Summary
Tumors are typically sequenced to depths of 75–100× (exome) or 30–50× (whole genome). We demonstrate that current sequencing paradigms are inadequate for tumors that are impure, aneuploid or clonally heterogeneous. To reassess optimal sequencing strategies, we performed ultra-deep (up to ~312×) whole genome sequencing (WGS) and exome capture (up to ~433×) of a primary acute myeloid leukemia, its subsequent relapse, and a matched normal skin sample. We tested multiple alignment and variant calling algorithms and validated ~200,000 putative SNVs by sequencing them to depths of ~1,000×. Additional targeted sequencing provided over 10,000× coverage and ddPCR assays provided up to ~250,000× sampling of selected sites. We evaluated the effects of different library generation approaches, depth of sequencing, and analysis strategies on the ability to effectively characterize a complex tumor. This dataset, representing the most comprehensively sequenced tumor described to date, will serve as an invaluable community resource (dbGaP accession id phs000159).
Introduction
Previous studies have estimated that sequencing human genomes to 30–40× mean coverage is sufficient to accurately detect germline heterozygous SNVs in 95% of the genome (Ajay et al., 2011). Despite additional complexity from factors like contamination of non-tumor cells, tumor heterogeneity, and aneuploidy, cancer genomes have generally been sequenced to comparable depths, typically between 30× and 50× mean coverage (Borad et al., 2014; Mardis, 2012). While this is enough coverage to discover SNVs in the founding clones of high purity tumors, most tumors are not pure, with estimated tumor cellularities ranging from 90–95% in many hematologic cancers, such as acute myeloid leukemia (AML) (Cancer Genome Atlas Research Network, 2013), to 25–75% in some solid tumors such as breast (Yuan et al., 2012), and to as low as 5% in some pancreatic cancer samples (Biankin et al., 2012). Most tumors also contain subclonal populations, some of which may contribute to therapeutic resistance (Ding et al., 2012; Ma et al., 2015). Increased sequencing depth has the potential to enable sensitive detection of mutations corresponding to these subclonal populations and improve inference of a tumor’s clonal architecture.
Although interrogating tumors with targeted approaches (including exome capture) can provide greater sequencing depth, these assays do not allow for complete characterization of a tumor genome. Whole genome sequencing (WGS) is necessary for comprehensive detection of many relevant classes of mutations, including structural variants, copy number alterations, non-protein-coding mutations with regulatory significance, and viral integration sites. In tumors with a low mutational load, WGS increases the number of detectable variants and is essential for accurately outlining subclonal architecture (Miller et al., 2014). In general, detecting low frequency mutations and characterizing the clonal structure of tumors may require greater depth than that afforded by current WGS strategies, and greater breadth than that afforded by current exome or gene panel sequencing strategies.
The reluctance to sequence cancer samples to high depth has been driven by financial constraints, but as the cost of sequencing has continued to drop, there have been a few studies that have generated large amounts of sequence data from single samples. Lam, et al. sequenced a normal individual to ~300× in order to compare the characteristics of multiple sequencing chemistries and instruments (Lam et al., 2012). Rieber, et al. generated ~145× coverage of a pair of medulloblastomas (when combining data across platforms) and used that data to evaluate coverage biases and variant calling performance (Rieber et al., 2013). One of the most deeply sequenced tumors to date is an ER+ breast cancer that was sequenced to ~188× mean coverage, which allowed for identification of subclonal populations as low as ~5% variant allele frequency (VAF) (Nik-Zainal et al., 2012). Though each study offered valuable insights, none examined the impact of sample and library preparation or attempted to quantify the lower limits of detection using current sequencing technologies.
While a growing body of evidence suggests that current sequencing paradigms are inadequate for evaluating relevant characteristics of tumors, consensus on optimal methodologies and depth of coverage targets is lacking. The International Cancer Genome Consortium (ICGC) has published anticipated standards, recommending that "80% of the somatic alterations should be identified in each sample and that coverage calculations on each sample should be based on this expectation” (https://icgc.org/icgc/goals-structure-policies-guidelines/e8-genome-analyses). At this point, it is unclear how such coverage depths can be estimated, especially when the number of mutations present at a variant allele frequency (VAF) of below ~15% is essentially unknown for all cancers sequenced to date.
In this study, we explore the value of sequencing genomes well beyond 100× by studying an individual with AML, referred to throughout this work as ‘AML31’. This patient was diagnosed with AML at age 55 and a primary tumor sample (from the bone marrow) was obtained at this timepoint. The patient achieved clinical remission following standard induction chemotherapy with cytarabine and idarubicin, and then received four rounds of high dose cytarabine consolidation therapy. The patient relapsed 16 months later, at which point a relapse sample was obtained (clinical features of this case have been described in detail in previous publications (Cancer Genome Atlas Research Network, 2013; Ding et al., 2012; Klco et al., 2014)). Previous genomic analyses of both the de novo and relapsed samples revealed common founding clone mutations between the two populations, as well as subclonal populations specific to the primary tumor (responsive to therapy) and to the relapse tumor (that became the dominant clone) (Ding et al., 2012). Although WGS was performed for these samples to typical coverage levels (25× and 38×), this depth was insufficient to identify the provenance of most mutations observed only in the relapse sample. The initial discovery sequencing for this case was also complicated by a normal skin sample that was heavily contaminated by tumor cells. In the current study we sequenced a second skin normal sample obtained when the patient was in a deep remission.
By whole genome sequencing this tumor to a depth of greater than 300× and applying comprehensive analysis strategies, we demonstrate that: 1) The commonly utilized sequencing depths of ~100× for exomes and 30–50× for WGS are inadequate when there exists even moderate heterogeneity, impurity, contamination, aneuploidy, or combinations thereof; 2) Current analysis strategies relying on a single alignment algorithm and variant caller, using data from a single DNA library, suffer from poor sensitivity and specificity; 3) Deep sequencing substantially improves the discovery of variants across a range of variant allele frequencies and allows for a more definitive model of tumor clonal architecture; and 4) This dataset, generated from the most deeply sequenced and cross-validated tumor described to date, will serve as a valuable community resource for improving tools and algorithms by providing an extremely high confidence set of low frequency mutations.
Results
Whole genome sequencing of an AML primary tumor and relapse specimen
Samples from a single patient (“AML31”) were originally sequenced to standard depths (24× tumor, 32× normal, 38× relapse), which allowed for the identification of 118 variants, including a DNMT3A R882H mutation and NPM1 W288fs in the founding clone (Ding et al., 2012). Nineteen sites, including FLT3 D835H and IDH1 R132H, comprised a subclone that was apparently eliminated by chemotherapy. An additional 42 sites were categorized as relapse-specific, though a few were found at measurable (but not statistically significant) depths in the primary tumor. Follow-up work further established that AML31 had multiple subclones at presentation (Klco et al., 2014), with at least three populations that responded differently to engraftment in immunodeficient mice, and confirmed that many mutations driving the relapse tumor were present at low levels in the primary tumor. That study also identified a presumed driver IDH2 R140Q mutation that was in less than 2% of the tumor, but present in the dominant subclone of the relapse. A total of 149 somatic SNVs and indels were discovered in these two paired samples.
To enable discovery of additional somatic mutations, we took samples from the primary and relapse tumors and matched them with a normal skin sample (Figure 1 and Table S1). We performed WGS for the primary tumor and normal samples, using multiple independent libraries to 312× and 121× coverage, respectively. Existing sequence data for the relapse tumor provided 38× coverage of that sample (Table 1 and Figure S1–S3). Using a combination of WGS and custom capture data (described below), we estimated the tumor cell purity of primary and relapse tumors as 90.7% and 36.2% respectively (Figure S4, Supplemental Experimental Procedures). Purity is the proportion of tumor cells derived from the initiating tumor cell and the remaining proportion represents putatively normal cells. Tumor contamination in the normal skin was low (likely because the sample was obtained while the patient was in remission), with a conservative upper estimate of 0.35% (Figure S4), based on the 95th percentile VAF value from selected somatic sites in the tumor (see Supplemental Experimental Procedures for details). This level may include sequencing errors and technical errors, but we were unable to find strong evidence for either at rates that would exceed 0.35%. Sequencing errors were examined using homozygous sites (as determined by SNP genotyping microarrays); the upper estimate of signal we expect from sequencing errors in this data is 0.23–0.35%, depending on the base identity (Figure S5 and Table S2). Much of the following analysis relies on accurate estimation and interpretation of tumor VAFs. These values can be substantially complicated by large-scale copy number variants (CNVs) that are common in many tumors. However, neither the primary tumor nor the relapse contained any detectable CNVs (Figure S6).
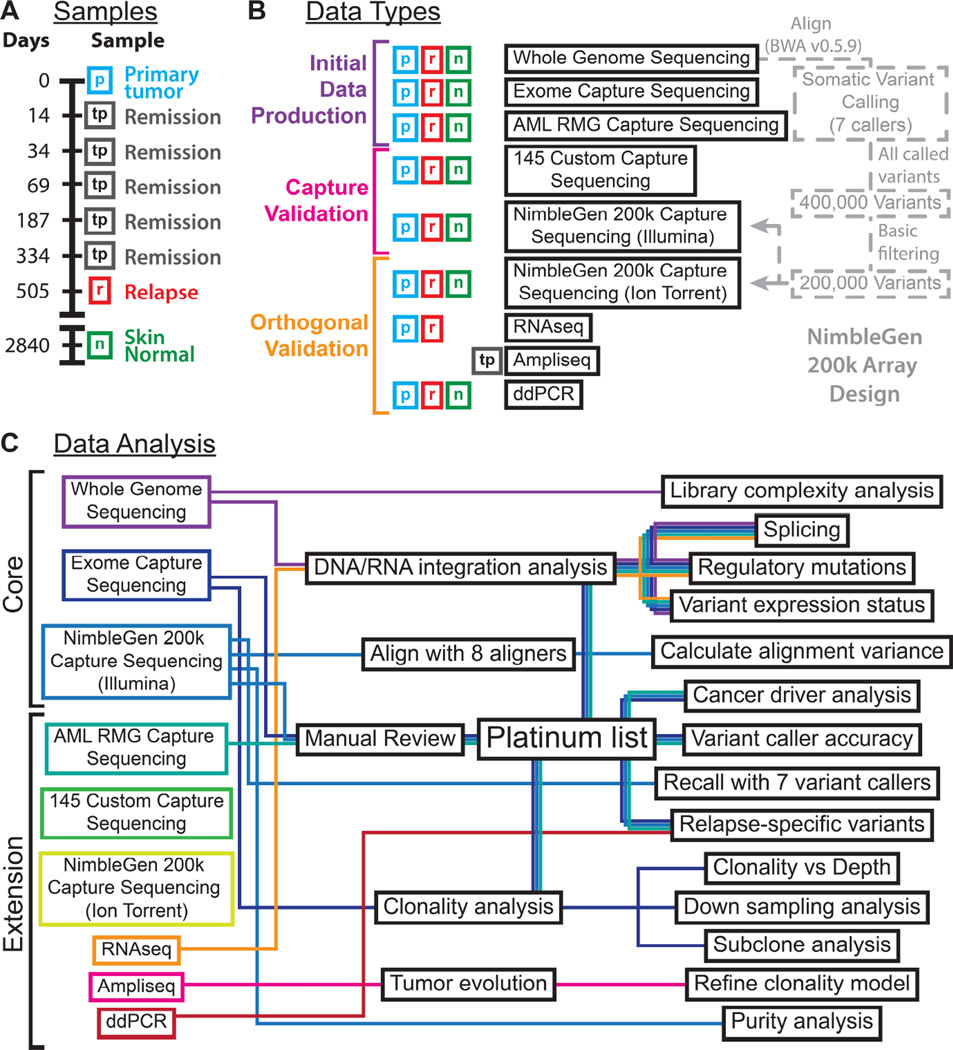
Figure 1. Experimental overview.
A) Samples from this study are depicted along a timeline with day 0 representing the day of AML diagnosis. B) Data types generated for each sample are indicated along with a basic summary of their dependencies. C) A depiction of the analysis strategies employed, their outputs and the datasets they rely on are depicted as a schematic ‘subway map’. Refer to Supplemental Experimental Procedures for additional methods and results associated with each.
Table 1.
Summary of data produced.
| Data type | Normal sample | Primary tumor sample (90.7% estimated purity) |
Relapse tumor sample (36.2% estimated purity) |
|---|---|---|---|
| Total number of DNA libraries | 12 libraries as 15 fractions | 13 libraries as 23 fractions | 8 libraries as 10 fractions |
| Illumina whole genome sequencing (WGS) * | 1 library, 4 reps; 422 Gbp; 121× coverage; 2,798 Mbp breadth |
4 libraries; 12 reps 1,167 Gbp; 312× coverage; 2,813 Mbp breadth |
1 libraries, 2 reps 153 Gbp; 38× coverage; 2,566 Mbp breadth |
| Illumina exome data (NimbleGen SeqCap EZ v3.0 exome) * | 1 library, 1 reps; 57 Gbp; 263× coverage; 149 Mbp breadth |
9 libraries 100 Gbp; 433× coverage; 174 Mbp breadth |
3 libraries; 77 Gbp; 251× coverage; 221 Mbp breadth |
| NimbleGen custom capture (200k sites) sequenced with Illumina * | 7 libraries, 7 reps; 99 Gbp; 1,130× coverage; 238 Mbp breadth |
6 libraries, 6 reps; 128 Gbp; 1,500× coverage; 359 Mbp breadth |
2 libraries, 2 reps; 29 Gbp; 280× coverage; 71 Mbp breadth |
| NimbleGen custom capture (200k sites) sequenced with Ion Torrent | 1 library; 6.1 Gbp; 43× coverage; 25 Mbp breadth |
1 library; 6.1 Gbp; 49× coverage; 31 Mbp breadth |
1 library; 6.6 Gbp; 45× coverage; 36 Mbp breadth |
| IDT custom capture (AML RMG set) sequenced with Illumina | 4 libraries; 3 Gbp; 270× coverage; 5.8 Mbp breadth |
10 libraries; 7 Gbp; 1,209× coverage; 7.3 Mbp breadth |
N/A |
| IDT custom capture (145 target sites) sequenced with Illumina | 4 libraries, 4 reps; 14 Gbp; 6,994 ×coverage; 2.8 Mbp breadth |
3 libraries, 3 reps; 9 Gbp; 6,939× coverage; 2.6 Mbp breadth |
3 libraries, 3 reps; 8 Gbp; 9,713× coverage; 2.4 Mbp breadth |
| ddPCR sequencing (15 targets) | N/A | 6,109× coverage (valid droplets) | 5,619× coverage (valid droplets) |
| PCR Amplicon Ion Torrent Sequencing (11 cancer driver targets). |
6,846× coverage; | 12,299× coverage; | 13,725× coverage; |
| Illumina RNA-seq | N/A | 8 libraries; 542 Gbp; |
1 library; 32 Gbp; |
An asterisk (*) denotes membership in the ‘core’ dataset.
‘Coverage’ refers to the median per base coverage. ‘Breadth’ refers to the number of sites sequenced to >= 20× coverage where only bases with >= 20 base quality score and reads with mapping quality >= 20 are considered. Refer to Supplemental Experimental Procedures for details of each sequence capture approach.
SNV detection and validation
Seven different SNV callers were run on the WGS data for the primary and relapse tumors (both compared to the normal skin sample). Application of these callers to the unusually high tumor WGS coverage resulted in 371,976 unique somatic variants called by one or more callers (Table S3 and Dataset S1, the complete call set is available in dbGaP accession id phs000159). A low stringency filter was applied to all of these variants to remove those with low read coverage in the tumor WGS data or unusually high evidence for the putative variant base in the normal sample (see Experimental Procedures for details). Only 3.3% of all variants were removed by this filter, leaving 359,619 calls. From the 359,619 remaining variants, we selected 198,814 for targeted re-sequencing using a custom NimbleGen capture reagent (Dataset S2, see Experimental Procedures for details). Oligonucleotide probes for 191,988 (96.6%) of these sites were successfully designed and synthesized. 3.4% of sites failed the design process because the resulting oligonucleotide sequences did not satisfy the manufacturer’s specified parameters with respect to GC content, melting temperature, repetitive sequence, or other factors. Capture hybridization followed by sequencing yielded adequate coverage for 164,090 sites (82.0%), with at least 20× coverage in the tumor, relapse and normal (median depth of 1,500× in the primary tumor sample, 280× in the relapse, and 1,130× in the normal skin). 135,788 of these sites (82.7%) contained at least one variant supporting read. Using these results along with data from deep exome sequencing (coverage of 263× normal, 433× tumor, and 251× relapse) as input (the “core” dataset), we performed extensive filtering and manual review to produce a ‘platinum’ variant list that contained 1,343 high quality validated sites (Table S4, Dataset S3, and Supplemental Experimental Procedures). The relapse data served as a useful control for this filtering process, since it contained many very-low frequency sites that expanded to higher VAFs in the relapse: 63 sites in the list with relapse VAF > 10% have a primary tumor VAF between 0.05 and 2%. This represents a greater than 11-fold increase in the number variants over the previous ~30× tumor with a tumorcontaminated normal (Ding et al., 2012).
The platinum variant list includes previously unidentified SNVs in a number of cancer-related genes, including FOXP1 (e11+1) and TP53 (G266R) (Table S5). The coverage for platinum variants in the core dataset was 4,182–11,603× in the primary sample and 3,392–10,557× in the relapse. We also identified 38 putative relapse-specific variants in this list, where the VAF observed in the primary tumor was less than 0.23% (the 95th percentile of what we might expect by random sequencing errors) and the VAF was greater than 5% in the relapse tumor (5–20% range, 7.2% median) (Table S6 and Figure S7). The relapse-specific variants included four predicted to affect protein sequence: MIB2 (G514S), CXCL17 (N83D), LMBR1 (R215H), and the TP53 (G266R) variant mentioned above. The TP53 site was covered by 9,292 reads across all sequence datasets with only 0.01% VAF in the primary tumor (Table S5), below that expected by sequencing error alone (Figure S5). To visualize support for these, along with an additional 228 putative relapse-specific events that did not meet the platinum criteria, we plotted the reference and variant read counts, coverage levels, and VAFs for both the primary and relapse sample (Figure S8). Many of these candidates had ~1,000–4,000× coverage with 0–2 variant-supporting reads in the primary tumor, close to what could be expected by sequencing errors alone. However, it was difficult to conclude that they were truly new mutations acquired at relapse, rather than mutations that were present at very low levels in the primary tumor. A subset of these events was therefore assayed by amplicon sequencing and ddPCR to assess their relapse-specific status (Figure S8, Tables S7 and S8). We obtained up to ~239,000× coverage in these assays (using up to 2 µg of input DNA, or ~580,000 haploid copies of the genome per assay). No variants assayed in this manner were truly relapse-specific. Some were present at low levels and were difficult to detect with the ~2,000–10,000× coverage from the custom capture assays, but upon deeper sequencing, all were found to be present in the primary tumor (Tables S8 and S9).
WGS offers the additional benefit of interrogating the regulatory regions of the genome. Out of the 1,343 variants in the platinum list, 8 occurred within putatively important non-coding regions defined by version 1 of our regulatory database, the ‘Regulome’, further described in the Supplemental Experimental Procedures (Table S10). FPKM expression values of the nearest neighboring gene were compared between RNA-seq data from the primary and relapse samples (4 variants were within gene introns, 3 were within 400 bp of a gene, and 1 was within the untranslated region of a pseudogene). Only 2 variants occurred within or adjacent to genes with >1 FPKM RNA-seq expression (RGS9 and ANKS6), both of which showed mild increases in FPKM values between primary and relapse (0.003 to 1.86 and 1.15 to 4.57, respectively).
Detection of other somatic events
Although the primary focus of this study was SNVs, we also examined other types of somatic variation. Detection of small insertions and deletions (indels) was performed using six different algorithms, yielding 667 indels within known Ensembl exons, of which only 23 (3.4%) remained after filtering and manual review (See Supplemental Experimental Procedures, Figure S9). Driver indels include FLT3 ITD, RUNX1 P339fs, and the previously described NPM1 W288fs (Table S11) (Ding et al., 2012). This tumor genome was exceptionally copy neutral, and even with the high resolution afforded by deep WGS, we were unable to detect any somatic copy number (Figure S6) or structural variants.
Clonal inference and tumor evolution
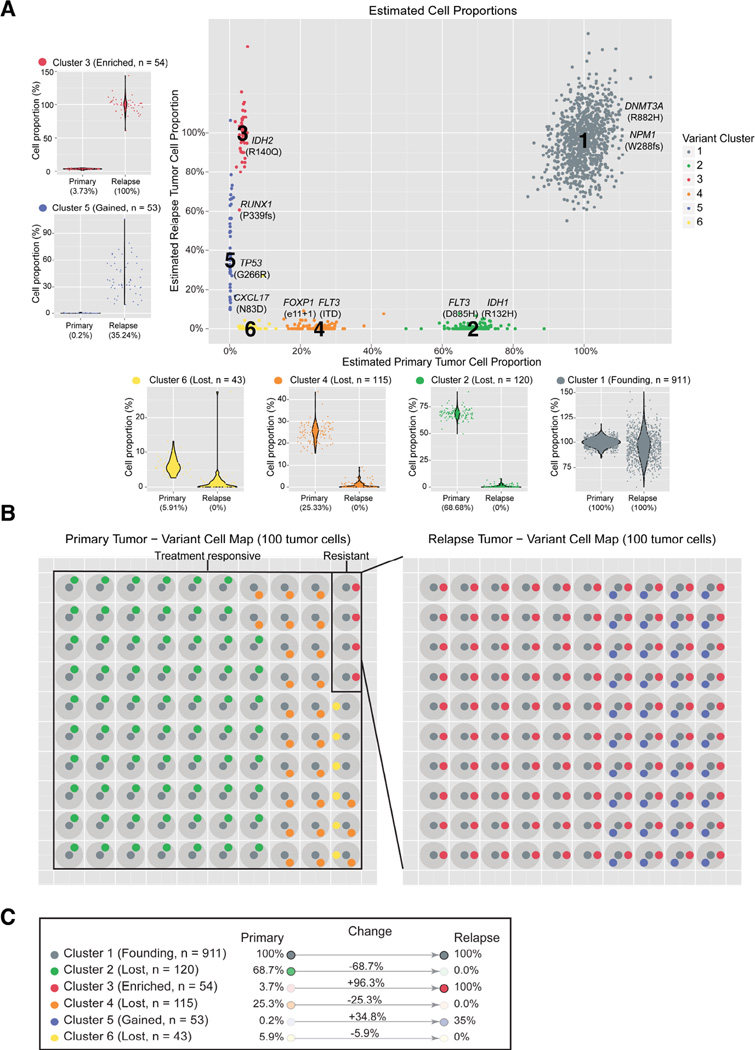
Using the variant allele frequencies of SNVs from the platinum list, we utilized the SciClone algorithm to infer the clonal structure of this tumor (Figure 2) (Miller et al., 2014). It detected 6 distinct clusters (Figure 2), with a founding clone (cluster 1), three tumor-specific subclones (clusters 2, 4, and 6), and two subclones enriched in the relapse sample (clusters 3 and 5). With the exception of the very-low VAF cluster 6, each cluster contained at least one potential driver mutation that may be relevant for that subclone’s growth advantage.
Figure 2. Cancer driver variants and their clonal identities in primary and relapse.
A) VAFs derived from the core dataset for all platinum variants are plotted for primary and relapse with colors assigned to variant clusters identified by SciClone. Each subpanel shows the change in VAF distributions for a single cluster from primary to relapse. Key AML-related variants are highlighted. B) A model of clonal heterogeneity within a theoretical 100 primary and relapse cells. C) Summary statistics and classification of each cluster with respect to clonal evolution.
Using the full clustering data as input, we generated possible evolutionary trees, based on relationships between subclones in the primary tumor and relapse (Figure S10). Though 5 structures are initially possible in the tumor, single cell sequencing data conclusively established the independence of clusters 2 and 4 (Klco et al., 2014). This eliminated all but two structures that differed only in assigning the parent of cluster 6 (Figure S10A,B).
Tracking clonal evolution over time
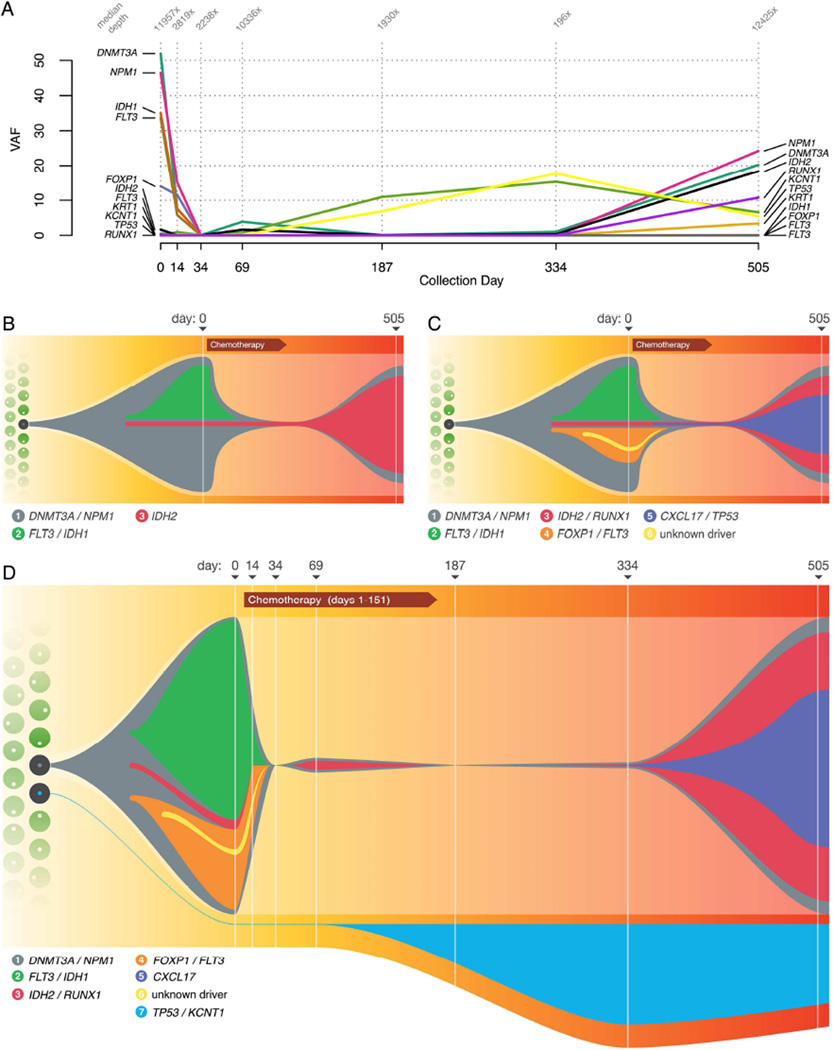
To add additional temporal resolution between presentation and relapse, we sequenced 11 variants drawn from several different subclonal populations to a depth of 6,463× in FFPE bone marrow biopsy samples collected at days 0 (primary tumor), 14, 34, 69, 187, 334, and 505 (relapse) using targeted amplicon sequencing (Figure 3A and Table S7). At day 14, the VAFs of all variants were diminishing and almost completely disappeared at day 34, with only 2 reads out of 2,878 (0.07%) supporting the presence of the DNMT3A founding clone variant. By day 69, however, the founding clone mutations had returned and were detected with VAFs between 1 and 3% until day 505. At this point the relapse tumor had emerged, containing the cluster 3 IDH2 mutation (18.64% VAF), as well as the founding clone DNMT3A and NPM1 mutations, all with VAFs of approximately 20%.
Figure 3. Tracking tumor evolution and refining a model of clonal architecture.
A) Variant allele frequency (VAF) of key mutations, at diagnosis (day 0), during progression through treatment, and at relapse (day 505). The median depth of coverage obtained at each variant position for each timepoint is indicated at top. Samples for intermediate timepoints between day 0 and 505 were obtained from FFPE blocks and in some cases were heavily degraded, leading to lower yields and sequence depth for some timepoints. B) Model of clonal architecture and tumor evolution, inferred from the original ~30× sequencing data. C) Ultra-deep sequencing and validation revealed additional subclonal complexity. D) Incorporating the results of single-cell sequencing and intermediate timepoints allows for refinements to the model, including establishing an independent origin for the TP53-mutant clonal population. Numbers in legend refer to cluster assignments. The ‘Chemotherapy’ label includes induction chemotherapy (day 1), and four rounds of consolidation chemotherapy at days 47, 81, 116, and 151.
Though the TP53 variant had a coverage of 7,664 reads and only 0.01% VAF at day 0, its proportion increased post-chemotherapy, with a VAF of 6.84% by day 187 and 17.65% by day 334. The other assayed variants all had VAFs below 1.02% at these timepoints. This indicates that the population of cells containing TP53 and KCNT1 mutations was derived from a hematopoietic stem/progenitor cell that was different from the one that produced the AML founding clone. This observation resulted in further modification of our model of tumor evolution (Figure 3B–D). This population had a selective advantage during consolidation chemotherapy, which lasted until day 150 (consistent with a previous report of TP53 as a driver of resistance to chemotherapy and expansion post-treatment (Wong et al., 2015)). Post-therapy, the tumor once again outcompeted the TP53-containing clone, which receded to a VAF of 5.43% at relapse.
RNA sequencing
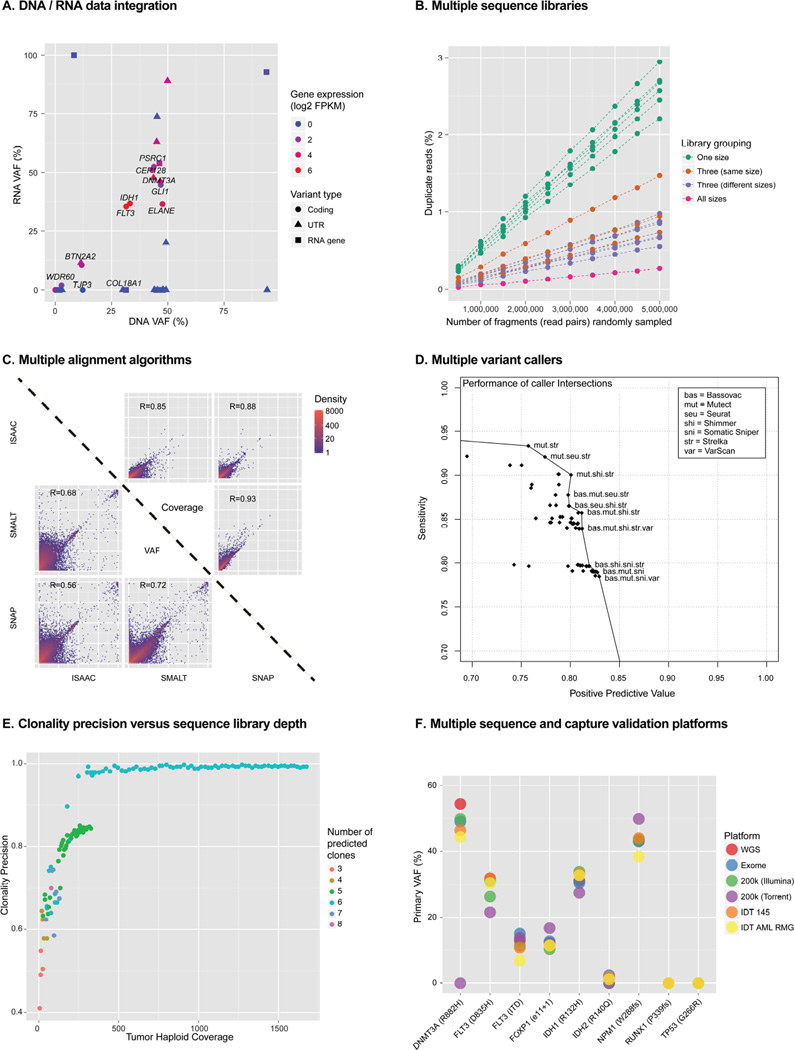
In addition to ultra-deep DNA sequencing of the primary tumor and relapse tumor genomes, we also conducted ultra-deep RNA-seq of the same two samples, producing 21 lanes of RNA-seq data from 10 individual libraries prepared using 8 different RNA-seq library construction strategies. We generated a total of 542 Gbp (2.7 billion 2×100 bp reads) of RNA-seq data for the primary tumor sample (Table 1). A detailed analysis of these data is being prepared for submission but we discuss here the integration of the RNA-seq data with respect to somatic SNV status. We examined all ~200k candidate somatic SNVs in the RNA-seq data. 74,897 variants (38%) predicted by the WGS were detected by >1 read in the RNA-seq data. 14,621 (7%) were detected with >100 reads. We observed the highest coverage for variants occurring within exons, UTRs, and RNA genes, followed by splice sites and intronic sites, and finally, "non-genic" variants had the least coverage. Despite substantial methodological differences across the 8 types of RNA-seq libraries sequenced, they were remarkably consistent with respect to RNA-seq VAF of driver AML variants. For example, VAFs obtained for the canonical IDH2 (R140Q) variant ranged from 33–43%. By considering the VAF estimates from RNA and DNA, along with gene level FPKM values from the RNA, we were able to categorize all 1,343 platinum variants according to their variant expression patterns (Supplemental Experimental Procedures). 43 were associated with a known or predicted Ensembl gene, 12 were confirmed as expressed and all but one of these were expressed in an allele-balanced fashion. ELANE Q194H exhibited wild type-biased expression, based on >9,000× coverage in both the DNA and RNA data (Figure 4D and Figure S11).
Figure 4. Selected findings from comprehensive data analyses.
A) Correlation of DNA and RNA VAFs for variants within exons. B) The effect of using multiple sequence libraries on library complexity. C) Representative comparisons of the effect of alignment algorithm on variant allele frequency estimation. D) The performance of variant callers when used in all possible combinations (intersections). E) The effect of increasing depth on the accuracy of clonal inference. F) Comparison of VAF estimation across six sequence platforms/datasets. Refer to the Supplemental Experimental Procedures and Supplemental Results for more details of each of these analyses.
Starting with the platinum variant list, we reviewed six putative somatic splicing mutations and found three with an observable effect on splicing in the RNA-seq data (Table S12 and Figure S12). Two resulted in exon skipping and one resulted in intron retention. FOXP1 had a donor site mutation that caused skipping of the associated exon, resulting in a frameshift. This FOXP1 splice site mutation was observed in a subclone of the primary tumor sample that was cleared at relapse. The level of aberrant splicing in the primary was consistent with the level expected for the subclone and at relapse, no exon skipping was observed. GGA2 also had a donor site mutation that resulted in skipping of the associated exon. This event appeared to be in the dominant clone of the primary and was maintained at relapse. The exon skipping pattern in the RNA-seq data for primary and relapse samples appeared consistent.
A comprehensive and integrative analysis strategy is critical
In addition to better describing the AML31 primary tumor and relapse, we used this dataset to perform an evaluation of sequencing and analysis methods and explored ways in which they could be optimized for genomic characterization of cancer samples.
Sample preparation
Generating multiple DNA libraries for WGS requires extra labor and expense, but starting with a larger pool of molecules should increase the complexity and coverage of the resulting sequence data. We generated data from 11 distinct libraries with insert sizes that were small (~407 bp), intermediate (~530 bp), or large (~792 bp) (Figures S1 and S2). We assessed uniqueness as a function of coverage depth by sampling from these libraries (either in isolation or pooled), then calculated the read duplication rate (Figures 4A and Figure S3). Combining libraries with different insert sizes added uniqueness, reducing the duplication rate an average of 3.6 fold, and combining libraries with similar insert sizes did almost as well, with 2.7 fold lower accumulation of duplicate reads (Figure S3). This is due to the fact that duplicates are marked only within a library, even if mapped to the same coordinates. The amount of input DNA (and thus the number of unique molecules) influenced duplication rates more than the library preparation parameters that we tested. Using all available data, we observed a 10.7 fold lower duplication rate than in individual libraries. We did not approach saturation of any library, and based on downsampling experiments using all libraries, we estimate that we could produce over 16-fold more WGS coverage (approaching 5,000×) before the overall duplication rate would exceed 50% (Supplemental Experimental Procedures).
A second benefit of multiple libraries is that it allows for a smaller number of amplification cycles, thereby reducing the technical substitution rate by restricting PCR errors to a smaller fraction of the total reads. We were unable to detect any significant library-specific artifacts in our data (see Supplemental Results). Previous studies indicate that such errors are likely to be more of an issue in samples with a limited quantity of nucleic acid, where more PCR cycles are required during library generation (Brodin et al., 2013). It seems likely that the large amount of input DNA used, coupled with our strategies of independent PCR amplifications and multiple libraries resulted in PCR error rates below those detectable from the depth of sequencing that was performed (<0.23–0.35%). We were also unable to evaluate the potential benefits of multiple library sizes on SV detection, since this genome is remarkably free of such events.
Comparison of sequencing assays
To assess the amount of information gained from deep sequencing and comprehensive variant calling, we compared our final results to those obtained from more typical sequencing assays and coverage levels (Figure S13). In each case, we called SNVs with a ‘standard’ calling pipeline implemented in the Genome Modeling System (GMS) (Griffith et al., 2015) consisting of SomaticSniper, VarScan, Strelka, and a number of additional filters (Supplemental Experimental Procedures). The data from targeted capture of 264 AML-related genes sequenced to ~1,200× depth revealed only 4 of 1,343 platinum SNVs. The sensitivity was good within this limited space, and only the very-low VAF (<2%) TP53 and IDH2 mutations were missed. Exome sequencing with a NimbleGen V3 reagent to a high depth of 433× found 30 SNVs from the platinum list, and only 6 that were below 15% VAF. 17 of these were protein-coding mutations, and only 4 of the 9 AML-related gene SNVs were discovered. Exome sequencing failed to detect 37% of the coding SNVs that were detectable in the tumor (mostly at low VAF) and missed 3 SNVs that were found to be expressed in corresponding RNA sequence data. In addition, none of the 9 candidate regulatory mutations we identified were called from the exome data (Table S10). The exome sensitivity within its target space was high, with 30 of 31 platinum mutations with VAFs of >= 2% in the tumor detected (and 30 of 41 overall).
To compare to ‘standard’ depths of 30× and 50× coverage, we randomly downsampled the WGS data to each depth 10 times. (Figure S14). SNVs were called with the same standard pipeline as the exome data, and the platinum variant list was used as the truth set. For this exercise, we considered a variant ‘consistently detected’ if it was identified in at least 50% of the 10 downsampled bam files. At 50× coverage, 95% of SNVs with a VAF of 15% or higher could be consistently detected, but only 10% of SNVs with less than 15% VAF could be consistently found. At 30× coverage, those numbers dropped to 94% (with a >= 15% VAF) and 3% (with a < 15% VAF). The positive predictive values (PPV) were between 63 and 68%, which corresponds to more than 500 false-positive calls in every case.
The above analyses examined variants originally discovered in the ultra-deep WGS data. We also performed de novo somatic variant calling using the 200k Illumina capture data and the same seven SNV callers. (Tables 1, S13, and S14 and Figure S2). Figure S15 and Figure S16 show the overlap between variants from this de novo calling compared to calls from the WGS data and the subset of these making up the platinum list. No variant caller was able to detect all of the platinum variants, even when considering only regions with >50× coverage in both the WGS and capture data. Furthermore, additional de novo variant calling on deeper data revealed a continuing accumulation of variant predictions. Most are expected to be false positives, but a small subset that might be true positives were discovered.
Orthogonal sequencing platforms
We used the Ion Torrent PGM sequencing platform and our custom NimbleGen capture reagent to generate additional sequence data from the normal (6.1 Gbp; 34×), primary tumor (6.1 Gbp; 36× coverage), and relapse tumor (6.6 Gbp; 40× coverage) samples (Tables S15 and S16). Of the platinum SNVs, 76.9% were adequately covered for variant confirmation in the tumor and 47.4% in the relapse samples, defined here as a 95% probability of detecting at least four variant-supporting reads, given the VAF of the variant (using the assumption of a binomial distribution). Using the criteria of at least four variant-supporting reads in the tumor and no more than one in the normal, 98.5% of covered sites in the tumor validated, as did 98.9% of sites in the relapse (Figure S17 and Table S16). None of the sites provided adequate evidence to suggest that any of the Illumina-derived platinum calls were the result of platform-specific artifacts. VAF estimates were consistent between the Illumina and Ion Torrent platforms (Figure S17). We also used a digital droplet PCR (ddPCR) platform to validate the presence and VAF of 15 somatic variants (Figure S8 and Table S9). VAFs from ddPCR and sequencing were highly correlated across a range of VAFs from ~1% to 47% (r2 = 0.992; p-value = 3.553e−15 based on Pearson's product moment correlation coefficient).
Algorithmic assessment
We hypothesized that comprehensive approaches to computational analysis might improve the quality of mutation calling. To evaluate this, we performed alignments of the WGS data with seven different algorithms (Supplemental Experimental Procedures). We then evaluated the coverage and VAF at 198,814 putative SNV sites to identify aligner-specific bias (Figure 4B and Figure S18). Positions with high variance in VAF reported by different aligners revealed regions of the genome that were difficult to characterize by short read sequencing and that represent a potential source of false-positive variant calls from misaligned reads.
Variant calling was also assessed, and sensitivity and PPV were calculated for each of the seven SNV callers, using BWA 0.5.9 aligned reads and the platinum variants as a truth set (Table S13 and S14). The algorithms had sensitivities ranging from 0.798 to 0.958 and PPVs ranging from 0.029 to 0.529 (Figure S13 and Supplemental Results). Notably, several variant callers appeared to miss many hemizygous variants on chromosome X (AML31 is male) that were of high quality by manual review. All callers performed reasonably well on variants at high VAFs, but for variants with less than 10% VAF, no caller had a PPV above 10% (Figure S19). This may be explained by the fact that none of these algorithms were designed specifically for ultra-deep sequence data. Furthermore, we used the authors’ recommended parameters for the evaluation and did not extensively explore parameter optimization (Supplemental Experimental Procedures).
In addition to benchmarking SNV-calling algorithms in isolation, we generated lists using the union and intersection of calls from multiple algorithms to determine whether such an approach could enhance the strengths and hide the weaknesses of particular variant detectors. Though union-based approaches depressed PPVs to unreasonable levels, intersecting multiple callers generally resulted in higher overall performance than using any single caller in isolation (Supplemental Results, Figure S20). The best performing combinations still had considerable room for improvement, with sensitivities of 80–95% and PPV of 75–85%. By plotting all possible combinations of algorithms, we create a ‘best performance’ curve that gives guidelines on which callers may be intersected to give a particular balance of sensitivity and PPV (Figure 4C, Figure S20, Table S17 and S18). We did not investigate more complicated schemes for merging variant calls, but believe that more complex and statistically rigorous methods for doing so could result in better overall performance.
Clonal inference
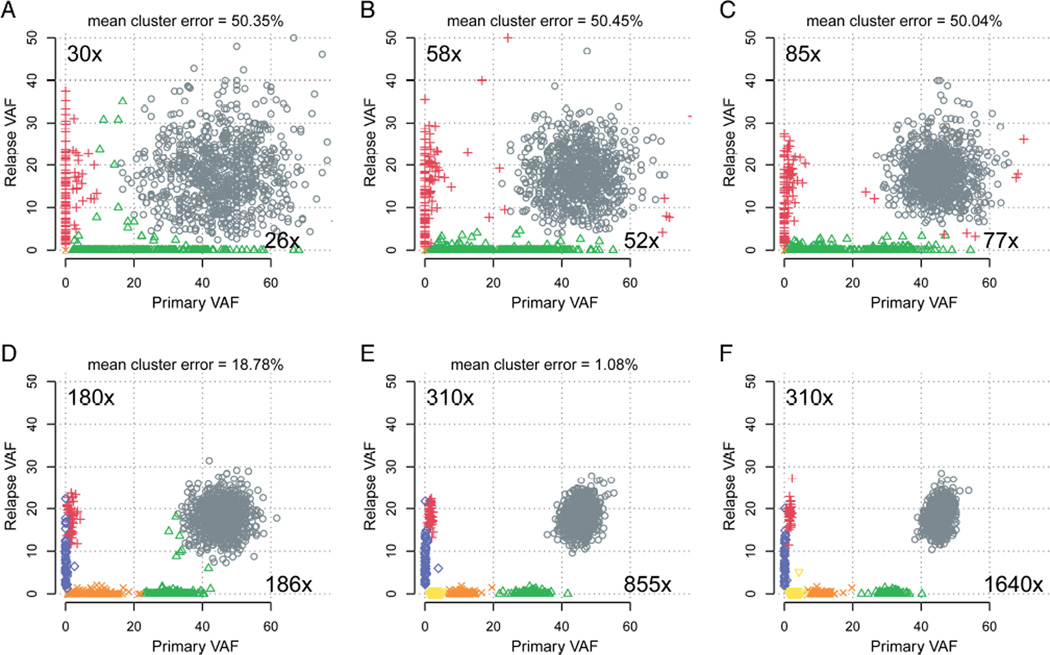
To systematically evaluate the effect of increasing sequencing depth on VAF estimation and inference of clonal architecture, we downsampled all variants from the platinum list to values between 10× and the maximum coverage available (1,640× for primary and 310× for relapse) (Figure 4E, Figure 5, Figure S21, and Video S1). VAFs were calculated for all sites, independently of whether a site could conceivably be called as a variant from that depth. Increased coverage provided more accurate VAFs, due to reduction in sampling error. This resulted in more compact clusters of variants and allowed distinct subclonal populations to be more easily resolved. At 30× depth, SciClone detected only four clusters, with a mean cluster error of over 50% (Miller et al., 2014). While 100× coverage improved the clustering, we did not reliably detect the fifth cluster until 200× coverage, and even then, the mean cluster error is 19.24%, owing largely to the absence of the sixth cluster. To resolve all six clusters, ~850× coverage depth in the primary tumor, and ~310× depth in the relapse was required.
Figure 5. The effect of coverage depth on subclonal inference.
Clustering was performed using read counts from downsampled sequence data at all platinum SNVs. Panels A-E show the results of lower coverage, while panel F uses the ‘core’ validation data. For each cluster in the ‘truth set’ (panel F), the percentage of that cluster’s points correctly assigned was calculated. One minus the mean of these values gives the mean cluster error. An animated version of these plots with additional coverage levels is available as Video S1.
We also explored the ability of different sequencing assays to reconstruct the clonal architecture of this tumor. Figure S13 shows that variants called from deep sequencing of a targeted RMG panel or a very deep exome did not provide enough data points to resolve the clonal architecture of this tumor. While 50× WGS sequencing represented an improvement, it lacked the power to detect the lowest-VAF subclones, and did not unambiguously separate the variant clusters. Though single-cell sequencing is generally limited by throughput, cost, and reduced accuracy due to errors and allelic dropout during amplification, using it in a limited way proved valuable for reconstructing the phylogeny of this tumor. We also performed extensive downsampling experiments to study the effect of depth on the accurate assignment of each individual variant to each subclone (Supplemental Experimental Procedures). At least ~500× coverage was needed before the majority of variants could be reliably assigned to the correct subclone (Figure 5D and Figure S21).
Since 5 of 6 tumor subclones contained mutations in genes with known relevance to AML, we examined the ability of each alternative sequencing and capture platform to detect them (Figure 4F and Figure S22). Some level of detection was generally observed for all nine potential driver mutations in all six sequencing platforms examined. However, poor detection was associated with the low coverage levels achieved by the Ion Torrent platform. Furthermore, low coverage from some other platforms for certain variants, especially subclonal variants and indels resulted in highly variable VAF estimates that would have hindered our ability to assign mutations in key AML genes to specific subclones without combining data across all platforms.
A rich resource for method development and algorithm evaluation
In addition to the data described in our analyses above, we have generated exome and custom targeted sequence data that represents an additional 126 billion bases (Gbp). In sum, we are releasing almost 3 terabases (Tbp) of sequence, representing over 10,000× coverage of some regions of these matched genomes. These raw data are being coupled with a database of analysis results that includes an extensively validated list of somatic variants. The platinum list has been carefully curated to incorporate a balance of information from ultra-deep validation data, multiple alignment algorithms, and manual review. This list is not intended to be comprehensive, as we believe that even more sequence data would uncover additional low-frequency variants, with the logical maximum being detection of mutations unique to a single cell. Though we do not approach this threshold of sensitivity, this list is very specific and includes very high-confidence mutations at variant allele frequencies of 1% or lower. We also provide a ‘gold’ list that includes sites that meet lower-stringency criteria for inclusion, but cannot definitively be ruled out as false positives (Dataset S4). We anticipate that these resources will be used to train the next generation of somatic variant callers, and to facilitate this, we have created an R package that allows users to input a list of variants and receive metrics and visualizations of their tool’s performance. This tool, links to the datasets, and a host of other analysis results can be accessed at http://aml31.genome.wustl.edu/ and as Dataset S5.
Discussion
Comprehensive characterization of tumors is challenging when a tumor has low-cellularity, polyploidy, and/or significant clonal heterogeneity. The sample that we have sequenced in this study represents a best-case scenario, with high purity and nearly no copy number alterations. Despite this, standard sequencing assays failed to capture many of the variants and most of the clonal complexity of this tumor. This lack of power is partially attributable to lack of sequencing depth - with 50× WGS coverage, detecting variants that exist in <= 1% of cells is essentially impossible. Resolving these rare variants is not as simple as simply adding coverage, though, as a number of technical issues contribute to the problem. Difficult to sequence or align regions result in poorly covered regions, including many that have been implicated in disease, such as the CEBPA gene and the TERT promoter (Horn et al., 2013; Huang et al., 2013). Library preparation can also introduce errors, and sequencing instruments do not “read” DNA with perfect fidelity. Though higher depth is sometimes obtained with targeted assays, these often miss key variants in genic or regulatory regions, have limited power to detect structural events or viral integrations, and may not provide enough variants to confidently segregate subclonal populations.
Even when high depth WGS data are obtained, significant challenges remain for effectively analyzing and interpreting these data. We show that there are significant portions of the genome where multiple aligners produce highly discordant results. These may represent problems with the reference genome or algorithmic limitations, but it is clear that they impact variant calling. Though some approaches, often based on a ‘panel of normals’ make help to ameliorate the issues of false positives in these regions, better strategies for identifying and handing these appropriately will be needed. Ultimately, improvements in longer-read sequencing technologies and assembly-based variant calling may help address these challenges.
We also demonstrate that while detecting most low-frequency SNVs is impossible in standard-depth genomes, it remains challenging even with very high depth sequencing. Though simulations based on in-silico mixing have provided some insight (Wang et al., 2013; Xu et al., 2014), such datasets do not represent the true spectrum of biases and experimental noise. By testing variant callers on the real ultra-deep sequence generated here, we find that the parameters and filtering strategies seem to have been heavily over-trained on the expectations of a 30–40× tumor/normal pair. Most tools still perform poorly when trying to identify SNVs in less than 30% of the cells sequenced. As we studied a single sample, and have not comprehensively explored the parameter space of each algorithm, we refrain from describing any SNV caller as ‘best’. Selection of the appropriate algorithm should be driven by each experiment’s design and should include factors like tolerance for false positives and false negatives. Intersecting calls from multiple tools does provide better results and more intelligent merging strategies may offer additional improvement. Additional efforts similar to the ICGC-TCGA DREAM Somatic Mutation Calling Challenge (Boutros et al., 2014), that utilized a large number of deeply sequenced samples to robustly quantify performance under a variety of conditions, might prove useful and spur efforts to optimize existing algorithms and develop new ones. This dataset should provide a rich substrate for those same efforts, by providing a very deeply sequenced, high-confidence set of mutation calls.
Though we discovered many somatic SNVs missed by more typical sequencing approaches, we also identified several thousand putative variants where even more sequence coverage would be necessary to make a confident SNV call. If confirmed, interpreting these low-VAF events will be a significant challenge. Similarly, even more coverage in the primary tumor would be useful for determining whether variants from the relapse were preexisting or acquired after the start of treatment. Given the low background mutation rate and the relatively short time frame, though, it seems quite possible that every mutation we detected was present in the original leukemia. Certainly, all identifiable subclonal driver mutations existed in the original AML sample, suggesting that selection—not chemotherapy-induced mutations—was the primary contributor to the patient’s relapse.
Many factors come into play when deciding how deeply to sequence a tumor, and making a ‘one-size-fits-all’ recommendation would be unwise. However, after comprehensive analysis of the rich and ultradeep resource dataset provided here, we believe we can provide some guidance to those seeking to optimize their tumor sequencing projects (Table 2). WGS is by far the best and most comprehensive strategy available, but the cost of achieving 500–1,000× coverage remains prohibitive with current platforms. A suitable intermediate-term compromise may be to increase the standard depth of tumor WGS to at least 200–300×, which would facilitate discovery of clinically relevant variants at sub-10% VAF. In tandem, it seems prudent to apply targeted sequencing of mutational ‘hotspots’ specific to cancer (or a particular tumor type) to achieve at least 1,000× coverage on potential drivers of therapeutic resistance. The data should be analyzed with a multi-caller, highly sensitive variant discovery pipeline, followed by custom capture of predicted variants and deep sequencing (1,000–10,000×). The complexity and time to produce data for this method would present significant challenges for clinical implementation though, and we suggest that matching the sequencing and analysis approaches to the experimental question are of paramount importance.
Table 2.
Key findings and recommendations for tumor genome sequencing and analysis.
| Sample and case selection |
|
| Matched normal samples |
|
| Library construction |
|
| Sequencing platform |
|
| Sequencing depth |
|
| Whole genome sequencing |
|
| Targeted Sequencing |
|
| Sequence alignment |
|
| Variant calling |
|
| Subclonal inference |
|
| RNA sequencing |
|
| Overall recommended strategy | Sequencing strategy will always be dependent on the goals of the project and budget, but an ideal tumor profiling study might include:
|
Regardless of which sequencing approach is used, it is clear that given the propensity of rare subclones to harbor mutations that contribute to therapy resistance there is an urgent need to become more adept at discovering low-frequency events at presentation. This requires pushing the boundaries of today’s sequencing platforms, developing a better understanding of technical details like error rates and lower bounds of detection, and developing new sets of best practices. The analyses and resources we present here represent a significant step in that direction, and thus will contribute to making targeted cancer therapies guided by genomics more effective.
Experimental Procedures
Sample collection and nucleic acid isolation
Bone marrow biopsy specimens and normal skin samples (Figure 1 and Table S1 for timepoints) were obtained from a single subject who provided written informed consent on a form that contained specific language authorizing WGS and data sharing. This consent procedure was approved by the Washington University School of Medicine Human Research Protection Office (HRPO) on 10/23/06 and renewed annually thereafter. Genomic DNA was prepared by column purification (Qiagen DNeasy).
Discovery sequencing
WGS for the primary AML tumor, relapse tumor and matched normal skin samples was performed using the Illumina HiSeq platform with paired 2×100 bp reads. Each sample was also subjected to exome sequencing using the Roche NimbleGen SeqCap EZ Human Exome Library v3.0. The resulting captured DNA was also sequenced by Illumina 2×100 bp sequencing. Each sample was also captured using a custom set of capture probes (manufactured by IDT) that were designed to tile across 264 genes found to be recurrently mutated in AML (Cancer Genome Atlas Research Network, 2013). Refer to Table 1 and Figure S2 for sequence coverage levels achieved and Figure 1 for an overview of the experimental design.
Sequence alignment and somatic variant calling
WGS, exome and custom capture reads were aligned to the human reference genome (NCBI build 37, ‘GRCh37’) using BWA (v0.5.9) (Li and Durbin, 2009). All reads obtained by custom capture of ~200,000 putative somatic variant sites were re-aligned using additional aligners (Supplemental Experimental Procedures). Prediction of somatic variants was performed primarily using the ultra-deep WGS data from the primary and relapse tumors (bone marrows) compared to a skin normal. Single nucleotide variants, small insertions and deletions, structural variants, and copy number variants were called with multiple algorithms designed for each of these variant types (Supplemental Experimental Procedures).
Design of a custom NimbleGen capture reagent
A total of 371,976 variants were called from the primary tumor or relapse WGS data by at least one of the seven variant callers described in the Supplement. Variants were removed from consideration for design of a capture validation reagent if (a) the minimum WGS coverage in the primary tumor was < 20× or (b) normal coverage was > 50× and the normal VAF was > 20%. This left 359,619 (96.7%) candidate somatic variants. All variants from this list were selected except for those called by Seurat where a random subset (~one-third) were selected (Table S3). A final list of 198,814 unique sites were sent to NimbleGen for design of a validation capture reagent. NimbleGen was able to design targeted probes for 191,988 (96.6%) of these sites. Capture followed by sequencing produced 135,731 (68.3%) sites with coverage > 100× coverage in the primary tumor.
Validation sequencing and analysis (Illumina capture, IDT, RMG)
Validation of predicted somatic variants from the primary and relapse tumors was conducted across multiple independent DNA samples, using multiple capture reagents, orthogonal sequencing platforms, and analysis strategies (Table 1 and Figure 1). Sequencing libraries were created in replicate from multiple aliquots of DNA as well as distinct DNA isolation events. Regions harboring putative somatic variants were enriched for deep validation by use of commercial exome reagents, design of a large custom NimbleGen capture panel (~200k sites), more focused capture reagents made by Integrated DNA Technologies, amplicon sequencing, and digital droplet PCR. Sequencing was performed on the Illumina HiSeq and Ion Torrent platforms. Analysis involved use of comprehensive downsampling experiments, integration of multiple sequence libraries and platforms, integration of DNA and RNA data, integration of data spanning across seven disease timepoints, and extensive manual review of raw data (Supplemental Experimental Procedures).
Sequence data is available through dbGaP accession id phs000159, and are described in more detail on the resource site: http://aml31.genome.wustl.edu
Refer to Supplemental Experimental Procedures for additional methods details.
Supplementary Material
Highlights.
Current sequencing strategies are inadequate given the complexity of most tumors
Current analysis strategies perform poorly, missing rare clinically relevant variants
A comprehensive strategy allows for a more definitive model of clonal architecture
We present a comprehensively sequenced and validated case as a community resource
Acknowledgements
We thank the McDonnell Genome Institute’s LIMS, Analysis Pipeline, and Systems groups for developing and maintaining the automated sequence analysis pipelines. Malachi Griffith was supported by a National Institutes of Health (NIH), National Human Genome Research Institute (NHGRI) grant (K99HG007940). Obi L. Griffith was supported by a National Cancer Institute (NCI) grant (K22CA188163). Christopher A. Maher was supported by NCI grants (R21CA185983 and R00CA149182). This work was funded by grants to Timothy J. Ley from the NCI (PO1CA101937) and to Richard K. Wilson from the NHGRI (U54HG003079).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
M.G. and C.A. Miller led the data analysis. M.G., C.A. Miller, O.L.G., K.K., Z.L.S., A.R., J.R.W., H.X.D., L.T., M.C.W., and D.E.L. performed data analysis and prepared figures and tables. R.T.D., R.E.A., V.M., S.D.M., S.K, M.G.C., C.C.F., A.L, and R.S.F. developed laboratory methods and performed laboratory experiments. J.F.M. contributed ideas and created figures. L.D. contributed ideas. T.J.L. and J.M.K. provided samples and clinical data. M.G., C.A. Miller, C.A. Maher, D.E.L., E.R.M., T.J.L., and R.K.W. developed the project concept and experimental design. D.E.L. provided informatics support. C.A. Miller and M.G. wrote the manuscript with input from O.L.G., K.K., Z.L.S., A.R., D.E.L., J.M.K, and T.J.L.
References
- Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011;21:1498–1505. doi: 10.1101/gr.123638.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, Bryce AH, McCullough AE, Barrett MT, Hunt K, Patel MD, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10:e1004135. doi: 10.1371/journal.pgen.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros PC, Ewing AD, Ellrott K, Norman TC, Dang KK, Hu Y, Kellen MR, Suver C, Bare JC, Stein LD, et al. Global optimization of somatic variant identification in cancer genomes with a global community challenge. Nat Genet. 2014;46:318–319. doi: 10.1038/ng.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin J, Mild M, Hedskog C, Sherwood E, Leitner T, Andersson B, Albert J. PCR-induced transitions are the major source of error in cleaned ultra-deep pyrosequencing data. PLoS One. 2013;8:e70388. doi: 10.1371/journal.pone.0070388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Smith SM, Ramu A, Callaway MB, Brummett AM, Kiwala MJ, Coffman AC, Regier AA, Oberkfell BJ, et al. Genome Modeling System: A Knowledge Management Platform for Genomics. PLoS Comput Biol. 2015;11:e1004274. doi: 10.1371/journal.pcbi.1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin M, Fronick C, Magrini V, Demeter RT, Fulton RS, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25:379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O'Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. doi: 10.1038/ncomms7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Applying next-generation sequencing to pancreatic cancer treatment. Nat Rev Gastroenterol Hepatol. 2012;9:477–486. doi: 10.1038/nrgastro.2012.126. [DOI] [PubMed] [Google Scholar]
- Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, Vij R, Tomasson MH, Graubert TA, Walter MJ, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10:e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieber N, Zapatka M, Lasitschka B, Jones D, Northcott P, Hutter B, Jager N, Kool M, Taylor M, Lichter P, et al. Coverage bias and sensitivity of variant calling for four whole-genome sequencing technologies. PLoS One. 2013;8:e66621. doi: 10.1371/journal.pone.0066621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Jia P, Li F, Chen H, Ji H, Hucks D, Dahlman KB, Pao W, Zhao Z. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 2013;5:91. doi: 10.1186/gm495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, et al. Role of TP53 mutations in the origin and evolution of therapyrelated acute myeloid leukaemia. Nature. 2015;518:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, DiCarlo J, Satya RV, Peng Q, Wang Y. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genomics. 2014;15:244. doi: 10.1186/1471-2164-15-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin SF, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4:157ra143. doi: 10.1126/scitranslmed.3004330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.