Abstract
ALK is a receptor tyrosine kinase with an oncogenic role in various types of human malignancies. Despite constitutive activation of the kinase through gene alterations, such as chromosomal translocation, gene amplification or mutation, treatments with kinase inhibitors invariably lead to the development of resistance. Aiming to develop new tools for ALK targeting, we took advantage of our previous demonstration identifying ALK as a dependence receptor, implying that in the absence of ligand the kinase-inactive ALK triggers or enhances apoptosis. Here, we synthesized peptides mimicking the proapoptotic domain of ALK and investigated their biological effects on tumor cells. We found that an ALK-derived peptide of 36 amino acids (P36) was cytotoxic for ALK-positive anaplastic large-cell lymphoma and neuroblastoma cell lines. In contrast, ALK-negative tumor cells and normal peripheral blood mononuclear cells were insensitive to P36. The cytotoxic effect was due to caspase-dependent apoptosis and required N-myristoylation of the peptide. Two P36-derived shorter peptides as well as a cyclic peptide also induced apoptosis. Surface plasmon resonance and mass spectrometry analysis of P36-interacting proteins from two responsive cell lines, Cost lymphoma and SH-SY5Y neuroblastoma, uncovered partners that could involve p53-dependent signaling and pre-mRNA splicing. Furthermore, siRNA-mediated knockdown of p53 rescued these cells from P36-induced apoptosis. Finally, we observed that a treatment combining P36 with the ALK-specific inhibitor crizotinib resulted in additive cytotoxicity. Therefore, ALK-derived peptides could represent a novel targeted therapy for ALK-positive tumors.
Designing targeted therapy for cancer has been a major goal of the last decade. Oncogenic tyrosine kinases have raised early interest, because elucidation of their structure facilitated the development of small-molecule inhibitors with therapeutic efficiency.1 The pioneer BCR-ABL inhibitor molecule imatinib was approved for therapeutic use as early as 2001 to treat chronic myeloid leukemia and Ph1-positive acute lymphoblastic leukemia.2 Later on, inhibitors targeting receptors for epidermal growth factor or vascular endothelial growth factor were approved for treatment of solid tumors, such as lung and breast cancer. To date, many tyrosine kinase inhibitors (TKIs) are used in the clinic.3 However, cancers treated by TKIs invariably become resistant to therapy and relapse. Acquired resistance develops through various mechanisms including secondary mutations of the targeted oncogene or activation of alternative proliferative signaling pathways.4 It seems thus necessary to invent new strategies designed to attack the tumor on multiple fronts.
ALK (anaplastic lymphoma kinase) is an oncogenic receptor tyrosine kinase associated with many tumor types. ALK was first identified in 1994 as a rearranged gene fusion (NPM–ALK) resulting from the t(2;5)(p23;q35) translocation occurring in 75% human anaplastic large-cell lymphomas (ALCLs).5, 6 Other translocations or gene inversions involving ALK were later described in solid tumors including 50–60% inflammatory myofibroblastic tumors, and a small proportion of diffuse large B-cell lymphomas, breast and renal carcinomas.7, 8 Recently, 4–8% non-small-cell lung cancer (NSCLC) were found to harbor an echinoderm microtubule-associated protein-like 4 (EML4)–ALK fusion.7, 9 Resulting fusion proteins associate the N-terminal portion of a protein partner (containing in most cases a dimerization domain) to the entire intracellular portion of ALK, including its tyrosine kinase domain. Subsequent dimerization of this fusion protein leads to constitutive activation of ALK kinase, resulting in enhanced signaling for cell proliferation, survival and oncogenicity.10
The full-length ALK receptor cDNA codes for a transmembrane receptor tyrosine kinase of the insulin receptor superfamily, which is essentially expressed in the developing nervous system.11, 12 Some authors proposed the two heparin-binding factors pleiotrophin (PTN) and midkine as ligands for ALK.10 However, their binding to ALK is controversed and might be indirectly mediated by heparin.13 ALK kinase signaling most likely involves co-receptors and/or co-signaling molecules such as the transmembrane receptor tyrosine phosphatase beta/zeta (RPTPb/z), a receptor for PTN and midkine. In the absence of ligand, RPTPb/z dephosphorylates ALK, whereas PTN and midkine direct binding to RPTPb/z inactivates its phosphatase activity.14 Expression of the full-length ALK receptor was also observed in neuroblastoma, a pediatric tumor derived from the neural crest affecting the peripheral nervous system. The ALK kinase in neuroblastoma is most often constitutively active as a result of gain-of-function mutations or protein overexpression, due to ALK gene amplification or copy number increase.10, 15
ALK appears therefore as an interesting therapeutic target to treat ALK-positive tumors. Indeed, since the identification of NPM–ALK and other ALK fusions as oncogenes for ALCL and inflammatory myofibroblastic tumors,6, 16, 17 several pharmaceutical companies developed ALK-specific TKIs. In 2010, a TKI targeting ALK and c-MET, crizotinib18 (also called PF-02341066), was authorized in clinical trials as a second-line therapy for advanced stage NSCLC harboring EML4–ALK. The initial clinical responses were so encouraging that crizotinib is currently tested in a growing number of advanced ALK-positive tumors (clinicaltrials.gov). Nevertheless, the tumors invariably develop resistance to the inhibitor, mostly through mutations of the kinase active site.19, 20 Therefore, it appears necessary to design alternate treatments or to associate TKIs with other molecules. One promising strategy would be to impair distinct functions of the oncogenic tyrosine kinase through targeting different sites of the ALK protein.
We recently demonstrated that the ALK receptor tyrosine kinase belongs to the functional family of so-called ‘dependence receptors'.21, 22 Such dependence receptors function with a dual signaling: in the presence of ligand (or a situation mimicking a ligand, e.g., inducing receptor dimerization and activation), the receptor exerts a prosurvival/antiapoptotic effect on the cell; in contrast, in absence of ligand and when the cell is submitted to environmental or genotoxic stress, a dependence receptor becomes proapoptotic. The proapoptotic effect is mediated by caspase-dependent cleavage of the receptor, either releasing or exposing a proapoptotic domain/sequence (called ‘addiction/dependence domain' or ADD), thus amplifying the apoptotic process.23 Molecular analysis of ALK deletion mutants allowed us to map the ADD domain of ALK to a 36-amino-acid (aa) stretch located within the juxtamembrane intracytoplasmic region of ALK. The ADD of ALK lacks homology with any known protein motif implicated in apoptotic processes and is necessary for ALK proapoptotic function.22 The purpose of the present study was to design a novel targeted therapy, taking advantage of the proapoptotic function of ALK.
Our hypothesis was that a synthetic peptide could mimic the proapoptotic function of ALK. Therefore, we synthesized several peptides whose sequence reproduced the entire ADD domain (36 aa) of ALK or part of it (12 aa) to assay their effects on various tumor cell lines. We show that several of these ALK-derived peptides are proapoptotic for ALK-expressing, but not ALK-negative, tumor cells. In addition, the ALK-derived 36-aa peptide (P36) enhanced the cytotoxic effect of the ALK kinase inhibitor crizotinib in ALK-positive ALCL and neuroblastoma cell lines. Thus our results uncover a new strategy for targeting ALK-expressing tumors.
Results
Design of peptides derived from the ADD of ALK
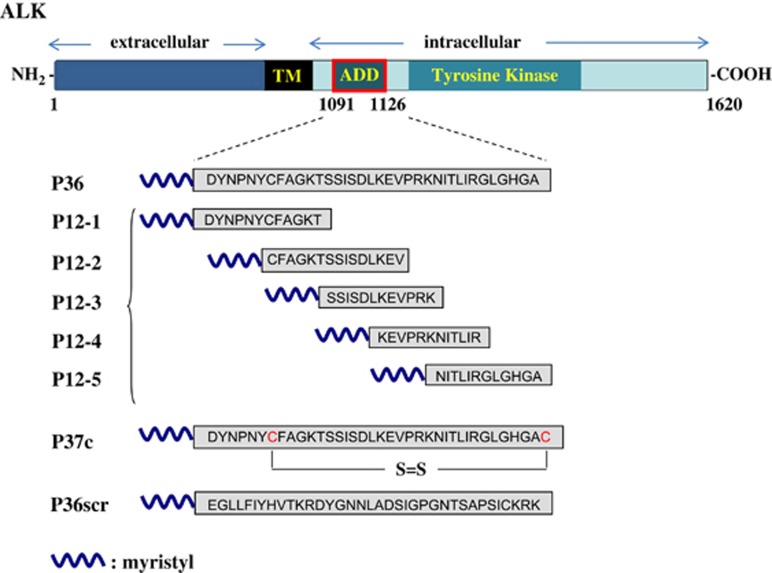
All the peptides are schematically represented in Figure 1. We first designed a 36-aa peptide (P36), mimicking the whole ADD domain of ALK,22 to assay its potential to kill tumor cells. P36 was identical to the original ALK sequence 1091–1126 (accession: NP_004295.2), whereas P36scr (for ‘scrambled') served as a control. To stabilize P36 structure, a cyclic peptide (P37c) was also designed. It comprised P36 sequence with an additional C-terminal cysteine creating a disulfide bridge with cysteine 1097 within the sequence. Finally, to try and identify smaller active molecules, five overlapping peptides of 12 aa spanning the P36 sequence (P12-1 to P12-5) were synthesized. All the peptides were myristylated at the N terminus to facilitate cell penetration.24
Figure 1.
Design of peptides derived from the ADD (addiction/dependence domain) domain of ALK. The ALK receptor is composed of an extracellular, a transmembrane (TM) and an intracellular region with a tyrosine kinase domain. ALK has been identified as a dependence receptor, with both antiapoptotic and proapoptotic functions. An ADD has been localized in the juxtamembrane portion of the intracellular region (aa 1091–1126). Peptides of 36, 12 or 37 aa (circularized through an S–S bond) that mimicked the whole or partial sequence of the ADD domain were synthesized with a myristyl at their N terminus
The ALK-derived peptide P36 is proapoptotic in ALK-positive ALCL and neuroblastoma cell lines
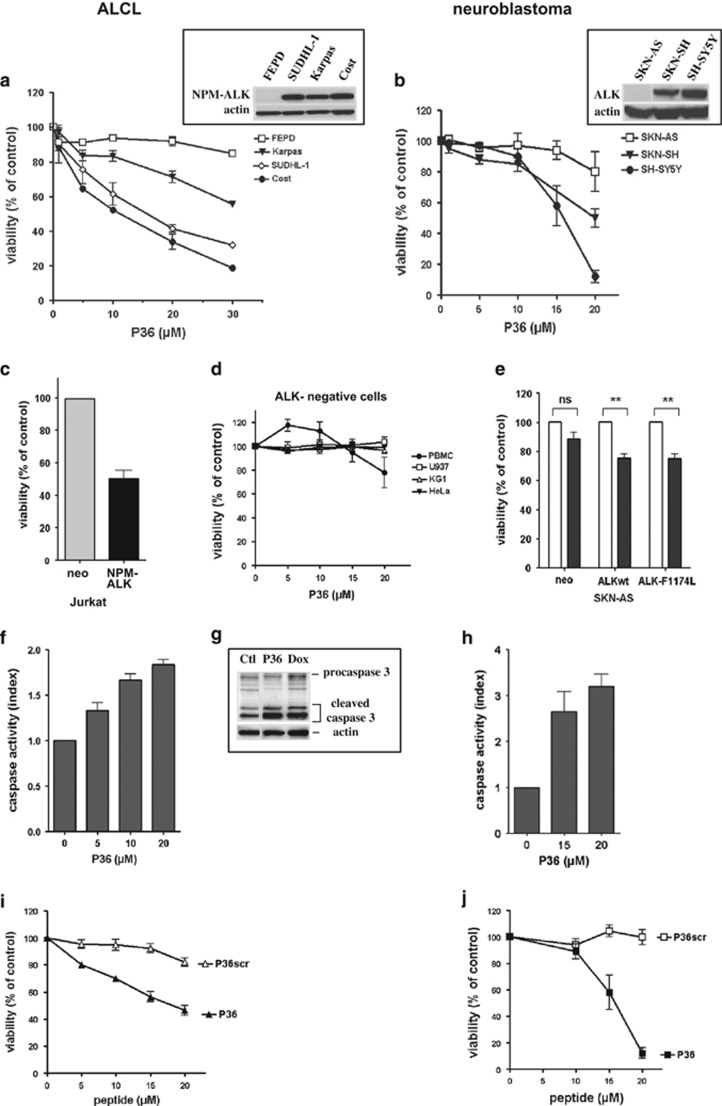
The effect of P36 was first assayed on ALCL and neuroblastoma, which harbor cases with and without oncogenic ALK. Three ALCL lines carrying the NPM–ALK fusion (Karpas 299, SUDHL-1 and Cost) and one ALK-negative ALCL (FEPD) were incubated with increasing doses of P36 for 48 h. Similarly, two neuroblastoma lines carrying the ALK-F1174L mutation (SKN-SH, SH-SY5Y) and one ALK-negative neuroblastoma (SKN-AS) were treated with P36. In contrast to ALK-negative cells, ALK-positive ALCL and neuroblastoma cells responded to P36 in a dose-dependent manner, showing a decrease of the number of viable cells compared with their untreated control counterpart (Figures 2a and b). This ALK dependence of P36-induced cytotoxicity was further demonstrated in oncogenic ALK-transfected Jurkat25 and SKN-AS cells (Supplementary Figure 1), and other ALK-negative normal or tumor cells (Figures 2c–e).
Figure 2.
A peptide (P36) derived from the ADD domain of ALK specifically induces apoptosis in ALK-positive tumor cells. Cells were incubated with various concentrations of P36 for 48 h, or with culture medium without peptide for the control. Top panels: effect of P36 on the viability of ALK-positive and -negative ALCL (a) and neuroblastoma (b) cell lines. Insets: western blot analysis of ALK expression in the cell lines. Actin detection was used as a loading control. (c–e) Specificity of P36-induced apoptosis for ALK-expressing cells. ALK-negative normal (PBMC) or tumor (U937, KG1, HeLa) cells are insensitive to P36 (d). ALK-negative cells stably transfected with ALK variants (NPM–ALK in Jurkat (c), ALKwt or mutant ALK-F1174L in SKN-AS (e)) or an empty vector (neo) control were incubated with 5 μM (Jurkat) or 15 μM (SKN-AS) P36 and their viability assessed at 48 h. (f–h) P36 induces caspase-dependent apoptosis in ALK-positive cells. Cost ALCL (f) or SH-SY5Y neuroblastoma (h) cells were incubated with various doses of P36 for 48 h. Caspase 3/7 activity was measured and expressed as an index relative to control untreated cells (f and h). An increased caspase 3 cleavage was detected by western blot in Cost cells (g) treated for 6 h with P36 (10 μM) or doxorubicin (Dox, 2 μM) compared with culture medium (ctl). (i and j) Specificity of P36 cytotoxic effect on Cost ALCL (i) and SH-SY5Y neuroblastoma (j) compared with a scrambled version (P36scr) of P36. Values in all graphs represent the mean ±S.E.M. from at least three independent experiments. NS, not significant, **P<0.01, statistical test: two-way analysis of variance (using Graphpad Prism 4 software)
Next, we sought to determine whether the decrease of viable cells induced by P36 was due to apoptosis. Incubation of Cost ALCL and SH-SY5Y neuroblastoma cells with P36 resulted in increased caspase 3/7 activity and cleaved forms of caspase 3, indicating that P36 induced caspase-dependent apoptosis in these cells (Figures 2f–h). P36 proapoptotic effect was sequence specific, as P36scr failed to affect P36-responsive cells (Figures 2I and j).
Taken together, our results indicate that the ALK-derived P36 peptide specifically induces apoptosis in ALK-expressing tumor cells.
Effect of different ALK-derived peptides on cell viability
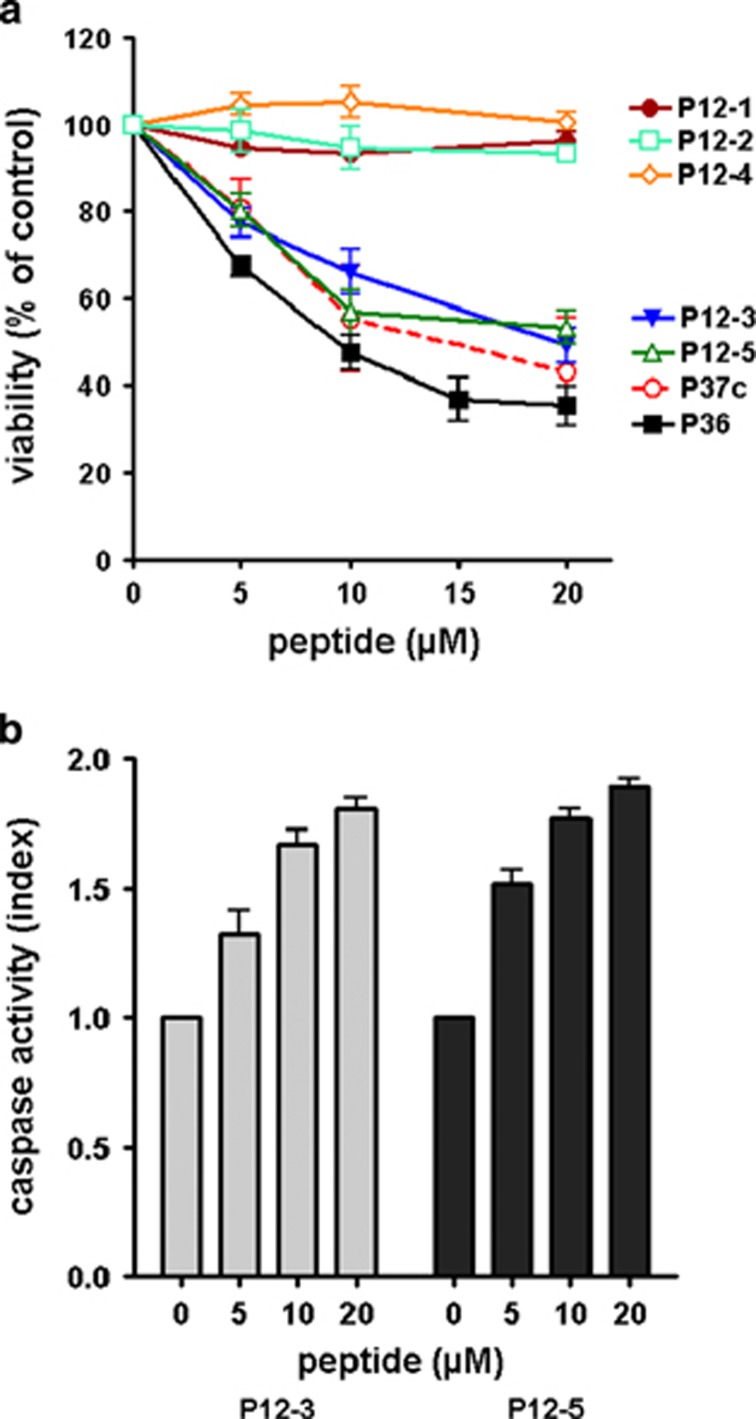
To determine whether a smaller motif within the ADD of ALK could be proapoptotic, we tested the effect of five overlapping peptides of 12 aa (P12-1 to P12-5, Figure 1) on Cost cells. P12-3 and P12-5 displayed a dose-dependent cytotoxicity, whereas P12-1, P12-2 and P12-4 had no effect. The cyclic peptide, P37c, was also cytotoxic for Cost cells (Figure 3a). Similarly to P36, P12-3 and P12-5 increased caspase-dependent apoptosis (Figure 3b). However, we found P36 was slightly but significantly more effective than P12-3, P12-5 or P37c, respectively (P<0.05, Student's t-test, Figure 3a). This result led us to preferentially use P36 in further work.
Figure 3.
Cytotoxicity due to caspase-dependent apoptosis is induced by different peptides derived from the ADD domain of ALK. (a) Dose–response curves of different peptides on Cost cell viability. (b) Caspase 3/7 activity of P12-3 or P12-5-treated Cost cells. Results are the mean ±S.E.M. from at least three independent experiments
P36 peptide proapoptotic effect depends on myristoylation
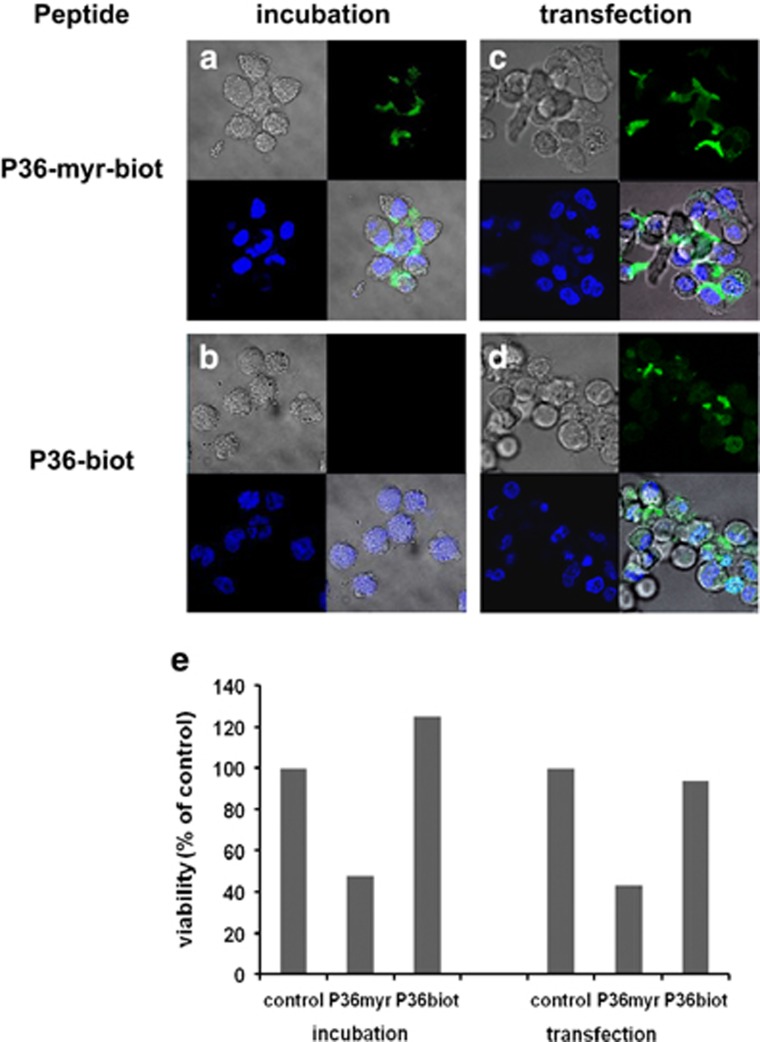
To determine the mechanisms implicated in P36-induced apoptosis, we first investigated whether P36 was internalized. Living cells were incubated for 1 h at 37 °C with a biotin-labeled N-myristylated P36 (P36-myr-biot), then fixed, permeabilized and stained with fluorescent streptavidin. A non-biotinylated peptide was used as negative control. P36-myr-biot was detected in the cytoplasm of both Cost ALCL (Figure 4a) and SH-SY5Y neuroblastoma cells (Supplementary Figure 2), but not in the nucleus. In contrast, cells incubated with a biotin-tagged non-myristylated P36 (P36-biot) did not display any fluorescence (Figure 4b), suggesting the myristyl was necessary for cell membrane insertion and penetration. To address this hypothesis, Cost cells were transfected with the biotin-coupled peptides using an Amaxa Nucleofector device and examined after 15 min incubation. With this alternate method, the non-myristylated P36-biot was delivered intracellularly as efficiently as P36-myr-biot (Figures 4c and d). To elucidate the respective roles of cell internalization and peptide myristoylation in the proapoptotic effect of P36, we compared the viability of cells treated with different peptides for 48 h following incubation or transfection. Results show that solely the myristylated P36 peptide was proapoptotic in both incubation and transfection conditions (Figure 4e). Of note, smaller myristylated ALK-derived peptides P12-3 (active) and P12-1 (inactive) were also internalized by cells (Supplementary Figure 2 and data not shown). These data demonstrate that the proapoptotic effect of active ALK-derived peptides depends on their myristoylation.
Figure 4.
Myristoylation is required for P36 proapoptotic effect in ALK-positive tumor cells. Cost ALCL cells were either incubated 1 h at 37 °C (a and b) or transfected using an Amaxa Nucleofector device (c and d) with biotin-coupled P36, either N-myristylated (P36-myr-biot) or unmyristylated (P36-biot). The peptides were visualized with streptavidin-FITC after cell fixation and permeabilization. Four-panel composites show Nomarski interference contrast (gray), peptide (green), cell nuclei (DAPI, blue) or a merge of all three (bottom right panels). (e) Cell viability following incubation or transfection with peptides was assayed after 48 h of treatment. The graph represents one of two independent experiments
P36 peptide proapoptotic effect is p53 dependent
To further explore the mechanism of P36-induced apoptosis, we conducted a search for P36 interaction partner proteins using surface plasmon resonance followed by mass spectrometry analysis. Biotinylated P36 was fixed onto a Biacore streptavidin-coated gold chip and cell protein extracts from Cost or SH-SY5Y peptide responder cells were allowed to bind onto the chip. Eluted bound proteins were analyzed by mass spectrometry. We identified a total of 13 proteins from Cost ALCL (Supplementary Table S1) and 179 proteins from SH-SY5Y neuroblastoma (Supplementary Table S2) that bound to P36, respectively. Assuming peptide cytotoxicity in ALCL and neuroblastoma cells might share a common mechanism, it seemed relevant to explore which P36-bound proteins were common to Cost and SH-SY5Y cells (Table 1). Six proteins were identical in both cell lines (i.e., SUB1, SSBP1, HRNR, XRCC5, EEF1A1 and LUC7L2). Interestingly, five of these structurally unrelated proteins have been reported to interact with the p53 tumor suppressor gene or protein.
Table 1. List of P36-interacting proteins that are identical or similar in Cost ALCL and SH-SY5Y neuroblastoma cell lines.
| Cell lines | Protein symbol | Cell process and function |
|---|---|---|
| Interaction with the p53 gene/protein or p53-dependent signaling pathways | ||
| Cost and SH-SY5Y | SUB1 (PC4) | Transcriptional coactivator for p53, neural gene silencing |
| Cost and SH-SY5Y | SSBP1 | p53-dependent DNA damage |
| Cost and SH-SY5Y | HRNR | Skin protein, member of S100 family, elevated in various tumors |
| Cost and SH-SY5Y | XRCC5 (KU80) | RNA helicase, repair of DNA ds breaks by NHEJ and V(D)J recombination |
| Cost and SH-SY5Y | EEF1A1 | Elongation factor, protein biosynthesis |
| Regulation of pre-mRNA splicing | ||
| Cost and SH-SY5Y | LUC7L2 | Pre-mRNA splicing, recognition of non-consensus splice donor sites |
| Cost | LUC7L3 | Pre-mRNA splicing, formation of spliceosome |
| Cost | SFRS3 | Pre-mRNA splicing, neural differentiation, proto-oncogene |
| Cost | SFRS11 | Pre-mRNA splicing, neural differentiation |
| Cost | DDX46 | RNA helicase, pre-mRNA splicing |
| SH-SY5Y | DDX5 | RNA helicase, p53-dependent DNA damage and apoptosis, neural differentiation |
| SH-SY5Y | DDX39 | RNA helicase, pre-mRNA splicing |
| SH-SY5Y | SFRS2 | Pre-mRNA splicing, cisplatin-mediated apoptosis |
| SH-SY5Y | SFRS7 | Pre-mRNA splicing, neural gene silencing |
| SH-SY5Y | U2AF1 | Pre-mRNA splicing, spliceosome |
| SH-SY5Y | U2AF2 | pre-mRNA splicing, spliceosome |
P36-interacting proteins were classified in two functional families: proteins potentially interfering with p53 tumor suppressor, and mRNA splicing factors. Information on genes' function was retrieved from Genecards (www.genecards.org)
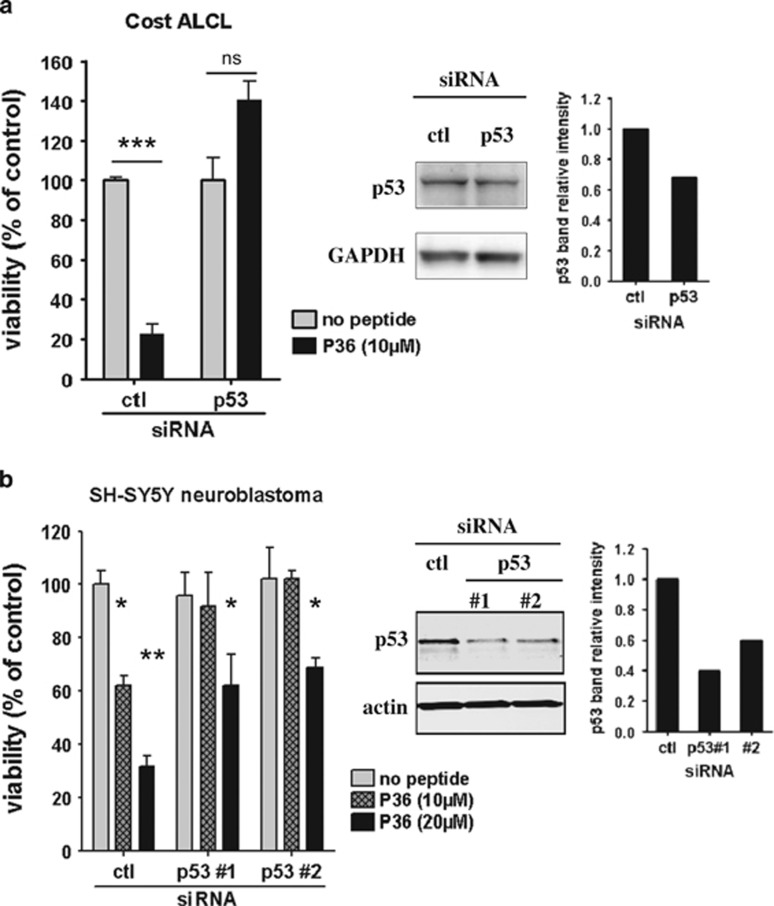
We next asked whether p53 knockdown would affect P36-induced apoptosis in ALK-positive cells. Cost and SH-SY5Y were preincubated with p53-specific or control siRNAs for 48 h and then treated with P36 for a further 48 h. Preincubation of Cost ALCL with a pool of four p53-specific siRNAs only achieved a 30% reduction of p53 protein expression (Figure 5a, right panel), whereas in SH-SY5Y cells, two distinct siRNAs reduced the p53 expression level to 40% and 60% of control, respectively (Figure 5b, right panel). Although partial, p53 downregulation clearly rescued both Cost ALCL and SH-SY5Y neuroblastoma cells from P36-mediated cytotoxicity (Figures 5a and b). These results demonstrate that the proapoptotic activity of P36 is p53 dependent.
Figure 5.
P36-induced apoptosis in ALK-positive tumor cells is p53 dependent. Cost ALCL (a) or SH-SY5Y neuroblastoma (b) cells were incubated with p53-specific (a pool of four siRNAs for Cost or two distinct siRNAs for SH-SY5Y) or non-targeted control pool (ctl) siRNAs for 48 h before addition of P36 or culture medium (no peptide) for another 48 h. Cell viability (left) and western blot analysis of p53 expression (middle) were assayed at that time. The intensity of p53 bands relative to loading controls (GAPDH, actin) was quantified by image analysis (graphs on the right). Results display one of two representative experiments with triplicate samples. Statistical test: Student's t-test. NS, non significant; *P<0.05; **P<0.01; ***P<0.001
Combined effects of P36 and crizotinib in ALK-positive tumor cells
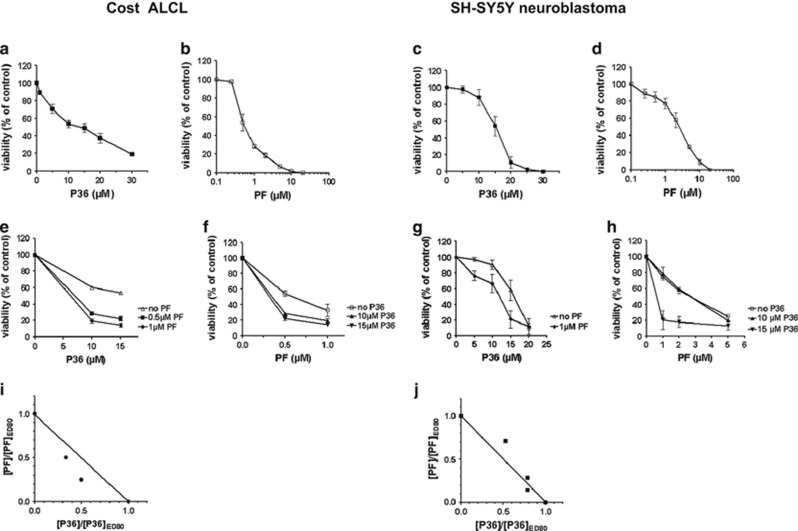
A promising therapeutic approach for ALK-expressing tumors is a targeted therapy using ALK kinase inhibitors. Crizotinib (PF-2341066, abbreviated ‘PF' in the text) is a dual kinase inhibitor for ALK and c-MET that is highly cytotoxic for ALK-positive ALCL cells in vitro and in vivo.18 To determine whether crizotinib (PF) and P36 could synergize in ALK-positive tumor cells, we examined the effect of single drug and combination treatments on the viability of Cost ALCL and SH-SY5Y neuroblastoma cells after 48 h exposure. To ensure an equivalent contributing effect from each drug, a dose–response was established for P36 and PF separately and the IC50 for each drug was determined (Cost cells: 10 μM for P36 and 0.5 μM for PF, respectively; SH-SY5Y cells: 16 μM for P36 and 2.5 μM for PF, respectively; Figures 6a–d). We then used a constant dose of either P36 or PF close to the IC50 while varying the dose of the other drug. Figures 6e–h show the combination of PF and P36 was always more efficient than each drug alone within the dose range assayed. To determine whether the effect of combined P36 and PF was synergistic or additive, normalized isobolograms were drawn (Figures 6i and j). In this type of analysis, points located on the straight line represent an additive effect for a given effective dose (ED), thus experimental data points that are above or under the straight line indicate an antagonist or synergistic effect, respectively.26 Figures 6i and j show at combined doses eradicating 80% of the viable cells (ED80), the killing effect of P36 and PF was synergistic in Cost ALCL and additive in SH-SY5Y neuroblastoma cells, respectively.
Figure 6.
Combined cytotoxic effect of P36 and PF on ALK-positive tumor cells. Cost ALCL (left) or SH-SY5Y neuroblastoma (right) cells were incubated for 48 h with increasing doses of P36 (0–30 μM) or PF (0–20 μM) or a combination of both drugs as indicated. (a–d) Dose–response curves of P36 and PF on Cost (a and b) and SH-SY5Y (c and d) cell viability, respectively. (e–h) Viability curves for Cost (e and f) and SH-SY5Y (g and h) treated with a constant dose of either P36 or PF close to the IC50 while varying the dose of the other drug. (i and j) Normalized isobolograms drawn for ED80 indicate a synergistic trend (i, Cost) or an additive effect (j, SH-SY5Y) of P36 and PF combination
P36 does not affect ALK phosphorylation
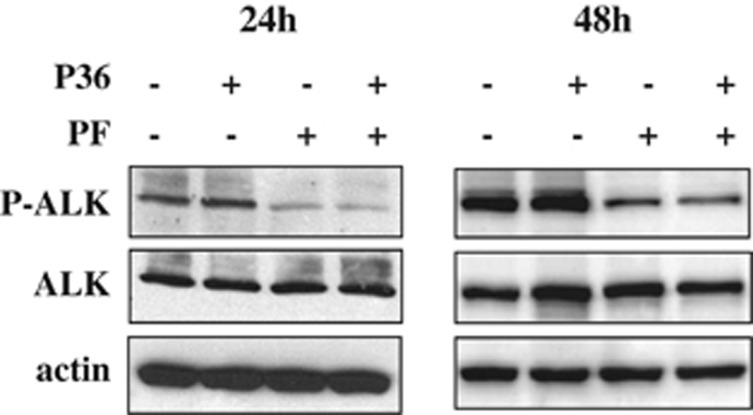
The finding that P36/PF combination induced at least additive cytotoxicity in both ALCL and neuroblastoma cells suggested the two agents could act on different pathways. We thus examined the level of ALK tyrosine phosphorylation of Cost cells incubated for 24 or 48 h with P36, PF or both drugs (Figure 7). As expected, the kinase inhibitor PF decreased ALK phosphorylation. In contrast, cell treatment with P36 did not affect the level of ALK phosphorylation. Moreover, combining PF and P36 did not enhance PF-induced dephosphorylation. Note that the expression level of total ALK protein during P36 treatment did not vary, indicating the protein was not degraded. Taken together, our results indicate the mechanism of P36 cytotoxicity is not mediated by kinase inhibition.
Figure 7.
Different effects of P36 and PF on ALK phosphorylation. Western blot analysis of ALK phosphorylation in Cost cells treated for 24 or 48 h with P36 (10 μM), PF (0.5 μM) or both
Discussion
ALK drives tumorigenesis in several types of cancers, including ALCL and neuroblastoma. ALK-targeted therapy using specific inhibitors such as crizotinib has shown promising results, but inevitably associates with resistance. To overcome this issue, we devised a strategy based on the dependence receptor properties of ALK, implying that in the absence of ligand or kinase activation, ALK is proapoptotic.21, 22 Using synthetic peptides to mimic the ALK proapoptotic domain (ADD),22 we showed human ALCL and neuroblastoma tumor cells were killed in a dose-dependent manner following incubation with an ADD-like peptide (P36). Cell death was due to caspase-dependent apoptosis. Cell sensitivity to the peptide was dependent on ALK expression by tumor cells as: (i) ALK-negative tumor cells failed to respond and (ii) expressing either NPM–ALK or full-length ALK restored P36-responsiveness of lymphoma or neuroblastoma cells, respectively. Unfortunately a loss-of-function approach, that is, silencing ALK by siRNA to confirm the requirement of ALK expression for P36-mediated apoptosis was not applicable. Indeed, ALK-expressing tumors present an oncogene addiction and die by apoptosis following ALK downregulation.27, 28 Moreover, despite different subcellular localization of oncogenic ALK forms (nuclear and cytoplasmic for NPM–ALK,25 cytoplasmic and at the plasma membrane for ALK,29 respectively) (Supplementary Figure 1), P36 killed both ALCL and neuroblastoma cells. Worthy of note, P36 was not toxic for normal peripheral blood mononuclear cells (PBMCs), which do not express ALK, making P36 a potential therapeutic agent.
Peptides are an attractive therapy due to relatively easy synthesis. To minimize protease degradation, cyclization can be used to strengthen peptide structure. Here, we showed P37c, like P36, was proapoptotic. Another challenge resides in delivering peptides at the right site, that is, tumor cells. We chose to add an N-myristyl tag because it reportedly facilitates insertion in the plasma membrane and subsequent peptide translocation into cells.24, 30 Surprisingly, the N-myristyl was not only necessary for cellular uptake but also for the cytotoxic activity of ALK-derived peptides. Note that many cell signaling molecules are N-myristylated. Myristoylation directs these proteins to specific cell compartments (e.g., the Golgi apparatus, endoplasmic reticulum or mitochondria), impacting their function.31 Moreover, the proapoptotic domain of ALK receptor is located close to the plasma membrane. Thus, N-myristoylation likely has a role in peptide subcellular localization and proapoptotic signaling. However, in vivo peptide delivery may require additional coupling, for example, to nanoparticles,32 and we may consider this approach in further work.
To our knowledge, this is the first time that synthetic peptides derived from the ADD domain of a dependence receptor are used to target and kill tumor cells. Alternative approaches using the proapoptotic potency of dependence receptors are also currently developed. In aggressive tumors coexpressing netrin-1 and its receptors DCC and UNC5H, a double therapeutic approach was designed: (i) overexpression of the intracellular domain (including the ADD) of the receptors, and/or (ii) synthesis of soluble decoy receptors used as ligand traps for autocrinally secreted netrin.33, 34, 35 Both strategies interfere with netrin-1 ability to inhibit DCC- or UNC5H-induced cell death. These treatments killed netrin-1-expressing tumor cells both in vitro and in vivo. However, the latter approach is only feasible for a receptor expressed on the cell surface, which is not the case for NPM–ALK or other ALK-associated fusions, therefore justifying our approach to use peptides designed to enter tumor cells.
Some authors have designed hybrid molecules coupling penetrating peptides derived from tat or antennapedia to proapoptotic nonspecific peptides such as BCL-2 family domain 3-mimetics.36 The originality of ALK-derived peptides described in this report is their specificity for ALK-expressing tumors. It would certainly be interesting to develop ALK-ADD peptide-mimetic small molecules in the future.
The fact that P36 only killed ALK-expressing cells suggested the peptide would likely interfere with ALK signaling. P36 may either antagonize ALK dependence receptor antiapoptotic or enhance proapoptotic signaling. Perhaps P36-mediated cytotoxicity requires binding of endogenous ALK. The chaperone Hsp90 was reported to protect ALK from degradation by the proteasome.37, 38 On the basis of the detection of Hsp90 in the proteomic analysis (Supplementary Table S2), P36 might sequester Hsp90 thus favoring ALK degradation and apoptosis. However, the lack of evidence for ALK degradation upon P36 cell treatment (Figure 7) did not support this hypothesis. A more likely mechanism would imply the titration of regulatory adaptors bound to the proapoptotic ALK domain. These propositions need to be further elucidated.
Cross-analysis of the P36 interactome of ALCL and neuroblastoma cells yielded six proteins susceptible to interact with the tumor suppressor p53, whereas the remaining proteins were involved in pre-mRNA splicing.
The presence of a significant number of pre-mRNA splicing factors among P36-interacting proteins was striking. Alternative splicing is an important regulator of apoptosis, as it determines the balance of antiapoptotic and proapoptotic isoforms of numerous gene products including caspases and members of the BCL-2 and p53 families.39 Therefore, sequestration of splicing factors through interaction with P36 could possibly impair the balance of antiapoptotic and proapoptotic signals.
Another feature was the ability of several P36-interacting proteins to interact with the tumor suppressor p53 or regulate its expression.40, 41, 42, 43, 44, 45, 46, 47 These interactions could modulate p53-dependent apoptosis or DNA damage response at the mitochondrial and/or nuclear level, respectively. Our finding that p53 downregulation (although partial) could rescue ALK-positive cells validated the role of p53 in P36-induced apoptosis. Moreover, P36 (in the absence of siRNA) did not increase p53 protein expression (data not shown), suggesting P36-induced apoptosis may involve a p53-dependent mitochondrial pathway.48
An interesting protein in our proteomic analysis was SUB1(PC4), a multifunctional nuclear protein having important roles in DNA transcription, replication and repair.49 PC4 interacts with p53 and is also regulated by p53. A recent study reported PC4 protein was upregulated in NSCLC carcinoma tissues. Moreover, PC4 knockdown induced growth arrest and apoptosis of NSCLC cell lines.50 Thus PC4 could act as an antiapoptotic protein. In the present report, we speculate P36 could sequester PC4 in the cytoplasm thus preventing it to interact with its nuclear target p53.
The recent development of ALK-specific TKIs brought hope for chemotherapy-resistant or relapsing ALK-expressing tumors. Crizotinib treatment could extend disease-free survival for ALK-positive NSCLC, ALCL and neuroblastoma.51, 52, 53 However, TKI resistance developed mostly due to secondary mutations.4, 54 Interestingly, acquired mutations affected the same hotspots (e.g., F1174) as those described in primary neuroblastomas.19, 20 Herein, we showed the F1174L-expressing SH-SY5Y and transfected SKN-AS neuroblastoma cells were sensitive to P36. This is in contrast with the reported resistance of this mutant to crizotinib.20, 55 Along the same line, we observed a fivefold difference between the IC50 of PF in SH-SY5Y neuroblastoma versus Cost ALCL, whereas the IC50 of P36 was in the same range for both cell lines (Figure 6).
Taken together, these observations brought evidence to associate two ALK-specific targeted drugs, P36 and PF, to enhance cytotoxicity. Whereas TKI-mediated killing implied inhibition of ALK phosphorylation, P36 cytotoxic effect involved a different, although presently unknown, mechanism. Accordingly, we observed an additive effect of TKI/ P36 combination in both ALK-positive ALCL and neuroblastoma.
Collectively our findings are novel and should contribute to better understand the mechanism of P36 proapoptotic activity. The proteomic analysis uncovered a role for p53, and further studies are in progress to establish which proteins have a direct or indirect role in the proapoptotic effect of P36 and how they act.
In conclusion, peptides derived from the ADD domain of ALK may represent a new therapeutic approach in ALK-expressing tumors. These peptides could potentially be useful both in tumors expressing ALK fusion proteins and the full-length ALK receptor. The enhancement of P36-mediated cytotoxicity when in association with an ALK TKI could be especially valuable in targeting tumors bearing ALK rearrangements or activating mutations acquired following a first-line treatment.
Materials and Methods
Reagents
All chemicals were from Sigma (www.sigmaaldrich.com) and culture reagents from Gibco, (www.invitrogen.com).
Peptides
Peptides of 36 (P36 and P36scr), 37 (P37c) and 12 (P12-1, P12-2, P12-3, P12-4, and P12-5) aa, respectively, were synthesized by 9-fluorenylmethyloxycarbonyl (Fmoc) solid-phase chemistry by Covalab (Villeurbanne, France) and purified by reverse-phase high-performance liquid chromatography (HPLC). Their purity (75–95%) was verified by HPLC and mass spectrometry. All the peptides (unless mentioned) were synthesized with a myristyl group at the N terminus and their C terminus was amidated. For visualization studies, peptides were coupled to biotin at the C terminus. Stock solutions of peptides were prepared in distilled water at 2.5 mM (P36, P36scr, P37c), 5 mM (P12-3, P12-4, P12-5) or in DMSO at 20 mM (P12-1, P12-2), sonicated, aliquoted and stored at −80 °C.
ALK inhibitor
Racemic PF-2341066 [3-[1-(2,6-dichloro-3-fluoro-phenyl)-ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)-pyridin-2-ylamine] (crizotinib) was synthesized according to the method described in the patent international application WO 2006/021881.
Cell culture
Cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. SU-DHL1, Karpas 299, Cost and FEPD ALCL cell lines56, 57, 58, 59 were maintained in Iscove's modified Dulbecco's medium (IMDM) (Gibco, www.invitrogen.com), 15% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, and 1 mM sodium pyruvate. The Jurkat (clone E6.1; TIB-152 from ATCC, Rockville, MD, USA) human T-lymphoblastic, KG1 and U937 myeloblastic leukemia cell lines were maintained in RPMI 1640 containing 10% FBS, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1 mM sodium pyruvate. Transfected Jurkat cells, that is, Jurkat/neo and Jurkat/NPM–ALK cells25 were cultured in the continuous presence of 2 mg/ml G418. The HeLa uterine carcinoma cell line was maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco), 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, and 1 mM sodium pyruvate. Neuroblastoma cell lines SKN-SH (ATCC: HTB-11), SHSY-5Y (ATCC: CRL-2266) and SKN-AS (ATCC: CRL-2137) were cultured in RPMI 1640, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, and 1 mM sodium pyruvate. PBMC from five healthy donors isolated using a Ficoll gradient were kindly provided by Dr Nabila Jabrane-Ferrat (CPTP, INSERM U1043, Toulouse, France) and used as normal lymphoid cell controls.
Cell transfection
SKN-AS cells were stably transfected using Fugene HD (Promega, www.promega.com) with pcDNA3.1 (empty vector), pcDNA-ALKwt or pcDNA-ALK-F1174L,15 cloned by limiting dilution and cultured in the continuous presence of 0.5 mg/ml G418.
Western blot
Cells were washed once with PBS, pelleted, and extracted in RIPA buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.7, 4 mM EDTA, 0.5% Triton X-100) containing 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1 mM 4-2-aminoethyl-benzenesulfonyl fluoride, 1 mM sodium orthovanadate, and 4 mM sodium fluoride for 30 min on ice. Cell extracts were either centrifuged at 10 000 × g for 20 min at 4 °C and the supernatant recovered, or sonicated for better extraction of nuclear proteins. The protein content was quantified using the Bio-Rad (www.biorad.com) Bradford protein assay. For western blotting, total cell proteins were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in a 4–12% bis/tris acrylamide (Novex NuPAGE, Invitrogen) gradient gel using the Nupage system with Mops SDS buffer under reducing conditions and transferred for 3 h at 60 V onto a nitrocellulose membrane. The blots were hybridized with the following primary antibodies:
ZAL4 anti-human ALK rabbit monoclonal antibody (1/200, Invitrogen), phospho-ALK (Y1604) (1/1000, #3341, Cell Signaling Technology, www.cellsignal.com) rabbit antiserum, mouse monoclonal anti-caspase 3 (1/200, sc-7148, Santa Cruz Biotechnologies, www.scbt.com), rabbit polyclonal anti-p53 (1/500, #554167, BD Pharmingen, www.bdbiosciences.com) and rabbit polyclonal anti-GAPDH (1/2000, Millipore, www.millipore.com) antibodies. Secondary antibodies were horseradish peroxidase-coupled anti-rabbit (1/1000) or anti-mouse (1/1000) Ig antiserum from Cell Signaling. For beta-actin detection, a peroxidase-coupled mouse monoclonal antibody (1/7500, Sigma) was used. Signal detection was performed with an enhanced chemoluminescence kit (ECL, Amersham, www.gelifesciences.com).
Cell viability assay
Exponentially growing cells were incubated in 96-well white microplates with a clear bottom at 105 cells/ml with peptide, PF, both, or culture medium for control, for 48 h at 37 °C. For adherent cell lines, the microwells were pre-coated with 50 μg/ml poly-l-lysine for better adhesion and cells were plated 24 h prior to treatment. At the end of incubation, cell viability was assessed by measuring the ATP content of wells using the luminescence-based CellTiterGlo assay (Promega) with a Berthold Mithras (http://www.berthold.com) plate reader. Each culture condition was assayed in triplicate.
Apoptosis assay
Exponentially growing cells were incubated in 96-well microplates at 105 cells/ml with peptide, PF or both, or culture medium for control, for 48 h at 37 °C. Apoptosis was assessed by measuring the relative caspase activity of treated versus control cells using the luminescence-based Caspase 3/7 Glo assay (Promega). Each culture condition was assayed in triplicate.
Peptide penetration assay
To enable visualization, we used C-terminal biotin-tagged peptides. Exponentially growing Cost cells were incubated for 1 h at 37 °C with 10 μM peptide. Alternatively, the peptides were electroporated into Cost cells with an Amaxa Nucleofector II device using program A30 and Nucleofector solution V (Lonza, www.lonza.com), according to a customized manufacturer protocol60 after which the cells were incubated in IMDM+1% FBS for 15 min at 37 °C. Then cells were centrifuged at 300 × g for 5 min, fixed, permeabilized and incubated with fluorescein isothiocyanate-coupled streptavidin (BD Pharmingen, www.bdbiosciences.com, 1/400) at room temperature using the Dako (www.dako.com) Intrastain kit. After washing and a second fixation in PBS-formaldehyde 1%, cells were mounted on slides and examined using an LSM 510 confocal microscope (Zeiss Axiovert 100, Carl Zeiss France, Marly le Roi, France).
Detection of surface plasmonic resonance and mass spectrometry
Real-time binding experiments were performed with a BIAcore 3000 biosensor instrument (www.gelifesciences.com) and quantified in terms of resonance units (RU) (1000 RU=1ng of protein bound/mm2 of flow cell surface).61 Synthetic biotinylated peptides were immobilized onto streptavidin-coated carboxymethylated dextran Biacore Sensor chips (www.gelifesciences.com). Cell extracts (100 μg/ml) were injected in the running buffer (HEPES 10 mM, NaCl 150 mM, EDTA 3 mM, polysorbate 0.005%). For recovery experiments, an optimized protocol was applied.62, 63 Synthetic P36 biotinylated peptide was immobilized at a level of 2000 RU. Cell extracts from Cost or SH-SY5Y were injected at 500 μg/ml. Five recovery cycles were performed for each cell line to reach a total amount of 2000 RU of recovered proteins. Further analysis of the recovered proteins was performed by nano-LC–MS/MS using an Ultimate3000 system (Dionex, Amsterdam, The Netherlands) coupled to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany).62, 63
RNAi transfection
RNAi transfection was optimized for Cost and SH-SY5Y cells therefore achieved by different methods and reagents. Cost cells were incubated for 48 h with 1 μM Accell Human TP53 siRNA SMARTpool or control Accell Non-targeting Pool siRNAs (Dharmacon, http://dharmacon.gelifesciences.com) in a 1 : 1 vol/vol mix of Accell siRNA Delivery Media and IMDM containing 1% FBS, according to the manufacturer's protocol. SH-SY5Y cells were seeded in poly-l-lysine pre-coated six-well plates and preincubated overnight in RPMI containing 1% FBS in the absence of antibiotics. The next day, SH-SY5Y cells were transfected using Dharmafect-1 transfection reagent (Dharmacon) with one of two siRNAs targeting p53 (Dharmacon, siGenome si-p53 #1: D-003329-05, si-p53 #2: D-003329-07) at 50 nM or an siRNA control (Qiagen, www.qiagen.com: Allstar negative control, cat# SI03650318). For both Cost and SH-SY5Y, cell incubation with siRNAs lasted for 48 h to allow p53 gene downregulation. Then P36 was added at 10 or 20 μM for the further 48 h. Cell viability and p53 expression (western blot) were assessed at that time.
Statistical analysis
Statistical analysis was done using the Student's t-test and two-way analysis of variance using GraphPad Prism 4 software (www.graphpad.com/scientific-software/prism/).
Acknowledgments
This work was supported by INSERM and Université Paul Sabatier and by a grant (R07055BB) from the Fondation de France (MA). We thank Dr Frédérique Gaits for helpful suggestions, Marie Terville, the GenoToul Proteomic facility of Toulouse, and the cellular imaging facility of INSERM UMR 1043 of Toulouse for technical assistance.
Glossary
- ALK
anaplastic lymphoma kinase
- TKI
tyrosine kinase inhibitor
- NPM
nucleophosmin
- ALCL
anaplastic large-cell lymphoma
- NSCLC
non-small-cell lung cancer
- EML4
echinoderm microtubule-associated protein-like 4
- PTN
pleiotrophin
- RPTPb/z
receptor tyrosine phosphatase beta/zeta
- ADD
addiction/dependence domain
- aa
amino acid
- PBMC
peripheral blood mononuclear cells
- PF
PF-02341066 or crizotinib
- ED
effective dose
- HPLC
high-performance liquid chromatography
- IMDM
Iscove's modified Dulbecco's medium
- FBS
fetal bovine serum
- DMEM
Dulbecco's modified Eagle's medium
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- RU
resonance units
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Oberst
Supplementary Material
References
- Williams DH, Mitchell T. Latest developments in crystallography and structure-based design of protein kinase inhibitors as drug candidates. Curr Opin Pharmacol 2002; 2: 567–573. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med 2001; 344: 1038–1042. [DOI] [PubMed] [Google Scholar]
- Uitdehaag JC, Verkaar F, Alwan H, de Man J, Buijsman RC, Zaman GJ. A guide to picking the most selective kinase inhibitor tool compounds for pharmacological validation of drug targets. Br J Pharmacol 2012; 166: 858–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch-Bentov R, Sauer K. Mechanisms of drug resistance in kinases. Expert Opin Investig Drugs 2011; 20: 153–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamant L, Meggetto F, Saati T, Brugieres L, de Paillerets BB, Dastugue N et al. High incidence of the t(2;5)(p23;q35) translocation in anaplastic large cell lymphoma and its lack of detection in Hodgkin's disease. Comparison of cytogenetic analysis, reverse transcriptase-polymerase chain reaction, and P-80 immunostaining. Blood 1996; 87: 284–291. [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non- Hodgkin's lymphoma. Science 1994; 263: 1281–1284. [DOI] [PubMed] [Google Scholar]
- Allouche M. ALK (anaplastic lymphoma receptor tyrosine kinase). Atlas Genet Cytogenet Oncol Haematol 2010; 14: 1059–1064. [Google Scholar]
- Minoo P, Wang HY. ALK-immunoreactive neoplasms. Int J Clin Exp Pathol 2012; 5: 397–410. [PMC free article] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448: 561–566. [DOI] [PubMed] [Google Scholar]
- Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J 2009; 420: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara T, Fujimoto J, Wen D, Cupples R, Bucay N, Arakawa T et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 1997; 14: 439–449. [DOI] [PubMed] [Google Scholar]
- Morris SW, Naeve C, Mathew P, James PL, Kirstein MN, Cui X et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin's lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) [published erratum appears in Oncogene 1997;15(23):2883]. Oncogene 1997; 14: 2175–2188. [DOI] [PubMed] [Google Scholar]
- Murray PB, Lax I, Reshetnyak A, Ligon GF, Lillquist JS, Natoli EJ Jr. et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal 2015; 8: ra6. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Zhang W, Chang Y, Vega JA, Deuel TF. Anaplastic lymphoma kinase is activated through the pleiotrophin/receptor protein-tyrosine phosphatase beta/zeta signaling pathway: an alternative mechanism of receptor tyrosine kinase activation. J Biol Chem 2007; 282: 28683–28690. [DOI] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 2008; 455: 967–970. [DOI] [PubMed] [Google Scholar]
- Armstrong F, Duplantier MM, Trempat P, Hieblot C, Lamant L, Espinos E et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene 2004; 23: 6071–6082. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer 2008; 8: 11–23. [DOI] [PubMed] [Google Scholar]
- Christensen JG, Zou HY, Arango ME, Li Q, Lee JH, McDonnell SR et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther 2007; 6: 3314–3322. [DOI] [PubMed] [Google Scholar]
- Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010; 363: 1734–1739. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res 2010; 70: 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allouche M. ALK is a novel dependence receptor: potential implications in development and cancer. Cell Cycle 2007; 6: 1533–1538. [DOI] [PubMed] [Google Scholar]
- Mourali J, Benard A, Lourenco FC, Monnet C, Greenland C, Moog-Lutz C et al. Anaplastic lymphoma kinase is a dependence receptor whose proapoptotic functions are activated by caspase cleavage. Mol Cell Biol 2006; 26: 6209–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P, Thibert C. Dependence receptors: between life and death. Cell Mol Life Sci 2004; 61: 1854–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AR, Borland L, Allbritton NL, Sims CE. Myristoyl-based transport of peptides into living cells. Biochemistry 2007; 46: 14771–14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenland C, Touriol C, Chevillard G, Morris SW, Bai R, Duyster J et al. Expression of the oncogenic NPM-ALK chimeric protein in human lymphoid T-cells inhibits drug-induced, but not Fas-induced apoptosis. Oncogene 2001; 20: 7386–7397. [DOI] [PubMed] [Google Scholar]
- Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 2006; 58: 621–681. [DOI] [PubMed] [Google Scholar]
- Di Paolo D, Ambrogio C, Pastorino F, Brignole C, Martinengo C, Carosio R et al. Selective therapeutic targeting of the anaplastic lymphoma kinase with liposomal siRNA induces apoptosis and inhibits angiogenesis in neuroblastoma. Mol Ther 2011; 19: 2201–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FY, Zhao Y, Anderson WF, Johnston PB. Downregulation of NPM-ALK by siRNA causes anaplastic large cell lymphoma cell growth inhibition and augments the anti cancer effects of chemotherapy in vitro. Cancer Invest 2007; 25: 240–248. [DOI] [PubMed] [Google Scholar]
- Mazot P, Cazes A, Boutterin MC, Figueiredo A, Raynal V, Combaret V et al. The constitutive activity of the ALK mutated at positions F1174 or R1275 impairs receptor trafficking. Oncogene 2011; 30: 2017–2025. [DOI] [PubMed] [Google Scholar]
- Goldenberg-Furmanov M, Stein I, Pikarsky E, Rubin H, Kasem S, Wygoda M et al. Lyn is a target gene for prostate cancer: sequence-based inhibition induces regression of human tumor xenografts. Cancer Res 2004; 64: 1058–1066. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Gouda M, Novatchkova M, Schleiffer A, Schneider G, Sirota FL et al. MYRbase: analysis of genome-wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins. Genome Biol 2004; 5: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agemy L, Friedmann-Morvinski D, Kotamraju VR, Roth L, Sugahara KN, Girard OM et al. Targeted nanoparticle enhanced proapoptotic peptide as potential therapy for glioblastoma. Proc Natl Acad Sci USA 2011; 108: 17450–17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delloye-Bourgeois C, Fitamant J, Paradisi A, Cappellen D, Douc-Rasy S, Raquin MA et al. Netrin-1 acts as a survival factor for aggressive neuroblastoma. J Exp Med 2009; 206: 833–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P, Guenebeaud C. Netrin-1 and its dependence receptors as original targets for cancer therapy. Curr Opin Oncol 2010; 22: 46–54. [DOI] [PubMed] [Google Scholar]
- Mille F, Llambi F, Guix C, Delloye-Bourgeois C, Guenebeaud C, Castro-Obregon S et al. Interfering with multimerization of netrin-1 receptors triggers tumor cell death. Cell Death Differ 2009; 16: 1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JK, Choi KH, Peng J, He F, Zhang Z, Weisleder N et al. Amphipathic tail-anchoring peptide and Bcl-2 homology domain-3 (BH3) peptides from Bcl-2 family proteins induce apoptosis through different mechanisms. J Biol Chem 2011; 286: 9038–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvini P, Dalla Rosa H, Vignes N, Rosolen A. Ubiquitination and proteasomal degradation of nucleophosmin-anaplastic lymphoma kinase induced by 17-allylamino-demethoxygeldanamycin: role of the co-chaperone carboxyl heat shock protein 70-interacting protein. Cancer Res 2004; 64: 3256–3264. [DOI] [PubMed] [Google Scholar]
- Sang J, Acquaviva J, Friedland JC, Smith DL, Sequeira M, Zhang C et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov 2013; 3: 430–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell 2005; 19: 1–13. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Andreeva A, Teufel DP, Freund SM, Fersht AR. Interaction between the transactivation domain of p53 and PC4 exemplifies acidic activation domains as single-stranded DNA mimics. J Biol Chem 2009; 284: 21728–21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Kundu TK. Activation of p53 function by human transcriptional coactivator PC4: role of protein-protein interaction, DNA bending, and posttranslational modifications. Mol Cell Biol 2007; 27: 7603–7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Gadad SS, Kundu TK. Human positive coactivator 4 controls heterochromatinization and silencing of neural gene expression by interacting with REST/NRSF and CoREST. J Mol Biol 2010; 397: 1–12. [DOI] [PubMed] [Google Scholar]
- Blanch A, Robinson F, Watson IR, Cheng LS, Irwin MS. Eukaryotic translation elongation factor 1-alpha 1 inhibits p53 and p73 dependent apoptosis and chemotherapy sensitivity. PLoS One 2013; 8: e66436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kulesz-Martin M. p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis 2001; 22: 851–860. [DOI] [PubMed] [Google Scholar]
- Wong TS, Rajagopalan S, Townsley FM, Freund SM, Petrovich M, Loakes D et al. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic Acids Res 2009; 37: 568–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Wu Y, Chen Q, Cao J, Hu K, Tang J et al. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res 2013; 23: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol SM, Bray SE, Black HD, Lorimore SA, Wright EG, Lane DP et al. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene 2013; 32: 3461–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta 2009; 1787: 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa C, Acker J. Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA Biol 2010; 7: 287–290. [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang J, Zhang E, Sun H, Wang Q, Wang T et al. Human positive coactivator 4 is a potential novel therapeutic target in non-small cell lung cancer. Cancer Gene Ther 2012; 19: 690–696. [DOI] [PubMed] [Google Scholar]
- Malik SM, Maher VE, Bijwaard KE, Becker RL, Zhang L, Tang SW et al. U.S. Food and Drug Administration approval: crizotinib for treatment of advanced or metastatic non-small cell lung cancer that is anaplastic lymphoma kinase positive. Clin Cancer Res 2014; 20: 2029–2034. [DOI] [PubMed] [Google Scholar]
- Gambacorti-Passerini C, Farina F, Stasia A, Redaelli S, Ceccon M, Mologni L et al. Crizotinib in advanced, chemoresistant anaplastic lymphoma kinase-positive lymphoma patients. J Natl Cancer Inst 2014; 106: djt378. [DOI] [PubMed] [Google Scholar]
- Mosse YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children's Oncology Group phase 1 consortium study. Lancet Oncol 2013; 14: 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardini E, Galvani A. ALK Inhibitors, a Pharmaceutical Perspective. Front Oncol 2012; 2: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler SC, Wood AC, Haglund EA, Courtright J, Belcastro LT, Plegaria JS et al. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci Transl Med 2011; 3: 108ra114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mistro A, Leszl A, Bertorelle R, Calabro ML, Panozzo M, Menin C et al. A CD30-positive T cell line established from an aggressive anaplastic large cell lymphoma, originally diagnosed as Hodgkin's disease. Leukemia 1994; 8: 1214–1219. [PubMed] [Google Scholar]
- Epstein AL, Kaplan HS. Biology of the human malignant lymphomas. I. Establishment in continuous cell culture and heterotransplantation of diffuse histiocytic lymphomas. Cancer 1974; 34: 1851–1872. [DOI] [PubMed] [Google Scholar]
- Fischer P, Nacheva E, Mason DY, Sherrington PD, Hoyle C, Hayhoe FG et al. A Ki-1 (CD30)-positive human cell line (Karpas 299) established from a high-grade non-Hodgkin's lymphoma, showing a 2;5 translocation and rearrangement of the T-cell receptor beta-chain gene. Blood 1988; 72: 234–240. [PubMed] [Google Scholar]
- Lamant L, Espinos E, Duplantier M, Dastugue N, Robert A, Allouche M et al. Establishment of a novel anaplastic large-cell lymphoma-cell line (COST) from a 'small-cell variant' of ALCL. Leukemia 2004; 18: 1693–1698. [DOI] [PubMed] [Google Scholar]
- Duplantier MM, Lamant L, Sabourdy F, de Reynies A, Delsol G, Espinos E. Serpin A1 is overexpressed in ALK+ anaplastic large cell lymphoma and its expression correlates with extranodal dissemination. Leukemia 2006; 20: 1848–1854. [DOI] [PubMed] [Google Scholar]
- Ferjoux G, Lopez F, Esteve JP, Ferrand A, Vivier E, Vely F et al. Critical role of Src and SHP-2 in sst2 somatostatin receptor-mediated activation of SHP-1 and inhibition of cell proliferation. Mol Biol Cell 2003; 14: 3911–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez F, Pichereaux C, Burlet-Schiltz O, Pradayrol L, Monsarrat B, Esteve JP. Improved sensitivity of biomolecular interaction analysis mass spectrometry for the identification of interacting molecules. Proteomics 2003; 3: 402–412. [DOI] [PubMed] [Google Scholar]
- Martinez LO, Jacquet S, Esteve JP, Rolland C, Cabezon E, Champagne E et al. Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nature 2003; 421: 75–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.