Abstract
Background
Autosomal recessive cerebellar ataxia (ARCA) comprises a large and heterogeneous group of neurodegenerative disorders. We studied 3 families diagnosed with ARCA.
Methods
To determine the gene lesions responsible for their disorders, we performed high density SNP genotyping and exome sequencing.
Results
We identified a new mutation in the SACS gene and a known mutation in SPG11. Notably we also identified a homozygous variant in APOB, a gene previously associated with ataxia.
Conclusions
These findings demonstrate that exome sequencing is an efficient and direct diagnostic tool for identifying the causes of complex and genetically heterogeneous neurodegenerative diseases, early stage disease or cases with limited clinical data.
Keywords: Exome sequencing, ataxia, SPG11, APOB, SACS, mutation
Introduction
The terminology ataxia covers a large spectrum of heterogeneous neurological disorders mainly characterized by cerebellar and/or extracerebellar damage [1]. Several classifications have been made to better understand its various mechanisms [2–4]. Hereditary ataxias include X-linked ataxia, autosomal dominant cerebellar ataxia such as spinocerebellar ataxia and autosomal recessive cerebellar ataxia such as spastic ataxia of Charlevoix-Saguenay (ARSACS) [1]. Despite these classification schemes, the diagnosis of ataxia is still challenging due to phenotypic variability and a great deal of genetic heterogeneity. This is apparent not only for the pure ataxia disorders, but also where ataxia may be a prominent, and sometimes presenting, symptom of other diseases such as hereditary spastic paraplegia (HSP).
Exome sequencing is a relatively new but increasingly popular technique that has been successfully used to identify novel genes causing rare disorders [5–7]. The power of this approach lies in the comprehensive nature of the data produced; with most exome sequencing approaches providing good coverage sequence on the vast majority of the protein coding genome. Thus in essence, this method skips linkage, and goes straight to association and segregation. While gene discovery is a powerful and useful application for exome sequencing, this method is increasingly used as a screening tool, to identify mutations in genes already implicated in disease. This is a cost and time efficient approach, particularly in diseases with a high degree of genetic heterogeneity [5].
In this study, we describe the successful application of exome sequencing in 3 unrelated Tunisian families each containing members who present with apparent autosomal recessive cerebellar ataxia (ARCA).
Patients and methods
Patients
This work was approved by the local ethics committee and by the Office of Human Subjects Research at the National Institutes of Health.
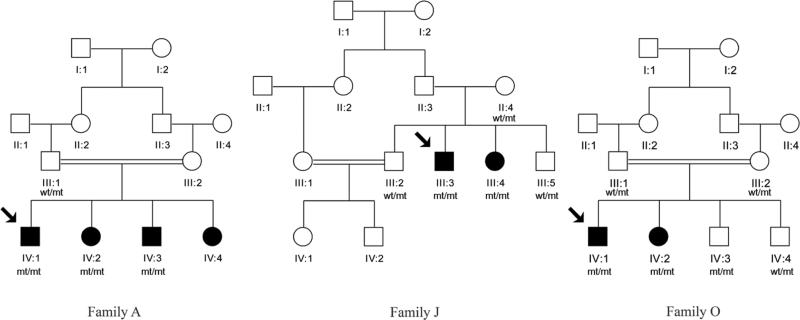
Three unrelated consanguineous families (A, J and O) of Tunisian decent consisting of 8 affected and 6 apparently unaffected individuals were studied (figure 1). Patients were selected according to the following criteria: 1) Clinical phenotype associating a cerebellar ataxia with knee and/or ankle retained tendon reflexes in at least the index patient, 2) Childhood or juvenile onset, and 3) Apparent autosomal recessive inheritance. A neurologist in the Department of Neurology at the National Institute of Neurology, Tunis, Tunisia performed a detailed phenotypic assessment of the patients.
Figure 1.
Family pedigrees
Molecular analysis
After obtaining informed consent from all study participants, blood was collected and genomic DNA was extracted from peripheral blood lymphocytes using standard protocols.
A linkage study on these 3 families was previously performed using microsatellite polymorphic fluorescent markers spanning 16 loci linked to cerebellar ataxia: FRDA (9q13 and 9p23), ARSACS (13q12), AVED (8q13), AOA (9p13 and 9q43), IOSCA (10p23), HLOA (6p21), AT (11q22), ATLD (11q21), SCAN1 (14q31), ATCAY (19p13), CAMOS (9q34), Salla syndrome (6q14), Marinesco Marinesco-Sjögren (5q31) and the Childhood spinocerebellar ataxia linked to 11p15. Linkage to all tested loci was excluded in this study [6].
To search for the disease locus, genome-wide SNP genotyping of all affected and unaffected individuals was conducted using the genome-wide human SNP array OmniExp-12,v1.0 DNA Analysis BeadChip according to the manufacturer's instructions (Illumina Inc., San Diego, CA). Because we believed the cause of disease in these families was a homozygous mutation driven by parental consanguinity, SNP array data were subjected to homozygosity mapping with the HomozygosityMapper software (http://www.homozygositymapper.org/) using only homozygous stretches 15 alleles or longer and identical homozygous regions segregating with disease within each family [7].
To find a gene mutation within the candidate loci, whole exome sequencing was performed on DNA extracted from each of the 8 affected individuals. Three micrograms of genomic DNA was processed according to the Nimblegen protocol (Nimblegen v2.0, Roche Nimblegen, Indianapolis, IN). Prior to sequencing, DNA templates were bridge amplified to form clonal clusters inside a flowcell via the cBot cluster generation process. The flowcell was then loaded into the next generation sequencer Illumina HiSeq™ 2000.
Paired end sequence reads were aligned using BWA against the reference human genome (UCSC hg18) [8]. Duplicate read removal, format conversion and indexing were performed with Picard (picard.sourceforge.net/index.shtml). The Genome Analysis Toolkit was used to recalibrate base quality scores, perform local re-alignments around possible indels and to call and filter the variants [9,10]. Variants previously identified in the 1000 Genomes project or within neurological normal controls exome sequence within the same laboratory were excluded. Vcftools was used to annotate gene information for the remaining novel variants [9]. Single nucleotide variants that are non-synonymous, stop loss or gain or within essential splice sites and indels that were non-reference homozygous and shared among affected family members were prioritized as potential candidates for causal variants. These prioritized, potentially novel shared protein coding variants, were then visually inspected using the Integrative Genomics Viewer (IGV) [10].
To verify that variants were not an artifact of the sequencing process and to test segregation of candidate variants within each family, Sanger sequencing was performed for all families’ members, using ABI BigDye Terminator Cycle Sequencing Kit on an ABI 3730 sequencer. Sequence traces were analyzed using Sequencher (version 4.2 Gene Codes Corporation).
Nucleotide and protein positions of identified variants are based on the following accession numbers from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). APOB, NM_000384.2 and NP_000375.2; SACS NM_014363.4 and NP_055178.3; SPG11 NM_001160227.1 and NP_001153699.1. Variant positions within the cDNA are numbered using the A of the translation initiation codon as position 1.
Results
Clinical study
We studied three families (A, J and O) composed of 8 affected individuals with cerebellar ataxia (Figure 1). The clinical findings for all patients are summarized in Table 1.
Table 1.
Summary of clinical findings of the families A, J and O
| Family ID | A | J | O | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient ID on pedigree | IV:1 | IV:2 | IV:3 | IV:4 | III:3 | III:4 | IV:1 | IV:2 | |
| Gender | M | F | M | F | M | F | M | F | |
| Initial signs | GA/D | GA | GA/D | GA | GA | GA | GA | GA | |
| Age at onset (y) | 21 | 21 | 16 | 12 | 1 | 5 | 19 | 13 | |
| Age at last examination (y) | 23 | 22 | 29 | 26 | 24 | 28 | 30 | 27 | |
| DD (y) | 2 | 1 | 7 | 14 | 23 | 23 | 11 | 14 | |
| ICARS score /100 | 55 | 32 | 56 | 44 | 34 | 36 | 50 | 19 | |
| ICARS score/DD | 27.5 | 32 | 8 | 3.1 | 1.5 | 1.6 | 4.5 | 1.4 | |
| Cereblellar syndrome | S/ K | S/ K | S/ K | S/ K | S/ K | S/ K | S/ K | S/ K | |
| Ocular signs | Nystg | Nystg/ Dip | Nystg | Nystg/ Dip | Nystg | Nystg | Nystg | Nystg/ Dip | |
| TR | Knee | Brisk | Brisk | Brisk | P | Brisk | Brisk | Brisk | Brisk |
| Ankle | A | Weak | A | A | A | A | Brisk | Brisk | |
| Pyr S | Babinski | P | P | P | P | P | P | P | P |
| Spasticity | P++ | P++ | P+ | P+ | P+ | P+ | P++ | P+ | |
| DSD | PE | A | A | A | A | P | A | P | P |
| VS | P | P | A | N/A | P | P | P | P | |
| Skel.def | PC/HT | A | A | A | A | PC/HT | PC/HT | A | A |
| Scoliosis | A | A | A | A | A | A | P | A | |
| MRI/CT | N TDM | N TDM | N TDM | N/A | N/A | TDM : Vermis/Cerebellar atrophy | N/A | N MRI | |
| Muscle biopsy | N/A | N/A | Neurogenic Atrophy± | Neurogenic Atrophy± | Neurogenic Atrophy± | N/A | Subnormal | Subnormal | |
| Nerve biopsy : loss of large myelinated fibers | N/A | N/A | P± | P± | N/A | P++ | A | A | |
| EMG | N/A | N/A | Mo Ax neur ± | Mo Ax neur ± | SM axono myelinic neur ++ | SM Ax neur ++ | N | N | |
| EP | S EP | Abn | N | Abn | Abn | Abn | Abn | Abn | Abn |
| VEP | N | N | N | N | N/A | N | N | N | |
| AEP | Abn | N | N | N | N/A | Abn | Abn | N | |
| MEP | N/A | N | Abn | N/A | N/A | N/A | Abn | Abn | |
| Sphincter disturbance | P | P | A | P | A | P | P | A | |
| Other | None | None | None | None | Mental retardation +/ Epilepsy | Epilepsy/ | Headaches | Headaches/Hypoaes thesia of right hemibody | |
A:absent Abn: Abnormal AEP:Auditory Evoked Potentials Ax: Axonal CT:Computed Tomography D:Dysarthria DD:Disease Duration Dip:Diplopia EP:Evoked Potentials F:Female GA:Gait Ataxia HT:Hammer Toes ICARS score: International Cooperative Ataxia Rating Scale/ 100 {Postural walking score/34+ Coordination score/52+D/8+ Oculomotor signs/6} K:Kinetic M:Male MEP: Motor Evoked Potentials Mo: Motor MRI:Magnetic Resonance Imaging N:Normal Neur:Neuropathy Nystg:Nystgmus P:Present Par:Paresthesia PC:Pes Cavus PE:Position Error Pyr S:Pyramidal Sign S:Static SM:Sensorimotor Skel.def.:Skeletal deformities TR:Tendon Reflexes SEP: Somatosensory Evoked Potentials VEP:Visual Evoked Potentials Y:Years ±: Mild +: Moderate ++: Severe
The four patients in Family A began the disease at age 12 to 21 years with walking instability. Gait ataxia and dysarthria were initial signs in two of them. Pyramidal syndrome with spasticity and Babinski sign was present in all patients. Static and Kinetic cerebellar syndrome and nystagmus were a constant finding. We noted that the evaluation of the dysmetria in lower limbs was distorted by the spasticity which was moderate to severe. Dysathria was absent in only one patient (IV2) where the ICARS score was the lowest. Intrafamilial variability, measured by the ICARS score divided by the disease duration, was between 3.1 and 27.5. Tendon reflexes were present or brisk in knee and absent or weak in ankles. There were no superficial sensory disturbances. Deep sensory disturbances were highlighted in two patients by a decrease in toe vibration sense. Urinary urgency was noted in three patients. There were no skeletal deformities. Cerebral TDM, which was assessed in three of four affected family members, was normal. Nerve conduction studies (NCS) in two patients found a mild axonal motor neuropathy with a decrease in the peroneal nerve amplitude confirmed by muscle biopsy showing neurogenic fascicular atrophy. There was a slight reduction of sural nerve velocity demonstrated by nerve biopsy showing rare axons with segmental demyelination and degeneration. Evoked potentials were variable between patients. Motor evoked potentials (MEP) recorded in one patient confirmed the injury of the pyramidal pathway.
The two patients of Family J had very early onset (<5 years), gait ataxia as initial sign and ICARS score divided by disease duration between 1.5 and 1.6. Cerebellar syndrome was static and kinetic. Nystagmus and dysarthria were present. Reflexes were brisk and ankle reflexes were abolished by the age of 23 years. Pyramidal syndrome with spasticity and Babinski sign was a main feature. Deep sensory disturbances were constant. Both patients had pes cavus and hammer toes. The brother (III:3) had epilepsy and died at age of 32 years by intracerebral hematoma. The sister (III4) had also epilepsy and had surgery for a malignant tumor of the ovary. Marked vermis and hemispheric cerebellar atrophy were noted in one patient. In both patients, there was severe sensori-motor neuropathy, mainly axonal as described previously [11], with mild neurogenic atrophy in peroneus brevis muscle biopsy. Nerve biopsy showed marked loss of large myelinated fibers. The two patients had abnormal SEP. One patient had an abnormal AEP and a normal VEP.
Both patients of Family O had onset in the second decade, with gait ataxia as initial symptoms. ICARS score was variable. Cerebellar syndrome was not associated with dysarthria in both patients. Nystagmus was multidirectional. Tendon reflexes were brisk. Pyramidal syndrome was obvious with positive Babinski sign and moderate to severe spasticity. Deep sensory disturbances were noted by impaired position and vibration sense. Scoliosis was present in one patient and urinary urgency in the other patient. Normal cerebral MRI was noted in one patient. Nerve conduction studies were normal. Muscle biopsy showed slight neurogenic atrophy. Both semi-thin sections and teased nerve fibers from nerve biopsy were normal. Evoked potential studies were variable. MEP were abnormal in both patients.
Molecular study
Comprehensive homozygosity mapping was performed in 8 affected and 7 unaffected individuals from the three families. For each family, we searched for homozygous regions shared among the affected individuals and not shared with the unaffected individuals. The homozygosity mapping revealed a total of 3 regions for family A and only one region for family O. The software revealed 4 regions for family J; including the region spanning chromosome 13 which contains SACS, linked to ARSACS. This region had been previously excluded by linkage [6].
Whole exome sequencing was performed on the DNA of 7 affected individuals. Overall, more than 200 million sequencing reads were produced for each sample covering more than 10 billion bases. Approximately, 98% of these were aligned to the human reference genome (hg 18). On average 94% of exome capture baits had at least 10X depth and 88% at least 30X depth. After filtering to identify variants not reported in dbSNP or 1000 genomes (April 2009 release), less than 10 homozygous coding variants and less than 15 homozygous indels were left per family.
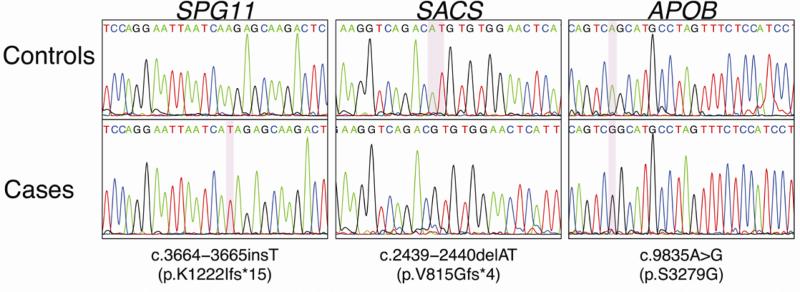
In family A, a homozygous insertion was found in SPG11, c.3664-3665insT leading to a premature stop codon (p.K1222Ifs*15). This mutation had been previously reported in a heterozygous state in an Italian family with spastic paraplegia [12]. In family J, a novel homozygous mutation was identified in SACS; a two base pair deletion in exon 9, c.2439-2440delAT predicted to produce a frameshift and a premature stop codon at 817 (p.V815Gfs*4). The sacsin protein is consequently truncated by 82%, likely leading to a complete loss of function. In family O, a homozygous missense mutation c. 9835A>G (S3279G) was found in APOB, this was the only novel homozygous coding variant within our identified runs of homozygosity in both affected family members. This mutation has been previously reported in a genome wide association study performed on 555 individuals as rare variant contributing to the heritability of hypertriglyceridemia [12].
Analysis of the runs of homozygosity detected by homozygosity mapper showed that these variants were in genetic segments inherited in a manner consistent with the apparent transmission of the disease; and suggesting that each of the variants segregated with disease perfectly. Sanger sequencing confirmed that the variants found in families A and J were homozygous in the patients and heterozygous in both parents. In family O, Sanger sequencing showed that the missense mutation c.9835A>G of APOB was homozygous in the affected siblings and heterozygous in both parents; notably one of the apparently healthy siblings was also homozygous for this variant. Unfortunately this family is lost to follow up.
Discussion
In this study, we used exome sequencing to identify the genetic causes of cerebellar ataxia in three Tunisian families. The technique proved to be advantageous in comparison to fluorescent marker linkage, which yielded misleading data that excluded SACS as a disease-causing gene in family J. While it is not clear why this discrepancy occurred, it is likely that the indirect nature of linkage contributed to this error, and this highlights the power of direct, comprehensive screening.
The first mutation we identified (in family A) was an insertion located in SPG11. SPG11 mutations are associated with a severe and complex form of autosomal recessive HSP with thin corpus callosum. HSP are a large group of neurological disorders that are both clinically and genetically heterogeneous. HSP are characterized mainly by slowly progressive spasticity affecting the lower limbs. In the group of autosomal recessive HSP, mutations in SPG11 seem to account for the majority of cases with thinning of the corpus callosum.
There are reported cases with complex spastic paraplegia exhibiting cerebellar ataxia and classified as such, especially in the absence of the MRI or the absence of an obvious thinning of the callosum on MRI [13,14]. In Tunisia, autosomal recessive HSP is frequent, with SPG11 mutations as a major cause of this entity (62%) [15]. The mutation we identified was not previously reported in Tunisia but has been reported in Italy. Since Italy is one of the closest Mediterranean countries to Tunisia, genetic similarities between the two populations would not be unexpected.
The second mutation we found was in APOB (in family O). Previous studies have reported that APOB mutations are associated with hypobetalipoproteinaemia, normotriglyceridemic hypobetalipoproteinemia, hypercholesterolemia and HTG. HTG is a complex polygenic disease defined by fasting plasma triglyceride concentrations above the ninety-fifth percentile [16]. Although secondary factors are important for clinical expression, susceptibility to HTG has a strong genetic component [17]. Our mutation was previously identified in a GWAS performed by Johansen et al., 2010, on 555 individuals with HTG [12]. In this study the authors described the variant occurring in less than 1% of cases (4 of 438) with HTG, and absent from 327 controls. Notably, the authors only observed this variant as a heterozygous alteration. In contrast, the variant was identified as a homozygous change in the family studied by us. There are a small number of affected family members in the current pedigree, and thus we have limited ability to show convincing segregation or reevaluate the unaffected family member who is also homozygous for the mutation. While ataxia has been previously reported as a presenting feature of APOB mutations, we believe the current finding should be interpreted with caution, particularly when considering exome sequencing does not capture every exon, or every type of mutation [19][5].
The third mutation we identified was in SACS (in family J). ARSACS was first identified in 1978 by Bouchard in the small region of Charlevoix-Saguenay in French Canada [20]. Clinical features include cerebellar syndrome, dysarthria, spasticity, distal muscle wasting, nystagmus and peripheral neuropathy [21]. Mutations causing ARSACS have been identified in SACS, located on chromosome 13q12.12, which encodes the large protein; sacsin [22]. More than 70 different SACS mutations have been identified worldwide [21]. Most of these mutations are frameshift or nonsense mutations located in the large exon 9 spanning ~12000 bp. In Tunisia, seven different mutations have been previously described [21]. The mutation we identified in our study is an insertion located in exon 9 that is predicted to lead to a truncated sacsin protein, which very likely leads to a loss of function.
In conclusion, our findings demonstrate that exome sequencing could be an efficient diagnostic tool for identifying genes that are implicated in complex neurodegenerative diseases, early stages of diseases or cases with limited clinical data. The continuous decrease of the cost and time needed for the technology will make it affordable even for laboratories without large budgets.
Figure 2.
Chromatograms showing the different mutations identified
Table 2.
Regions of homozygosity
| Family | Chromosome | Start SNP | End SNP | Start position | End position |
|---|---|---|---|---|---|
| A | 6 | rs9376622 | rs6928093 | 96396321 | 103010466 |
| 15 | rs7178596 | rs8028106 | 36966613 | 44783815 | |
| 17 | rs1005651 | rs2286674 | 61868473 | 62231014 | |
| J | 8 | rs11780869 | rs9773025 | 154984 | 6661868 |
| 8 | rs4841462 | rs2173117 | 10841027 | 11642539 | |
| 11 | rs11263623 | rs11237348 | 69394552 | 77382240 | |
| 13 | rs9552800 | rs17083511 | 22497673 | 25729419 | |
| O | 2 | rs2075007 | rs205618 | 74323087 | 74799693 |
Acknowledgments
The authors thank Jeffrey Hammer and Marguerite Meitzler for their contribution in the correction of the manuscript.
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, part of the Department of Health and Human Services. Project Number ZIA AG000958-09.
Footnotes
Disclosure of conflict of interest
The authors declare no financial or other conflict of interests.
References
- 1.Manto M, Marmolino D. Cerebellar ataxias. Curr Opin Neurol. 2009;22:419–429. doi: 10.1097/WCO.0b013e32832b9897. doi:10.1097/WCO.0b013e32832b9897. [DOI] [PubMed] [Google Scholar]
- 2.Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;1:1151–1155. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- 3.De Michele G, Coppola G, Cocozza S, Filla A. A pathogenetic classification of hereditary ataxias: is the time ripe? J Neurol. 2004;251:913–922. doi: 10.1007/s00415-004-0484-2. doi:10.1007/s00415-004-0484-2. [DOI] [PubMed] [Google Scholar]
- 4.Klockgether T. Parkinsonism & related disorders. Ataxias. Parkinsonism Relat Disord. 2007;13(Suppl 3):S391–394. doi: 10.1016/S1353-8020(08)70036-1. doi:10.1016/S1353-8020(08)70036-1. [DOI] [PubMed] [Google Scholar]
- 5.Singleton AB. Exome sequencing: a transformative technology. Lancet Neurol. 2011;10:942–946. doi: 10.1016/S1474-4422(11)70196-X. doi:10.1016/S1474-4422(11)70196-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouhlal Y, El-Euch-Fayeche G, Amouri R, Hentati F. Distinct phenotypes within autosomal recessive ataxias not linked to already known loci. Acta Myol. 2005;24:155–161. [PubMed] [Google Scholar]
- 7.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–599. doi: 10.1093/nar/gkp369. doi:10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. doi:10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. doi:10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. doi:10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermeer S, Meijer RPP, Pijl BJ, Timmermans J, Cruysberg JRM, et al. ARSACS in the Dutch population: a frequent cause of early-onset cerebellar ataxia. Neurogenetics. 2008;9:207–214. doi: 10.1007/s10048-008-0131-7. doi:10.1007/s10048-008-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denora PS, Schlesinger D, Casali C, Kok F, Tessa A, et al. Screening of ARHSP TCC patients expands the spectrum of SPG11 mutations and includes a large scale gene deletion. Hum Mutat. 2009;30:E500–519. doi: 10.1002/humu.20945. doi:10.1002/humu.20945. [DOI] [PubMed] [Google Scholar]
- 13.Rajakulendran S, Paisán-Ruiz C, Houlden H. Thinning of the corpus callosum and cerebellar atrophy is correlated with phenotypic severity in a family with spastic paraplegia type 11. J Clin Neurol. 2011;7:102–104. doi: 10.3988/jcn.2011.7.2.102. doi:10.3988/jcn.2011.7.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depienne C, Stevanin G, Brice A, Durr A. Hereditary spastic paraplegias: an update. Curr Opin Neurol. 2007;20:674–680. doi: 10.1097/WCO.0b013e3282f190ba. doi:10.1097/WCO.0b013e3282f190ba. [DOI] [PubMed] [Google Scholar]
- 15.Boukhris A, Stevanin G, Feki I, Denis E, Elleuch N, et al. Hereditary spastic paraplegia with mental impairment and thin corpus callosum in Tunisia: SPG11, SPG15, and further genetic heterogeneity. Arch Neurol. 2008;65:393–402. doi: 10.1001/archneur.65.3.393. doi:10.1001/archneur.65.3.393. [DOI] [PubMed] [Google Scholar]
- 16.Yuan G, Al-Shali KZ, Hegele RA. Hypertriglyceridemia: its etiology, effects and treatment. CMAJ. 2007;176:1113–1120. doi: 10.1503/cmaj.060963. doi:10.1503/cmaj.060963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansen CT, Hegele RA. Genetic bases of hypertriglyceridemic phenotypes. Curr Opin Lipidol. 2011;22:247–253. doi: 10.1097/MOL.0b013e3283471972. doi:10.1097/MOL.0b013e3283471972. [DOI] [PubMed] [Google Scholar]
- 18.Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. doi:10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homer VM, George PM, du Toit S, Davidson JS, Wilson CJ. Mental retardation and ataxia due to normotriglyceridemic hypobetalipoproteinemia. Ann Neurol. 2005;58:160–163. doi: 10.1002/ana.20531. doi:10.1002/ana.20531. [DOI] [PubMed] [Google Scholar]
- 20.Bouchard JP, Barbeau A, Bouchard R, Bouchard RW. Autosomal recessive spastic ataxia of Charlevoix-Saguenay. Can J Neurol Sci. 1978;5:61–69. [PubMed] [Google Scholar]
- 21.Bouhlal Y, El Euch-Fayeche G, Hentati F, Amouri R. A novel SACS gene mutation in a Tunisian family. J Mol Neurosci. 2009;39:333–336. doi: 10.1007/s12031-009-9212-9. doi:10.1007/s12031-009-9212-9. [DOI] [PubMed] [Google Scholar]
- 22.Bouhlal Y, Amouri R, El Euch-Fayeche G, Hentati F. Autosomal recessive spastic ataxia of Charlevoix-Saguenay: an overview. Parkinsonism Relat Disord. 2011;17:418–422. doi: 10.1016/j.parkreldis.2011.03.005. doi:10.1016/j.parkreldis.2011.03.005. [DOI] [PubMed] [Google Scholar]