Abstract
Background & Aims
Bile acids are physiologic detergents that also activate nuclear receptors to regulate glucose and lipid homeostasis. Cholesterol 7α-hydroxylase (Cyp7a1), the rate-limiting enzyme that converts cholesterol to bile acids, is transcriptionally regulated by bile acids and circadian rhythms. Fasting, nutrients, and the circadian clock critically control hepatic bile acid and lipid homeostasis, and circadian misalignment is associated with the metabolic syndrome in humans. To delineate these interactions, we employed a sleep disruption model to induce circadian disruption and examined hepatic metabolism with respect to bile acids, lipids, and clock gene expression.
Methods
B6xC57 mice were maintained on chow or Western diet and were sleep disrupted for 6 hours/day for 5 days. Mice were sacrificed at 4-hour intervals over 24 hours. Hepatic metabolic genes were examined, and bile acid pool and lipid profiles were measured over 24 hours.
Results
Sleep disruption significantly suppressed circadian expression of core clock genes, genes involved in lipid metabolism, and key regulators of Cyp7a1 as well as Cyp7a1 expression itself. Sleep disruption abolished the peak in serum cholesterol and increased liver and serum free fatty acids. Bile acid pool size was increased while liver bile acids were decreased. Chromatin immunoprecipitation assay revealed that hepatocyte nuclear factor 4α (HNF4α) and D-site binding protein (Dbp) occupancies were suppressed at the Cyp7a1 promoter in sleep-disrupted mice. When coupled with Western diet, sleep disruption abolished liver clock rhythms and elevated free fatty acids.
Conclusions
Even short-term circadian disruption dramatically alters hepatic clock gene expression, bile acid metabolism, and lipid homeostasis to contribute to dyslipidemia.
Keywords: Bile Acid Synthesis, Circadian Rhythm, Lipid Metabolism
Abbreviations used in this paper: BMAL1, brain and muscle Arnt-like 1; CCG, clock-controlled genes; ChIP, chromatin immunoprecipitation; CLOCK, circadian locomotor output cycles kaput; CYP7A1, cholesterol 7α-hydroxylase; DBP, D-site binding protein; HNF4α, hepatocyte nuclear factor 4α; Per, period; Rev-erbα, reverse-erythroblastosis α; RORα, retinoic acid-related orphan receptor α; Shp, small heterodimer partner; Srebp-1, sterol regulatory element-binding protein-1; ZT, Zeitgeber time
Summary.
Short-term circadian disruption disturbs bile acid and lipid homeostasis in mice, in part via transcriptional occupancy of the Cyp7a1 promoter by HNF4α and Dbp. Coupled with Western diet, free fatty acids are increased, and hepatic clock gene expression is altered.
Many cellular and physiologic reactions occur at specific times of the day and follow a circadian rhythm (circa: approximate; diem: day), which allows for timing of biochemical and behavioral processes that are synchronized to the external environment. The environmental light/dark cycle is the predominant Zeitgeber, or entraining agent, to the hypothalamic biological clock (the suprachiasmatic nucleus), which is the master synchronizer that coordinates timing of central and peripheral rhythms, including the sleep-wake cycle, the fasting-feeding cycle, and body temperature.1 On a cellular level, organisms are met with the need to preserve glucose and lipid homeostasis over 24 hours. In peripheral organs such as the liver and white adipose tissue, neural and hormonal circadian signals serve to temporally segregate metabolic reactions. This, coupled with circadian regulation of activity, allows for advantageous metabolic responses to changes in the external environment such that individual organs and tissues are operating under maximal efficiency.
In mammals, core molecular clockwork is present in nearly all cell types and consists of rhythmically expressed clock genes that produce an autoregulatory feedback loop. The products of these clock genes transcriptionally activate and repress clock gene activity, such that the core molecular clock oscillates with a period of ∼24 hours. Among these genes, circadian locomotor output cycles kaput (Clock) and brain and muscle Arnt-like 1 (Bmal1) represent the forward limb of the regulatory loop, whose proteins heterodimerize and drive transcription of period (Per) and cryptochrome (Cry) genes. PER and CRY enter the nucleus and subsequently inhibit CLOCK/BMAL1 activity, thus reducing their own transcription. The molecular clock is further regulated by an additional feedback loop in which the nuclear receptors reverse-erythroblastosis α (Rev-erbα) and retinoic acid-related orphan receptor α (RORα) negatively and positively, respectively, regulate Bmal1 transcription by competing for retinoic acid-related orphan receptor response elements in the Bmal1 promoter.2 These interlocking regulatory loops allow for temporal management of many metabolic processes, including glucose and cholesterol homeostasis.
Recent studies indicate approximately 10% of genes within the liver transcriptome are rhythmically expressed,3 up to 20% of the mouse liver proteome is under circadian control,4 and the phases of gene and protein expression can be used as reporters of liver clock time. One such gene is the rate-limiting enzyme involved in the conversion of cholesterol to bile acids, cholesterol 7α-hydroxylase (Cyp7a1), which exhibits a circadian rhythm of expression of mRNA, protein, and activity in rodents and humans.5, 6 Bile acids are amphipathic molecules that aid in absorption of dietary lipids and regulate the gut microbiome population. They are also natural ligands for several nuclear receptors that regulate metabolic pathways. Conversion to bile acids represents the main catabolic pathway for cholesterol in humans;7 thus, bile acids and Cyp7a1 are crucial to homeostatic maintenance of lipid metabolism.
Growing research evidence suggests disruptions in circadian rhythms negatively impact human health. Disturbances such as sleep deprivation, shift work, and the 20th-century phenomenon of increased exposure to light at night are associated with increased incidence of cardiovascular events,8 gastrointestinal disorders,9 cancer,10 and metabolic syndrome.11, 12 Synchronization between internal physiology and external environment is crucial, but in modernized populations this fundamental relationship may be increasingly disturbed. Recently, it was reported that sleep restriction altered transcriptional regulation of white adipose tissue in mice13 and the ClockΔ19 mutation in mice may contribute to the deregulation of lipid accumulation or mobilization.14 Studies in humans demonstrate an association between shift work or chronodisruption and the development of obesity.15, 16
Given these facts and the implications for bile acids in the regulation of glucose and lipid homeostasis, we employed a sleep-deprivation protocol in mice to examine the circadian and epigenetic regulation of bile acid and liver metabolism under disrupted circadian conditions.
Materials and Methods
Animals and Diets
Cohorts of 2- to 4-month-old female wild-type mice (B6xC57 background) were bred in-house, maintained on a standard chow diet and water ad libitum, and were housed in a temperature-controlled room with a 12/12 light/dark cycle, Zeitgeber time (ZT) 12 = lights off (6:00 pm). Mice were individually housed, and sleep-disruption techniques were performed using gentle stimulation (physical contact with a soft artist’s paintbrush and light puffs of air) for 6 hours/day for 5 days during the middle of the light phase (ZT 2 – ZT 8). Body weight and daily food intake were recorded, and the mice were visually monitored continuously for signs of sleep during the procedure. The control mice were monitored and were not manipulated. After the sleep-disruption procedure had been completed on day 5, the mice were deeply anesthetized with isoflurane vapor and were sacrificed (in the fed-state) by cervical dislocation in 4-hour intervals (ZT 2, 6, 10, 14, 18, 22; n = 5–6 mice per time point) followed by rapid tissue extraction. The samples were stored at −80°C until analysis.
Additional cohorts of female wild-type mice were maintained on a high-fat, high-cholesterol Western diet (42% kcal from fat, 0.2% cholesterol, cat. no. TD.88137; Harlan Laboratories; Indianapolis, IN), and their body weight growth was monitored. Growth plateaued after approximately 4 months of dietary treatment, after which the mice were subjected to the previously described sleep-disruption protocol for 5 days (n = 4 mice per time point) and were sacrificed at the previously mentioned intervals.
All animal procedures followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All procedures were approved by the Institutional Animal Care and Use Committee of Northeast Ohio Medical University (protocol #13-023).
Experimental Procedures
Quantitative Polymerase Chain Reaction
Total RNA was isolated using TRI-Reagent (Sigma-Aldrich, St. Louis, MO) from mouse livers harvested at the described ZTs, followed by centrifugation with chloroform and isopropanol. Reverse transcription and quantitative polymerase chain reaction (qPCR) were performed using Taqman probes and primers (Life Technologies, Carlsbad, CA) according to manufacturer’s instructions. Relative mRNA levels were calculated using the comparative cycle threshold (2−ΔΔCt) method with values normalized to mouse glyceraldehyde 3-phosphate dehydrogenase expression as an internal control.
Lipid Analysis
Lipids were isolated by homogenizing liver tissue in 7:11:0.1 chloroform/isopropanol/NP-40 followed by evaporation at 60°C. Commercially available kits and reagents were used to quantify the total cholesterol (BioVision Incorporated, Milpitas, CA), triglycerides (Sekisui Diagnostics, Lexington, MA), and free fatty acids (Wako Chemicals, Richmond, VA) in liver and serum.
Bile Acid Analysis
Bile acids were isolated from 100-mg liver samples or whole intestine and gallbladder by a series of extractions and centrifugations in ethanol followed by incubation overnight at 60°C. Content was analyzed by kit (Diazyme, Poway, CA) according to the manufacturer’s instructions. The bile acid pool size was calculated by summing the bile acid content in the liver, gallbladder, and intestine.
Chromatin Immunoprecipitation Assay
Effects of sleep disruption on transcription factor recruitment of Cyp7a1 promoter chromatin were investigated using a commercially available chromatin immunoprecipitation (ChIP) kit (EMD Millipore, Billerica, MA). Briefly, nuclei were isolated from 200 mg of pooled frozen liver tissue at each time point over 24 hours. Formaldehyde (37%) was used to crosslink protein to DNA, after which the nuclei were centrifuged and washed. Approximately 1 × 106 nuclei were used per time point. The samples were lysed, sonicated, and precleared with agarose beads for 1 hour, after which the samples were incubated in 10 μg of antibody against hepatocyte nuclear factor 4α (HNF4α) (cat. no. C11F12; Cell Signaling Technology, Danvers, MA) or D-site binding protein (DBP) (cat. no. sc-98411X; Santa Cruz Biotechnology, Dallas, TX) at 4°C overnight. The antibody/protein complex was precipitated with agarose beads, and the complex was washed in a series of salt buffers, finishing with DNA elution buffer.
The crosslinks were reversed by incubating samples with 5 M NaCl at 65°C for 4 hours. The DNA was purified with 25:24:1 phenol/chloroform/isoamyl alcohol followed by precipitation with glycogen. Quantitative polymerase chain reaction was performed using 25 μg of DNA, and SYBR primers were used to detect mouse Cyp7a1 promoter at -1 kB region: forward primer-219ACCTTCGGCTTATCGACTATTGC, reverse primer-163TATCTGGCCTTGAACTAAGTCCATCT. Input samples were 10% of total DNA, and negative controls were not incubated in primary antibody. SYBR primers were designed to amplify a fragment in intron 5 of the Cyp7a1 gene and were used to detect off-site binding (data not shown): forward primer 162TGTTTGGACACTTTGGGTAGAG; reverse primer 261TGAGGATCTGGGAGAAGGTATT.
Statistical Analysis
Data were analyzed using Sigma Plot 11.0 computer software (Systat Software, San Jose, CA), and the results were expressed as mean ± standard error of the mean (SEM). For each gene, 24-hour mRNA data were normalized to total wild-type mRNA expression for ease of comparison. Data were analyzed by Student t test or two-way analysis of variance (ANOVA) followed by a post hoc Holm-Sidak test where appropriate. P < .05 was considered statistically significant. All authors had had access to the study data and reviewed and approved the final manuscript.
Results
Sleep Disruption and Western Diet Alter Food Intake and Liver and White Adipose Tissue Weight Without Changing Body Weight
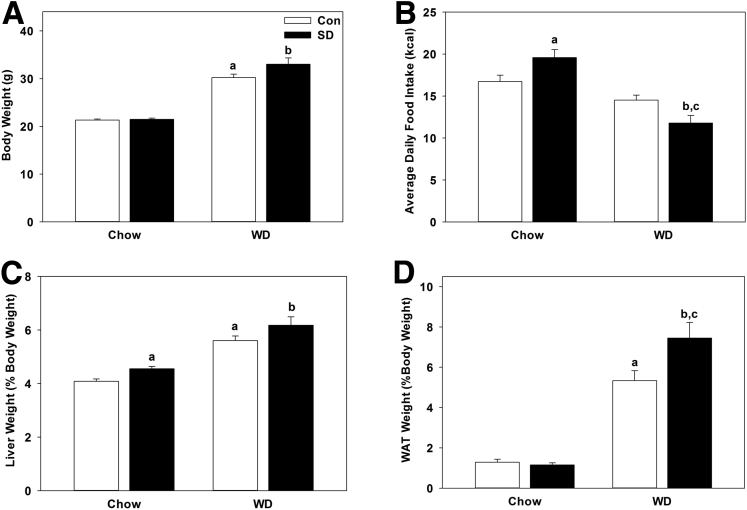
Wild-type mice maintained on normal chow diet underwent sleep disruption for 5 consecutive days in the middle of the light phase (ZT 2 – ZT 8) using gentle handling and stimulation techniques. To determine the effects Western diet and diurnal disruption on metabolism, we coupled sleep disruption with a high-calorie, high-cholesterol Western diet in a separate group of mice. Mice were then sacrificed at 4-hour intervals over 24 hours corresponding to ZT 2, ZT 6, ZT, 10, ZT 14, ZT 18, and ZT 22. Wet liver and white adipose tissue weights were normalized and expressed as a percentage of total body weight. Sleep disruption did not statistically significantly affect total body weight across chow-fed (P = .660) or Western diet-fed mice (P = .089) although Western diet statistically significantly increased body weight in both control and sleep-disturbed groups (P = 2.88E-10; Figure 1A). Average daily caloric consumption was statistically significantly increased during sleep disruption in chow-fed mice (P = .039) and was significantly decreased in Western diet-fed mice undergoing sleep disruption compared with sleep disruption under chow-fed conditions (P = 5.36E-5) and compared with Western diet-fed control mice (P = .01; Figure 1B). The liver-to-body weight ratio was statistically significantly increased in chow-fed mice undergoing sleep disruption (P = .014) and by Western diet treatment alone in both control (P = 1.26E-5) and sleep-disturbed mice (P = .001; Figure 1C). White adipose tissue weight did not change with sleep disruption (P = .540) in chow-fed mice and was statistically significantly increased by Western diet feeding (P = 1.49E-5); it was further increased in Western diet-fed mice undergoing sleep disruption (P = .046; Figure 1D). This suggests even short-term rhythm disruption (as little as 5 days) is sufficient to induce physiologic changes in mice.
Figure 1.
Sleep disruption and Western diet alter food intake and liver and white adipose tissue weight. (A) Average body weight of control and sleep-disrupted (SD) mice on chow or Western diet (WD). (B) Average daily food intake was significantly increased during sleep disruption in chow-fed mice and was reduced in Western diet–fed mice undergoing sleep disruption. (C) Wet liver weight from each mouse was normalized to total body weight. Sleep disruption significantly increased liver weight in mice maintained on both chow and Western diet. (D) White adipose tissue weight (normalized to body weight) did not differ significantly due to sleep disruption in mice maintained on chow diet, but was significantly increased in mice maintained on Western diet. Data were pooled from chow-fed control (n = 36) and sleep-disrupted (n = 34) mice, and from Western diet-fed control (n = 24) and sleep-disrupted (n = 24) mice across all time points measured. White bar, control; black bar, sleep disrupted. Data were analyzed by one-way analysis of variance. aP < .05 within control treatment; bP < .05 with Western diet; cP < .05 within sleep disruption treatment.
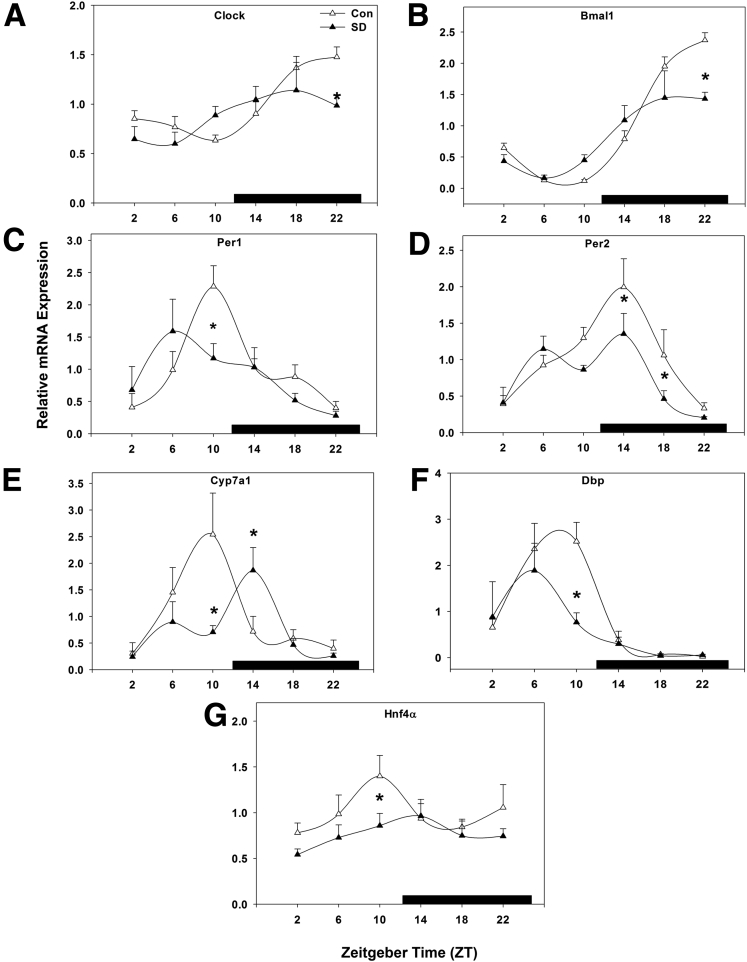
Sleep Disruption Suppresses Hepatic Molecular Core Clock Gene Expression
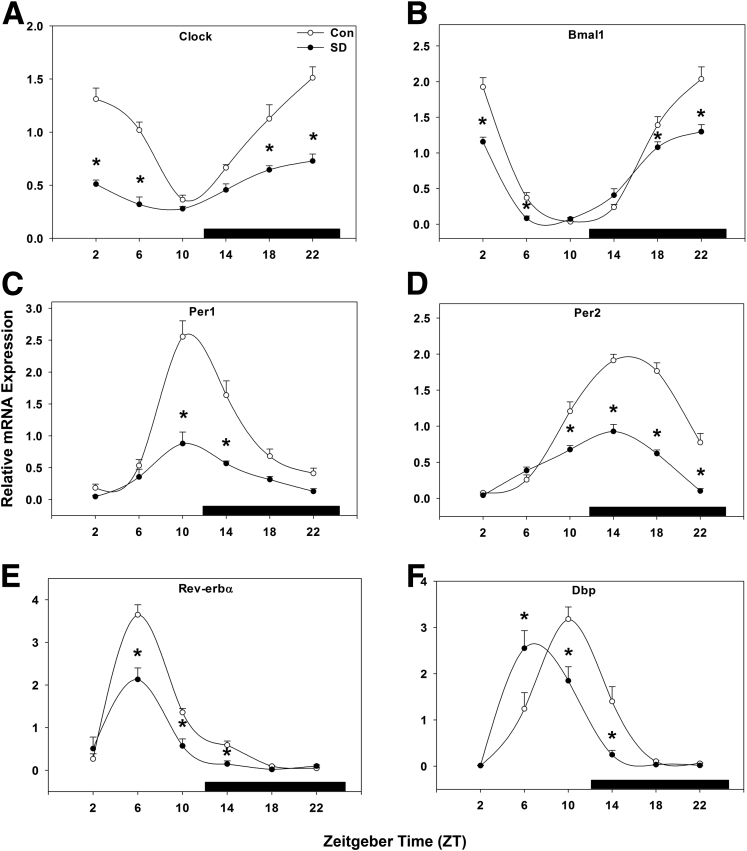
Core clock gene mRNA expression was statistically significantly suppressed across the majority of time points examined. In control mice, peak Clock gene expression occurred from mid-night to early morning (ZT 22–2; Figure 2A), and sleep disruption caused a statistically significant suppression of Clock gene expression levels and reduced expression at nearly all times (P < .001). Peak Bmal1 expression occurred between ZT 22 and ZT 2 (Figure 2B), and was suppressed by sleep disruption (P < .001), while the phase of peak expression was not changed.
Figure 2.
Sleep disruption suppresses hepatic molecular core clock gene expression. Messenger RNA rhythms of (A) Clock, (B) Bmal1, (C) Per1, and (D) Per2, (E) Rev-erbα, and (F) Dbp, mean ± standard error of the mean after 5 days of sleep disruption. ○, control; ●, sleep disrupted; n = 5–6 mice. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT), black bar indicates the dark phase.
Hepatic Per1 peaked at ZT 10 and exhibited a nadir in late-night to early morning. Five days of sleep disruption resulted in approximately a 2.5-fold decrease in Per1 peak amplitude compared with control mice, though peak expression similarly occurred at ZT 10 (P < .001; Figure 2C). In control mice, Per2 peaked near the middle of the dark phase (ZT 14–18) with a nadir in early morning. Sleep disruption statistically significantly suppressed Per2 gene expression from late-day throughout the night by twofold, but did not change the phase of peak expression (P < .001; Figure 2D). These data suggest that both the forward limb of the molecular circadian clock (Clock and BMAL) and the repressive limb (Per1/2) are affected by short-term diurnal disruption.
Next, we examined the molecular clock feedback regulators Rev-erbα and the clock-controlled gene (CCG) D-site binding protein (Dbp), an oscillatory transcription factor shown to bind to and induce transcription of Per117 in mice and implicated in the regulation of Cyp7a1.18 Rev-erbα and RORα are negative and positive regulators, respectively, of the circadian rhythms of many liver genes. Interestingly, Rev-erbα is a positive regulator of the Cyp7a1 gene,19 and RORα is a positive regulator of the sterol 12α-hydroxylase (Cyp8b1) gene in cholic acid synthesis.20 Rev-erbα expression peaked in mid-day (ZT 6) with a nadir in late-night. Sleep disruption suppressed Rev-erbα expression, resulting in decreased amplitude during the day hours (P < .001; Figure 2E). Dbp expression was unchanged in overall gene expression level (P = .094; Figure 2F) but was phase-advanced about 4 hours by sleep disruption, resulting in amplitude changes at corresponding times (P < .001).
Sleep Disruption Suppresses Hepatic Genes Involved in Bile Acid and Lipid Metabolism
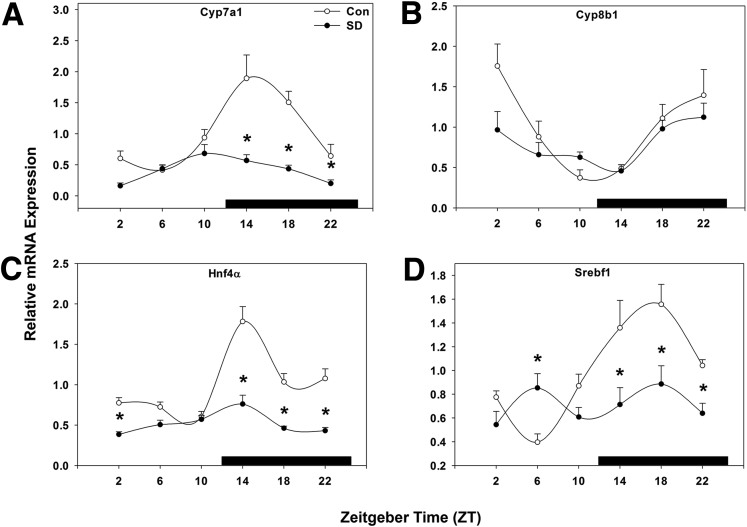
Cyp7a1 gene expression peaked at the beginning of the dark phase in control mice, and this peak was statistically significantly reduced by sleep disruption from ZT 14–22, and peak expression was advanced by approximately 4 hours (P < .001; Figure 3A). Conversely, Cyp8b1 was not altered at any time point by sleep disruption (P = .061; Figure 3B). Hepatocyte nuclear receptor 4α (Hnf4α), an important transcriptional regulator of Cyp7a1, exhibited a peak at ZT 14 coinciding with the Cyp7a1 peak, which was markedly reduced by sleep disruption (P < .001; Figure 3C). Finally, sterol regulatory element-binding protein 1c (Srebf1), which regulates fatty acid synthesis in hepatocytes, was rhythmically expressed in wild-type mice and was statistically significantly suppressed by sleep disruption across most time points, resulting in arrhythmic expression (P < .001; Figure 3D).
Figure 3.
Sleep disruption suppresses hepatic genes involved in bile acid and lipid metabolism. Messenger RNA rhythms of (A) Cyp7a1, (B) Cyp8b1, (C) Hnf4α, and (D) Srebf1, mean ± standard error of the mean after 5 days of sleep disruption. ○, control; ●, sleep disrupted; n = 5–6 mice. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT), and black bar indicates the dark phase.
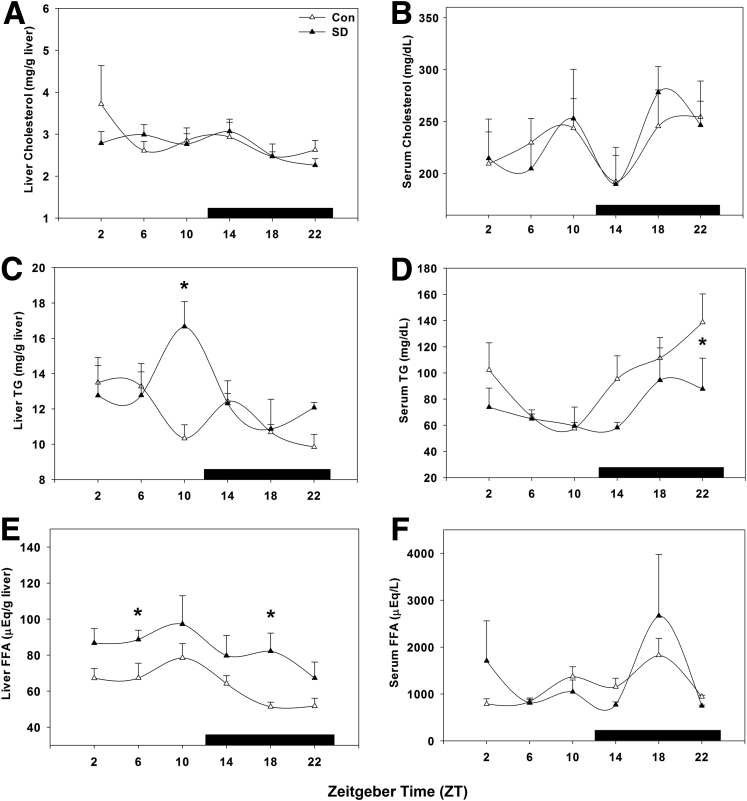
Short-Term Sleep Disruption Altered Hepatic and Serum Lipid Profiles
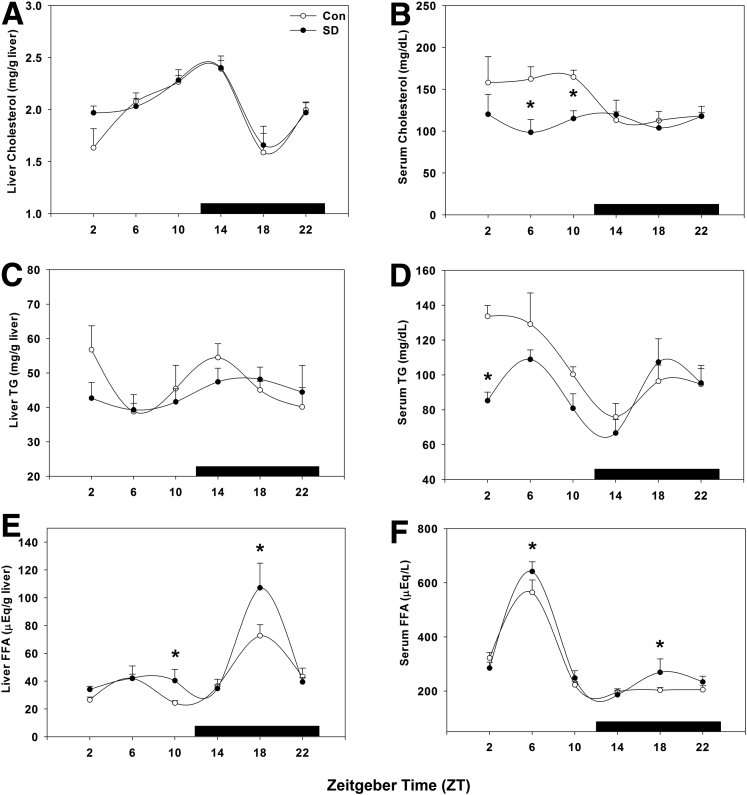
Hepatic and serum samples were analyzed for cholesterol, triglycerides, and free fatty acids to determine the metabolic effect of short-term sleep disruption. Hepatic cholesterol levels were unchanged, peaking near the light/dark transition in both treatment groups (P = .375; Figure 4A), whereas the diurnal daytime elevation in serum cholesterol was statistically significantly lower in sleep-disturbed mice (P = .004; Figure 4B), resulting in near-total suppression of rhythmicity.
Figure 4.
Hepatic and serum profiling indicates short-term sleep disruption affects lipid metabolism. Lipid content was profiled over 24 hours by quantifying (A) liver cholesterol, (B) serum cholesterol, (C) liver triglycerides (TG), (D) serum TG, (E) liver free fatty acids (FFA), and (F) serum FFA. ○, control; ●, sleep disrupted; n = 5–6 mice. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT), and black bar indicates the dark phase.
Similarly, hepatic triglyceride content was not statistically significantly altered by sleep disruption at any time point compared with that of wild-type mice (P = .421; Figure 4C), whereas serum triglyceride levels were statistically significantly suppressed by sleep disruption early in the day at ZT 2 (P = .018; Figure 4D). Free fatty acids were elevated in both hepatic tissue at ZT 10 and ZT 18 (P = .017; Figure 4E) and in serum only at ZT 6 (P = .044; Figure 4F). Also of note, liver and serum free fatty acid content are rhythmic with opposing phases of peak content.
Sleep Disruption Suppresses Key Genes Involved in Lipid Metabolism
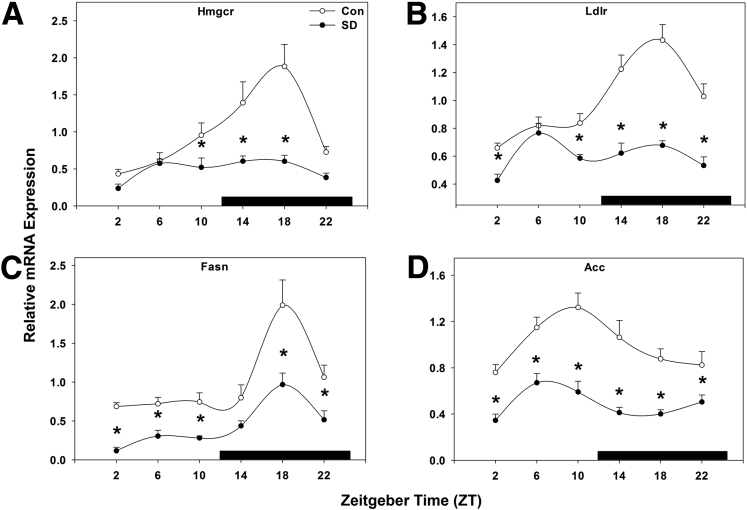
To delineate the mechanism by which sleep disruption could result in altered lipid metabolism, we next examined the expression of several key genes involved in lipid metabolism. HMG-CoA reductase (3-hydroxy-3-methylglutaryl-coenzyme A reductase) is the rate-limiting enzyme in the cholesterol production pathway and exhibits a nighttime peak in gene expression in the liver.21 Sleep disruption abolished this peak and suppressed HMG-CoA reductase expression (P < .001; Figure 5A) compared with control mice. Likewise, low-density lipoprotein receptor under normal conditions exhibits a nighttime peak in gene expression in liver tissue;13 here, sleep disruption rendered low-density lipoprotein receptor expression arrhythmic and suppressed its expression (P < .001; Figure 5B). Fatty acid synthase, which codes for the enzyme that catalyzes fatty acid synthesis, exhibited a peak in gene expression in mid-night (ZT 18), and this peak was statistically significantly decreased, but not abolished, by sleep disruption (P < .001; Figure 5C). Related to this, acetyl-CoA carboxylase, which converts acetyl-CoA to malonyl-CoA and thus provides substrate for fatty acid synthesis, exhibited a rhythmic pattern of gene expression, peaking in late day at ZT 10. Sleep disruption suppressed this expression, though the phase of peak expression remained unaffected at ZT 10 (P < .001; Figure 5D). These data support the notion that short-term sleep disruption significantly alters genes involved in fatty acid synthesis, which may lead to alteration of triglyceride and free fatty acid homeostasis in liver and serum.
Figure 5.
Sleep disruption suppresses key genes involved in lipid metabolism. Messenger RNA rhythms of (A) HMG-CoA reductase (Hmgcr), (B) low-density lipoprotein receptor (Ldlr), (C) fatty acid synthase (Fasn), and (D) acetyl CoA carboxylase (Acc), mean ± standard error of the mean after 5 days of sleep deprivation. ○, control; ●, sleep disrupted; n = 5–6 mice. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT), and black bar indicates the dark phase.
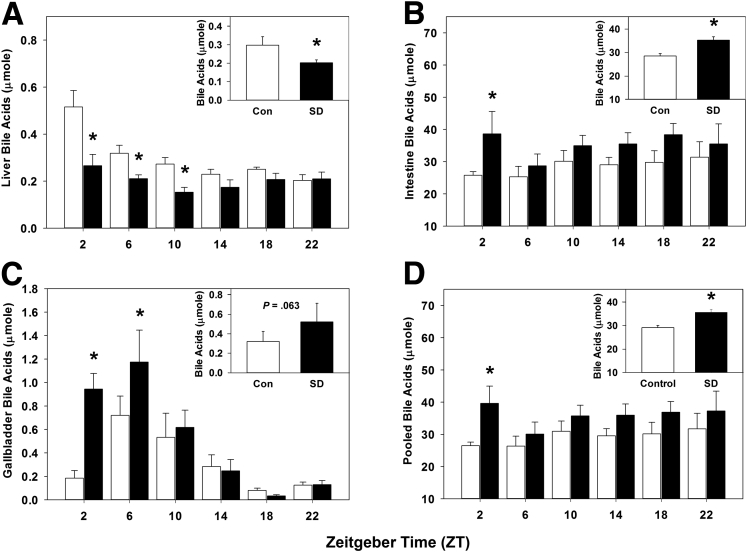
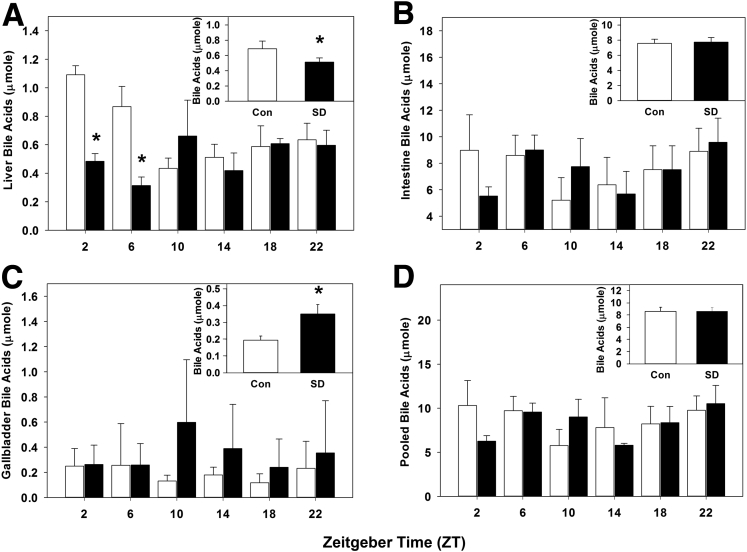
Sleep Disruption Alters Bile Acid Homeostasis
We quantified the liver, intestinal, and gallbladder bile acids after sleep disruption, as circadian disturbances have been shown to influence bile acid metabolism.22 Liver bile acids were statistically significantly decreased by sleep disruption during the daytime from ZT 2–10 (Figure 6A), and total liver bile acids were statistically significantly decreased across pooled times measured (P < .001; Figure 6A, inset). This finding is consistent with the observed decrease in Cyp7a1 expression and rhythmicity and is the result of decreased de novo synthesis.
Figure 6.
Sleep disruption alters bile acid homeostasis. Bile acid quantification over 24 hours (summed over 24 hours, inset) in (A) liver, (B) intestine, (C) gallbladder, and (D) total pool size, from wild-type mice after 5 days of sleep disruption. White bar, control; black baŗ sleep disrupted; n = 5–6 mice per time point. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT).
Conversely, intestinal bile acids were nearly doubled by sleep disruption in early day at ZT 2, and they trended toward increasing at all other times measured (Figure 6B); total intestinal bile acids across all times were statistically significantly increased (P = .005; Figure 6B, inset). Interestingly, gallbladder bile acid content at ZT 2 and ZT 6 was increased approximately 5 times and 2 times, respectively, by sleep disruption (Figure 6C), though when pooled over all times measured, the gallbladder bile acid content did not statistically significantly differ but did trend toward an increase (P = .063; Figure 6C, inset). Bile acid pool size, defined as the total amount of bile acids in the gallbladder, intestine, and liver, was statistically significantly increased at ZT 2 in sleep-disturbed mice and trended toward an increase at all other times measured. When the data were pooled across all times, the bile acid pool size was statistically significantly increased by sleep disruption (P = .003; Figure 6D, inset). These data suggest that sleep disruption altered bile acid homeostasis and enterohepatic circulation of bile acids.
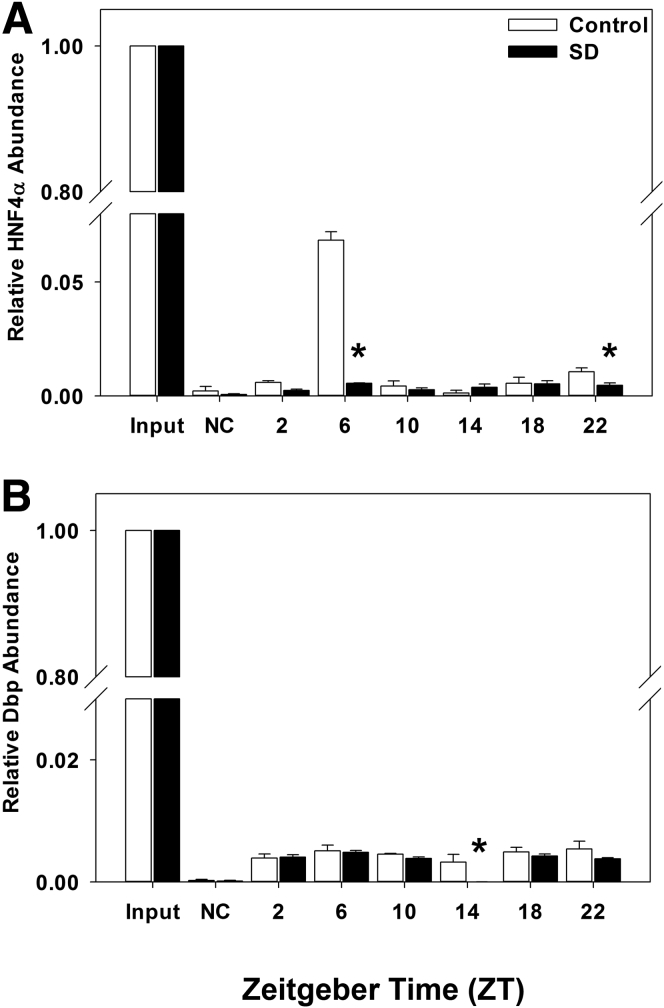
Sleep Disruption Alters Transcription Factor Occupancy at the Cyp7a1 Gene Promoter
HNF4α plays a major role in the liver-specific transcription of Cyp7a1 gene expression. We performed ChIP assays using nuclear extracts from mouse liver to demonstrate that sleep disruption significantly reduced HNF4α occupancy at the Cyp7a1 gene promoter.
In control mice, HNF4α occupancy at the Cyp7a1 promoter chromatin is greatest in mid-day at ZT 6 (Figure 7A), which is 8 hours in advance of the peak expression of both Cyp7a1 and HNF4α mRNA at ZT 14 (Figure 3A and C). In sleep-disturbed mice, HNF4α occupancy at the Cyp7a1 promoter was markedly reduced at ZT 6 and again at ZT 22 (P < .001; Figure 7A). Dbp occupancy was also significantly reduced by sleep disruption at ZT 14 (P = .022; Figure 7B). It is noted that DBP occupancy at the Cyp7a1 promoter is very low and nonrhythmic. DBP is a major CCG regulating Cyp7a1 mRNA rhythmicity, but its half-life is short and may contribute to weak binding to the Cyp7a1 promoter. These results, combined with gene expression profiles (Figure 3A and C), indicate that disruption of the HNF4α circadian rhythm may be the major cause of altered Cyp7a1 rhythmicity.
Figure 7.
Sleep disruption alters transcription factor occupancy at the Cyp7a1 gene promoter. Chromatin immunoprecipitation (ChIP) assay depicting (A) HNF4α and (B) Dbp occupancy at the Cyp7a1 gene promoter over 24 hours. Samples were pooled from 5–6 mice per time point; white bar, control; black bar, sleep disrupted. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT).
Sleep Disruption Coupled With Western Diet Alters Hepatic Molecular Core Clock and Bile Acid Gene Expression
We next examined mRNA gene expression of molecular core clock and bile acid genes in the liver, as consumption of high-calorie diet is known to alter liver gene expression in mice.22, 23 In sleep-disturbed mice fed a Western diet, the pattern of Clock gene expression was inverted compared with the control mice though the amplitude remained nearly unchanged except at ZT 22 (P = .143; Figure 8A); Bmal1 mRNA expression remained unchanged, with a suppression at ZT 22 (P = .103; Figure 8B).
Figure 8.
Sleep disruption coupled with Western diet alters hepatic molecular core clock and bile acid gene expression. Messenger RNA rhythms of (A) Clock, (B) Bmal1, (C) Per1, (D) Per2, (E) Cyp7a1, and (F) Dbp, mean ± standard error of the mean after 5 days of sleep deprivation. △, control + Western diet; ▲, sleep disrupted + Western diet; n = 4 mice. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT), and black bar indicates the dark phase.
Peak Per1 gene expression was suppressed at ZT 10 (P = .003) in sleep-disturbed mice fed a Western diet (Figure 8C). Per2 gene expression peaked later at ZT 14 in control mice, and the rhythm was suppressed in sleep-disturbed mice (P = .032), rendering the pattern arrhythmic (Figure 8D). Cyp7a1 mRNA expression exhibited an advanced peak at ZT 10 in Western diet-fed control mice, while in sleep-disturbed mice the expression was suppressed at ZT 10 (P = .002) and shifted to ZT 14 (Figure 8E).
Dbp mRNA expression in Western diet-fed mice displayed a similar pattern to its expression in chow-fed mice (Figure 8F, compared with chow-fed mice in Figure 2F), with peak expression occurring near the end of the day at ZT 10; sleep disruption advanced and suppressed the peak expression to ZT 6 (P = .048; Figure 8F). Peak HNF4α mRNA expression was advanced by Western diet to ZT 10 in control mice, and in sleep-disturbed mice the expression pattern was suppressed and resembled that of sleep-disturbed chow-fed mice (P = .024; Figure 8G). We also noticed that the effect of sleep disruption on rhythmicity of Cyp7a1 is similar to that of HNF4α in both chow diet (Figure 3A versus C) and Western diet (Fig 8E versus G), but not Dbp. These results further support our conclusion that HNF4α is a major regulator of Cyp7a1 gene transcription. It appears that Dbp gene expression is more susceptible to disruption of circadian systems than to a high-fat, high-cholesterol diet, whereas HNF4α expression may be more susceptible to diet-induced disruption.
Lipid and Bile Acid Profiling of Sleep-Disturbed Mice Fed a Western Diet
Hepatic and serum cholesterol, triglycerides, and free fatty acids were profiled in Western diet-fed mice to determine the metabolic effects of a high-calorie diet and circadian system disruption. Liver and serum cholesterol content were rendered arrhythmic in both groups of Western diet-fed mice, and sleep disruption did not cause statistically significant changes in liver (P = .508) or serum (P = .915) cholesterol levels (Figure 9A and B), though the serum cholesterol levels fluctuated dramatically across all times measured. Liver triglycerides after Western diet treatment were increased only at ZT 10 in sleep-disturbed mice (P = .001; Figure 9C), whereas serum triglycerides were suppressed due to sleep disruption (P = .031; Figure 9D). Liver free fatty acids were statistically significantly elevated by sleep disruption and Western diet compared with Western diet alone (P < .001; Figure 9E) while serum free fatty acids remained unchanged (P = .618; Figure 9F).
Figure 9.
Hepatic and serum profiles of sleep-disturbed mice fed Western diet. Lipid content was profiled over 24 hours by quantifying (A) liver cholesterol, (B) serum cholesterol, (C) liver triglycerides (TG), (D) serum TG, (E) liver free fatty acids (FFA), and (F) serum FFA. △, control; ▲, sleep disrupted; n = 4 mice. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate, *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT), and black bar indicates the dark phase.
We next profiled bile acid content over 24 hours in sleep-disrupted and control mice fed a Western diet. Liver bile acid content was statistically significantly suppressed by sleep disruption (P = .018; Figure 10A), similar to the effects of sleep disruption under normal diet conditions. Intestinal bile acid content was unchanged (P = .875; Figure 10B), while gallbladder bile acid content was statistically significantly increased by sleep disruption (P = .026; Figure 10C). When analyzed as total pool size, these effects resulted in no difference in bile acid pool size (P = .974; Figure 10D). However, taken together these data indicate a shift in hepatobiliary metabolism toward decreased de novo synthesis under conditions of sleep disruption and Western diet feeding.
Figure 10.
Sleep disruption coupled with Western diet alters bile acid homeostasis. Bile acid quantification over 24 hours (summed over 24 hours, inset) in (A) liver, (B) intestine, (C) gallbladder, and (D) total pool size from wild-type mice after 5 days of sleep deprivation. White bar, control + Western diet; black bar, sleep disrupted + Western diet; n = 4 mice. Data were analyzed by two-way analysis of variance followed by Holm-Sidak post hoc comparison where appropriate. *P < .05 within time point. The x-axis indicates Zeitgeber time (ZT).
Discussion
Circadian organization represents a means by which internal biological processes can be synchronized to the timing of the external environment to maximize physiologic efficiency. Coordination of this timing is regulated by the suprachiasmatic nucleus master clock, which directs neurochemical and hormonal signals to synchronize the core clock gene expression in peripheral tissues to maintain homeostasis. Although circadian disruption (mainly in the form of shift work) is strongly correlated to metabolic syndrome in humans, little evidence is available regarding the molecular and biochemical mechanisms by which metabolism and human health are negatively affected.
Genetic mutation or ablation of molecular clock genes in mice results in wide-ranging metabolic disturbances. ClockΔ19 mutant mice exhibit hyperphagia, obesity, hepatic steatosis, and impaired glucose tolerance,24 and whole-body or tissue-specific ablation of Bmal1 causes impaired glucose tolerance24, 25 in mice. In our study, we demonstrate that a different form of circadian system disturbance, sleep disruption, resulted in altered hepatic gene expression and lipid homeostasis reflected in the hepatobiliary system, a critical component of lipid homeostasis.
Short-term diurnal disruption in wild-type female mice resulted in widespread suppression of core clock gene expression in the liver compared with control mice. This study shows that sleep disruption markedly altered the circadian rhythms of Clock, Per1, and Per2, but not Bmal1 gene expression in mice. Rhythms of several liver clock genes and CCGs, including Rev-erbα and Dbp, and transcription factors Hnf4α and Srebp-1c involved in the regulation of Cyp7a1 and lipid metabolism, were altered by just 5 days of circadian system disruption. In Rev-erbα-deficient mice, the Cyp7a1 rhythm in gene expression was not affected.19 However, activation of Rev-erbα by a synthetic agonist has been shown to increase Cyp7a1 expression.
HNF4α and DBP are major positive regulators of CYP7A1 expression, and key regulators of the circadian rhythm of CYP7A1.26, 27 HNF4α is one of the most abundant nuclear receptors in the liver and participates in glucose, cholesterol, bile acid, and xenobiotic metabolism.28 Interestingly, it was shown that liver-specific HNF4α-null mice did not exhibit the nightly peak in Cyp7a1 gene expression. Instead, nighttime Cyp7a1 expression in these mice was more than 50% reduced and was comparable to low levels detected during the day.29 It is possible that the suppression in HNF4α occupancy at the Cyp7a1 promoter demonstrated in this study, combined with reduced Hnf4α gene expression, contributes to the suppression in Cyp7a1 gene expression induced by sleep disruption, in turn leading to disrupted bile acid homeostasis.
The phase advance in Dbp peak gene expression may contribute to the advance of the Cyp7a1 peak by sleep disruption. Additionally, we demonstrate that Dbp presence at the Cyp7a1 gene promoter is reduced in early night at ZT 14, though binding did not appear to be rhythmic. Still, reduced Rev-erbα and HNF4α expression caused by sleep disruption may concurrently influence the reduction in Cyp7a1 gene expression, resulting in a suppressed and shifted gene expression pattern that is ultimately reflected in the bile acid pool. We speculate that HNF4α is the main driving factor in this scenario, as gene expression was reduced to basal levels by sleep disruption and peak HNF4α abundance at the Cyp7a1 promoter was also abolished. However, Dbp, Rev-erbα or other transcription factors may still play additional roles in the rhythmic regulation of Cyp7a1 gene expression.
It has been shown that genetic ablation of Per1 and Per2 results in increased serum bile acids and suppression of Cyp7a1.22 Similarly, we demonstrate that sleep disruption markedly decreased Per1 and Per2 expression, which may contribute to reduction of the peak in Cyp7a1 expression in the dark phase and may also contribute to its phase shift. Significantly reduced liver bile acid content (specifically during the day) coincides with reduced Cyp7a1 levels, which suggests that sleep disruption may increase intestinal bile acid reabsorption to account for increased bile acid pool size. It is known that bile acids inhibit apical sodium-dependent bile salt transporter (ASBT) expression in the intestine.30 Reduced bile acid synthesis may increase apical sodium-dependent bile salt transporter to stimulate bile acid reabsorption and increase the bile acid pool size under conditions of sleep disruption/disturbed rhythms. Overall, sleep disruption increased food intake and may partially account for the increased bile acid pool size and altered bile acid homeostasis and enterohepatic circulation of bile acids, which plays a critical role in regulation of hepatic lipid metabolism. However, Western diet-fed mice consumed less food under conditions of sleep disruption but still exhibited altered bile acid homeostasis.
Both liver and serum free fatty acids were increased by sleep disruption in the middle of the night in the liver (ZT 18) and in the middle of the day in serum (ZT 6), whereas serum triglyceride content was decreased in early day (ZT 2), possibly indicating a shift toward increased triglyceride breakdown. Serum free fatty acid is rhythmic in content and peaks during mid-day,23 and we demonstrate that short-term sleep disruption is capable of increasing free fatty acids after only 5 days. Rhythmic triglyceride content was shown to be increased in Clock mutant mice via decreased interaction of Clock with small heterodimer partner (Shp), a nuclear receptor that represses Cyp7a1 gene expression, which resulted in sustained microsomal triglyceride transfer protein expression.31, 32 Liver gene expression of both Shp and microsomal triglyceride transfer protein were reduced after sleep disruption (data not shown), indicating differential gene expression changes in response to distinct types of circadian disruption (genetic or behavioral). Interestingly, Shp−/− mice are protected against fatty liver disease, and knockdown of the clock gene neuronal PAS domain protein 2 (Npas2) reversed this phenotype and resulted in steatosis.33
We next examined bile acid content in the liver, intestine, and gallbladder of sleep-disturbed mice. The data show a significant trend toward decreased de novo synthesis of bile acids (demonstrated by suppressed Cyp7a1 gene expression and decreased liver bile acid content) yet an overall increased pool size. After Western diet treatment, liver bile acids were again suppressed by sleep disruption while gallbladder content was increased, though the total pool size was unchanged. This may indicate an underlying diet-independent mechanism by which the hepatobiliary system responds to circadian disruption. Few studies show the effects of circadian system disruption on bile acids, specifically. Ma et al22 demonstrated that in Per1/2 double-knockout mice hepatic bile acids were transiently increased, though Cyp7a1 gene expression was suppressed. Our laboratory has previously shown that the fasting-to-refeeding transition also plays an important role in bile acid metabolism; experimentally manipulating fasting and refeeding in mice results in a drastic suppression and then induction, respectively, of Cyp7a1 gene expression.34 Clearly, the hepatobiliary system is susceptible to both endogenous circadian control and mismanagement due to perturbation.
After Western diet treatment, hepatic clock gene expression was severely altered, with the exception of Bmal1. The pattern of Clock gene expression was inverted, and Per1 and Per2 expression was rendered arrhythmic by sleep disruption. We demonstrate that Western diet-feeding itself appears to mildly suppress core clock gene expression (Clock, Per1, Per2; Figures 2 and 8) in control mice alone; when coupled with sleep disruption, these rhythms were further suppressed or shifted.
It has been reported that high-fat feeding can increase core clock gene expression and induce phase delays in expression of metabolic genes in mouse liver23, 35 though at least one study refutes this observation.36 With respect to bile acid metabolism, Cyp7a1 peak gene expression in mice fed a Western diet was advanced approximately 4 hours compared to the well-established early nighttime peak, and sleep disruption delayed Cyp7a1 gene expression. Dbp gene expression was again advanced approximately 4 hours (from ZT 10 to ZT 6) in sleep-disturbed mice, which was similar to the expression profile seen in their chow-fed counterparts. We speculate that the advance in Cyp7a1 gene expression in Western diet-fed control mice may be mainly due to diet-induced changes in HNF4α gene expression. Interestingly, it was shown that feeding a high-fat diet to mice resulted in a 5-hour advance in clock gene rhythmicity cultured hepatocyte in vitro.37
Although we did not find a diet-induced difference specifically in Per2 gene expression, it is possible the in vivo manifestation of dietary effects on liver rhythms is reflected by phase shifting of key hepatic genes, including HNF4α. Additionally, Matsumoto et al38 demonstrated that alterations in nutrients (high-fat, high-carbohydrate, or high-protein diets fed to mice) dramatically advanced the mRNA rhythm of Srebp-1, a transcription factor critically involved in cholesterol and fatty acid synthesis, while liver clock gene and CCG expression (including Dbp) were only modestly affected by dietary changes. The interactions of excess nutrient conditions and peripheral rhythms, and the downstream effects on hepatobiliary physiology, appear to be independently linked, supported by our data and others, but definite conclusions cannot yet be drawn.
Conclusion
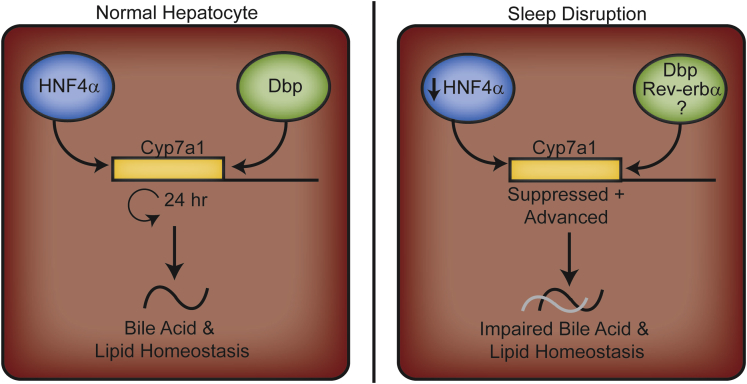
Research evidence continues to demonstrate multiple links between rhythms and metabolism, both under physiologic and disturbed circadian conditions. We have presented evidence that even very short-term circadian system disruption is capable of inducing measurable and significant physiologic changes with respect to lipids and bile acids, which were further modulated by nutrient conditions. We demonstrate that even a short-term disruption of rhythms markedly reduced Hnf4α expression and binding, which in part may contribute to suppression and phase shifting of Cyp7a1 expression, and result in reduced hepatic bile acid synthesis and altered lipid homeostasis in mice (Figure 11). However, the precise mechanisms by which the clock controls cellular metabolism and how altered nutrient condition affects the clock within different tissue types have yet to be fully understood.
Figure 11.
Schematic diagram representing the effects of short-term sleep disruption in the liver. Under normal conditions, the transcription factors HNF4α and Dbp regulate the circadian rhythm expression of the Cyp7a1 gene, which regulates bile acid synthesis and homeostasis (left panel). Under conditions of short-term sleep disruption demonstrated here, HNF4α gene expression and abundance at the Cyp7a1 promoter was suppressed, which may partially contribute to the suppression of Cyp7a1 gene transcription. This ultimately leads to altered bile acid pool size and homeostasis (right panel). Additional circadian-regulated components, including Dbp, Rev-erbα, and others, may also contribute to phase shifting by unknown mechanisms.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded by the National Institutes of Health grants DK44442 and DK58379 (to J.Y.L.C.) and DK096784 (to J.M.F.).
References
- 1.Moore R.Y. Organization and function of a central nervous system circadian oscillator: the suprachiasmatic hypothalamic nucleus. Fed Proc. 1983;42:2783–2789. [PubMed] [Google Scholar]
- 2.Reppert S.M., Weaver D.R. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 3.Panda S., Antoch M.P., Miller B.H. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 4.Reddy A.B., Karp N.A., Maywood E.S. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 5.Gälman C., Angelin B., Rudling M. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology. 2005;129:1445–1453. doi: 10.1053/j.gastro.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Noshiro M., Nishimoto M., Okuda K. Rat liver cholesterol 7 alpha-hydroxylase. Pretranslational regulation for circadian rhythm. J Biol Chem. 1990;265:10036–10041. [PubMed] [Google Scholar]
- 7.Chiang J.Y.L. Regulation of bile acid synthesis. Front Biosci. 1998;3:176–193. doi: 10.2741/a273. [DOI] [PubMed] [Google Scholar]
- 8.Scheer F.A.J.L., Hilton M.F., Mantzoros C.S., Shea S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nojkov B., Rubenstein J.H., Chey W.D., Hoogerwerf W.A. The impact of rotating shift work on the prevalence of irritable bowel syndrome in nurses. Am J Gastroenterol. 2010;105:842–847. doi: 10.1038/ajg.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis S., Mirick D.K., Stevens R.G. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–1562. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 11.Pietroiusti A., Neri A., Somma G. Incidence of metabolic syndrome among night-shift healthcare workers. Occup Environ Med. 2010;67:54–57. doi: 10.1136/oem.2009.046797. [DOI] [PubMed] [Google Scholar]
- 12.Esquirol Y., Bongard V., Mabile L. Shift work and metabolic syndrome: respective impacts of job strain, physical activity, and dietary rhythms. Chronobiol Int. 2009;26:544–559. doi: 10.1080/07420520902821176. [DOI] [PubMed] [Google Scholar]
- 13.Husse J., Hintze S.C., Eichele G. Circadian clock genes Per1 and Per2 regulate the response of metabolism-associated transcripts to sleep disruption. PLoS One. 2012;7:e52983. doi: 10.1371/journal.pone.0052983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shostak A., Meyer-Kovac J., Oster H. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes. 2013;62:2195–2203. doi: 10.2337/db12-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F., Zhang L., Zhang Y. Meta-analysis on night shift work and risk of metabolic syndrome. Obes Rev. 2014;15:709–720. doi: 10.1111/obr.12194. [DOI] [PubMed] [Google Scholar]
- 16.McFadden E., Jones M.E., Schoemaker M.J. The relationship between obesity and exposure to light at night: cross-sectional analyses of over 100,000 women in the Breakthrough Generations Study. Am J Epidemiol. 2014;180:245–250. doi: 10.1093/aje/kwu117. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi S., Mitsui S., Yan L. Role of DBP in the circadian oscillatory mechanism. Mol Cell Biol. 2000;20:4773–4781. doi: 10.1128/mcb.20.13.4773-4781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavery D.J., Schibler U. Circadian transcription of the cholesterol 7a-hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- 19.Duez H., van der Veen J.N., Duhem C. Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology. 2008;135:689–698.e685. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 20.Pathak P., Li T., Chiang J.Y.L. Retinoic acid-related orphan receptor α regulates diurnal rhythm and fasting induction of sterol 12α-hydroxylase in bile acid synthesis. J Biol Chem. 2013;288:37154–37165. doi: 10.1074/jbc.M113.485987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solt L.A., Wang Y., Banerjee S. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma K., Xiao R., Tseng H.-T. Circadian dysregulation disrupts bile acid homeostasis. PLoS One. 2009;4:e6843. doi: 10.1371/journal.pone.0006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohsaka A., Laposky A.D., Ramsey K.M. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Marcheva B., Ramsey K.M., Buhr E.D. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudic R.D., McNamara P., Curtis A.-M. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:1893–1899. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroup D., Chiang J.Y.L. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7α-hydroxylase gene (CYP7A1) J Lipid Res. 2000;41:1–11. [PubMed] [Google Scholar]
- 27.Lavery D.J., Schibler U. Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- 28.Hwang-Verslues W.W., Sladek F.M. HNF4α—role in drug metabolism and potential drug target? Curr Opin Pharmacol. 2010;10:698–705. doi: 10.1016/j.coph.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue Y., Yu A.-M., Yim S.H. Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4α. J Lipid Res. 2006;47:215–227. doi: 10.1194/jlr.M500430-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neimark E., Chen F., Li X., Shneider B.L. Bile acid–induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 2004;40:149–156. doi: 10.1002/hep.20295. [DOI] [PubMed] [Google Scholar]
- 31.Pan X., Zhang Y., Wang L., Hussain M.M. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12:174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussain M.M., Pan X. Clock regulation of dietary lipid absorption. Curr Opin Clin Nutr Metab Care. 2012;15:336–341. doi: 10.1097/MCO.0b013e3283548629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S.M., Zhang Y., Tsuchiya H. Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology. 2015;61:497–505. doi: 10.1002/hep.27437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T., Francl J.M., Boehme S. Glucose and insulin induction of bile acid synthesis: mechanisms and implications in diabetes and obesity. J Biol Chem. 2011;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barnea M., Madar Z., Froy O. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology. 2009;150:161–168. doi: 10.1210/en.2008-0944. [DOI] [PubMed] [Google Scholar]
- 36.Yanagihara H., Ando H., Hayashi Y. High fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol Int. 2006;23:905–914. doi: 10.1080/07420520600827103. [DOI] [PubMed] [Google Scholar]
- 37.Pendergast J.S., Branecky K.L., Yang W. High-fat diet acutely affects circadian organisation and eating behavior. Eur J Neurosci. 2013;37:1350–1356. doi: 10.1111/ejn.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumoto E., Ishihara A., Tamai S. Time of day and nutrients in feeding govern daily expression rhythms of the gene for sterol regulatory element-binding protein (SREBP)-1 in the mouse liver. J Biol Chem. 2010;285:33028–33036. doi: 10.1074/jbc.M109.089391. [DOI] [PMC free article] [PubMed] [Google Scholar]