Abstract
Epidermal keratinocyte differentiation on the body surface is a carefully choreographed process that leads to assembly of a barrier that is essential for life. Perturbation of keratinocyte differentiation leads to disease. Activator protein 1 (AP1) transcription factors are key controllers of this process. We have shown that inhibiting AP1 transcription factor activity in the suprabasal murine epidermis, by expression of dominant-negative c-jun (TAM67), produces a phenotype type that resembles human keratoderma. However, little is understood regarding the structural and molecular changes that drive this phenotype. In the present study we show that TAM67-positive epidermis displays altered cornified envelope, filaggrin-type keratohyalin granule, keratin filament, desmosome formation and lamellar body secretion leading to reduced barrier integrity. To understand the molecular changes underlying this process, we performed proteomic and RNA array analysis. Proteomic study of the corneocyte cross-linked proteome reveals a reduction in incorporation of cutaneous keratins, filaggrin, filaggrin2, late cornified envelope precursor proteins, hair keratins and hair keratin-associated proteins. This is coupled with increased incorporation of desmosome linker, small proline-rich, S100, transglutaminase and inflammation-associated proteins. Incorporation of most cutaneous keratins (Krt1, Krt5 and Krt10) is reduced, but incorporation of hyperproliferation-associated epidermal keratins (Krt6a, Krt6b and Krt16) is increased. RNA array analysis reveals reduced expression of mRNA encoding differentiation-associated cutaneous keratins, hair keratins and associated proteins, late cornified envelope precursors and filaggrin-related proteins; and increased expression of mRNA encoding small proline-rich proteins, protease inhibitors (serpins), S100 proteins, defensins and hyperproliferation-associated keratins. These findings suggest that AP1 factor inactivation in the suprabasal epidermal layers reduces expression of AP1 factor-responsive genes expressed in late differentiation and is associated with a compensatory increase in expression of early differentiation genes.
The epidermis is a highly differentiated structure that acts as a barrier to reduce fluid and nutrient loss, and prevent infection. Assembly of the barrier requires that keratinocytes undergo a complex terminal differentiation process that results in the conversion of proliferating basal cells to suprabasal differentiated keratinocytes. During this process, nuclei, organelles and other cellular structures are destroyed. The ultimate fate of these cells is formation of the stratum corneum, which is comprised of covalently cross-linked proteins, lipids and keratin bundles, and functions as a barrier. Activator protein one (AP1) transcription factors are essential regulators of this process.1 These factors form homo- and heterodimers that bind DNA response elements to regulate gene expression.2 An example is mutation of a single AP1 transcription factor binding site in the distal regulatory region of the involucrin gene promoter results in a complete loss of involucrin expression in epidermis.3 Moreover, selective inactivation of AP1 factor function in epidermis produces phenotypes that mimic human epidermal disease.4, 5 To investigate the role of AP1 factors in epidermis, we utilized TAM67, a dominant-negative form of c-jun. TAM676 dimerizes with other AP1 transcription factors. These complexes bind to DNA, but this interaction does not activate transcription and this reduces AP1-mediated gene expression. We targeted TAM67 to the suprabasal epidermis to inhibit AP1 factor-related transcription in this compartment.4, 5 Our findings show that TAM67-dependent inactivation of AP1 factor function in the suprabasal epidermis results in increased cell proliferation and delayed differentiation and that this is associated with extensive epidermal hyperkeratosis and formation of constriction rings on the tail and digits5 to produce a phenotype that resembles human keratoderma.4, 5
However, the structural and biochemical changes underlying this phenotype are not well understood. To assess the biochemical impact of suprabasal AP1 factor inactivation, we analyzed epidermal structure and the corneocyte cross-linked proteome and RNA expression profile. We show that the cornified envelope (CE) cross-linked proteome in AP1 factor-deficient (TAM67-positive) mice is enriched for early envelope precursors, and that late envelope precursors and filaggrin-related proteins are reduced in level. In many cases, these changes are reflected in parallel changes in gene expression. This phenotype is associated with reduced cornified envelope formation, reduced formation of filaggrin-type keratohyalin granules and keratin filaments, and abnormal desmosome formation, lipid processing and desquamation.
Results
TAM67-rTA mice develop a unique phenotype
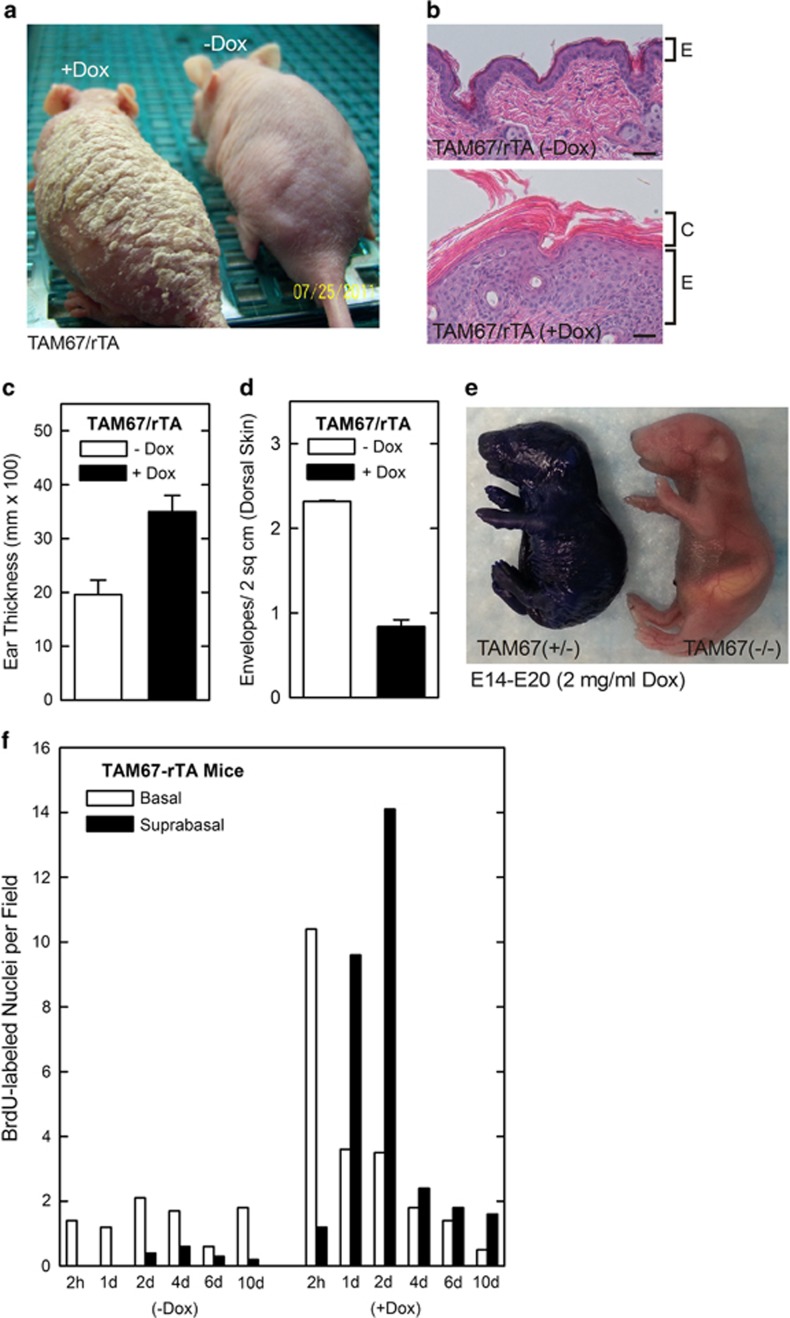
At 2 weeks after induction of TAM67 expression in the suprabasal epidermis, TAM67-rTA mice are covered with scale over the trunk and appendages (Figure 1a).5 Microscopic examination reveals that the epidermis is four times thicker than in TAM67-negative mice, and the histology suggests there is enhanced proliferation and cornification (Figure 1b) which is also reflected in increased ear thickness (Figure 1c). However, although extensive hyperkeratosis is evident (Figure 1b), the number of CEs, as counted following boiling in SDS and reducing agent, is reduced (Figure 1d), indicating that the envelopes are not well formed. In addition, there is a 13% reduction in body weight in adult TAM67-positive mice that we believe is caused by increased evaporative water loss due to compromised barrier function. To assess the impact of loss of AP1 factor functional inactivation on barrier integrity, TAM67 expression was induced on E14 and embryos were collected on the day before birth (E20) to assess barrier integrity. Figure 1e shows that the TAM67-expressing littermate has a clear barrier defect as evidenced by enhanced epidermal dye uptake.
Figure 1.
Impact of suprabasal epidermal AP1 factor inactivation on epidermal phenotype . (a and b) TAM67-rTA mice were treated with 0 or 2 mg/ml doxycycline for 7 days and the mice were photographed and skin sections were processed and stained with hematoxylin and eosin. E indicates the epidermis and C the cornified layers. (c) Increased ear thickness in TAM67-FLAG-positive epidermis. Ear thickness was monitored using calipers at 7 days after initiation of doxycycline treatment. (d) Reduced cornified envelope number in TAM67–FLAG-positive epidermis. Epidermis scale was collected from TAM67-rTA mice treated as above. The scale was then boiled in detergent and reducing agent and surviving envelope structures were counted. The values are mean±S.E.M. of three separate experiments. Envelope number is significantly reduced (P<0.001, n=3). (e) TAM67 expression compromises barrier function. Pregnant female mice were treated with 2 mg/ml doxycycline beginning on E14 and the embryos were removed and stained with toluidine blue at E20. All TAM67-positive mice displayed compromised barrier function. (f) TAM67 expression is associated with increased cell proliferation. Adult TAM67-rTA mice were treated with 0 or 2 mg/ml doxycycline and on day 4 were injected IP with 50 mg of BrdU per kg body weight. At 2 h to 10 days animals were killed and the epidermis was sectioned and stained with anti-BrdU. The number of basal and suprabasal BrdU-positive cells were counted at each time point. Similar findings were observed in each of two experiments
To measure changes in keratinocyte proliferation, 8-week-old adult female TAM67-rTA mice were treated with 0 or 2 mg/ml doxycycline for 4 days.4 Mice were then injected with bromodeoxyuridine (BrdU) and the fate of the BrdU-labeled cells was assessed by sectioning epidermis and staining with anti-BrdU. Counts of labeled cells at 2 h suggest that seven times more cells are dividing in the TAM67-positive epidermis and that the majority of the cells are in the basal layer (Figure 1f). In control (-Dox) epidermis the number of labeled cells remains relatively stable and most labeled cells localize to the basal layer. In contrast, by 24 and 48 h, the majority of the labeled cells are suprabasal in the TAM67-positive epidermis; moreover, at day 4 the number of labeled cells is reduced, and by 10 days 80% of the labeled cells have been released from the surface. In contrast, in TAM67-negative epidermis only 1% of labeled cells have been released by day 10.
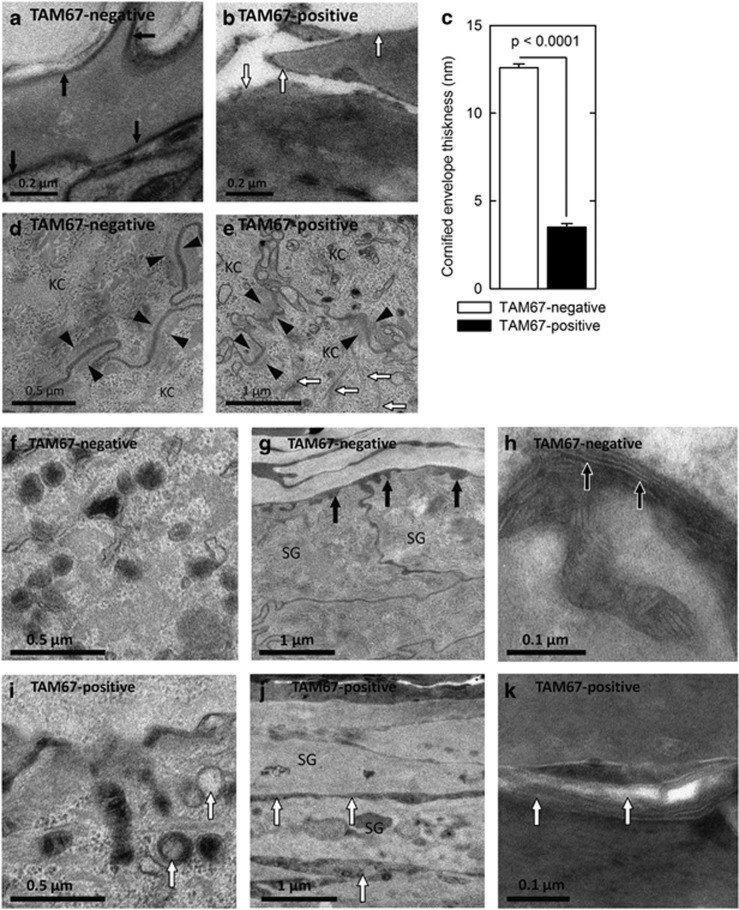
EM images of TAM67-positive epidermis demonstrate reduced cornified envelope thickness (Figures 2a, b and c), and abnormal desmosomes and keratin filaments (Figures 2d and e). TAM67-positive epidermis exhibits abnormal lamellar body contents (Figures 2f/i), premature lamellar lipid secretion (Figures 2g/j) and incomplete processing of secreted lipid (Figures 2h/k).
Figure 2.
Attenuated cornified envelope, keratin filament and desmosome formation and altered lipid secretion in TAM67-positive epidermis. (a and b) Electron microscopy reveals uniform cornified envelopes (black arrows) in stratum corneum of TAM67-negative as compared with less-developed envelopes (white arrows) in TAM67-positive epidermis. (c) Quantification of the cornified envelope thickness. The values are mean+S.E.M. (d and e) TAM67-negative epidermis demonstrates compact desmosomes flanked with keratin filaments (black arrowheads) compared with disorganized desmosomes (black arrowheads) and haphazardly arranged keratin filaments (white arrows) in TAM67-positive epidermis. KC indicates keratinocyte. Epidermis from TAM67-negative mice is characterized by lamellar bodies of uniform size filled with stacked membranous lipid contents (f), lipid secretion at the SG–SC junction (g, black arrows) and processing of secreted lipid into extended membrane arrays (h, ruthenium stained, black arrows). In contrast, TAM67-rTA epidermis displays abnormal lamellar body contents (i, white arrows), premature lipid secretion in the middle SG layers (j, white arrows) and incomplete post-secretory processing of secreted lipid (k, white arrows). SG, stratum granulosum; SC, stratum corneum
The cross-linked proteome
To understand the biochemical basis for these changes, we examined the composition of epidermal scale from TAM67-positive and -negative epidermis using proteomic methods.7, 8 Cornified material was collected and extracted with SDS to remove soluble proteins and fatty acids. The residual was protease digested and analyzed by mass spectrometry.
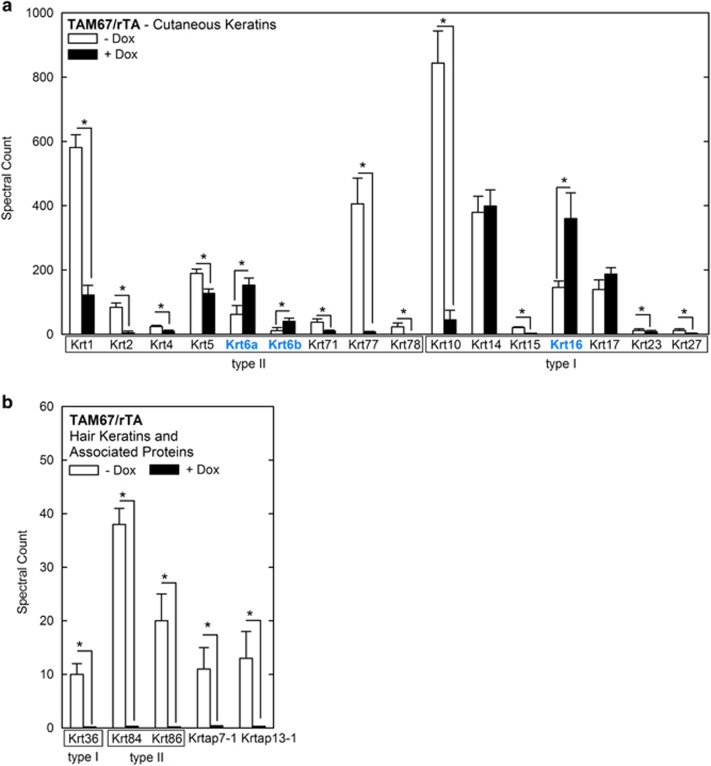
Analysis of keratin composition in TAM67-positive epidermis reveals a decrease in Krt1, 2, 4, 5, 71, 77 and 78 (type II) and Krt10, Krt15, Krt23 and Krt27 (type I; Figure 3a) and an increase in the hyperproliferation-associated keratins, Krt6a, Krt6b and Krt16 (Figure 3a). We also explored the impact on hair keratins and hair keratin-associated proteins. Figure 3b shows that Krt36, 84 and 86 are decreased, as are the Krtap7-1 and Krtap13-1 hair keratin-associated proteins.
Figure 3.
Keratins and keratin-associated protein levels in the cross-linked proteome. (a and b) Epidermal scale and cornified material were analyzed from three TAM67-positive and three TAM67-negative mice. Samples were processed for mass spectrometry as outlined in Materials and Methods.8 The values are weighted spectral counts shown as the mean±S.D.69 The asterisks indicate a significant difference (P<0.001, n=3)
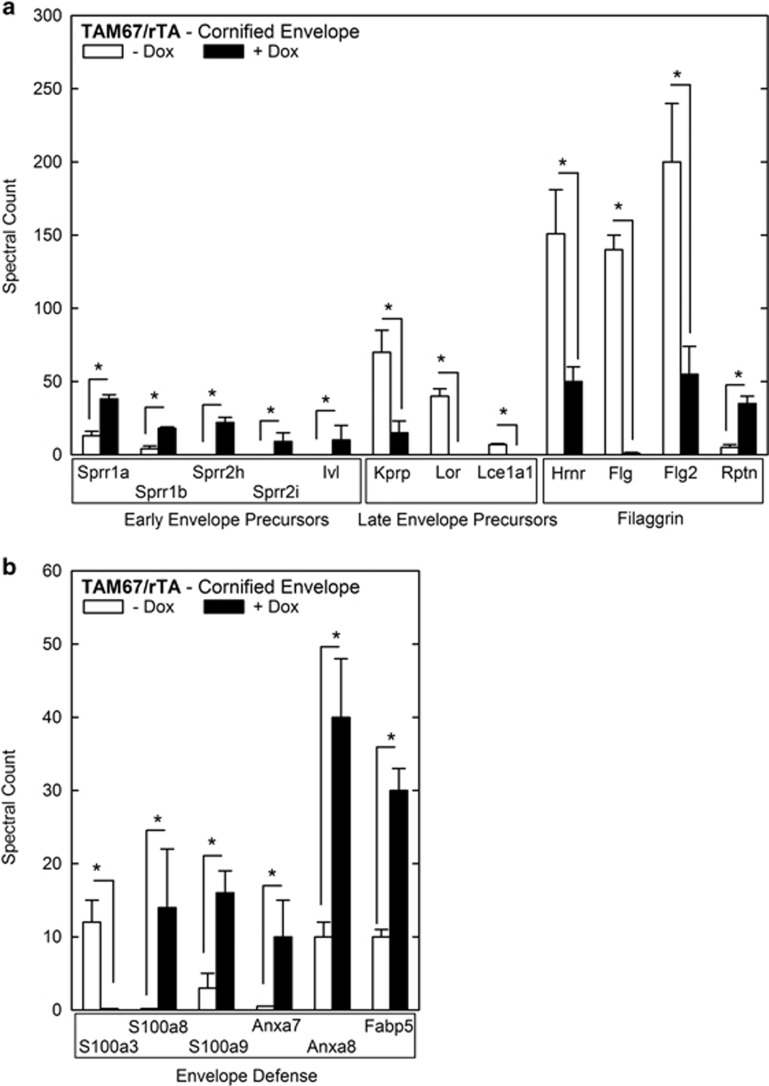
We detected significant changes in the level of 12 CE precursors (Figure 4a). Precursors expressed early in normal differentiation are increased in AP1 factor-deficient mice. In contrast, late envelope precursors (keratinocyte proline-rich proteins, loricrin, Lce1a1) are reduced in level, as are most of the S100 fused-type proteins (filaggrin related; Figure 4a). It is interesting that many of these genes are positively regulated by AP1 transcription factors.9
Figure 4.
Envelope precursor and defense levels in cross-linked proteome. (a and b). Level of early envelope, late envelope, filaggrin-related proteins and envelope defense proteins in the cross-linked proteome. Epidermal scale and cornified material was collected and processed as described in the legend to Figure 3. The values are spectral units shown as the mean±S.D. The asterisks indicate a significant difference (P<0.001, n=3)
Annexin, fatty-acid binding and S100 proteins form functional complexes that are involved in response to pathogen invasion and inflammation.10, 11, 12 S100a3 is reduced, but S100a8 and S100a9 are increased (Figure 4b). In addition, the S100 interacting proteins, annexin 7, annexin 8 and Fabp5 are increased. This suggests a tissue inflammatory response.
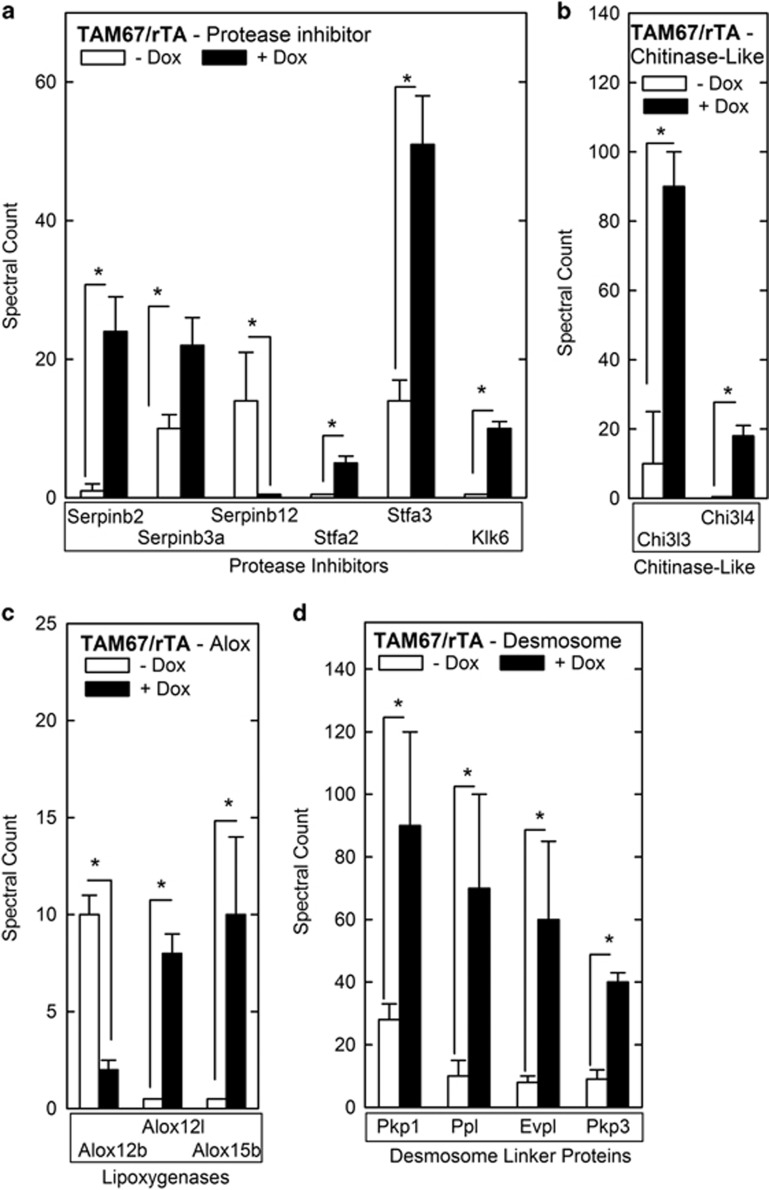
Proteolysis is an important event in the epidermis.13 Composition analysis reveals enhanced serpin and stefin levels (Figure 5a). Serpins are irreversible serine protease inhibitors14, 15 and serpin2 and serpin3a are increased in cross-linked material from TAM67-expressing epidermis, while serpinb12 level is reduced. Stefin 2 and 3 (Stfa2 and Stfa3) are members of the cystatin family and inhibit lysosomal cysteine proteinases including cathepsin B, H, K, L and S.16, 17 Stfa2 and Stfa3 are increased in cross-linked material from TAM67-expressing epidermis. In addition, kallikrein-related peptidase 6 (Klk6), which has trypsin-like properties, is elevated in level (Figure 5a).
Figure 5.
Protease inhibitor, chitinase-like protein, lipoxygenase and desmosome proteins in cornified envelope. (a–d) Level of the indicated proteins in epidermal scale and cornified material collected and processed as described in the legend to Figure 3. The values are spectral units shown as the mean±S.D. The asterisks indicate a significant difference (P<0.001, n=3)
Chitinases hydrolyze chitin.18 Mice express two catalytically inactive chitinase-like genes, Chi3l3 (Ym1) and Chi3l4 (Ym2).19 Increased Chi3l3 and Chi3l4 are observed in the epidermis of mouse models of chronic proliferative dermatitis and are derived from macrophages, dendritic cells and mast cells.19 We observed increased Chi3l3 and Chi3l4 content in the CE of TAM67-expressing mice (Figure 5b). 12/15-Lipoxygenases catalyze the oxidation of free and esterified fatty acids to generate bioactive lipids that form ceramides, which are required for formation of the lipid barrier.20 Alox12b, Alox12l and Alox15b, are murine lipoxygenase genes,20 and these proteins are increased in TAM67-expressing epidermis (Figure 5c).
Desmosomes are essential barrier components,13 and are altered in TAM67-positive epidermis (Figure 2e). The desmosome includes membrane-spanning core proteins, including desmocollins (DSCs) and desmoglein (DSGs),21 and linker protein components, including envoplakin (Evpl), periplakin (Ppl), plakophilin 1 (Pkp1) and plakophilin 3 (Pkp3), which tether these core proteins to the actin and keratin intermediate filaments.22 Analysis reveals no change in the level of desmosomal core proteins in TAM67-positive cross-linked protein preparations; however, there is substantial increase in the level of tethering proteins (Pkp1, Ppl, Evpl and Pkp3; Figure 5d).
RNA expression
We next used gene array to compare mRNA expression (Table 1). TAM67-negative epidermis, which differentiates normally, is enriched in mRNA encoding cutaneous keratins, hair keratins and hair keratin associated proteins, and expression of these genes is reduced in the epidermis of TAM67-expressing epidermis. Cornified envelope precursors also change. Sprr4, an early-expressed envelope marker, and a host of late envelope precursor genes, including loricrin and the late cornified envelope (Lce) genes, are enriched in TAM67-negative epidermis. Filaggrin-related proteins, including filaggrin2, trichohyalin and cornulin, are also enriched in TAM67-negative versus -positive epidermis.
Table 1. Gene array analysis.
| Ratio: (log2)a TAM67+/TAM67− | Gene name | Entrez gene ID | Description | Category |
|---|---|---|---|---|
| Gene expression elevated in TAM67-negative epidermis | ||||
| Cutaneous keratins | ||||
| −4.08 | Krt71 | 56735 | Keratin 71 | |
| −3.64 | Krt25 | 70810 | Keratin 25 | |
| −3.47 | Krt27 | 16675 | Keratin 27 | |
| −3.47 | Krt26 | 320864 | Keratin 26 | |
| −3.04 | Krt15 | 16665 | Keratin 15 | |
| −3.02 | Krt28 | 70843 | Keratin 28 | |
| −2.88 | Krt72 | 105866 | Keratin 72 | |
| −2.41 | Krt77 | 406220 | Keratin 77 | |
| −2.37 | Krt73 | 406220 | Keratin 73 | |
| −2.32 | Krt23 | 94179 | Keratin 23 | |
| −2.13 | Krt78 | 332131 | Keratin 78 | |
| Hair keratins and associated proteins | ||||
| −3.66 | Krt33a | 71888 | Keratin 33a | |
| −3.19 | Krt86 | Keratin 86 | ||
| −2.47 | Krt31 | 16660 | Keratin 31 | |
| −4.43 | Krtap7-1 | 71363 | Keratin associated protein 7-1 | |
| −4.39 | Krtap8-1 | 16703 | Keratin associated protein 8-1 | |
| −3.46 | Krtap3-3 | 66380 | Keratin associated protein 3-3 | |
| −3.27 | Krtap1-5 | 69664 | Keratin associated protein 1-5 | |
| −3.04 | Krtap6-1 | 100040214 | Keratin associated protein 6-1 | |
| −3.03 | Krtap16-8 | 68484 | Keratin associated protein 16-8 | |
| −2.92 | Krtap4-16 | 435285 | Keratin associated protein 4-16 | |
| −2.91 | Krtap4-1 | 665891 | Keratin associated protein 4-1 | |
| −2.77 | Krtap8-2 | 16704 | Keratin associated protein 8-2 | |
| −2.67 | Krtap13-1 | 268905 | Keratin associated protein 13-1 | |
| −2.64 | Krtap6-3 | Keratin associated protein 6-3 | ||
| −2.57 | Krtap4-7 | 76444 | Keratin associated protein 4-7 | |
| −2.23 | Krtap1-4 | 629873 | Keratin associated protein 1-4 | |
| −2.03 | Krtap4-2 | 68673 | Keratin associated protein 4-2 | |
| −1.83 | Krtap3-1 | 69473 | Keratin associated protein 3-1 | |
| −1.83 | Krtap16-7 | 170656 | Keratin associated protein 16-7 | |
| −1.80 | Krtap3-2 | 66380 | Keratin associated protein 3-2 | |
| −1.77 | Krtap1-3 | 435273 | Keratin associated protein 1-3 | |
| −1.70 | Krtap16-5 | 77918 | Keratin associated protein 16-5 | |
| Early envelope | ||||
| −1.8 | Sprr4 | 229562 | Small protein-rich protein 4 | |
| Late envelope | ||||
| −3.97 | Lce1m | 66203 | Late cornified envelope 1M | |
| −3.83 | Lce1d | 69611 | Late cornified envelope 1D | |
| −3.79 | Lce1a2 | 73722 | Late cornified envelope 1A2 | |
| −3.73 | Lce6a | 78382 | Late cornified envelope 6A | |
| −3.56 | Lce1f | 67828 | Late cornified envelope 1F | |
| −3.51 | Lce1e | 68694 | Late cornified envelope 1E | |
| −3.40 | Lce1b | 68720 | Late cornified envelope 1B | |
| −3.37 | Kprp | 433619 | Keratinocyte proline-rich protein | |
| −3.23 | Lce1l | 73730 | Late cornified envelope 1L | |
| −2.79 | Lce1c | 73719 | Late cornified envelope 1C | |
| −2.48 | Lce1a1 | 67127 | Late cornified envelope 1A1 | |
| −2.29 | Lce1h | Late cornified envelope 1H | ||
| −2.16 | Lce1j | Late cornified envelope 1J | ||
| −2.16 | Lce1g | 66195 | Late cornified envelope 1G | |
| −2.67 | Lor | 16939 | Loricrin | |
| Filaggrin-related proteins | ||||
| −5.55 | Flg2 | 229574 | Filaggrin2 | |
| −3.53 | Tchh | 99681 | Trichohyalin | |
| −2.26 | Crnn | 381457 | Cornulin | |
| Proteases and protease inhibitors | ||||
| −3.68 | Tmprss11f | 243083 | Transmembrane protease, serine 11f | |
| −3.02 | Serpina12 | 68054 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 | |
| Lipoxygenases | ||||
| −3.67 | Alox12e | 11685 | Arachidonate lipoxygenase, epidermal | |
| Gene expression elevated in TAM67-positive epidermis | ||||
| Cutaneous keratins | ||||
| +1.75 | Krt6a | 16687 | Keratin 6a | |
| +1.68 | Krt16 | 16666 | Keratin 16 | |
| +1.60 | Krt17 | 16667 | Keratin 17 | |
| Early envelope | ||||
| +2.40 | Sprr2b | 20756 | Small proline-rich protein 2B | |
| +2.14 | Tgm3 | 21818 | Transglutaminase type III | |
| +2.00 | Sprr2d | 20758 | Small proline-rich protein 2D | |
| +1.62 | Sprr2e | Small proline-rich protein 2E | ||
| Proteases and protease inhibitors | ||||
| +4.19 | Serpinb3a | 20248 | Serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A | |
| +2.51 | Serpinb3d | 394252 | Serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3D | |
| +1.80 | Serpinb6c | Serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 6C | ||
| +2.07 | Spink12 | 78242 | Serine peptidase inhibitor, Kazal type 12 | |
| +2.04 | Klk13 | 626834 | Kallikrein related-peptidase 13 | |
| +1.89 | Serpinb2 | 18788 | Plasminogen activator inhibitor 2 | |
| +1.84 | Capn2 | 12334 | Calpain 2 | |
| Envelope defense | ||||
| +3.82 | S100a8 | 20201 | S100A8 | |
| +3.72 | Defb3 | 27358 | Defensin beta 3 | |
| +3.19 | S100a9 | 20202 | S100A9 | |
| +2.00 | Defb4 | 56519 | Defensin beta 4 | |
| Chitinase-like proteins | ||||
| +4.18 | Chi3l4 | 104183 | Chitinase-3-like protein 3 | |
| Desmosome | ||||
| +1.99 | Dsc2 | 13506 | Desmocollin 2 | |
| +1.91 | Dsg3 | Desmoglein 3 | ||
Larger negative or positive numbers indicate greater enrichment
Negative log2 values indicate relative enrichment in TAM67-negative epidermis and positive numbers indicate relative enrichment in TAM67-positive epidermis
In contrast, TAM67-positive mice display increased expression of mRNA encoding hyperproliferation-associated keratins (Krt6a, Krt16 and Krt17), early envelope precursors (Sprr2b, Sprr2d, Sprr2e) and type III transglutaminase. We also observed a marked enrichment of mRNA encoding serine protease inhibitors (Serpins, Spink12), proteins involved in inflammation/bacterial defense (S100 proteins, defensins) and chitinase-3-like protein 4 (Chi3l4). We also observed an impact on desmosomal protein expression. DSC 2 and DSG 3, which constitute the core of the desmosome,21 were increased.
Time course of change in expression
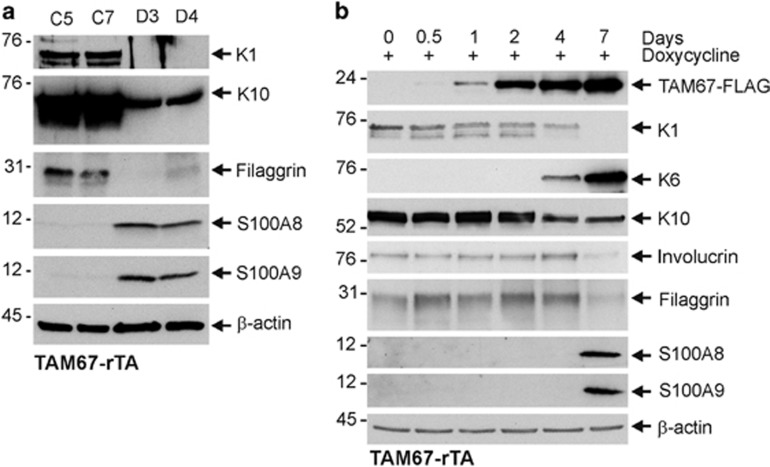
We next assayed selected proteins to confirm expression changes. Figure 6a compares protein level in epidermal extracts prepared from TAM67-negative (C5, C7) and positive (D3, D4) mice. This reveals reduced levels of Krt1 and Krt10 and filaggrin, and increased S100A8 and S100A9 in TAM67-positive epidermis. Time course studies (Figure 6b) reveal TAM67-FLAG expression at 1 day after addition of doxycycline. Krt1, Krt10, involucrin and filaggrin levels are reduced and Krt6 and S100A8 and S100A9 levels increased at days 4 and 7.
Figure 6.
Protein levels in TAM67-negative versus -positive epidermis. (a) Suprabasal AP1 factor inactivation alters epidermal protein level. TAM67-rTA mice were treated for 7 days with 0 (C5, C7) or 2 (D3, D4) mg/ml doxycycline administered in drinking water. The epidermis was then separated from the dermis using dispase and equivalent protein amounts were electrophoresed on denaturing and reducing 10% polyacrylamide gel. The proteins were transferred to nitrocellulose for incubation with appropriate antibodies.70 (b) Protein level as a function of time after AP1 factor inactivation. TAM67-rTA mice were treated for 0–7 days with 0 or 2 mg/ml doxycycline administered in drinking water. The level of the indicated proteins was then detected by immunoblot. Similar results were observed in each of three experiments
Discussion
We are interested in control of epidermal gene expression by the AP1 family of transcriptional regulators.23 This family includes c-jun, junB, junD, c-fos, Fra-1 and Fra-2.1 We recently showed that inactivation of suprabasal AP1 factor function in epidermis causes a phenotype characterized by hyperplasia, hyperkeratosis, parakeratosis and pseudoainhum that resembles human keratoderma.5 The present study expands on these findings and examines the impact of AP1 factor inactivation on the structure and composition of the CE, and on epidermal keratinocyte gene expression. We show that the keratoderma-like phenotype is associated with defective CE formation. Although the epidermis is thicker and the cornified layer is expanded,5 the number of mature envelopes per unit area of skin is markedly reduced, and barrier function is compromised. In addition, electron microscopic examination of the epidermis reveals reduced CE formation, as well as abnormalities at each step of lipid barrier formation: lamellar body loading, lipid secretion and secreted lipid processing. Defective lipid barrier formation likely contributes to the defective skin barrier in these mice and may induce expression of hyperproliferative proteins including Krt6 and Krt16. The premature lipid secretion observed in TAM67-positive epidermis (e.g., in the lower layers of stratum granulosum rather than chiefly at the stratum granulosum-stratum corneum) is also seen in Par2 knockout mice24 and other models of abnormal cornification, and may result from reduced entombment of lamellar bodies in the setting of the delayed apoptosis that occurs with epidermal hyperproliferation.
Keratins, envelope precursors and desmosomes
The keratoderma-like phenotype we observe is associated with reduced expression of differentiation-associated cutaneous keratins, and increased expression of hyperproliferation-associated (Krt6a, Krt16 and Krt17) keratins. Analysis of the cross-linked proteome in these mice reveals a reduction in differentiation-associated type I and type II cutaneous keratins (Krt1, Krt2, etc.) coupled with increased levels of Krt6a, Krt6b and Krt16. First, this is consistent with the likelihood that keratins which are available during CE formation are used in proportion to their abundance in keratinocytes. Second, these finding are consistent with reduced keratinocyte differentiation. Third, it appears that the change in keratin composition is associated with altered keratin filament formation, as the EM images suggest a decrease in keratin filaments in TAM67-positive epidermis. This may also have to do with the substantial reduction in the level of keratin organizing proteins, such as filaggrin.
Interestingly, we also observe a dramatic reduction in hair keratin and hair keratin-associated protein level in cross-linked proteome preparations derived from TAM67-positive epidermis (Figure 3b). Expression of a large number of genes encoding this class of proteins is reduced in TAM67-positive epidermis (Table 1). The hINV promoter,23 which we used to target TAM67 to epidermis, is expressed in the hair follicle.25 This suggests that hair follicle-localized AP1 factor function is required for expression of these hair follicle keratins.
We also observe increased expression of early CE (sprr proteins and involucrin) and reduced expression of late CE (Kprp, Lor, Lce1a1) precursors and filaggrin-related proteins (hornerin, filaggrin, filaggrin2). Many Lce genes are reduced in expression, as are genes encoding loricrin, filaggrin2, trichohyalin and cornulin. Here again, this finding suggests a key role for AP1 transcription factors in maintaining appropriate expression. Considering the marked reduction in filaggrin level, it is interesting that the level of bleomycin hydrolase, a neutral cysteine protease that breaks down filaggrin into amino acids,26 is not altered in expression or in cross-linked proteome content (not shown).
Desmosome protein expression is also altered. Desmoplakin is an abundant desmosomal protein and epidermal genetic disorders are associated with mutation of the desmoplakin gene.27 Desmoplakin haploinsufficiency is associated with striate palmoplantar keratoderma27, 28 and a desmoplakin mutant causes autosomal recessive cardiomyopathy-associated PPK.28 Plakoglobin mutations are associated with autosomal recessive Naxos Syndrome29 and plakoglobin null mice display a severe skin phenotype.30 DSCs and DSGs are core components to the desmosome that are expressed in epidermis. DSG1 and DSC1 are expressed in the suprabasal, whereas DSG2, DSG3 and DSC2 are expressed in the basal epidermis.29 Consistent with reduced differentiation/enhanced proliferation of TAM67-positive epidermis, the basal layer DSCs, Dsc2 and Dsg3, are increased in expression. It is interesting that the cornified layer has abnormal desmosomes. In this context, we note that caspase 14, which localizes at desmosomes in epidermis,31 and may impact desmosome processing, is markedly reduced in preparations derived from TAM67-expressing epidermis (not shown). In addition, cathepsin D that degrades desmosomes during desquamation,32 is 50% reduced in TAM67-positive envelopes (not shown). Moreover, the level of mRNA encoding the hair follicle DSG, Dsg4,29 is reduced in TAM67-positive epidermis (not shown). These findings are consistent with a general reduction in expression of hair follicle associated genes (e.g., hair keratins and associated proteins).
As loss of suprabasal AP1 factor function results in loss of differentiation-associated keratin, envelope precursor and desmosomal protein expression, it is not surprising that the epidermis, as shown in Figures 1 and 2, is visually, histologically and functionally impaired. To better understand this, we monitored markers associated with epidermal response to stress.
Epidermal defense
S100 proteins and annexins interact to from macromolecular complexes33, 34 that are incorporated as components of the CE where they have inflammatory and bacterial defense actions.12, 34 S100A8 and S100A9, which are markedly enriched in TAM67-positive epidermis, are frequently highly expressed in hyperproliferative diseases.35 S100A8 and S100A9 mRNA levels are among the most increased in TAM67 epidermis. Expression of two defensin genes, Defb3 and Defb4, is also markedly increased. Taken together, these results suggest a response to impending or active infection.
Lipoxygenases
Lipoxygenases are a class of ubiquitous nonheme iron containing dioxygenases that catalyze the stereo- and regiospecific incorporation of molecular oxygen into polyunsaturated fatty acids containing a (cis,cis)-1,4-pentadiene structure. The products are hydroperoxides that are rapidly transformed into other compounds.36 The mouse genome encodes seven lipoxygenase genes on mouse chromosome 11. The six human genes are on chromosome 17.37 Lipoxygenase genes are classified based on the positional specificity of arachidonic acid oxygenation (e.g., 5-, 8-, 12- and 15-LOX) and the tissue of discovery (i.e. platelet-type 12-LOX (p12-LOX)). Our analysis reveals that lipoxygenease gene expression and lipoxygenase incorporation into CEs is altered in TAM67-positive epidermis and suggests these changes may contribute to development of the abnormal epidermal phenotype.
The ALOX12B gene (ID 11686) product catalyzes oxidation of acyl-ceramide to form ox-acyl-ceramide during synthesis of the cornified lipid envelope.38 Our studies show that Alox12b levels are reduced in the cross-linked proteome of TAM67-positive epidermis. Mutations that inactivate the ALOX12B gene product are associated with autosomal recessive congenital ichthyosis (ARCI) types including harlequin ichthyosis, lamellar ichthyosis and congenital ichthyosiform erythroderma.39 These rare diseases are characterized by epidermal scaling and birth as collodion babies.40 After loss of the collodion membrane in the first weeks of life these patients exhibit generalized scaling, erythema, epidermal hyperplasia and hyperkeratosis. It is likely that this phenotype is a compensatory response to the loss of cutaneous barrier function. These patients may also display palmoplantar hyperlinearity with or without keratoderma. A recent study identified mutations in ALOX12B as a cause of a form of ARCI called ‘self-healing collodion baby'.41 A specific ALOX12B (p.Tyr521Cys) mutation is frequently present in these patients and small deletion, insertion and single-nucleotide mutations are also observed.39, 41, 42 Mice harboring a loss-of-function Alox12b mutation display an ARCI phenotype,43 and Alox12b knockout mice are born with red/shiny skin and rapidly desiccate and perish after birth.44 Our studies showing reduced Alox12b level in CE are consistent with reduced ceramide processing and reduced assembly of the cornified lipid envelope as a contributing factor in phenotype development. This is consistent with our EM micrographs showing a loss of lipid envelope formation.
The Alox15b gene (ID 11688) product converts 8(S)-hydroperoxyeicosatetraenoic acid (8S-HPETE) from arachidonic acid and also oxygenates α-linolenic acid and docosahexaenoic acid and converts 5-HPETE to LTA4.45 Alox15b is expressed in cutaneous adnexa and hair follicles, and is also abundant in sebaceous, eccrine and apocrine glands.46 The presence of ALOX15B in secretory glands suggests a role in regulating secretory processes.46 Alox15b is also expressed in mouse and human interfollicular epidermis. In mouse epidermis, expression is observed in the differentiated layers.47 Alox15b enhances keratinocyte differentiation and suppresses cell growth.48 Expression of Alox15b in cultured cells suppresses cell growth via effects on p38 mitogen-activated protein kinase signaling.49, 50 Similarly, transgenic mice overexpressing Alox15b display enhanced epidermal differentiation, and reduced tumor formation.48, 50 Likely as a compensatory response, Alox15b level is substantially increased in the granulosum of IκBα-deficient mice, which display a psoriatic phenotype.51 Alox15b is also induced following treatment of epidermis with phorbol ester.47 Thus, it has been proposed that Alox15b is a differentiation marker and may be involved in promoting terminal differentiation. We observed a substantial increase in Alox15b level in the cross-linked proteome prepared from TAM67-positive epidermis. We propose that this is a compensatory response as part of a program to restore normal differentiation.
The Alox12l gene (ID 11687) encodes leukocyte-type 12-lipoxygenase, which is produced by peritoneal macrophages, but is not produced in epidermis.37 Our finding that Alox12l is elevated in the CE in TAM67-positive mouse epidermis probably represents macrophage invasion into the epidermis. Finally, we observed a substantial increase in the level of mRNA encoded by the Alox12e gene (ID 11685). This lipoxygenase is predominantly expressed in the suprabasal epidermis and part of the hair follicle.52 We are not sure why this gene is increased in expression, except that it may be a compensatory mechanism associated with reduced differentiation.
Chitinase-like proteins
Chitinases are enzymes that breakdown chitin. Two active enzymes have been identified and several related proteins that lack enzymatic activity.19 It has been proposed that active chitinases are involved in defense in response to infection; however, the function of the inactive (chitin like) enzymes is not well understood. The level of these proteins is markedly increased in the chronic proliferative dermatitis mouse. This is associated with epidermal thickening, dermal neovascularization and fibrosis, and infiltration by eosinophils, macrophages and dendritic cells.53, 54 Chi3l3 and Chi3l4 accumulation in epidermis is localized to macrophages, dendritic cells and mast cells.19 This may suggest that the increase in these proteins in TAM67-expressing epidermis is owing to immune cell invasion.
Summary
Previous studies suggest that AP1 factors are important regulators of differentiation in the human epidermis.1, 2, 55 The present study expands the number of potential genes that may be regulated by AP1 factors. Moreover, altered expression of these genes is associated with other skin diseases. Some of these genes are candidates as direct targets of AP1 factor action (e.g., late envelope precursors, filaggrin, loricrin, etc.),9, 56, 57, 58, 59, 60, 61 whereas other genes likely change expression and envelope incorporation as a compensatory response to reduced differentiation or loss of barrier function. Additional studies will be necessary to sort out the regulatory arrangements. An important point is that TAM67-associated AP1 factor inactivation in the suprabasal epidermis produces a dramatic phenotype4, 5 (Figure 1), but TAM67 expression in the basal epidermis does not.62 This supports our contention that perturbing transcription factor function can produce changes in phenotype in an epidermal localization-specific manner.4
Materials and Methods
Animals
The TAM67-rTA mice used in this study are the offspring of a cross of hINV-rTA +/− mice and TetO-TAM67-FLAG mice and are maintained in a hairless SKH1 background.5 The hINV-rTA mice express the reverse-tetracycline activator (rTA) protein targeted to the suprabasal epidermis. Interaction with doxycycline converts the rTA protein to an active conformation that binds to the TetO in the TetO-TAM67-FLAG cassette to drive TAM67 expression. Thus, treatment of TAM67-rTA mice with doxycycline in the drinking water results in suprabasal TAM67-FLAG expression.4, 5 The transgenes are maintained in the SKH-1 hairless genetic background to facilitate visualization of the epidermal phenotype.5 Mice were housed in the University of Maryland School of Medicine animal facility in compliance with NIH regulations. Eight-week-old female TAM67-rTA mice were treated with 0 or 2 mg/ml doxycycline in their drinking water for 0–2 weeks to induce the phenotype. At various time points, the mice were sedated, euthanized and the dorsal skin and epidermis were collected.
Barrier integrity, envelope counting and BrdU labeling assays
To assess the impact of loss of AP1 factor function on barrier integrity, a TetO-TAM67-FLAG male was bred with a hINV-rTA +/− female. At E14, the pregnant female was treated with 2 mg/ml doxycycline in the drinking water and embryos were collected on the day before birth (E20). Pregnant females were anesthetized and killed by cervical dislocation and the embryos were removed, washed with phosphate-buffered saline and treated sequentially with 25, 50, 75 and 100% methanol. The mice were washed with phosphate-buffered saline and incubated for 10 min in a 0.1% toluidine blue solution. The pups were thoroughly washed with phosphate-buffered saline and photographed.
To count cornified envelopes, epidermis from dorsal skin (2 cm2) was collected, minced and boiled in 2% SDS containing 5 mM β−mercaptoethanol. Under these conditions, cells are solubilized but CEs survive. An aliquot of the samples was diluted 1 : 10 in sterile water and counted.33
For BrdU labeling of epidermis, eight week old female TAM67-rTA SKH1 mice were intraperitoneally injected with 50 mg BrdU per kg body weight. At 0–10 days thereafter, mice were collected and paraffin-embedded/formalin-fixed epidermal sections were prepared and stained with mouse monoclonal anti-BrdU clone BU-33 (Sigma, B8434, St. Louis, MO, USA). Primary antibody binding was visualized using biotin-labeled anti-mouse IgG in the Mouse on Mouse detection kit from Vector Labs (Burlingame, CA, USA) (PK-2200).
Electron microscopy
Skin biopsies were taken for electron microscopy.63 Briefly, samples were minced to <0.5 mm3, fixed in modified Karnovsky's fixative overnight and postfixed in 1% aqueous osmium tetroxide containing 1.5% potassium ferrocyanide or in 0.2% ruthenium tetroxide (the latter only where indicated in figure legends). After fixation, all samples were dehydrated in a graded ethanol series and embedded in an Epon-epoxy mixture. Ultrathin sections were examined, with or without further contrasting with lead citrate, in a JEOL electron microscope (JEOL USA, Inc., Peabody, MA, USA), operated at 60 kV. Cornified envelope thickness was determined by three independent measurements of each envelope present on three fields for each genotype using Gatan Imaging software (Pleasanton, CA, USA).
Immunoblot protein detection
Equivalent amounts of protein were electrophoresed on denaturing and reducing polyacrylamide gels and transferred to nitrocellulose. The nitrocellulose membrane was blocked by 5% nonfat dry milk for 1 hour and incubated with primary antibody (diluted 1 : 1000) in 5% nonfat dry milk followed by incubation with secondary antibody (diluted 1 : 5000) for 2 h at 37 °C. Secondary antibody binding was visualized with ECL Prime (Amersham) chemiluminescence detection reagent.
Mass spectrometry sample preparation
After the outer scale was removed and collected by scraping, the remaining epidermis was collected from the dermis by heating the excised skin in water at 52 °C for 1 min.64 The samples were then quick frozen at −70 °C. Scale samples were heated in 2% SDS −0.1 M sodium phosphate (pH 7.8) for 10 min in a 90 °C water bath and centrifuged for 5 min at 20 000 × g. Extraction of corneocytes with sodium dodecyl sulfate (SDS) yields proteins linked by disulfide and isopeptide bonding, nearly all of which are represented in the isopeptide cross-linked (envelope) fraction.8, 33 The cloudy supernatant was discarded, and the samples were washed four times with and then resuspended in 2% SDS −0.1 M sodium phosphate, pH 7.8. Samples (0.4 ml) were then reduced with 25 mM dithioerythritol and alkylated with 50 mM iodoacetamide at room temperature in the dark with stirring. The protein components were collected by addition of 1.1 ml ethanol followed by centrifugation, rinsed twice with 67% ethanol and once in 0.1 M ammonium bicarbonate and digested with reductively methylated bovine trypsin as previously described.8 The samples were then submitted for mass spectrometry.
Mass spectrometry
Sample digests (adjusted to equal peptide amounts by A280) were acidified with trifluoroacetic acid and directly loaded onto a nano LC column from which peptides were eluted with a formic acid-acetonitrile gradient and analyzed as previously described.65 The extraction of peptide spectra was performed using X! Tandem (The GPM, thegpm.org; version CYCLONE (2013.02.01.1)) to search the Uniprot mouse database (86 070 entries) and a database of reverse sequences for estimation of false discovery rate for trypsin digests. Peptide and protein (minimum of two peptides) identifications (0.2 and 3.6% false discovery rates, respectively) were verified by Scaffold version_4.2.1 (Proteome Software Inc., Portland, OR). Numbers of distributed spectral counts (called weighed spectral counts in Scaffold) were tabulated using experiment-wide grouping, which provides more accuracy than total spectral counts for proteins (e.g., certain keratins) with shared peptides.65
Gene array analysis
For gene array analysis, 8-week-old female TAM67-rTA SKH1 mice were treated for 8 days with 0 or 2 mg/ml doxycycline and the epidermis was recovered using dispase.4, 5 Approximately 5–8 μg of total RNA was reverse transcribed, amplified and labeled as described.66 Labeled cRNAs were hybridized to Affymetrix Mouse Gene 1.0 ST Arrays, which cover 28 853 genes. The arrays were washed, stained with anti-biotin streptavidin-phycoerythrin labeled antibody using fluidics station and then washed again according to the Affymetrix protocol. Data analysis was described previously.67 Briefly, intensity values were obtained using Microarray Suite version 5.0 (Affymetrix, Santa Clara, CA, USA), and scaled by calculating the overall signal for each array. Differential expression was defined as a twofold or greater difference in expression. For annotation we used the DAVID program [http://david.abcc.ncifcrf.gov].68 DAVID provides tables containing functional and ontological details of the genes in the uploaded list, charts containing ontological categories, pathways and so on, over-represented in the gene lists, as well as clusters of such ontological categories, which consolidates redundancies and overlaps, transcription factors over-represented in the promoters of the genes, as well as sublists of genes specific for each ontological category.
Acknowledgments
This work was supported by National Institutes of Health grants to RLE (R21 AR065266), RHR (P42 ES04699) and PME (AR061106). We thank Jan Babus for excellent technical assistance with animal breeding, characterization and care, and Brett S. Phinney for expert assistance with mass spectrometry.
Glossary
- CE
cornified envelope
- Krt
keratin
- AP1
activator protein 1
- SDS
sodium dodecyl sulfate
- ARCI
autosomal recessive congenital ichthyosis
- BrdU
bromodeoxyuridine
- DSC
desmocollin
- DSG
desmoglein
The authors declare no conflict of interest.
Footnotes
Edited by E Candi
References
- 1Eckert RL, Welter JF. Transcription factor regulation of epidermal keratinocyte gene expression. Mol Biol Rep 1996; 23: 59–70. [DOI] [PubMed] [Google Scholar]
- 2Angel P, Szabowski A, Schorpp-Kistner M. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene 2001; 20: 2413–2423. [DOI] [PubMed] [Google Scholar]
- 3Efimova T, Broome AM, Eckert RL. A regulatory role for p38 delta MAPK in keratinocyte differentiation. Evidence for p38 delta-ERK1/2 complex formation. J Biol Chem 2003; 278: 34277–34285. [DOI] [PubMed] [Google Scholar]
- 4Rorke EA, Adhikary G, Jans R, Crish JF, Eckert RL. AP1 factor inactivation in the suprabasal epidermis causes increased epidermal hyperproliferation and hyperkeratosis but reduced carcinogen-dependent tumor formation. Oncogene 2010; 29: 5873–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Rorke EA, Adhikary G, Young CA, Roop DR, Eckert RL. Suppressing AP1 Factor Signaling in Suprabasal Epidermis Produces A Keratoderma Phenotype. J Invest Dermatol 2014; 135: 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 6Brown PH, Kim SH, Wise SC, Sabichi AL, Birrer MJ. Dominant-negative mutants of cJun inhibit AP-1 activity through multiple mechanisms and with different potencies. Cell Growth Differ 1996; 7: 1013–1021. [PubMed] [Google Scholar]
- 7Rice RH, Bradshaw KM, Durbin-Johnson BP, Rocke DM, Eigenheer RA, Phinney BS et al. Distinguishing ichthyoses by protein profiling. PLoS One 2013; 8: e75355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Rice RH, Bradshaw KM, Durbin-Johnson BP, Rocke DM, Eigenheer RA, Phinney BS et al. Differentiating inbred mouse strains from each other and those with single gene mutations using hair proteomics. PLoS One 2012; 7: e51956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Jang SI, Steinert PM. Loricrin expression in cultured human keratinocytes is controlled by a complex interplay between transcription factors of the Sp1, CREB, AP1, and AP2 families. J Biol Chem 2002; 277: 42268–42279. [DOI] [PubMed] [Google Scholar]
- 10Schroder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci 2006; 63: 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Hattinger E, Zwicker S, Ruzicka T, Yuspa SH, Wolf R. Opposing functions of psoriasin (S100A7) and koebnerisin (S100A15) in epithelial carcinogenesis. Curr Opin Pharmacol 2013; 13: 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Eckert RL, Broome AM, Ruse M, Robinson N, Ryan D, Lee K. S100 proteins in the epidermis. J Invest Dermatol 2004; 123: 23–33. [DOI] [PubMed] [Google Scholar]
- 13Lin TK, Crumrine D, Ackerman LD, Santiago JL, Roelandt T, Uchida Y et al. Cellular changes that accompany shedding of human corneocytes. J Invest Dermatol 2012; 132: 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Heit C, Jackson BC, McAndrews M, Wright MW, Thompson DC, Silverman GA et al. Update of the human and mouse SERPIN gene superfamily. Hum Genomics 2013; 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Fischer J, Meyer-Hoffert U. Regulation of kallikrein-related peptidases in the skin - from physiology to diseases to therapeutic options. Thromb Haemost 2013; 110: 442–449. [DOI] [PubMed] [Google Scholar]
- 16Tsui FW, Tsui HW, Mok S, Mlinaric I, Copeland NG, Gilbert DJ et al. Molecular characterization and mapping of murine genes encoding three members of the stefin family of cysteine proteinase inhibitors. Genomics 1993; 15: 507–514. [DOI] [PubMed] [Google Scholar]
- 17Mihelic M, Teuscher C, Turk V, Turk D. Mouse stefins A1 and A2 (Stfa1 and Stfa2) differentiate between papain-like endo- and exopeptidases. FEBS Lett 2006; 580: 4195–4199. [DOI] [PubMed] [Google Scholar]
- 18Kzhyshkowska J, Gratchev A, Goerdt S. Human chitinases and chitinase-like proteins as indicators for inflammation and cancer. Biomark Insights 2007; 2: 128–146. [PMC free article] [PubMed] [Google Scholar]
- 19HogenEsch H, Dunham A, Seymour R, Renninger M, Sundberg JP. Expression of chitinase-like proteins in the skin of chronic proliferative dermatitis (cpdm/cpdm) mice. Exp Dermatol 2006; 15: 808–814. [DOI] [PubMed] [Google Scholar]
- 20Uderhardt S, Kronke G. 12/15-lipoxygenase during the regulation of inflammation, immunity, and self-tolerance. J Mol Med (Berl) 2012; 90: 1247–1256. [DOI] [PubMed] [Google Scholar]
- 21Nekrasova O, Green KJ. Desmosome assembly and dynamics. Trends Cell Biol 2013; 23: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Green KJ, Kowalczyk AP, Bornslaeger EA, Palka HL, Norvell SM. Desmosomes: integrators of mechanical integrity in tissues. Biol Bull 1998; 194: 374–376. [DOI] [PubMed] [Google Scholar]
- 23Crish JF, Howard JM, Zaim TM, Murthy S, Eckert RL. Tissue-specific and differentiation-appropriate expression of the human involucrin gene in transgenic mice: an abnormal epidermal phenotype. Differentiation 1993; 53: 191–200. [DOI] [PubMed] [Google Scholar]
- 24Demerjian M, Hachem JP, Tschachler E, Denecker G, Declercq W, Vandenabeele P et al. Acute modulations in permeability barrier function regulate epidermal cornification: role of caspase-14 and the protease-activated receptor type 2. Am J Pathol 2008; 172: 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Murthy S, Crish JF, Zaim TM, Eckert RL. A dual role for involucrin in the epidermis-ultrastructural localization in epidermis and hair follicle in humans and transgenic mice. J Struct Biol 1993; 111: 68–76. [DOI] [PubMed] [Google Scholar]
- 26Kamata Y, Taniguchi A, Yamamoto M, Nomura J, Ishihara K, Takahara H et al. Neutral cysteine protease bleomycin hydrolase is essential for the breakdown of deiminated filaggrin into amino acids. J Biol Chem 2009; 284: 12829–12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Lai Cheong JE, Wessagowit V, McGrath JA. Molecular abnormalities of the desmosomal protein desmoplakin in human disease. Clin Exp Dermatol 2005; 30: 261–266. [DOI] [PubMed] [Google Scholar]
- 28Armstrong DK, McKenna KE, Purkis PE, Green KJ, Eady RA, Leigh IM et al. Haploinsufficiency of desmoplakin causes a striate subtype of palmoplantar keratoderma. Hum Mol Genet 1999; 8: 143–148. [DOI] [PubMed] [Google Scholar]
- 29Brooke MA, Nitoiu D, Kelsell DP. Cell-cell connectivity: desmosomes and disease. J Pathol 2012; 226: 158–171. [DOI] [PubMed] [Google Scholar]
- 30Kottke MD, Delva E, Kowalczyk AP. The desmosome: cell science lessons from human diseases. J Cell Sci 2006; 119: 797–806. [DOI] [PubMed] [Google Scholar]
- 31Alibardi L, Dockal M, Reinisch C, Tschachler E, Eckhart L. Ultrastructural localization of caspase-14 in human epidermis. J Histochem Cytochem 2004; 52: 1561–1574. [DOI] [PubMed] [Google Scholar]
- 32Igarashi S, Takizawa T, Takizawa T, Yasuda Y, Uchiwa H, Hayashi S et al. Cathepsin D, but not cathepsin E, degrades desmosomes during epidermal desquamation. Br J Dermatol 2004; 151: 355–361. [DOI] [PubMed] [Google Scholar]
- 33Robinson NA, Lapic S, Welter JF, Eckert RL. S100A11, S100A10, annexin I, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J Biol Chem 1997; 272: 12035–12046. [DOI] [PubMed] [Google Scholar]
- 34Ruse M, Lambert A, Robinson N, Ryan D, Shon KJ, Eckert RL. S100A7 S100A10, and S100A11 are transglutaminase substrates. Biochemistry 2001; 40: 3167–3173. [DOI] [PubMed] [Google Scholar]
- 35van BL, Cossu M, Loof A, Gohar F, Wittkowski H, Vonk M et al. Proteomic analysis of plasma identifies the Toll-like receptor agonists S100A8/A9 as a novel possible marker for systemic sclerosis phenotype. Ann Rheum Dis 2014; 73: 1585–1589. [DOI] [PubMed] [Google Scholar]
- 36Haeggstrom JZ, Funk CD. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev 2011; 111: 5866–5898. [DOI] [PubMed] [Google Scholar]
- 37Krieg P, Furstenberger G. The role of lipoxygenases in epidermis. Biochim Biophys Acta 2014; 1841: 390–400. [DOI] [PubMed] [Google Scholar]
- 38Zheng Y, Yin H, Boeglin WE, Elias PM, Crumrine D, Beier DR, Brash AR. Lipoxygenases mediate the effect of essential fatty acid in skin barrier formation: a proposed role in releasing omega-hydroxyceramide for construction of the corneocyte lipid envelope. J Biol Chem 2011; 286: 24046–24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Eckl KM, de JS, Kurtenbach J, Natebus M, Lugassy J, Oji V et al. Molecular analysis of 250 patients with autosomal recessive congenital ichthyosis: evidence for mutation hotspots in ALOXE3 and allelic heterogeneity in ALOX12B. J Invest Dermatol 2009; 129: 1421–1428. [DOI] [PubMed] [Google Scholar]
- 40Oji V, Traupe H. Ichthyoses: differential diagnosis and molecular genetics. Eur J Dermatol 2006; 16: 349–359. [PubMed] [Google Scholar]
- 41Vahlquist A, Bygum A, Ganemo A, Virtanen M, Hellstrom-Pigg M, Strauss G, Brandrup F, Fischer J. Genotypic and clinical spectrum of self-improving collodion ichthyosis: ALOX12B, ALOXE3, and TGM1 mutations in Scandinavian patients. J Invest Dermatol 2010; 130: 438–443. [DOI] [PubMed] [Google Scholar]
- 42Jobard F, Lefevre C, Karaduman A, Blanchet-Bardon C, Emre S, Weissenbach J et al. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet 2002; 11: 107–113. [DOI] [PubMed] [Google Scholar]
- 43Moran JL, Qiu H, Turbe-Doan A, Yun Y, Boeglin WE, Brash AR, Beier DR. A mouse mutation in the 12R-lipoxygenase, Alox12b, disrupts formation of the epidermal permeability barrier. J Invest Dermatol 2007; 127: 1893–1897. [DOI] [PubMed] [Google Scholar]
- 44de JS, Epp N, Latzko S, Neumann M, Furstenberger G, Hausser I et al. Development of an ichthyosiform phenotype in Alox12b-deficient mouse skin transplants. J Invest Dermatol 2009; 129: 1429–1436. [DOI] [PubMed] [Google Scholar]
- 45Qiao N, Takahashi Y, Takamatsu H, Yoshimoto T. Leukotriene A synthase activity of purified mouse skin arachidonate 8-lipoxygenase expressed in Escherichia coli. Biochim Biophys Acta 1999; 1438: 131–139. [DOI] [PubMed] [Google Scholar]
- 46Shappell SB, Keeney DS, Zhang J, Page R, Olson SJ, Brash AR. 15-Lipoxygenase-2 expression in benign and neoplastic sebaceous glands and other cutaneous adnexa. J Invest Dermatol 2001; 117: 36–43. [DOI] [PubMed] [Google Scholar]
- 47Furstenberger G, Hagedorn H, Jacobi T, Besemfelder E, Stephan M, Lehmann WD, Marks F. Characterization of an 8-lipoxygenase activity induced by the phorbol ester tumor promoter 12-O-tetradecanoylphorbol-13-acetate in mouse skin in vivo. J Biol Chem 1991; 266: 15738–15745. [PubMed] [Google Scholar]
- 48Muga SJ, Thuillier P, Pavone A, Rundhaug JE, Boeglin WE, Jisaka M et al. 8S-lipoxygenase products activate peroxisome proliferator-activated receptor alpha and induce differentiation in murine keratinocytes. Cell Growth Differ 2000; 11: 447–454. [PubMed] [Google Scholar]
- 49Schweiger D, Furstenberger G, Krieg P. Inducible expression of 15-lipoxygenase-2 and 8-lipoxygenase inhibits cell growth via common signaling pathways. J Lipid Res 2007; 48: 553–564. [DOI] [PubMed] [Google Scholar]
- 50Kim E, Rundhaug JE, Benavides F, Yang P, Newman RA, Fischer SM. An antitumorigenic role for murine 8S-lipoxygenase in skin carcinogenesis. Oncogene 2005; 24: 1174–1187. [DOI] [PubMed] [Google Scholar]
- 51Schneider C, Strayhorn WD, Brantley DM, Nanney LB, Yull FE, Brash AR. Upregulation of 8-lipoxygenase in the dermatitis of IkappaB-alpha-deficient mice. J Invest Dermatol 2004; 122: 691–698. [DOI] [PubMed] [Google Scholar]
- 52Funk CD, Keeney DS, Oliw EH, Boeglin WE, Brash AR. Functional expression and cellular localization of a mouse epidermal lipoxygenase. J Biol Chem 1996; 271: 23338–23344. [DOI] [PubMed] [Google Scholar]
- 53HogenEsch H, Gijbels MJ, Offerman E, van HJ, van Bekkum DW, Zurcher C. A spontaneous mutation characterized by chronic proliferative dermatitis in C57BL mice. Am J Pathol 1993; 143: 972–982. [PMC free article] [PubMed] [Google Scholar]
- 54Aggarwal BB, Sethi G, Ahn KS, Sandur SK, Pandey MK, Kunnumakkara AB et al. Targeting signal-transducer-and-activator-of-transcription-3 for prevention and therapy of cancer: modern target but ancient solution. Ann N Y Acad Sci 2006; 1091: 151–169. [DOI] [PubMed] [Google Scholar]
- 55Zenz R, Wagner EF. Jun signalling in the epidermis: From developmental defects to psoriasis and skin tumors. Int J Biochem Cell Biol 2006; 38: 1043–1049. [DOI] [PubMed] [Google Scholar]
- 56Jang SI, Steinert PM, Markova NG. Activator protein 1 activity is involved in the regulation of the cell type-specific expression from the proximal promoter of the human profilaggrin gene. J Biol Chem 1996; 271: 24105–24114. [DOI] [PubMed] [Google Scholar]
- 57Rutberg SE, Adams TL, Glick A, Bonovich MT, Vinson C, Yuspa SH. Activator protein 1 transcription factors are fundamental to v-rasHa-induced changes in gene expression in neoplastic keratinocytes. Cancer Res 2000; 60: 6332–6338. [PubMed] [Google Scholar]
- 58Crish JF, Eckert RL. Synergistic activation of human involucrin gene expression by Fra-1 and p300—evidence for the presence of a multiprotein complex. J Invest Dermatol 2008; 128: 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59Kamioka N, Akahane T, Kohno Y, Kuroki T, Iijima M, Honma I, Ohba M. Protein kinase C delta and eta differently regulate the expression of loricrin and Jun family proteins in human keratinocytes. Biochem Biophys Res Commun 2010; 394: 106–111. [DOI] [PubMed] [Google Scholar]
- 60Navarro JM, Casatorres J, Jorcano JL. Elements controlling the expression and induction of the skin hyperproliferation-associated keratin K6. J Biol Chem 1995; 270: 21362–21367. [DOI] [PubMed] [Google Scholar]
- 61Phillips MA, Jessen BA, Lu Y, Qin Q, Stevens ME, Rice RH. A distal region of the human TGM1 promoter is required for expression in transgenic mice and cultured keratinocytes. BMC Dermatol 2004; 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62Young MR, Li JJ, Rincon M, Flavell RA, Sathyanarayana BK, Hunziker R, Colburn N. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA 1999; 96: 9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63Hou SY, Mitra AK, White SH, Menon GK, Ghadially R, Elias PM. Membrane structures in normal and essential fatty acid-deficient stratum corneum: characterization by ruthenium tetroxide staining and x-ray diffraction. J Invest Dermatol 1991; 96: 215–223. [DOI] [PubMed] [Google Scholar]
- 64Macdiarmid J, Wilson JB. Separation of epidermal tissue from underlying dermis and primary keratinocyte culture. Methods Mol Biol 2001; 174: 401–410. [DOI] [PubMed] [Google Scholar]
- 65Laatsch CN, Durbin-Johnson BP, Rocke DM, Mukwana S, Newland AB, Flagler MJ et al. Human hair shaft proteomic profiling: individual differences, site specificity and cuticle analysis. PeerJ 2014; 2: e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66Li D, Turi TG, Schuck A, Freedberg IM, Khitrov G, Blumenberg M. Rays and arrays: the transcriptional program in the response of human epidermal keratinocytes to UVB illumination. FASEB J 2001; 15: 2533–2535. [DOI] [PubMed] [Google Scholar]
- 67Mimoso C, Lee DD, Zavadil J, Tomic-Canic M, Blumenberg M. Analysis and meta-analysis of transcriptional profiling in human epidermis. Methods Mol Biol 2014; 1195: 61–97. [DOI] [PubMed] [Google Scholar]
- 68Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003; 4: P3. [PubMed] [Google Scholar]
- 69Nagele P. Misuse of standard error of the mean (SEM) when reporting variability of a sample. A critical evaluation of four anaesthesia journals. Br J Anaesth 2003; 90: 514–516. [DOI] [PubMed] [Google Scholar]
- 70Efimova T, Deucher A, Kuroki T, Ohba M, Eckert RL. Novel protein kinase C isoforms regulate human keratinocyte differentiation by activating a p38 delta mitogen-activated protein kinase cascade that targets CCAAT/enhancer-binding protein alpha. J Biol Chem 2002; 277: 31753–31760. [DOI] [PubMed] [Google Scholar]