Abstract
The fragmentation pattern of several protonated 1+ phosphatidylcholines (PCs) were studied using low energy collision induced dissociation (CID) and helium metastable atom-activated dissociation (He-MAD). He-MAD of the protonated compounds produced a dominant phosphocholine head group at m/z 184 as well as typical sn-1 and sn-2 glycerol fragments such as [M+H–Rx-1CHC=O]+ and [M+H-Rx-1CO2H]+. Within the aliphatic chain, He-MAD showed fragments consistent with high-energy collision induced dissociation (HE-CID) and products/pathways consistent with Penning ionization of the 1+ precursor ions to their respective radical dications. These Penning ionization products included both singly and doubly charged radical fragments, and the fragment ions are related to the number and position of double bonds in the acyl chains. Fragments created through HE-CID-like fragmentation followed classic charge remote fragmentation pathways including ladder-like fragmentation along the acyl chain, except for additional or missing peaks due to predictable rearrangement reactions. He-MAD therefore shows utility in being able to effectively fragment singly charged lipids into a variety of useful product ions using both radical and high-energy processes in the confines of a 3D ion trap.
Graphical abstract
Introduction
Recent advances in mass spectrometry have allowed scientists to probe different biological systems that were unattainable in the past. This increase in investigative power has contributed to the development of different fields of research including proteomics [1], genomics [2], and lipidomics [3, 4]. Lipidomics is defined as the full characterization of lipid molecular species and of their biological roles with respect to expression of proteins involved in lipid metabolism and function, including gene regulation [5]. This field is expansive and covers everything from identifying which lipids are localized in cells to the role those particular lipids plays in a metabolic cycle [4]. Mass spectrometry is an attractive technique for analyzing lipids due to its high selectivity and sensitivity, ability to quantify, abundance of structural information and ability to perform a variety of experiments [5, 6].
Mass spectrometric characterization of lipids began with electron ionization (EI) [7–9] via GC-interfaces, but most current research now relies on low-energy collisional induced dissociation (CID) [10–13] using matrix-assisted laser desorption ionization (MALDI) or atmospheric ion sources like electrospray ionization (ESI), APCI and atmospheric pressure (AP)-MALDI. The fragmentation products of glycerolipids are normally restricted to [M+Y-Rx-1CO2H]+, [M+Y-Rx-1CHC=O]+, where R corresponds to the sn-1 or sn-2 fatty acid chain from which the fragment originated, and Y is the charging adduct, such as Na+ or H+. The phosphocholine head group is another major fragment [12, 14, 15]. CID can provide useful information about a lipid by identifying its class, lipid chain lengths and the degree of unsaturation [16] but this does not represent all of the pertinent information about the structure/function of lipids. The position and isomeric form of double bonds is also important.
Researchers have developed a variety of tandem mass spectrometry approaches to interrogate the gas-phase structure of lipids in mass spectrometry. Some recently developed approaches includes multistage mass spectrometry [16, 17], post-source decay (PSD) matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF) [18], chemical IRMPD [19–21], and ion-molecule chemistry like Paternò-Büchi reactions [22], and ion-ion chemistry [23]. Blanksby’s group has provided a relatively straightforward and reliable method to determine the lipid class and double bond positions of unsaturated lipids using ozone-induced dissociation (OzID) of gas-phase ions [16, 24–27]. Although most lipids seem to fragment through even electron pathways using OziD, there is evidence that heavily conjugated lipids (e.g. fatty acid methyl esters, FAMES) can produce odd-electron fragments [28].
Although radical-induced dissociation of multiply-charged peptides and proteins is readily achievable via a variety of photon- or electron-based activation methods [29–34], radical ion chemistry is generally harder to drive when starting with a 1+ or 1− even-electron precursor ion, like most lipids. Recent examples of radical-induced fragmentation methods of lipids include ETD [35], UVPD [20], and electron-impact ionization excitation of ions from organics (EIEIO) [36]. These new methods have pros and cons and are still in their developing stages. They are likely to become more useful as research in these areas progresses.
An alternate activation method known as metastable atom-activated dissociation (MAD) [37–44] has been used to fragment peptides and small proteins via radical cation chemistry, and we believe this is the first report applying MAD to the fragmentation of gas-phase lipids. In contrast to CID, which almost exclusively involves even electron rearrangements, MAD causes fragmentation through radical-induced rearrangements that can induce fragmentation pathways unavailable through even electron mechanisms.
Methods and Instrumentation
Instrumentation
All experiments were performed on a modified Esquire-LC or amaZon QIT mass spectrometer (Bruker Daltronics, Bremen, Germany), the former of which has been described in a previous work.[41] Metastable atoms were generated with an Ion Tech FAB gun (P50, PSU, Teddington UK) and deflection electrodes were used to remove electrons and ions from the beam. The FAB gun was pulsed using custom electronics (described previously) to coincide with the fragmentation period in the scan function normally reserved for CID. The CID amplitude was set to 0 V during MAD so the ions are effectively just stored at a specified qz while the metastable atom source is pulsed on for ~300 ms. A visual schematic and description of the connections used in this process have been provided elsewhere [41].
Reagents
All the lipids used in this experiment were purchased from Avanti Polar Lipids (Alabaster, AL). Lipids used in this experiment included 1-hexadecanoyl-2-octadecanoyl-sn-glycero-3-phosphocholine PSPC (PSPC, 16:0/18:0), 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine (POPC, 16:0/18:1(9Z)), 1,2-di-(9E-octadecenoyl)-sn-glycero-3-phosphocholine (9E-DOPC, 18:1(9E)/18:1(9E)), 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine (9Z-DOPC, 18:1(9Z)/18:1(9Z)), 1,2-di-(5Z,8Z, 11Z, 14Z-eicosatetraenoyl)-sn-glycero-3-phosphocholine (DAPC, 20:4/20:4), and sphingomyelin (SM, d18:1/18:0). HPLCgrade methanol and glacial acetic acid were purchased from Sigma-Aldrich (St. Louis, MO). All lipids were reconstituted in a 9:1 mixture of methanol:water (with 1% acetic acid) to provide lipid solutions of approximately 60 µM for analysis. Ultra high purity helium (Airgas, Parkersburg, WV) was used with the FAB gun and further purified using a noble gas purifier (HP2, VICI, Houston, TX) to remove impurities that could otherwise prevent the formation of, or quench, metastable atoms.
Method
Singly charged lipid ions were generated through electrospray ionization (ESI) using an electronic syringe pump (74900, Cole-Parmer Instrument Company, Vernon Hills, IL) at a rate of 250 µL/h. After injection, precursor ions were isolated using a width of 1 to 4 Da before exposing them to the helium metastable atom beam at a low mass cut off (LMCO) of m/z 100. The metastable atom beam was typically pulsed on for 299 ms at an anode voltage of 6 kV. The vacuum chamber base pressure outside of the ion trap measured 1.68 × 10−5 mbar and was populated mostly by helium bath gas leaking out of the trap. After the addition of helium gas to the FAB gun, the vacuum pressure increased to 3.0 × 10−5 mbar. The holes in the ring electrode and the difference in bath gas pressure in the trap did not have any measurable effect on trap performance, but the instrument was re-tuned and re-calibrated prior to use anyway.
Collection of MAD data for each lipid took a approximately 10 minutes and consisted of: 1) 2 minutes of the full scan acquisition of the ESI spectrum of the sample; 2) 2 minutes of the isolated precursor ion; 3) 2 minutes of He MAD of the precursor with the deflection electrode on; 4) 2 minutes of He MAD with the deflection electrode off; and 5) 2 minutes of He MAD background signal (ESI off). These relatively long acquisition times enabled averaging many spectra together to improve the signal-to-noise ratio of low-abundance peaks. The MAD background is defined as the ion signal that is collected with the FAB gun on and the ESI source off, and consists of Penning ionization products of residual gases and pump oil. We expect a mixture of ion/ion and metastable atom chemistry with the deflection electrodes off, but a higher relative proportion of metastable atom-induced chemistry with the deflection electrodes on. MAD spectra shown in the figures have been background corrected using the average of the 2-minute MAD background signal. The magnitude of the background signal is roughly two orders of magnitude larger than the low-abundance fragment ions of the lipids. Because the absolute magnitude of the background various by a few percent, depending on the averaging, the variance is on the same order of magnitude as the low abundance fragment ions. It is for this reason that background subtraction in the low mass region does not result in a flat baseline
Collisional activation (CID) analysis was then performed following MAD acquisition and included: 1) 2 minutes of the full-scan ESI spectrum; 2) 2 minutes of the isolated precursor (no CID); 3) 2 minutes of CID of the precursor. Ion generation, accumulation, manipulation, and detection were optimized for each lipid to provide consistent precursor ion signals of approximately 1×106 AU. All fragments were identified manually based on the predicted masses and were found to be within m/z 0.3 of the observed masses.
CID
All lipids were fragmented with the “SmartFrag” option in the Bruker Esquire NT 4.5 software. SmartFrag exposes the precursor ions to a linear-amplitude-modulated-waveform from 30 to 200 percent of the selected CID amplitude. The fragmentation time was set to 25 ms in all the CID experiments. The 100% amplitude setting of the CID ranged from 0.85 to 1.40 V, depending on the isolated mass. A typical acquisition spanning about 2 minutes contained approximately 400 spectra (each an average of 5 scans), which were subsequently averaged for data analysis.
MAD-MS of protonated phospholipids
Penning ionization of precursor lipids in the 1+ charge state is expected to occur at sterically-exposed sites of highest electron density such as lone pairs on oxygen and π-electrons [45, 46]. Therefore, one would expect the new radical and charge to be initially located on carbonyl or phosphate oxygen atoms, as is the case for electron ionization (EI) of neutral lipids [7, 8]. Following long-established EI mechanisms, fragmentation adjacent to the site of Penning ionization can be charge directed, radical directed, or follow common rearrangements like γ-hydrogen shifts.
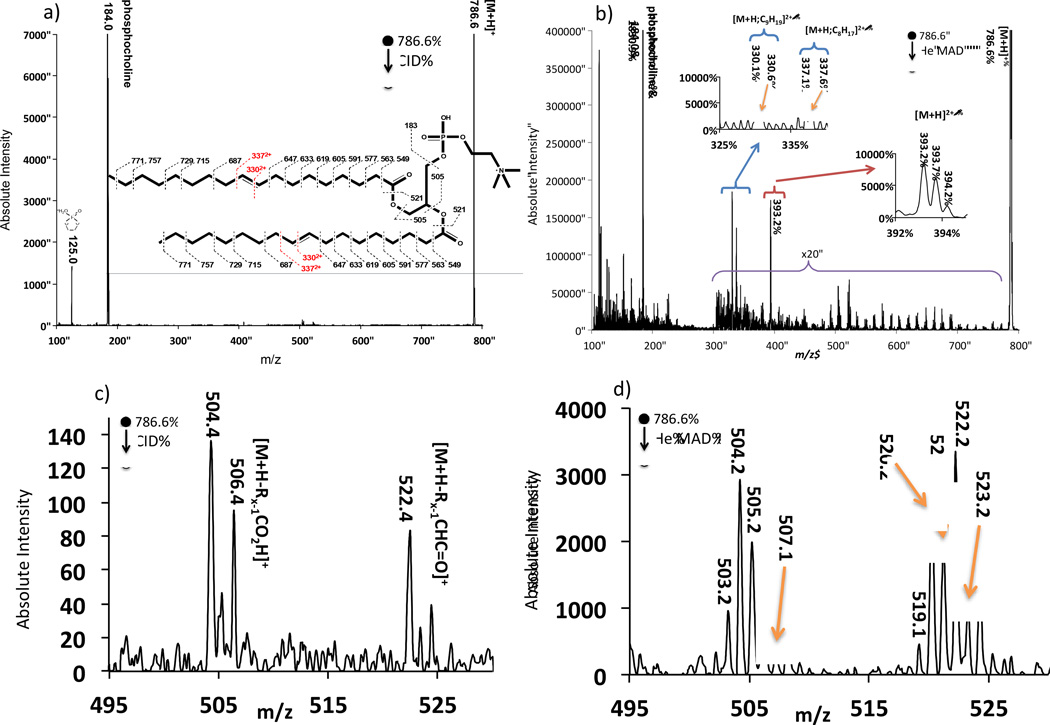
Figure 1 shows the comparison of the spectra obtained from the fragmentation of 9E-DOPC using He-MAD and CID. In the CID spectrum, the major fragments correspond to the phosphocholine head group and the complete loss of each fatty acid chain. The even electron, even mass losses of the fatty acid chains are typically referred to as the [M+H-Rx-1CO2H]+ and [M+H-Rx-1CHC=O]+ fragments, where R corresponds to the sn-1 or sn-2 fatty acid chain from which the fragment originated. For DOPC (18:1/18:1), R1 and R2 are identical, so cannot be distinguished. The CID fragmentation patterns observed for our lipids are consistent with low energy CID spectra obtained by others [10–12, 18, 47].
Figure 1.
Comparison spectra of a) even electron CID of [9E-DOPC+H]1+ (18:1/18:1) and b) odd-electron He-MAD of the same 1+ protonated lipid. c) and d) show expanded regions of the loss of sn-1 and sn-2 acyl chains for CID and He MAD, respectively. The inset in a) also shows possible cleavages and theoretical masses for fragmentation with no H-atom transfers. The masses in the insert may not agree with those shown in the spectra due to hydride shifts or rearrangements.
When MAD and CID fragmentation for 9E-DOPC is compared, MAD clearly produces several CID-type fragments, too. For example, Figure 1c highlights a region of even-electron products at m/z 504 and m/z 522 for conventional CID of 9E-DOPC. These fragments are typically low abundance or missing in low energy CID, but are more prominent in higher-energy CID. These fragments are ascribed to the sn-1 and sn-2 products [M+H-Rx-1CO2H]+ at m/z 504 and [M+H-Rx-1CHC=O]+ at m/z 522. We cannot assign R1 or R2 to the fragments for DOPC because the two R groups are identical in this instance. In fact, the identical nature of the two acyl chains of DOPC would normally be expected to produce only two peaks in this region using CID [12], not the 3 or 4 peaks observed here. We therefore question the purity of the lipid standard, which is also brought into question by the MAD results discussed later. All the lipids studied—including POPC, 9Z-DOPC and DAPC—showed similar losses of the acyl chains, although the sn-1 and sn-2 products are usually distinguished when the acyl chains have different masses (see Figure 2, for example).
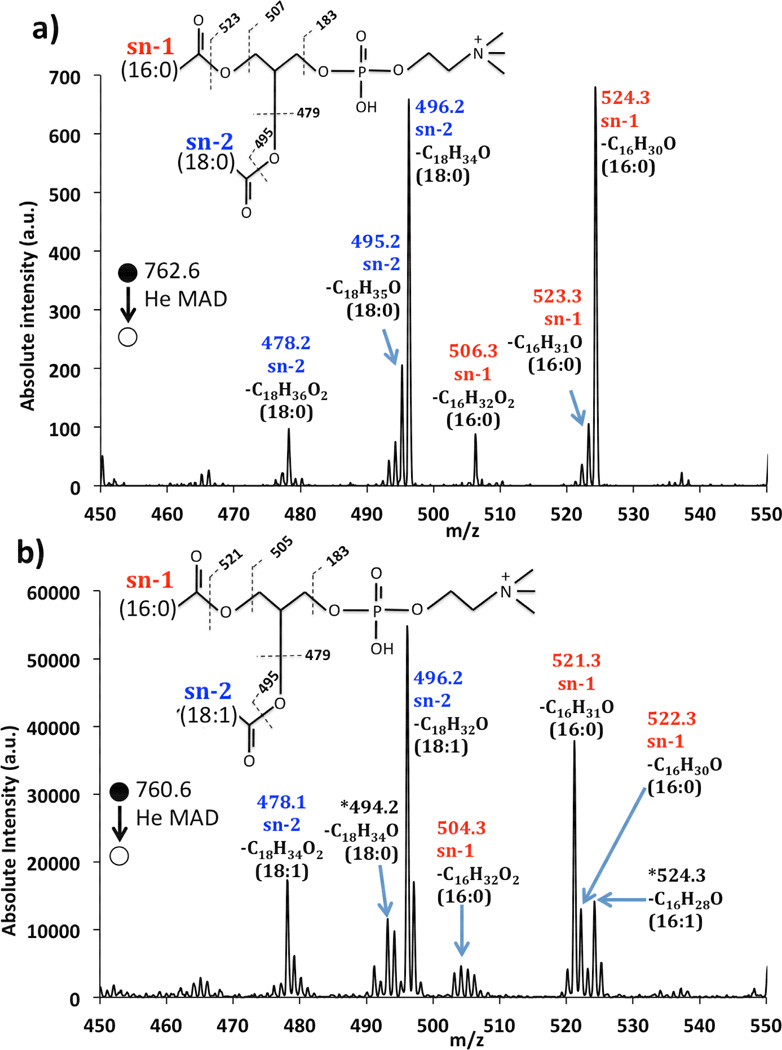
Figure 2.
Zoomed-in spectra of a) He-MAD of PSPC (16:0/18:0) and b) He-MAD of POPC (16:0/18:1) labeled to show the acyl chain identities. The asterisks in b) identify peaks suspected to come from an isobaric contaminant (16:1/18:0).
In contrast to the even-electron even-mass rearrangement products observed through conventional CID, He-MAD spectra of 9E-DOPC in Figure 1b and Figure 1d shows additional radical products not common in CID, such as the peaks at m/z 505 and m/z 521. The odd mass fragments observed in the MAD spectra could only be produced with the introduction of a radical, such as via Penning ionization. Odd mass fragments of this type have been observed in post-source decay, for example [18], which often proceeds through radical pathways.
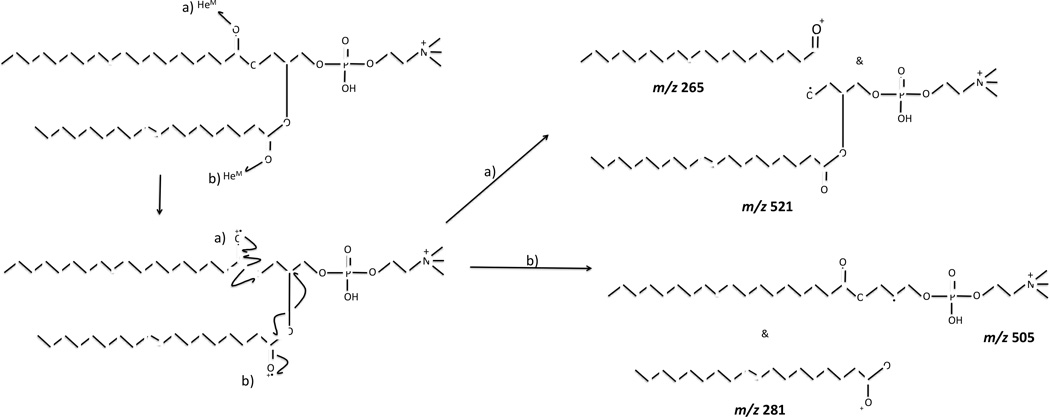
Although Penning ionization occurs through an oxidation (ionization) process that is somewhat similar to EI ionization, the fragmentation pattern observed for MAD of [DOPC+H]+ is vastly different from the EI-MS fragmentation pattern obtained for neutral or DOPC [7, 8], presumably because the charging proton significantly influences the site of ionization and pathways for fragmentation. However, the MAD spectrum of protonated DOPC appears to be very similar that of EIEIO of protonated DOPC, which indicates more-similar mechanisms between MAD and EIEIO [36]. For example, MAD and EIEIO products are analogous to common homolytic α- or β-cleavages observed in electron ionization of neutral esters [48], and are consistent with ionization of the carbonyl oxygen atoms. Two examples of homolytic cleavages originating from ionization of the carbonyl oxygen atoms are given in pathways a and b of Scheme 1, although these pathways are of course hypothetically possible on either acyl chain. The complementary ion pairs at m/z 265 and m/z 281 could not be confirmed in the MAD spectra of 9E or 9Z DOPC because the background noise is greater in this lower mass region and obscures their observation.
Scheme 1.
Proposed mechanism for the formation of odd-mass radical cation species at m/z 521 and m/z 505 through Penning ionization of carbonyl oxygen atoms of [9E-DOPC+H]+ precursor ion. When the acyl chains differ, homolytic bond cleavages in the α- and β-positions for sn-1 and sn-2 cleavages would each result in a unique product ion.
A more careful examination of the glycerol cleavages around m/z 500 is presented in Figure 2 for PSPC (16:0/18:0) and POPC (16:0/18:1). He-MAD of PSPC (Figure 2a) shows that rearrangements are dominant pathways for the common glycerol losses, giving of m/z 496 and m/z 478 for the loss of the C18:0 chain from the sn-2 position, and at m/z 524 and m/z 506 for the loss of the C16:0 chain from the sn-1 position (details shown in Figure 2). In addition to these common rearrangement losses, Figure 2 also shows evidence for radical cleavages, which result in product ions at m/z 495 and 523 for sn-2 and sn-1 losses, respectively. Unlike other methods [23, 36, 49–51], the sn-2 and sn-1 pathways in He-MAD seem equally favorable for PSPC. The general preference for sn-2 eliminations in CID is often used to help distinguish sn-1 from sn-2 products. In this regard, the apparent lack of preference for either pathway in He-MAD—which remains to be validated with other lipids—can be considered a possible detriment to structural elucidation of glycerolipids.
He-MAD of POPC (16:0/18:1) in Figure 2b also provides the expected sn-2 rearrangement products at m/z 496 and m/z 478, and sn-1 products at m/z 522 and m/z 504, respectively. For He-MAD of POPC, the radical-induced product at m/z 521 is considerably more abundant than the rearrangement product at m/z 522. The fragment ion containing the remaining acyl chain has the double bond, and it is possible that the presence of the double bond stabilizes or enhances radical fragmentation, as has been observed by others [28, 52]. There are several additional peaks in this vicinity that are difficult to assign to any simple fragmentation mechanisms besides a contaminant isobaric lipid. The presence of ~20% (by mass) of 16:1/18:0 in the POPC solution could easily explain the occurrence of fragments at m/z 524 and 492. We could not confirm the purity of each lipid by CID because signal-to-noise ratio for the low abundance fragment of the impurity was below the signal-to-noise threshold. However, impure lipid standards have been identified by others [36, 53] and are known to cause spectral interpretation problems.
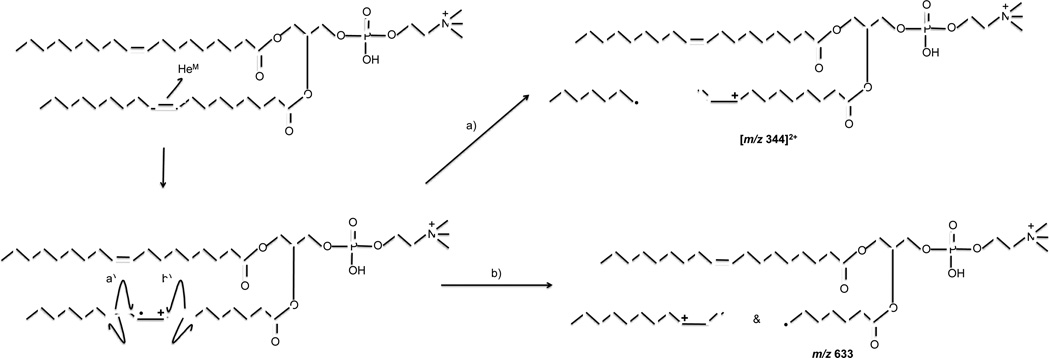
A series of MAD products of 9E-DOPC are also present in the range m/z 500 to 800 of Figure 1b that are not observed in the corresponding CID spectrum. Many of these MAD fragments are spaced in 14 Da increments, which is indicative of the aliphatic chain. These lipid chain fragments are also observed in the MAD spectra of other phosphoglycerolipids studied here, including PSPC, POPC, 9Z-DOPC and 9E-DOPC, examples of which are shown in Figure 3 and the supplemental material. High energy CID and EIEIO are known to produce similar ladder-like fragmentation along the lipid chain [36, 54, 55], but low-energy even-electron CID is not typically capable of producing such fragmentations [16, 17]. High-energy fragmentation processes like high energy CID (HE-CID) proceed through charge remote fragmentation (CRF) [6, 52, 56–59], the mechanism of which may proceed via 1,4 elimination reactions—as proposed by Gross et al. [56, 59]—or by homolytic carbon cleavage and hydrogen rearrangement reaction, as proposed by Wysocki and Ross [58]. In addition, Claeys et. al. suggested that a homolytic carbon-hydrogen cleavage may be another path to generate such fragments [52].
Figure 3.
He-MAD spectra of a) PSPC (16:0/18:0) and b) POPC (16:0/18:1) and c) 9Z-DOPC (18:1/18:1) showing the high energy or radical-induced cleavage of the acyl chains. Mass labels on the spectra show the most abundant masses in a cluster. Mass labels on the structures show expected fragments without H-transfers or rearrangements.
Shimma et. al. found that in addition to the typical low-energy CID fragments observed in low- and high-energy CID, HE-CID of protonated PC(18:1/18:0) produced a series of CRF fragments associated with the fatty acid chain [60]. A similar series of fatty acid fragments were also observed by Cheng and Gross when using HE-CID to study triacylglycerols [61]. The similarity between our results and those reported in the literature suggests HE-CID- or CRF-like fragmentation is occurring, although the mechanism might be different. This capability is interesting because HE-CID is normally performed in larger beam-type instruments like magnetic sectors, time-of-flight (TOF) or Fourier-transform ion cyclotron resonance (FT-ICR) mass spectrometers. MAD appears to provide similar capabilities in an ion trap instrument.
During MAD, metastable and ground state atoms have up to 6 keV of kinetic energy in the lab frame, which at first glance would appear to exceeds the ~1 keV (lab frame) threshold required for the onset of HE-CID. However, here, we are accelerating the neutral atoms into the precursor ions, instead of the ions into the neutral atoms, as typically occurs in HE-CID. Because the projectile (Hem) has considerably less mass than lipid ions, there is considerably less energy available in the center-of-mass (COM) frame. Previous work has shown through the study of different metastable atoms that the electronic energy available in the metastable atoms has a more pronounced effect on fragment ion types than the COM collision energy [37, 40, 41].
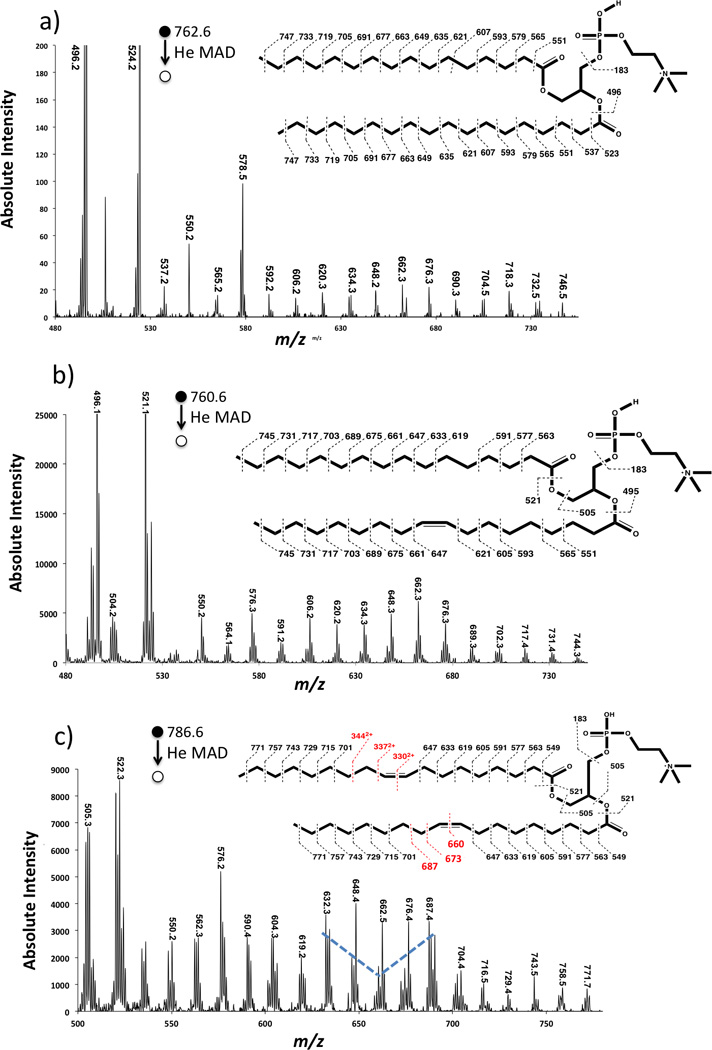
Figure 4 shows more detail in the He-MAD fragmentation of 9Z-DOPC. Figure 4b shows a region of the spectrum around the cleavage of the double bond. The even mass fragments labeled in red indicate common high energy or charge-remote fragmentation products, which are generally spaced by 14 Da and involve rearrangement looses of [CnH2n-2] from the precursor ion. In contrast, the fragments labeled in blue indicate fragments that do not involve hydrogen shifts across the cleaved C-C bonds. The peak at m/z 660.3 corresponds to cleavage at the double bond, which is admittedly quite unlikely and therefore a suspicious assignment. However, this fragment was observed on replicate trials of 9Z-DOPC, so is reliably present—even if the assignment is incorrect.
Figure 4.
a) He MAD spectrum of protonated (9Z) DOPC (18:1/18:1 PC). The insets and panel b) show zoomed in regions of the spectrum. The inset also shows possible cleavages and theoretical masses for fragmentation with no H-atom transfers. The masses in the insert may not agree with those shown in the spectra due to hydride shifts or rearrangements.
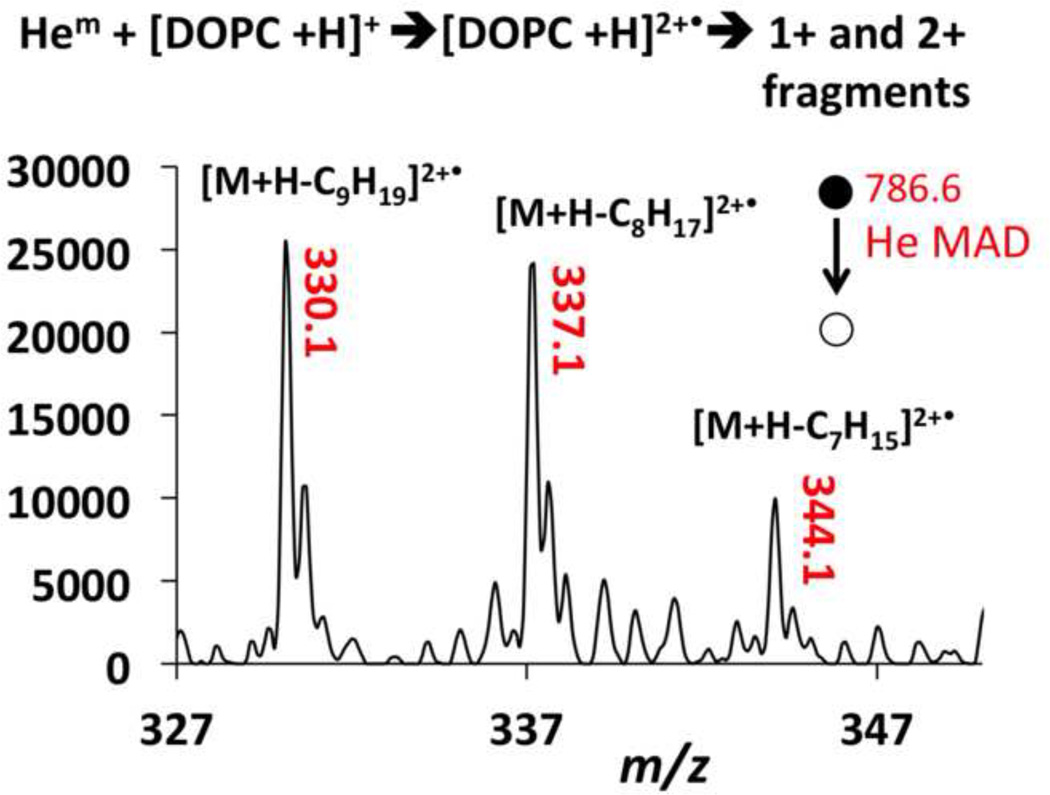
Another unique feature of MAD is the observation of 2+ product ions in Figures 1b and 4a, which are easily observed in the MAD spectra of 9E-DOPC and 9Z-DOPC. The 2+ fragment ions appear at m/z 330.1, m/z 337.1 and m/z 344.1 for 9Z-DOPC and m/z 330.1, m/z 337.1 for 9E-DOPC. We assume the only real difference was a difference in signal-to-noise ratio between the two spectra, as no other spectral differences could be found between the two geometric isomers.
The assignment of the 2+ product ions was confirmed by the presence of the 13C isomers at m/z 0.5 intervals. Doubly charged product ions are common features of MAD of 1+ peptide ions [38–41]. As shown in Scheme 2, if Penning ionization occurred at the site of unsaturation in the lipid, one would expect EI-like homolytic cleavages to produce a 1+ α-cleavage product at m/z 633.2 or a 2+ α-cleavage product at m/z 344.1, depending on which end of the ionized double bond the radical resides. Although the Coulombic repulsion exit-channel should presumably favor the formation of the m/z 633.2 product ion, both of these fragments are observed in similar abundances. Heterolytic α-cleavage on the distal side of the ionized double bond is responsible for the fragment at m/z 687.3, which is considerably more favorable than the non-observed α-cleavage on the proximal side, which would have given a 2+ product at m/z 316.5).
Scheme 2.
Proposed mechanism for the formation of complementary ion pairs at m/z 344 and m/z 633, which are both observed in the He MAD spectrum of [9Z-DOPC+H]+. In both pathways, fragmentation occurs via homolytic cleavage alpha to the radical formed by Penning ionization of the double bond. The intermediate shows the radical on C10 of the sn-2 acyl chain, but pathway b assumes the radical starts on C9.
EI spectra of olefins often contain evidence of γ-hydrogen shifts after ionizing at the double bond position [48]. For He-MAD of 9Z-DOPC, a γ-hydrogen shift from the distal side of the ionized double bond would be expected to result in cleavage of the β-carbon bond and provide product ions at either m/z 688.6 or m/z 632.3, both of which are observed. Whereas a classical or concerted McLafferty rearrangement results in the homolytic cleavage of the bond β to the original carbonyl group, other factors—like hetero-atoms—can encourage heterolytic
cleavage or step-wise radical migration away from this region [48]. In the case of MAD of 1+ lipid ions, the inductive acyl chain and the charge-repulsion exit channel both favor the heterolytic β-cleavage products, which has the only noticeable difference that the extra electron on the head group fragment results in 1+ product ions instead of 2+ product ions. These observed fragments, in conjunction with the presence of the in-tact radical cation [M+H]2+• at m/z 393.2 provide compelling evidence for radical-induced fragmentation through Penning ionization, as shown in Equation 1.
| [1] |
It is known that in the Penning ionization of gas-phase neutral organics, metastable atoms are attracted to, and therefore ionize at, areas of high electron density [45, 46, 62], which is based on dipole attraction and steric factors at close range [63–66]. The He-MAD spectra presented here indicate that Penning ionization of charged lipids also follows preferential oxidation/activation of the lone pairs on carbonyl or phosphate oxygen atoms and the π-electrons in the unsaturated C=C bonds.
The He-MAD spectrum in Figure 4a shows a 2+ product at m/z 330.1 as the most abundant doubly charged product. This product apparently requires cleavage across the double bond position with no hydrogen or hydride shifts. We cannot find any precedent for cleavage at a double bond position, and do not have a satisfactory mechanism at this time. One explanation for this peak could be the presence of an isobaric molecular ion with a different double bond position or acyl chains. Contamination of DOPC with an isobaric 18:0/18:2 lipid would actually help explain the fragment ions in the glycerol loss region at m/z 520 and m/z 524 in Figure 3c. These two fragments are difficult to explain based on known sn-1 and sn-2 glycerol or ketene cleavages of the 18:1/18:1 acyl chains.
In these experiments, we only observed the doubly-charged fragments in the spectra of 9E-DOPC and 9Z-DOPC. POPC does not produce 2+ fragments, even though one of the acyl chains has a double bond in the same position (C9) as DOPC. Presumably, the second double bond in DOPC must stabilize or enhance the formation of 2+ ions, or else provide an additional site for attack and therefore higher probability and better signal-to-noise in the product ion spectrum. Clearly, these first experiments raise a lot of questions about the ability to unequivocally identify the position of double bonds in acyl chains. However, additional studies—including the use of sodiated lipids and MS3 experiments–should help elucidate the potential of He-MAD for the structural interrogation of gas-phase lipids.
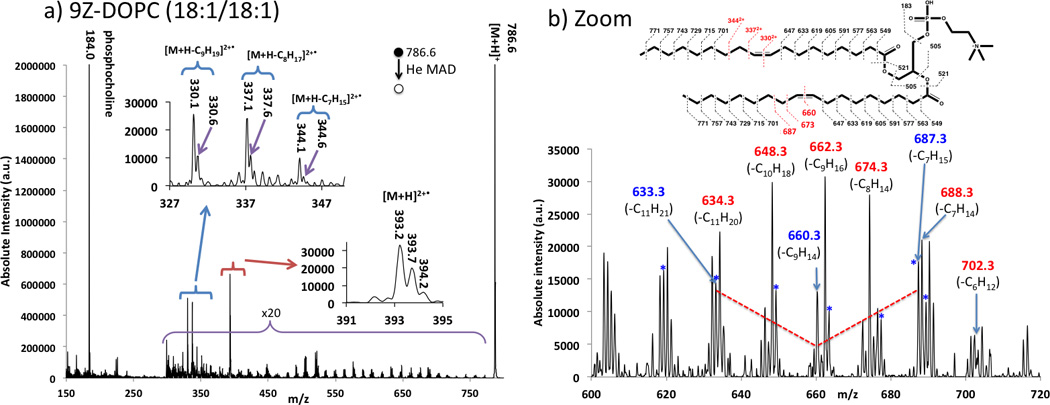
Two additional lipids, SM and DAPC, did not fragment as readily as the other lipids during He-MAD. In CID, SM typically produces the charged phosphocholine head group fragment and no other significant fragments [15, 67, 68]. Here, SM provides very similar MAD and CID spectra with the phosphocholine fragment being the dominant product in both cases (Figure 5). Small neutral losses are also apparent in the in-house CID spectrum of SM, such as a water loss at m/z 713 and the formation of the O, O’-dimethylenephosphoric acid at m/z 124. The O,O’-dimethylenephosphoric acid fragment at m/z 124 could also formed in the He-MAD spectrum, but this fragment is overwhelmed by the background signal and cannot be confirmed. He-MAD of DAPC provides more fragments than He-MAD of SM, but does not provide the extensive array of fragments observed for POPC, PSPC, 9E-DOPC, or 9Z-DOPC. We do not yet understand the factors that limit the MAD fragmentation of these two lipids.
Figure 5.
Comparison of fragmentation of sphingomyelin (SM) (d18:0/18:0): a) by He-MAD, and b) by low-energy CID.
DAPC contains four double bonds per acyl chain and these bonds are expected to impart some selectivity to the acyl chain fragmentation. The supplemental material shows a spectrum and fragmentation scheme of how some of fragments we observe in DAPC could be formed. DAPC provided low fragmentation efficiencies, which could be related to the large number of canonical forms that the molecule can adopt to stabilize intermediate radicals [69–71]. Another surprising feature of DAPC and sphingomyelin is the lack of Penning ionized peak, [M+H]2+•, which could be attributed to a combination of the ionization energy of the [M+H]+ precursor ion, competition between electronic and vibrational excitation, or the relative lability of the [M+H]2+• product ions. Although a Penning ionized [M+H]2+• is not observed in DAPC, there are many odd-electron fragment ions that are indicative of radical-induced dissociation. For example, the fragmentation spectra of DAPC in the supplemental material shows peaks corresponding to acyl chain cleavages and H-atom transfers.
Conclusion
MAD-MS was used to study several phosphatidylcholine lipids in the protonated form. MAD-MS was able to produce the phosphocholine head group, and sn-1 and sn-2 glycerol cleavages of [M+H-Rx-1CO2H]+ and [M+H-Rx-1CHC=O]+, which are commonly observed in other fragmentation methods such as CID. In addition to several examples of in-tact Penning ionized products, [M+H]2+•, several unsaturated lipids gave 2+ product ions corresponding to cleavage at or near the double bond position. He-MAD yielded several fragments associated with both radical-induced and HE-CID fragmentation pathways within the fatty acid chain of PSPC, POPC, 9E-DOPC and 9Z-DOPC, which is a notable achievement in a 3D ion trap. Although, MAD appears to be quite effective for fragmenting lipids through a variety of mechanisms, He-MAD spectra are quite complicated, have relatively poor signal-to-noise levels and does not seem to provide significantly more information then can be obtained using MSn, CID or HE-CID, or OziD. Additional experiments and refinement of MAD or the spectral interpretation could provide a more optimistic outlook for this novel radical-based fragmentation method.
Supplementary Material
Highlights.
Ability to form radical dications from protonated even electron lipids
Ability to produce high-energy fragmentation in an ion trap
Ability to produce radical-induced fragmentation in an ion trap
Ability to activate and fragment at double bonds position in acyl chains
Acknowledgments
Funding Sources
The authors acknowledge financial support form the National Institutes of Health (NIH) (1R01 GM114494-01). The opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of NIH.
ABBREVIATIONS
- CID
Collision induced dissociation
- CRF
Charge remote fragmentation
- DAPC
1,2-di-(5Z,8Z, 11Z, 14Z-eicosatetraenoyl)-sn-glycero-3-phosphocholine
- 9E-DOPC
1,2-di-(9E-octadecenoyl)-sn-glycero-3-phosphocholine
- 9Z-DOPC
1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine
- EI
Electron Ionization
- ESI
Electrospray Ionization
- FT-ICR
Fourier transform ion cyclotron resonance
- HE-CID
High-energy collision induced dissociation
- LMCO
Low mass cut-off
- MAD-MS
Metastable atom-activated dissociation
- PC
Phosphatidylcholine
- PI
Penning ionization
- POPC
1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine
- PSPC
1-palmitoyl-2-stearoyl-sn-glycero-3-phosphocholine
- SM
Sphingomyelin
- TOF
Time of flight
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Blackstock Wp Fau - Weir MP, Weir MP. Proteomics: quantitative and physical mapping of cellular proteins. Tibtech. 1999;17 doi: 10.1016/s0167-7799(98)01245-1. [DOI] [PubMed] [Google Scholar]
- 2.Kevin PW. Functional genomics and the study of development, variation and evolution. Nature Reviews Genetics. 2001;2:528–537. doi: 10.1038/35080565. [DOI] [PubMed] [Google Scholar]
- 3.Wenk MR. The emerging field of lipidomics. Nature Reviews Drug Discovery. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 4.Watson AD. Thematic review series: Systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: A global approach to lipid analysis in biological systems. J. Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Roberts LD, McCombie G, Titman CM, Griffin JL. A matter of fat: An introduction to lipidomic profiling methods. J. Chromatogr. B. 2008;871:174–181. doi: 10.1016/j.jchromb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Blanksby SJ, Mitchell TW. Advances in mass spectrometry for lipidomics. Annual Review of Analytical Chemistry. 2010;3:433–465. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- 7.Klein RA. Mass spectrometry of the phosphatidylcholines: dipalmitoyl, dioleoyl, and stearoyl-oleoyl glycerylphosphorylcholines. J. Lipid Res. 1971;12:123–131. [PubMed] [Google Scholar]
- 8.Klein RA. Mass spectrometry of the phosphatidylcholines: fragmentation processes for dioleoyl and stearoyl-oleoyl glycerylphosphorylcholine. J. Lipid Res. 1971;12:628–634. [PubMed] [Google Scholar]
- 9.Furlong ST, Leary JA, Costello CE, Dawidowicz EA. Isolation and identification of 1(3),2-diacylglyceryl-(3)-O-4'-(N,N,N-trimethyl)homoserine from the soil amoeba, Acanthamoeba castellanii. J. Lipid Res. 1986;27:1182–1189. [PubMed] [Google Scholar]
- 10.Hsu F-F, Turk J. Studies on phosphatidylglycerol with triple quadrupole tandem mass spectrometry with electrospray ionization: fragmentation processes and structural characterization. J. Am. Soc. Mass. Spectrom. 2001;12:1036–1043. [Google Scholar]
- 11.Ho Y-P, Huang P-C. A novel structural analysis of glycerophosphocholines as TFA/K+ adducts by electrospray ionization ion trap tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2002;16:1582–1589. doi: 10.1002/rcm.751. [DOI] [PubMed] [Google Scholar]
- 12.Hsu F-F, Turk J. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: The fragmentation processes. J. Am. Soc. Mass. Spectrom. 2003;14:352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 13.McLuckey SA, Goeringer DE. Special Feature: Tutorial slow heating methods in tandem mass spectrometry. J. Mass Spectrom. 1997;32:461–474. [Google Scholar]
- 14.Domingues P, Domingues MRM, Amado FML, Ferrer-Correia AJ. Characterization of sodiated glycerol phosphatidylcholine phospholipids by mass spectrometry. Rapid Commun. Mass Spectrom. 2001;15:799–804. doi: 10.1002/rcm.300. [DOI] [PubMed] [Google Scholar]
- 15.Murphy RC, Harrison KA. Fast atom bombardment mass spectrometry of phospholipids. Mass Spectrom. Rev. 1994;13:57–75. [Google Scholar]
- 16.Mitchell TW, Pham H, Thomas MC, Blanksby SJ. Identification of double bond position in lipids: From GC to OzID. J. Chromatogr. B. 2009;877:2722–2735. doi: 10.1016/j.jchromb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Hsu F-F, Turk J. Structural characterization of unsaturated glycerophospholipids by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J. Am. Soc. Mass. Spectrom. 2008;19:1681–1691. doi: 10.1016/j.jasms.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Saad K, Siems W, Hill HH, Zabrouskov V, Knowles NR. Structural analysis of phosphatidylcholines by post-source decay matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Am. Soc. Mass. Spectrom. 2003;14:373–382. doi: 10.1016/S1044-0305(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 19.Zehethofer N, Scior T, Lindner B. Elucidation of the fragmentation pathways of different phosphatidylinositol phosphate species (PIPx) using IRMPD implemented on a FT-ICR MS. Anal Bioanal Chem. 2010;398:2843–2851. doi: 10.1007/s00216-010-4184-8. [DOI] [PubMed] [Google Scholar]
- 20.Madsen JA, Cullen TW, Trent MS, Brodbelt JS. IR and UV photodissociation as analytical tools for characterizing lipid A structures. Anal. Chem. 2011;83:5107–5113. doi: 10.1021/ac103271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo HJ, Hakansson K. Determination of Phospholipid Regiochemistry by Ag(I) Adduction and Tandem Mass Spectrometry. Anal. Chem. (Washington, DC, U. S.) 2011;83:1275–1283. doi: 10.1021/ac102167q. [DOI] [PubMed] [Google Scholar]
- 22.Ma X, Xia Y. Pinpointing Double Bonds in Lipids by Paterno-Buechi Reactions and Mass Spectrometry. Angew. Chem., Int. Ed. 2014;53:2592–2596. doi: 10.1002/anie.201310699. [DOI] [PubMed] [Google Scholar]
- 23.Stutzman JR, Blanksby SJ, McLuckey SA. Gas-phase transformation of phosphatidylcholine cations to structurally Iinformative anions via ion/ion chemistry. Anal. Chem. 2013;85:3752–3757. doi: 10.1021/ac400190k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas MC, Mitchell TW, Blanksby SJ. Ozonolysis of phospholipid double bonds during electrospray ionization: A new tool for structure determination. JACS. 2006;128:58–59. doi: 10.1021/ja056797h. [DOI] [PubMed] [Google Scholar]
- 25.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Murphy RC, Blanksby SJ. Elucidation of double bond position in unsaturated lipids by ozone electrospray ionization mass spectrometry. Anal. Chem. 2007;79:5013–5022. doi: 10.1021/ac0702185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Nealon JR, Blanksby SJ. Ozone-induced dissociation: Elucidation of double bond position within mass-selected lipid ions. Anal. Chem. 2008;80:303–311. doi: 10.1021/ac7017684. [DOI] [PubMed] [Google Scholar]
- 27.Pham HT, Maccarone AT, Thomas MC, Campbell JL, Mitchell TW, Blanksby SJ. Structural characterization of glycerophospholipids by combinations of ozone- and collision-induced dissociation mass spectrometry: the next step towards "top-down" lipidomics. Analyst (Cambridge, U. K.) 2014;139:204–214. doi: 10.1039/c3an01712e. [DOI] [PubMed] [Google Scholar]
- 28.Pham HT, Maccarone AT, Campbell JL, Mitchell TW, Blanksby SJ. Ozone-induced dissociation of conjugated lipids reveals significant reaction rate enhancements and characteristic odd-electron product ions. J. Am. Soc. Mass. Spectrom. 2013;24:286–296. doi: 10.1007/s13361-012-0521-9. [DOI] [PubMed] [Google Scholar]
- 29.Zubarev RA, Haselmann KF, Budnik B, Kjeldsen F, Jensen F. Towards an understanding of the mechanism of electron-capture dissociation: a historical perspective and modern ideas. Eur. J. Mass Spectrom. 2002;8:337–349. [Google Scholar]
- 30.Zubarev RA. Electron-capture dissociation tandem mass spectrometry. Curr. Opin. Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Coon JJ, Shabanowitz J, Hunt DF, Syka JEP. Electron Transfer Dissociation of Peptide Anions. J. Am. Soc. Mass. Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Xia Y, Chrisman PA, Pitteri SJ, Erickson DE, McLuckey SA. Ion/Molecule Reactions of Cation Radicals Formed from Protonated Polypeptides via Gas-Phase Ion/Ion Electron Transfer. JACS. 2006;128:11792–11798. doi: 10.1021/ja063248i. [DOI] [PubMed] [Google Scholar]
- 33.Ly T, Julian RR. Residue-Specific Radical-Directed Dissociation of Whole Proteins in the Gas Phase. JACS. 2008;130:351–358. doi: 10.1021/ja076535a. [DOI] [PubMed] [Google Scholar]
- 34.Yoo HJ, Wang N, Zhuang S, Song H, Håkansson K. Negative-Ion Electron Capture Dissociation: Radical-Driven Fragmentation of Charge-Increased Gaseous Peptide Anions. JACS. 2011;133:16790–16793. doi: 10.1021/ja207736y. [DOI] [PubMed] [Google Scholar]
- 35.Liang X, Liu J, LeBlanc Y, Covey T, Ptak AC, Brenna JT, McLuckey SA. Electron transfer dissociation of doubly sodiated glycerophosphocholine lipids. J. Am. Soc. Mass. Spectrom. 2007;18:1783–1788. doi: 10.1016/j.jasms.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell JL, Baba T. Near-complete structural characterization of phosphatidylcholines using electron impact excitation of ions from organics. Anal. Chem. 2015 doi: 10.1021/acs.analchem.5b01460. [DOI] [PubMed] [Google Scholar]
- 37.Misharin AS, Silivra OA, Kjeldsen F, Zubarev RA. Dissociation of peptide ions by fast atom bombardment in a quadrupole ion trap. Rapid Commun. Mass Spectrom. 2005;19:2163–2171. doi: 10.1002/rcm.2038. [DOI] [PubMed] [Google Scholar]
- 38.Berkout VD. Fragmentation of protonated peptide ions via interaction with metastable atoms. Anal. Chem. 2006;78:3055–3061. doi: 10.1021/ac060069a. [DOI] [PubMed] [Google Scholar]
- 39.Berkout VD, Doroshenko VM. Fragmentation of phosphorylated and singly charged peptide ions via interaction with metastable atoms. Int. J. Mass Spectrom. 2008;278:150–157. doi: 10.1016/j.ijms.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berkout VD. Fragmentation of singly protonated peptides via interaction with metastable rare gas atoms. Anal. Chem. 2009;81:725–731. doi: 10.1021/ac802214e. [DOI] [PubMed] [Google Scholar]
- 41.Cook SL, Collin OL, Jackson GP. Metastable atom-activated dissociation mass spectrometry: leucine/isoleucine differentiation and ring cleavage of proline residues. J. Mass Spectrom. 2009;44:1211–1223. doi: 10.1002/jms.1598. [DOI] [PubMed] [Google Scholar]
- 42.Cook SL, Jackson GP. Characterization of tyrosine nitration and cysteine nitrosylation modifications by metastable atom-activation dissociation mass spectrometry. J. Am. Soc. Mass. Spectrom. 2011;22:221–232. doi: 10.1007/s13361-010-0041-4. [DOI] [PubMed] [Google Scholar]
- 43.Cook SL, Jackson GP. Metastable atom-activated dissociation mass spectrometry of phosphorylated and sulfonated peptides in negative ion mode. J. Am. Soc. Mass. Spectrom. 2011;22:1088–1099. doi: 10.1007/s13361-011-0123-y. [DOI] [PubMed] [Google Scholar]
- 44.Cook SL, Zimmermann CM, Singer D, Fedorova M, Hoffmann R, Jackson GP. Comparison of CID, ETD and metastable atom-activated dissociation (MAD) of doubly and triply charged phosphorylated tau peptides. J. Mass Spectrom. 2012;47:786–794. doi: 10.1002/jms.3023. [DOI] [PubMed] [Google Scholar]
- 45.Pasinszki T, Yamakado H, Ohno K. Penning ionization of CH3CN and CH3NC by collision with He (23S) metastable atoms. J. Phys. Chem. 1995;99:14678–14685. [Google Scholar]
- 46.Kishimoto N, Aizawa J, Yamakado H, Ohno K. Collision-energy-resolved penning ionization electron spectroscopy of nitriles: Conjugation effects on interactions with He*(23S) metastable atoms. J. Phys. Chem. A. 1997;101:5038–5045. [Google Scholar]
- 47.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 48.McLafferty F. Interpretation of Mass Spectra. New York City, New York: W.A. Benjamin; 1966. [Google Scholar]
- 49.Xu Y, Brenna JT. Atmospheric Pressure Covalent Adduct Chemical Ionization Tandem Mass Spectrometry for Double Bond Localization in Monoene- and Diene-Containing Triacylglycerols. Anal. Chem. 2007;79:2525–2536. doi: 10.1021/ac062055a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerwin JL, Tuininga AR, Ericsson LH. Identification of molecular species of glycerophospholipids and sphingomyelin using electrospray mass spectrometry. J. Lipid Res. 1994;35:1102–1114. [PubMed] [Google Scholar]
- 51.Ho Y-P, Huang P-C, Deng K-H. Metal ion complexes in the structural analysis of phospholipids by electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:114–121. doi: 10.1002/rcm.880. [DOI] [PubMed] [Google Scholar]
- 52.Claeys M, Nizigiyimana L, Van den Heuvel H, Derrick PJ. Mechanistic aspects of charge-remote fragmentation in saturated and mono-unsaturated fatty acid derivatives. Evidence for homolytic cleavage. Rapid Commun. Mass Spectrom. 1996;10:770–774. [Google Scholar]
- 53.Maccarone AT, Duldig J, Mitchell TW, Blanksby SJ, Duchoslav E, Campbell JL. Characterization of acyl chain position in unsaturated phosphatidylcholines using differential mobility-mass spectrometry. J. Lipid Res. 2014;55:1668–1677. doi: 10.1194/jlr.M046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams J, Gross ML. Charge-remote fragmentations of closed-shell ions. A thermolytic analogy. J. Am. Chem. Soc. 1989;111:435–440. [Google Scholar]
- 55.Deterding LJ, Gross ML. Tandem mass spectrometry for identifying fatty acid derivatives that undergo charge-remote fragmentations. Org. Mass Spectrom. 1988;23:169–177. [Google Scholar]
- 56.Jensen NJ, Tomer KB, Gross ML. Gas-phase ion decomposition occurring remote to a charge site. JACS. 1985;107:1863–1868. [Google Scholar]
- 57.Jensen N, Tomer K, Gross M. Fast atom bombardment and tandem mass spectrometry of phosphatidylserine and phosphatidylcholine. Lipids. 1986;21:580–588. doi: 10.1007/BF02534056. [DOI] [PubMed] [Google Scholar]
- 58.Wysocki VH, Ross MM. Charge-remote fragmentation of gas-phase ions: mechanistic and energetic considerations in the dissociation of long-chain functionalized alkanes and alkenes. Int. J. Mass Spectrom. Ion Processes. 1991;104:179–211. [Google Scholar]
- 59.Cheng C, Gross ML. Applications and mechanisms of charge-remote fragmentation. Mass Spectrom. Rev. 2000;19:398–420. doi: 10.1002/1098-2787(2000)19:6<398::AID-MAS3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 60.Shimma S, Kubo A, Satoh T, Toyoda M. Detailed structural analysis of lipids directly on tissue specimens using a MALDI-SpiralTOF-ReflectronTOF mass spectrometer. PLoS One. 2012;7:e37107. doi: 10.1371/journal.pone.0037107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng C, Gross ML, Pittenauer E. Complete structural elucidation of triacylglycerols by tandem sector mass spectrometry. Anal. Chem. 1998;70:4417–4426. doi: 10.1021/ac9805192. [DOI] [PubMed] [Google Scholar]
- 62.Madison TA, Siska PE. Penning ionization and ion fragmentation of formamide HCONH2 by He*, Ne*, and Ar* in molecular beams. J. Chem. Phys. 2009;131 doi: 10.1063/1.3242782. [DOI] [PubMed] [Google Scholar]
- 63.Siska PE. Molecular-beam studies of Penning ionization. Rev. Mod. Phys. 1993;65:337–412. [Google Scholar]
- 64.Ohno K, Okamura K, Yamakado H, Hoshino S, Takami T, Yamauchi M. Penning ionization of HCHO, CH2CH2, and CH2CHCHO by collision with He (23S) metastable atoms. J. Phys. Chem. 1995;99:14247–14253. [Google Scholar]
- 65.Yamakado H, Yamauchi M, Hoshino S, Ohno K. Penning ionization of CH3OH,(CH3) 2O, and (CH3CH2) 2O by collision with He (23S) metastable atoms. J. Phys. Chem. 1995;99:17093–17099. [Google Scholar]
- 66.Morgner H. The characterization of liquid and solid surfaces with metastable helium atoms. In: Benjamin B, Herbert W, editors. Advances In Atomic, Molecular, and Optical Physics. Academic Press; 2000. pp. 387–488. [Google Scholar]
- 67.Ann Q, Adams J. Collision-induced decomposition of sphingomyelins for structural elucidation. Biological Mass Spectrometry. 1993;22:285–294. [Google Scholar]
- 68.Hsu F-F, Turk J. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass. Spectrom. 2000;11:437–449. doi: 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- 69.Longuet-Higgins HC. Some Studies in Molecular Orbital Theory I. Resonance Structures and Molecular Orbitals in Unsaturated Hydrocarbons. The Journal of Chemical Physics. 1950;18:265–274. [Google Scholar]
- 70.Herndon WC. Resonance theory and the enumeration of Kekule structures. J. Chem. Educ. 1974;51:10. [Google Scholar]
- 71.Glendening ED, Weinhold F. Natural resonance theory: I. General formalism. J. Comput. Chem. 1998;19:593–609. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.