Abstract
Nucleotide excision repair (NER) protects against sunlight-induced skin cancer. Defective NER is associated with photosensitivity and a high skin cancer incidence. Some clinical treatments that cause photosensitivity can also increase skin cancer risk. Among these, the immunosuppressant azathioprine and the fluoroquinolone antibiotics ciprofloxacin and ofloxacin, interact with UVA radiation to generate reactive oxygen species (ROS) that diminish NER capacity by causing protein damage. The RPA DNA binding protein plays a pivotal role in DNA metabolism and is an essential component of NER. The relationship between protein oxidation and NER inhibition was investigated in cultured human cells expressing different levels of RPA. We show here that RPA is limiting for NER and that oxidative damage to RPA compromises NER capability. Our findings reveal that cellular RPA is surprisingly vulnerable to oxidation and we identify oxidized forms of RPA that are associated with impaired NER. The vulnerability of NER to inhibition by oxidation provides a connection between cutaneous photosensitivity, protein damage and increased skin cancer risk. Our findings emphasize that damage to DNA repair proteins, as well as to DNA itself is likely to be an important contributor to skin cancer risk.
Introduction
Skin cancer is one of the few human cancers that are associated with a well-defined and pervasive mutagen. Non-melanoma skin cancer genomes are dominated by the mutational signatures of sunlight-induced DNA lesions (Alexandrov et al., 2013; Jayaraman et al., 2014; South et al., 2014). The nucleotide excision repair (NER) system provides an important protection against skin cancer by removing potentially mutagenic DNA cyclobutane pyrimidine dimer (CPD) and pyrimidine (6-4) pyrimidone (6-4Py:Py) photolesions induced by solar UVB radiation. Inefficient NER in the genetic disorder xeroderma pigmentosum is associated with photosensitivity and a hugely increased skin cancer risk (Friedberg et al., 2006).
Photosensitivity and an increased susceptibility to sunlight-related malignancy are side effects of some medications. For example, patients prescribed azathioprine for inflammatory bowel disorders or to prevent organ transplant rejection are UVA photosensitive and have high rates of non-melanoma skin cancer (Peyrin-Biroulet et al., 2011; Ramiscal et al., 2013; Euvrard et al., 2003). Azathioprine metabolism culminates in the incorporation of 6-thioguanine (6-TG) into patients’ DNA where it acts as a potent UVA photosensitizer. In cultured human cells, the combination of DNA 6-TG and UVA generates reactive oxygen species (ROS). These cause widespread DNA and protein damage that is associated with inhibition of DNA repair (Gueranger et al., 2014). Other photosensitizing drugs include the fluoroquinolone antibiotics that are photocarcinogens in mice (Itoh et al., 2005) and may increase skin cancer risk in patients (Traianou et al., 2012).
Combinations of UVA and the fluoroquinolones ciprofloxacin or ofloxacin duplicate many of the effects of DNA 6-TG/UVA in cultured human cells. In particular, fluoroquinolone/UVA treatment causes DNA and protein damage and inhibits DNA repair, including NER (Peacock et al., 2014). Effective NER requires the coordinated activities of numerous proteins to recognise and process DNA lesions. The human replication protein A (RPA) is an abundant DNA binding protein complex that participates in almost all aspects of DNA metabolism and is essential for NER. Its high-affinity binding protects single-stranded DNA against nucleases and facilitates the correct processing of unpaired DNA regions generated during replication and repair (Binz et al., 2004). Recruitment of specific proteins to DNA-bound RPA is important in the activation of the cellular checkpoints that maintain genome stability in the face of arrested replication (Friedel et al., 2009). Consistent with its pivotal role in protecting DNA, genetic inactivation of murine RPA is lethal and haploinsufficiency is associated with cancer proneness and reduced lifespan (Wang et al., 2005). Reduced RPA expression in cultured human cells results in spontaneous DNA damage and activation of DNA damage-related checkpoints (Dodson et al., 2004). RFA, the yeast RPA homolog, is limiting for UV protection and hypomorphic RFA mutations confer UVC sensitivity (Umezu et al., 1998) and a mutator phenotype (Chen et al., 1998).
RPA is a heterotrimer (Wold, 1997). Its subunits of 70, 32 and 14 kDa (RPA70, RPA32 & RPA14) contain several DNA- and protein-interacting domains. The most important DNA binding motifs lie in the RPA70 subunit. Protein interacting domains are located in the N terminus of RPA70 and the C terminus of RPA32 (Binz et al., 2004). An N-terminal RPA32 domain also contains multiple conserved phosphorylation sites that control RPA function. RPA32 phosphorylation triggered by the presence of long single-stranded DNA tracts at arrested replication forks, may serve to regulate the distribution of limiting RPA resources between replication and repair (Reviewed in (Binz et al., 2004)). The role of RPA14 appears to be largely to stabilize the heterotrimer.
We previously showed that protein oxidation is associated with NER inhibition in human cells treated with 6-TG/UVA or fluoroquinolone/UVA (Gueranger et al., 2014; Peacock et al., 2014). Our results suggested that RPA damage might underlie the reduced NER capability. Here we demonstrate that the vulnerability of RPA to oxidation confers a risk of suboptimal NER under oxidative stress conditions. We describe oxidative RPA lesions in cultured human cells exposed to conditions that simulate events in the sun-exposed skin of patients taking azathioprine or fluoroquinolones. Our findings emphasize the importance of protein oxidation as a determinant of DNA repair efficiency and skin cancer risk in patients taking photosensitizing drugs.
Results
RPA expression, NER and toxicity
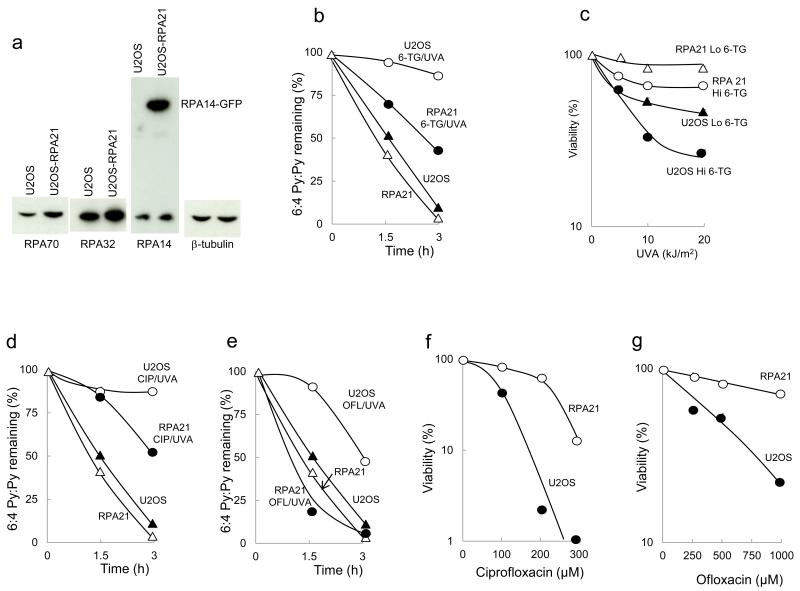
Treatment of human cells with 6-TG/UVA inhibits NER (Gueranger et al., 2014). To investigate how RPA influences NER efficiency, we compared the removal of UVC-induced 6-4 Py:Pys by U2OS and the related U2OS-RPA21 cells in which RPA is overexpressed (Toledo et al., 2013). Western blotting confirmed an approximately two-fold RPA overexpression in U2OS-RPA21 cells (Figure 1a). Increased RPA protected NER against inhibition by 6-TG/UVA. Whereas 6-TG/UVA treated U2OS cells excised < 20% of UVC-induced 6-4 Py:Pys in three hours, similarly damaged U2OS-RPA21 cells removed approximately 60% of these photoproducts in the same period (Figure 1b). U2OS and U2OS-RPA21 cells irradiated with UVC alone removed > 95% of 6-4 Py:Pys in three hours.
Figure 1.
RPA overexpression, NER and sensitivity
a) Western blotting for RPA14, RPA32 and RPA70.
b) 6:4 Py:Py ELISA of 6-TG-treated U2OS and U2OS-RPA21 cells irradiated with UVA (20 kJ/m2) + 10 J/m2 UVC. ( ) 10 J/m2 UVC alone. Mean values from ≥ 3 experiments.
) 10 J/m2 UVC alone. Mean values from ≥ 3 experiments.
c) Viability of UVA irradiated cells with Lo (~ 0.06%) or Hi (~ 0.14%) DNA 6-TG. Mean values from 2 experiments.
d) 6:4 Py:Py ELISA of cells treated with ciprofloxacin (CIP) (1 h; 0.5 mM) + UVA (20 kJ/m2) + UVC (10 J/m2). Mean values from ≥ 3 experiments.
e) As D). Cells treated with 1 mM ofloxacin, UVA + UVC.
f) Ciprofloxacin/UVA toxicity. Viability of cells treated for 1 h with ciprofloxacin and UVA (20 kJ/m2). Mean values from ≥ 3 experiments.
g) Ofloxacin/UVA toxicity. As F). Cells were treated with ofloxacin and UVA.
Increased RPA expression also conferred resistance to the toxicity of 6-TG/UVA. MTT assays revealed a UVA dose-dependent decrease in U2OS viability at two levels of DNA 6-TG substitution. At the higher DNA 6-TG level, approximately < 20% of U2OS cells survived after 20 kJ/m2 UVA. In contrast, DNA 6-TG-containing U2OS-RPA21 cells were largely insensitive to UVA and > 80% survived even at the highest UVA dose (Figure 1c). Treatment with 6-TG or UVA alone reduced viability by < 10%.
6-TG is an atypical photosensitizer in that it is DNA-embedded. The UVA absorbing fluoroquinolone antibiotics ciprofloxacin and ofloxacin replicate many of the photosensitizing effects of DNA 6-TG without incorporation into DNA. In particular, ciprofloxacin/UVA and ofloxacin/UVA induce oxidative stress and cause widespread protein damage that compromises NER (Peacock et al., 2014). 6-4Py:Py ELISA confirmed that ciprofloxacin/UVA and ofloxacin/UVA also inhibited NER in U2OS cells. Increased RPA expression in U2OS-RPA21 protected against inhibition. Figure 1d shows that whereas 500 μM ciprofloxacin/20 kJ/m2 UVA almost completely abolished 6-4Py:Py excision in U2OS cells, U2OS-RPA21 cells removed around 50% of these photoproducts within 3 hours under the same conditions. Higher RPA expression also protected against NER inhibition by ofloxacin/UVA (Figure 1e). 1 mM ofloxacin/20 kJ/m2 UVA that caused a significant inhibition of 6-4Py:Py removal in U2OS cells, did not detectably affect excision by U2OS-RPA21 cells. Repair inhibition was the result of UVA/drug interaction and ciprofloxacin, ofloxacin or these low UVA doses alone were without effect.
Increased RPA expression also protected against fluoroquinolone/UVA toxicity. Figure 1f shows that the viability of UVA-irradiated (20 kJ/m2) U2OS cells was compromised by ciprofloxacin and < 2% survived treatment with 200 μM. In contrast, > 80% of irradiated U2OS-RPA21 cells survived treatment with 200 μM ciprofloxacin. Ofloxacin/UVA was significantly less toxic to both U2OS and U2OS-RPA21 cells. More than 20% of U2OS cells remained viable following 1000 μM/20 kJ/m2 and the effect of RPA overexpression on U2OS-RPA21 viability was correspondingly more modest (Figure 1g).
RPA chromatinization
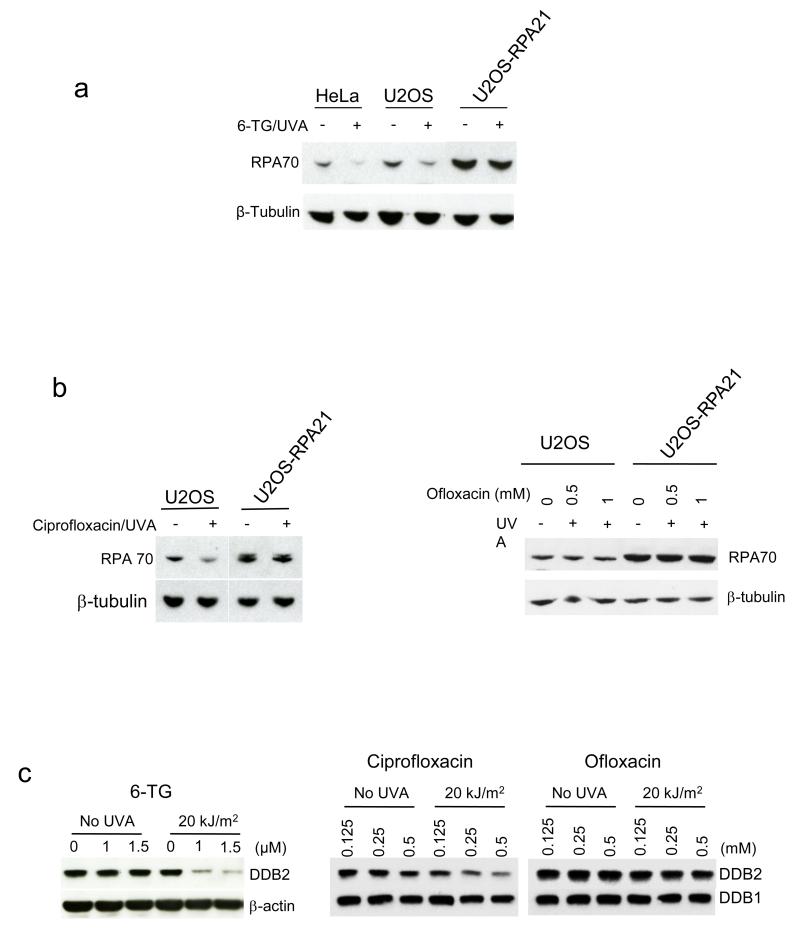
6-TG/UVA generates DNA lesions that are powerful replication blocks. UVA activation of certain fluoroquinolones induces T<>T CPDs (Lhiaubet-Vallet et al., 2009) that also arrest replication. Since RPA can become sequestered at arrested replication forks and no longer available for NER, we investigated recruitment of RPA to chromatin. 6-TG/UVA or ciprofloxacin/UVA reduced recovery of RPA in U2OS cell extracts (Figure 2a,b). Consistent with RPA chromatinization and the presence of replication-arresting DNA lesions, the missing RPA was recovered by digestion of the extracts with benzonase (Supplementary Figure 2). RPA recovery from U2OS-RPA21 cells was less affected owing to their higher RPA levels. As expected, ofloxacin/UVA did not detectably affect RPA recovery in U2OS extracts as it is a poor source of CPDs (Peacock et al., 2014) (Figure 2b). Chromatinization of the DDB2 component of the DDB1:DDB2 DNA damage recognition complex is another marker for the presence of NER substrates (Otrin et al., 1997). Figure 2c shows that 6-TG/UVA and ciprofloxacin/UVA but not ofloxacin/UVA provoked immediate relocalization of a substantial fraction of DDB2 in U2OS. This observation provides further confirmation that 6-TG/UVA and ciprofloxacin/UVA induce DNA damage that includes potential substrates for NER. Ofloxacin/UVA is, however, a poor source of such lesions.
Figure 2.
RPA and DDB2 chromatinization following treatment
a) 6-TG/UVA. HeLa, U2OS or U2OS-RPA21 cells grown for 48 h in 1 μM, 4 μM or 10 μM 6-TG, respectively to ensure equivalent DNA substitution were UVA irradiated (20 kJ/m2). Whole cell extracts were analysed by western blotting for RPA70.
b) Ciprofloxacin/UVA and Ofloxacin/UVA. U2OS or U2OS-RPA21 cells were treated with 0.5 mM ciprofloxacin or ofloxacin/UVA as indicated and irradiated with UVA (20 kJ/m2). Whole cell extracts were analysed as in A.
c) DDB2. U2OS cells treated with 6-TG for 24 h or fluoroquinolone for 1 h at the concentrations shown were UVA irradiated as indicated. Whole cell extracts prepared immediately after irradiation were analysed by western blotting for DDB2 or DDB1/ β-actin as loading controls.
RPA damage
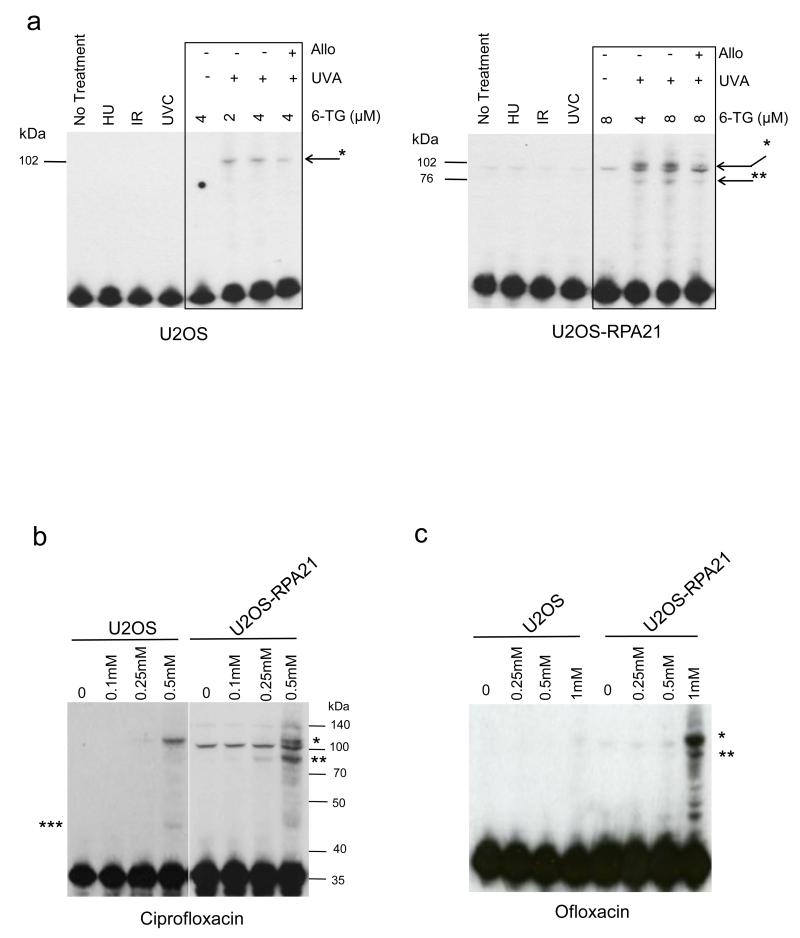
All three photosensitizers cause protein oxidation (Peacock et al., 2014). Since their effects on NER and survival did not correlate with the induction of DNA lesions, we examined whether RPA itself was damaged. Western blotting of extracts from 6-TG/UVA-treated U2OS cells revealed an RPA32 species of approximately 100 kDa (Figure 3a) that was absent from untreated cells. Western analysis of RPA32 in U2OS-RPA21 cells was complicated by the presence of additional constitutive RPA forms generated by incomplete processing of the full-length GFP-RPA polypeptide encoded by the construct (Supplementary Figure 1). In addition to the three correctly processed RPA proteins, U2OS-RPA21 extracts contained low but detectable levels of polypeptides corresponding to full length GFP/RPA14-RPA70-RPA32 (143 kDa) and RPA32-RPA70 fusion polypeptides (102 kDa) (Supplementary Figure 1). 6-TG/UVA treated U2OS-RPA21 extracts contained an RPA32 species, apparently identical to that induced in U2OS cells. This migrated slightly behind the incompletely processed constitutive 102 kDa RPA32-RPA70 product (Figure 3b). These extracts also contained an additional RPA32-reactive polypeptide of 70-100 kDa that was not present in extracts from untreated cells (Figure 3b). The antioxidant allopurinol suppressed the formation of both 100 and 70-100 kDa complexes, consistent with oxidation-dependent formation. Neither complex was detected after exposure to IR, UVC, or hydroxyurea all of which inhibited DNA replication by > 90% indicating that they are not generated in response to extensive DNA damage or replication arrest. Ciprofloxacin/UVA (Figure 3b) and ofloxacin/UVA (Figure 3c) generated an apparently identical 100 kDa RPA32 species. Consistent with the induction of a lower level of overall protein oxidation (Peacock et al., 2014), this complex was less prominent following ofloxacin/UVA treatment. The enrichment provided by nuclear extracts (see Supplementary Figures 3 & 4) confirmed its presence in ofloxacin/UVA treated cells. Additional complexes with apparent masses between 40 and 50 kDa and 70-100 kDa were observed in extracts from ciprofloxacin/UVA-treated U2OS and U2OS-RPA21 cells, respectively. These changes in RPA32 were not restricted to U2OS osteosarcoma cells and both ciprofloxacin/UVA and ofloxacin/UVA induced the 100 kDa and 40-50 kDa RPA species in untransformed GM8339 human fibroblasts (Supplementary Figure 3).
Figure 3.
Formation of RPA32 complexes
a) (Left panel). RPA32 western blots of extracts from U2OS cells either untreated or treated with hydroxyurea (HU; 3 mM, 3 h), IR (20 Gy), UVC (100 J/m2) or 6-TG/UVA (2 or 4 μM 6-TG, 24 h/20 kJ/m2). (Right panel) RPA32 western blots of U2OS-RPA21 cells treated identically except that 6-TG concentrations were 4 or 8 μM 6-TG/UVA to ensure similar levels of DNA 6-TG. Allopurinol (1 mM) was included during 6-TG treatment as indicated. RPA32 complexes are indicated (* and **).
b) RPA32 western blots of extracts from U2OS and U2OS-RPA21 cells treated for 1 h with ciprofloxacin/UVA (20 kJ/m2) as indicated. MW markers and RPA32 complexes (*, ** and ***) are indicated.
c) As Panel B. Cells treated with ofloxacin/UVA (20 kJ/m2).
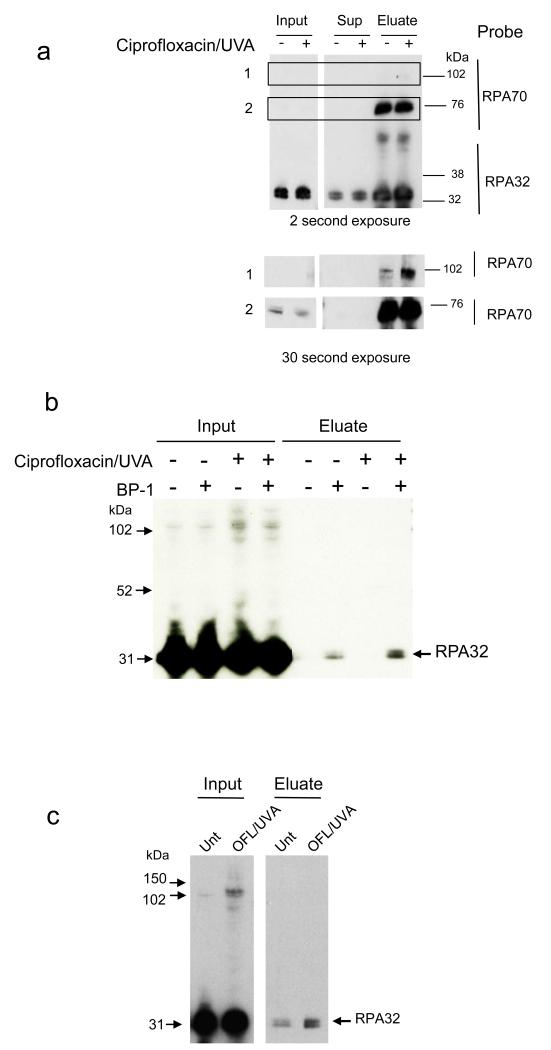
The sizes of the RPA photoproducts are consistent with covalent crosslinking of RPA32 to its RPA neighbours. The 100 kDa species is a potential RPA32:RPA70 dimer. RPA32-RPA14 dimerization is consistent with the 40-50 kDa species observed in U2OS and GM8339 cells. In U2OS-RPA21 cells, the GFP protein fused to RPA14 would contribute an additional 27 kDa to an RPA32:RPA14 complex to generate the 70-100 kDa species. To address these possibilities, RPA32 immunoprecipitated from ciprofloxacin/UVA-treated U2OS-RPA21 cells was probed for the presence of RPA70 by western blotting. Consistent with the presence of a partially processed GFP-RPA polypeptide, the material immunoprecipitated from untreated cells included an antiRPA70-reactive species of approximately 100 kDa (Figure 4a). Ciprofloxacin/UVA increased the amount of antiRPA70 reactive material at around 100 kDa, consistent with the formation of an RPA32:70 dimer. In Figure 4a, this photoproduct is not resolved from the endogenous unprocessed material. We conclude that one of the oxidation products of these photosensitizers is a covalent complex of RPA32 and RPA70 subunits.
Figure 4.
Identity of RPA oxidation products
a) Immunoprecipitation Anti-RPA32 immunoprecipitates from U2OS-RPA21 cells treated with ciprofloxacin (0.5 mM, 1 h) and UVA (20 kJ/m2) were analysed by western blotting for RPA32 and RPA70. The two lower panels represent longer exposure of the boxed areas 1 & 2. Positions of molecular weight markers are indicated.
b) RPA32 cysteine sufenate. U2OS-RPA21 cells that had been treated with ciprofloxacin (250 μM, 1 h) and UVA (20 kJ/m2) were lysed in the presence of the BP-1 biotinylated sulfenate-reactive probe. Derivatized proteins were recovered from streptavidin-linked beads (Eluate) and analysed by western blotting for RPA32. Samples prior to streptavidin enrichment (Input) were also analysed.
c) Extracts from U2OS-RPA21 cells were treated with ofloxacin (OFL 500 μM, 1 h) and UVA (20 kJ/m2) and analysed as in B).
Cysteine residues in RPA play important structural and functional roles. We therefore examined RPA cysteine thiol (-SH) oxidation to cysteine sulfenate (Cys-SOH). To do this, protein sulfenates in extracts of treated U2OS-RPA21 cells were selectively derivatized with a probe (BP-1) which comprises a sulfenate-reactive group attached to biotin (Qian et al., 2011) to enable their recovery via affinity binding to streptavidin. Western blotting confirmed that ciprofloxacin/UVA and ofloxacin/UVA induce the approximately 100 kDa RPA32:70 complex in U2OS-RPA21 cells (Input, Figure 4b,c). BP-1 derivatized RPA32 was present in untreated and treated U2OS-RPA21 cells and the level was increased by both ciprofloxacin/UVA and ofloxacin/UVA (Figure 4b,c). These findings demonstrate that fluoroquinolone/UVA treatment causes RPA cysteine thiol oxidation. They also suggest that some RPA may contain Cys-SOH under normal oxic growth conditions, at least when it is overexpressed. Both intersubunit crosslinking and cysteine sulfenation are potential contributors to RPA inactivation and NER inhibition in oxidatively stressed cells.
RPA damage and in vitro NER
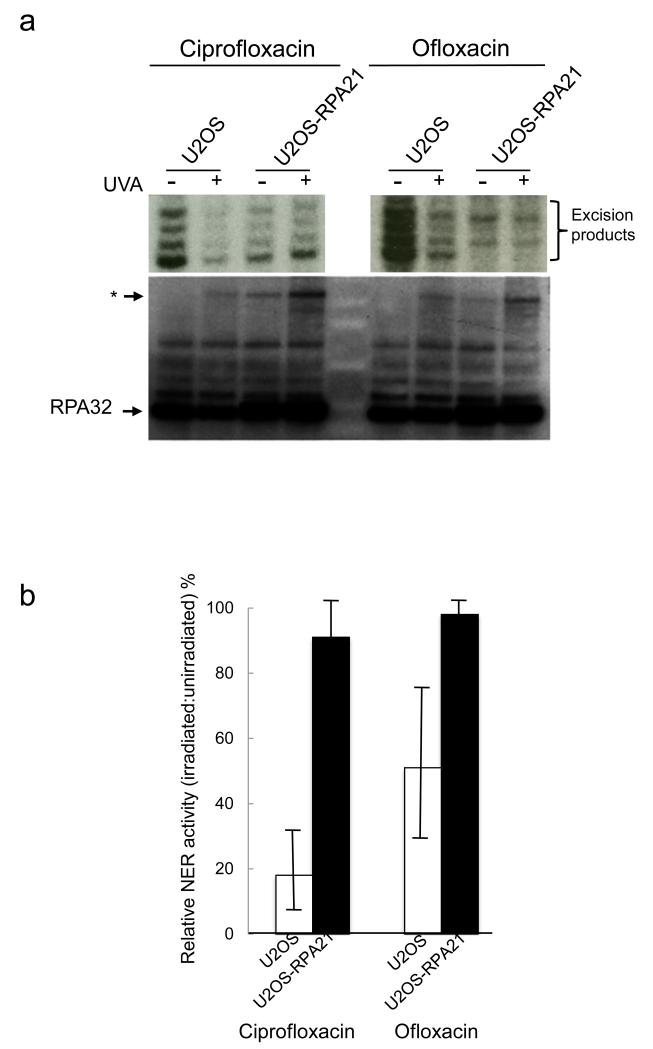
Protein damage underlies the inhibitory effects of fluoroquinolone/UVA on NER (Peacock et al., 2014). Consistent with previous observations, in vitro assays revealed that extracts from ciprofloxacin/UVA- or ofloxacin/UVA-treated U2OS cells were defective in NER. Similar treatments caused a more modest reduction in NER capacity of nuclear extracts from U2OS-RPA21 cells (Figure 5). Probing the extracts for RPA32 revealed that that impaired NER activity coincided with the appearance of an RPA32 species of approximately 100 kDa.
Figure 5.
NER in vitro
a) NER assays of nuclear extracts from U2OS or U2OS-RPA21 cells treated (1 h) with ciprofloxacin (0.5 mM) or ofloxacin (1 mM) and irradiated with 20 kJ/m2 UVA as indicated. Excision products (indicated) were radiolabelled and analysed by urea gel electrophoresis (upper panels). The same nuclear extracts were analysed by western blotting (lower panel). The position of the treatment-related RPA32 complex is indicated *.
b) Excision products were quantitated by GelDoc and NER efficiency by extracts from drug/UVA treated cells is expressed relative to that of extracts from cells treated with drug alone. Means of 3-5 assays of independently prepared extracts.
The relationship between the 100 kDa RPA32 species and NER capacity was investigated further in extracts of CCRF-CEM cells. NER inhibition by ciprofloxacin/UVA and ofloxacin UVA in CCRF-CEM cells was demonstrated previously (Peacock et al., 2014)(Supplementary Figure 4 upper panels). In agreement with the observations with U2OS, examination of NER-negative extracts from ciprofloxacin/UVA- or ofloxacin/UVA treated CCRF-CEM cells revealed novel RPA32 species of approximately 100 kDa and 40-50 kDa (Supplementary Figure 4 lower panels). Neither RPA32 complex was observed in NER-competent extracts from untreated, UVA-irradiated or drug-treated cells.
We conclude that these fluoroquinolone/UVA combinations cause sufficient oxidative RPA damage to compromise NER. Damage includes RPA subunit crosslinking and thiol oxidation. Modest RPA overexpression provides sufficient RPA to prevent NER becoming compromised.
Discussion
Our observations indicate that, despite its manifest importance in DNA processing, RPA is limiting for NER under oxidative stress conditions. Oxidative RPA damage compromises NER and modest RPA overexpression is sufficient to prevent this. RPA, and therefore NER, was shown to be surprisingly susceptible to inhibition under the oxidative stress conditions induced by photosensitizers. Diminished NER capacity was associated with readily detectable forms of damaged RPA.
RPA is essential for the NER of the UV-induced photolesions that are a major cause of skin cancer. In addition, it is also recruited to the extensive regions of exposed single-stranded DNA at replication forks stalled by UV-induced lesions. Cells exposed to UV radiation therefore experience conflicting demands on a limiting RPA pool. In conditions of extreme replication stress and in the absence of a functional S phase checkpoint to prevent replicon firing, RPA levels are insufficient to protect stalled replication forks from collapse and DNA breakage ensues (Toledo et al., 2013). This observation has prompted the suggestion that NER capacity is reduced by RPA sequestration at arrested replication forks (Tsaalbi-Shtylik et al., 2014) even in cells with a functional checkpoint. Whilst this appears to be the case under conditions of extreme replication stress induced by hydroxyurea, the extent to which these events occur following replication arrest by DNA photolesions is unclear.
Ciprofloxacin/UVA and 6-TG/UVA inhibit NER (Peacock et al., 2014). Consistent with enhanced removal of potentially lethal DNA lesions, improved NER in RPA overexpressing U2OS-RPA21 cells decreased their sensitivity to killing by ciprofloxacin/UVA and 6-TG/UVA.
Ciprofloxacin/UVA is a source of replication-arresting T<>T CPDs that are canonical NER substrates (Lhiaubet-Vallet et al., 2009). Chromatinization of both RPA and DDB2 (Otrin et al., 1997) after ciprofloxacin/UVA treatment is consistent with the presence of CPDs. 6-TG/UVA also induced RPA and DDB2 relocation indicating that it too generates replication-blocking NER substrates. Depletion of RPA by sequestration at arrested replication forks and competition by ciprofloxacin/UVA- and 6-TG/UVA-induced photolesions for NER proteins are therefore both potential contributors to a reduced NER capacity.
Several observations indicate that neither RPA sequestration nor the presence of competing substrates fully accounts for NER inhibition. Firstly, our measure of NER activity, 6-4Py:Py removal, is largely independent of DDB2 and proceeds much more rapidly than T<>T CPD excision (Hwang et al., 1999). DDB-recruiting lesions induced by 6-TG/UVA and ciprofloxacin/UVA induced T<>Ts by are therefore unlikely to be effective competitors of 6-4Py:Py NER. Secondly, RPA sequestration following replication arrest is S phase-specific. In our experiments, both 6-TG/UVA and ciprofloxacin/UVA effectively abolished 6-4Py:Py excision indicating that their inhibitory effects are not confined to S phase. Most pertinently, ofloxacin/UVA also inhibited NER and increased RPA expression prevented this inhibition. Ofloxacin/UVA is a very weak source of T<>T CPDs (Lhiaubet-Vallet et al., 2009; Peacock et al., 2014). Under conditions of NER inhibition it is relatively non-toxic and the induction of potentially lethal, replication-arresting DNA lesions cannot explain NER inhibition by ofloxacin/UVA. Thus, although these data do not exclude a contribution of DNA damage to NER inhibition, the effects of fluoroquinolone/UVA and 6-TG/UVA in U2OS and U2OS-RPA21 cells indicate additional or alternative events that compromise RPA function.
Our findings identify oxidation of RPA itself as an important determinant of NER efficiency both in vivo and in vitro. Inhibitory 6-TG/UVA, ciprofloxacin/UVA and ofloxacin/UVA treatments that severely compromised NER in U2OS cells induced covalent crosslinking between RPA subunits to generate RPA32:70 and most likely RPA32:14 dimers. RPA crosslinking was oxygen-dependent and occurred together with RPA cysteine oxidation to Cys-SOH. Crosslinked RPA was present in all NER-negative extracts of ciprofloxacin/UVA and ofloxacin/UVA treated U2OS, U2OS-RPA21 and CCRF-CEM cells but was not detected in NER-competent extracts. RPA is limiting for NER by cell extracts (Coverley et al., 1991) and our findings indicate that even the modest overexpression in U2OS-RPA21 cells provides significant protection against NER inhibition. Only a small fraction of the total RPA is present in crosslinked or sulfenated forms in NER-negative extracts. Although this is consistent with a powerful dominant negative effect, it seems more likely that the altered RPA species are markers for more widespread RPA damage.
RPA is redox sensitive and presents a significant target for oxidation (Park et al., 1999; Men et al., 2007). Eleven of its 15 cysteines are located in the 70 kDa subunit. Four in the C-terminal region form a zinc finger essential for DNA replication but apparently dispensable for NER (Lin et al., 1998). These two pairs of RPA70 cysteines are particularly sensitive to oxidant-induced disulfide formation (Men et al., 2007). The remaining 11 cysteines were unaffected and no cysteine sulfinic or sulfonic acid residues were observed. In our experiments, fluoroquinolone/UVA treatment increased the levels of RPA Cys-SOH - a reactive cysteine oxidation product and a marker for protein oxidation. Protein Cys-SOH can act as a redox switch to regulate redox homeostasis and signalling pathways (Groitl et al., 2014). These metastable intermediates can be reduced back to cysteine or undergo further and largely irreversible oxidation to more stable cysteine sulfinic and sulfonic acids. Importantly, they can react with other protein functional groups including amino groups to form sulfenamide crosslinks. The RPA32 complexes were resistant to the reducing conditions associated with western blotting, consistent with a crosslink of this nature. In addition to its two cysteines, RPA32 is rather methionine-rich and these residues are also plausible candidates for oxidation to sulfoxide (Men et al., 2007). Any of these oxidative changes to RPA might adversely affect its structure and are potentially deleterious for its function. In summary, RPA is particularly susceptible to oxidation damage and it is noteworthy in this regard that screens for protein carbonyls (Peacock, 2014) and DNA-protein crosslinks (M. Guven, PK, unpublished observations) both identified cellular RPA as a potential target for oxidative damage by 6-TG/UVA. RPA is of critical importance in almost all DNA transactions. It is therefore surprising that RPA levels are limiting and that its susceptibility to oxidative stress can compromise NER. Perhaps there are constraints on RPA expression. Based on the oxidation sensitivity of RPA both in vitro (Men et al., 2007) and in vivo as described here, it is tempting to speculate that a sensitivity to oxidative damage constrains its expression level. Protein oxidation is an unavoidable consequence of aerobic metabolism and oxidized proteins are generally detrimental to cellular fitness. If the multiple functions of RPA require it to be constructed in a way that renders it particularly vulnerable to oxidation damage, this requirement may determine its safe expression level. Limiting RPA may simply represent a trade-off between function and avoidance of unacceptable levels of protein oxidation.
The photosensitizers we use are associated with clinical photosensitivity. They amplify the effects of solar UVA which is an acknowledged generator of cellular ROS. Our demonstration that RPA is particularly susceptible to oxidation damage and that this compromises NER serves to emphasize the importance of protecting skin against the effects of the full spectrum of UV wavelengths in sunlight.
Materials & Methods
Chemicals
6-TG, ciprofloxacin and ofloxacin were obtained from Sigma Aldrich.
Cells and UV irradiation
HeLa, U2OS and U2OS-RPA21 cells (Toledo et al., 2013) kindly provided by Dr. J. Lucas, were grown in Dulbecco's MEM supplemented with 10% fetal calf serum. UVA irradiation was by a UVH 253 lamp (UV Light Technology Limited) with maximum emission at 365 nm. The dose rate was 0.1 kJ/m2/sec. UVC (254 nm) was delivered by a Stratalinker UV Crosslinker (Stratagene) at a dose rate of 10 J/m2/sec. For experiments that combined drug/UVA treatment with UVC radiation, drug/UVA treatment preceded UVC irradiation. Cell viability was determined by MTT assay on triplicate samples. All experiments were repeated at least twice.
6-TG determination
For initial incorporation measurements, HeLa, U2OS and U2OS-RPA21 cells were grown for 48 hours in medium containing 6-TG. DNA 6-TG was measured as previously described (Zhang et al., 2007). Initial determinations indicated that the more slowly growing U2OS-RPA21 cells incorporated 2-3 times less 6-TG at the same external 6-TG concentration. In subsequent experiments, 6-TG concentrations were adjusted to achieve similar 6-TG incorporation levels during 24 h growth. DNA 6-TG values (as % DNA G) were measured in each experiment to ensure comparability.
UVC 6:4 Py:Py measurements
DNA extracted by QIAamp DNA mini kit (Qiagen) from drug/UVA and UVC treated cells was analysed by ELISA according to the supplier’s instructions (Cosmo Bio Co). These experiments confirmed that none of the drug/UVA treatments induces detectable 6:4 Py:Pys (Peacock et al., 2014) and initial values for UVC-induced 6:4 Py:Py were in agreement within 10% independently of prior drug/UVA treatment. Repair data were therefore expressed as percentage of the initial lesions at various times after irradiation.
NER assays in vitro
NER by nuclear extracts of treated cells was assayed as described previously (Gueranger et al., 2014). Each assay was performed with least three independent extracts. Excision products were quantified by summing band intensities by GelDoc (Biorad). NER efficiency was calculated relative to extracts from cells treated with drug alone.
Immunoblotting
Whole cell extracts were prepared with radioimmunoprecipitation assay (RIPA) buffer. Proteins (20 μg) were separated on 10% polyacrylamide gels (Invitrogen). After transfer, membranes were probed with antibodies against RPA32, RPA70, DDB1 or DDB2 (Abcam). Antigen–antibody complexes were visualized using ECL blotting detection agent (GE Healthcare).
Immunoprecipitation
Cells were lysed in non-denaturing lysis buffer. RPA32 antibody was incubated with Dynabeads ProteinG (Invitrogen) prior to addition of protein extract (500 μg).
Sulfenate labelling and detection
Treated cells were lysed with RIPA buffer containing 1 mM biotin-1,3-cyclopentanedione (BP-1) probe (KeraFAST). N-ethylmaleimide 20 mM (Sigma) was immediately added and proteins precipitated with acetone. The dried protein pellet was dissolved in 2% SDS. Protein concentrations were determined by BCA assay (Thermo Scientific). Solubilized proteins were mixed with M280 Streptavidin Dynabeads (Invitrogen) which were washed three times with 4 volumes of 0.1% SDS. Following rotation overnight at 4°C beads were washed sequentially (30 min each) with 2 M urea, 1 M NaCl, 0.1% SDS/10 mM DTT, PBS. BP-1-derivatized proteins were recovered by boiling and analysed by western blotting.
Supplementary Material
Supplementary Figure 1
RPA in U2OS and overexpressing U2OS-RPA21 cells.
A RPA expression construct in U2OS-RPA21. The mRNA encodes all three RPA subunits translated as a single polypeptide in the order indicated. Individual subunits are linked by P2A sequences that permit processing into individual RPA proteins. RPA14 is also linked to GFP as indicated.
B Western blots of U2OS-RPA21 cell extracts reveal low levels of partially processed polypeptide.
RPA32:70 fusion (* left & centre panels);
Full length unprocessed polypeptide (*** right panel).
The correctly processed GFP-RPA14 fusion and endogenous RPA14 are also shown.
NS = non specific band
Supplementary Figure 2
RPA70 becomes chromatin-associated after 6-TG/UVA treatment and is recovered by benzonase digestion.
U2OS cells grown for 24 h in 4 μM 6-TG were irradiated with 20 kJ/m2 UVA. Whole cell extracts prepared immediately or 30 min after irradiation were analysed by western blotting for RPA70 with (+) or without (−) chromatin digestion with benzonase.
Supplementary Figure 3
RPA32:RPA70 complex formation in normal human fibroblasts.
GM8339 untransformed human fibroblasts were treated for 1 h with ciprofloxacin or ofloxacin at the concentrations shown and UVA irradiated. Extracts prepared immediately after irradiation were analysed by western blotting.
The positions of RPA32 and the putative RPA32:70 (*) and RPA32:14 (**) complexes are indicated.
Supplementary Figure 4
RPA32 photoproducts and NER in CCRF-CEM nuclear extracts.
Upper panels: NER by extracts prepared from ciprofloxacin/UVA or ofloxacin/UVA-treated CCRF-CEM cells.
Repair capability was determined by analysing the formation of oligonucleotides generated during repair of a cisplatin crosslink. These excision products (indicated) were radiolabelled and intrastrand separated by PAGE.
The NER assays were reported in (Peacock et al. Nucleic Acids Res., 42, 13714-13722)
Lower panels: Western blots of the same nuclear extracts were probed with antiRPA32.
The position of the putative RPA32:70 (*) and RPA32:14 (**) photoproducts are indicated.
Acknowledgements
We are grateful to Drs L. Toledo and J. Lucas for providing U2OS and U2OS-RPA21 cells and to Prof. C. Furdui for advice on BP-1 derivatization. The assistance of The Francis Crick Institute Cell Services department is gratefully acknowledged. Funding was provided by Cancer Research UK.
Footnotes
The authors state no conflict of interest.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–24. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Chen C, Umezu K, Kolodner RD. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Molecular Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- Coverley D, Kenny MK, Munn M, et al. Requirement for the replication protein SSB in human DNA excision repair. Nature. 1991;349:538–41. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- Dodson GE, Shi Y, Tibbetts RS. DNA replication defects, spontaneous DNA damage, and ATM-dependent checkpoint activation in replication protein A-deficient cells. J. Biol. Chem. 2004;279:34010–4. doi: 10.1074/jbc.C400242200. [DOI] [PubMed] [Google Scholar]
- Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N. Engl. J. Med. 2003;348:1681–91. doi: 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, et al. DNA Repair and Mutagenesis. ASM Press; Washington: 2006. [Google Scholar]
- Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 2009;21:237–44. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Groitl B, Jakob U. Thiol-based redox switches. Biochim. Biophys. Acta. 2014;1844:1335–43. doi: 10.1016/j.bbapap.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueranger Q, Li F, Peacock M, et al. Protein oxidation and DNA repair Inhibition by 6-thioguanine and UVA radiation. J. Invest. Dermatol. 2014;134:1408–17. doi: 10.1038/jid.2013.509. [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Ford JM, Hanawalt PC, et al. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA. 1999;19:424–8. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Miyauchi-Hashimoto H, Sugihara A, et al. The photocarcinogenesis of antibiotic lomefloxacin and UVA radiation is enhanced in xeroderma pigmentosum group A gene-deficient mice. J. Invest. Dermatol. 2005;125:554–9. doi: 10.1111/j.0022-202X.2005.23862.x. [DOI] [PubMed] [Google Scholar]
- Jayaraman SS, Rayhan DJ, Hazany S, et al. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J. Invest. Dermatol. 2014;134:213–20. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- Lhiaubet-Vallet V, Bosca F, Miranda MA. Photosensitized DNA damage: The case of fluoroquinolones. Photochem. & Photobiol. 2009;85:861–8. doi: 10.1111/j.1751-1097.2009.00548.x. [DOI] [PubMed] [Google Scholar]
- Lin Y-L, Shivji MKK, Chen C, et al. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J. Biol. Chem. 1998;273:1453–61. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- Men L, Roginskaya M, Zou Y, et al. Redox-dependent formation of disulfide bonds in replication protein A. Rapid Commun. Mass Spectrometry. 2007;21:2743–9. doi: 10.1002/rcm.3144. [DOI] [PubMed] [Google Scholar]
- Otrin VR, McLenigan M, Takao M, et al. Translocation of a UV-damaged DNA binding protein into a tight association with chromatin after treatment of mammalian cells with UV light. J. Cell Sci. 1997;110:1159–68. doi: 10.1242/jcs.110.10.1159. [DOI] [PubMed] [Google Scholar]
- Park J-S, Wang M, Park S-J, et al. Zinc finger of replication protein A, a non-DNA binding element, regulates its DNA binding activity through redox. J. Biol. Chem. 1999;274:29075–80. doi: 10.1074/jbc.274.41.29075. [DOI] [PubMed] [Google Scholar]
- Peacock M. UVA photosensitizers, protein oxidation and DNA repair. University College; London: 2014. PhD Thesis. [Google Scholar]
- Peacock M, Brem R, Macpherson P, et al. DNA repair inhibition by UVA photoactivated fluoroquinolones and vemurafenib. Nucleic Acids Res. 2014;42:13714–22. doi: 10.1093/nar/gku1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrin-Biroulet L, Khosrotehrani K, Carrat F, et al. Increased risk for nonmelanoma skin cancers in patients who receive thiopurines for inflammatory bowel disease. Gastroenterology. 2011;141:1621–8. doi: 10.1053/j.gastro.2011.06.050. [DOI] [PubMed] [Google Scholar]
- Qian J, Klomsiri C, Wright MW, et al. Simple synthesis of 1,3-cyclopentanedione derived probes for labeling sulfenic acid proteins. ChemComm. 2011;47:9203–5. doi: 10.1039/c1cc12127h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiscal JAB, Brewer JD. Thiopurines and risk of nonmelanoma skin cancer in inflammatory bowel disease. JAMA Dermatol. 2013;149:92–4. doi: 10.1001/2013.jamadermatol.616. [DOI] [PubMed] [Google Scholar]
- South AP, Purdie KJ, Watt SA, et al. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J. Invest. Dermatol. 2014;34:2630–8. doi: 10.1038/jid.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask M-B, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Traianou A, Ulrich M, Apalla Z, et al. Risk factors for actinic keratosis in eight European centres: a case-control study. Br. J. Dermatol. 2012;167(Suppl. 2):36–42. doi: 10.1111/j.1365-2133.2012.11085.x. [DOI] [PubMed] [Google Scholar]
- Tsaalbi-Shtylik A, Moser J, Mullenders LHF, et al. Persistently stalled replication forks inhibit nucleotide excision repair in trans by sequestering replication protein A. Nucleic Acids Res. 2014;42:4406–13. doi: 10.1093/nar/gkt1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Sugawara N, Chen C, et al. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Putnam CD, Kane MF, et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nature Genetics. 2005;37:750–5. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single stranded DNA-binding protein required for eukaryotic DNA metabolism. Ann. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jeffs G, Ren X, et al. Novel DNA lesions generated by the interaction between therapeutic thiopurines and UVA light. DNA Repair. 2007;6:344–54. doi: 10.1016/j.dnarep.2006.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
RPA in U2OS and overexpressing U2OS-RPA21 cells.
A RPA expression construct in U2OS-RPA21. The mRNA encodes all three RPA subunits translated as a single polypeptide in the order indicated. Individual subunits are linked by P2A sequences that permit processing into individual RPA proteins. RPA14 is also linked to GFP as indicated.
B Western blots of U2OS-RPA21 cell extracts reveal low levels of partially processed polypeptide.
RPA32:70 fusion (* left & centre panels);
Full length unprocessed polypeptide (*** right panel).
The correctly processed GFP-RPA14 fusion and endogenous RPA14 are also shown.
NS = non specific band
Supplementary Figure 2
RPA70 becomes chromatin-associated after 6-TG/UVA treatment and is recovered by benzonase digestion.
U2OS cells grown for 24 h in 4 μM 6-TG were irradiated with 20 kJ/m2 UVA. Whole cell extracts prepared immediately or 30 min after irradiation were analysed by western blotting for RPA70 with (+) or without (−) chromatin digestion with benzonase.
Supplementary Figure 3
RPA32:RPA70 complex formation in normal human fibroblasts.
GM8339 untransformed human fibroblasts were treated for 1 h with ciprofloxacin or ofloxacin at the concentrations shown and UVA irradiated. Extracts prepared immediately after irradiation were analysed by western blotting.
The positions of RPA32 and the putative RPA32:70 (*) and RPA32:14 (**) complexes are indicated.
Supplementary Figure 4
RPA32 photoproducts and NER in CCRF-CEM nuclear extracts.
Upper panels: NER by extracts prepared from ciprofloxacin/UVA or ofloxacin/UVA-treated CCRF-CEM cells.
Repair capability was determined by analysing the formation of oligonucleotides generated during repair of a cisplatin crosslink. These excision products (indicated) were radiolabelled and intrastrand separated by PAGE.
The NER assays were reported in (Peacock et al. Nucleic Acids Res., 42, 13714-13722)
Lower panels: Western blots of the same nuclear extracts were probed with antiRPA32.
The position of the putative RPA32:70 (*) and RPA32:14 (**) photoproducts are indicated.