Abstract
Activation of a telomere maintenance mechanism (TMM) is permissive for replicative immortality and a hallmark of human cancer. While most cancers rely on reactivation of telomerase, a significant fraction utilizes the recombination dependent alternative lengthening of telomeres (ALT) pathway. ALT is enriched in tumors of mesenchymal origin, including those arising from bone, soft tissue, and the nervous system, and usually portends a poor prognosis. Recent insights into the mechanisms of ALT are uncovering novel avenues to exploit vulnerabilities and may facilitate clinical development of ALT detection assays and personalized treatment decisions based on TMM status. Treatments targeting ALT may hold promise for a broadly applicable therapeutic modality specific to mesenchymal lineage tumors, something that has thus far remained elusive.
Keywords: telomere, alternative lengthening of telomeres, recombination, cancer
Telomere maintenance in cancer
Telomeres (see Glossary) are nucleoprotein structures that protect the ends of linear chromosomes. Composed of DNA repeats (5’-TTAGGG-3’) and specialized proteins that cooperatively prevent recognition as DNA double-strand breaks (DSBs), telomere structure is essential for genome stability [1,2]. Telomeres shorten with each cell cycle in somatic cells, which generally lack a telomere maintenance mechanism (TMM), eventually leading to senescence or crisis depending on the degree of telomere dysfunction. Therefore, in order to bypass such roadblocks to proliferation, continuously dividing cells need to figure out a means of maintaining their telomeres. Indeed, it is now well accepted that cancers must achieve a state of replicative immortality in order to form robust tumors and that activation of a TMM is central to this process [3]. Most human cancers reactivate the reverse transcriptase telomerase [4]. However, a significant proportion (~5-15%) utilize a homologous recombination (HR) based TMM known as alternative lengthening of telomeres (ALT) (Figure 1, Key Figure) [5,6].
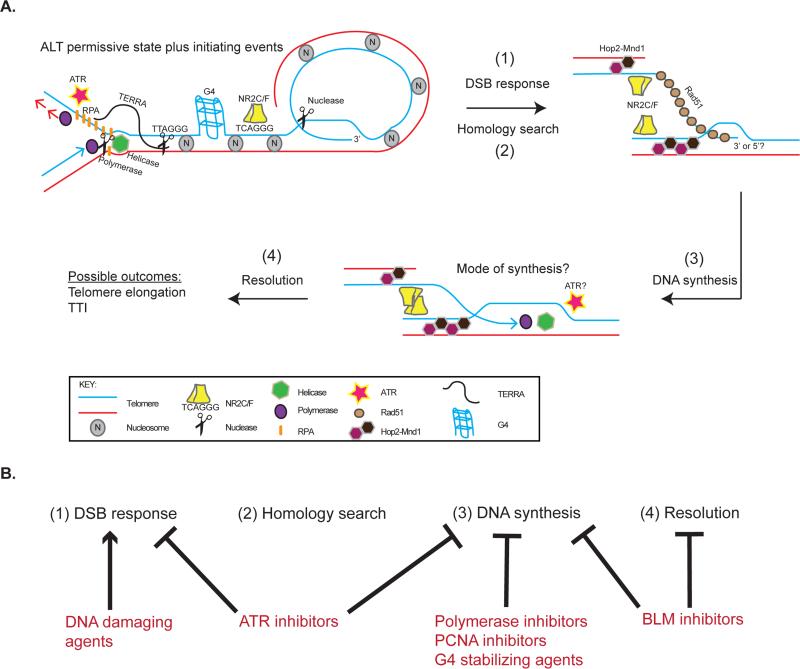
Figure 1, Key Figure.
Updated model of alternative lengthening of telomeres (ALT) (A). A permissive environment for telomere recombination likely occurs through telomeric chromatin changes, such as those mediated by mutations in ATRX/DAXX/H3.3, altered histone dynamics, changes in DNA and histone modifications, and decreased nucleosome density. In this permissive environment, DNA double strand breaks (DSBs) initiate ALT recombination. The source of these break signals may come from replication stress due to chromatin changes, TERRA mediated R-loops, or G-quadruplex (G4) structures. Endonuclease activity or telomere uncapping could also lead to DSB signals. Excessive RPA and ATR signaling may create a permissive environment for recombination and/or signal the presence of DSBs. DSB signals lead to a Rad51 and Hop2-Mnd1 mediated homology search and capture of a homologous sequence (telomere or internal genomic site) by the 3’ or 5’ end of the recipient telomere. The NR2C/F family of nuclear receptors can facilitate the proximity of homologous sequences. DNA synthesis and resolution of synapsed telomeres occur by unknown mechanisms. Ultimately, this process leads to elongation of telomeres or targeted telomere insertion (TTI) at genomic sites. (B) The model of ALT provides a framework for therapeutically targeting essential steps including: (1) DSB response, (2) Homology search, (3) DNA synthesis, and (4) Resolution. DNA damaging agents could cause hyperactivity of the DSB response and lead to toxic recombination. ATR inhibitors can function to inhibit DSB responses or DNA synthesis events. Other potential ways to attack synthesis include DNA polymerase, PCNA, and BLM inhibitors. G4 stabilizing agents could be used to cause replication stress.
ALT was originally discovered in immortalized cell lines and subsequently shown to occur in human tumors [7,8]. The first definitive evidence for a homology directed process between telomeres on different chromosomes came from a study showing that a telomere tag could be copied to multiple telomeres only in telomerase negative cells, indicating that ALT occurs via HR [9]. Indeed, a requirement for HR was predicted from studies in yeast definitively showing Rad52 recombination dependent telomere maintenance and survival of telomerase null yeast [10–13]. In the last two decades, tremendous progress has been made in characterizing the ALT phenotype. It is now widely accepted that ALT cells have highly heterogeneous, fluctuating telomere lengths [7,14], high levels of telomere sister chromatid exchanges (t-SCEs) [15], abundant extrachromosomal telomeric repeat DNA (ECTR) [16,17], and a specialized telomeric DNA nuclear structure termed ALT-associated promyelocytic leukemia (PML) bodies (APBs) [18]. Such characteristics have afforded opportunities to investigate the prevalence and prognostic consequences of ALT in cancer patients and explore potential targeted therapies for ALT-positive tumors, thereby attacking a crucial cornerstone of cancer cell autonomous proliferation.
Spectrum and characteristics of ALT-positive cancers
The recent application of tests for ALT phenotypic markers, including telomere length heterogeneity, APBs, and ECTR, to large sets of human tumor samples has begun to reveal the spectrum of cancers that utilize ALT [8,19–49]. The most comprehensive study to date analyzed APBs in over 6000 tumor specimens from a wide range of cancer subtypes and found ALT activity in just under 4% of the samples [20]. When combined with other more focused studies, the prevalence of ALT activity increases to 11% of all tumors (Table 1). However, there is no universal definition for ALT activity and most studies of patient samples investigate only one or two of the phenotypic markers. Additionally, tumors can have intratumoral heterogeneity in telomere lengths and TMM activity, with ALT and telomerase functioning in different cells within the same tumor [23,27]. Therefore, it is possible that the prevalence of ALT may be over- or under- represented by the markers currently used.
Table 1.
Summary of alternative lengthening of telomeres (ALT) prevalence in human cancer by tissue type.
| Tissue of origin | % ALT+ | References |
|---|---|---|
| Adrenal/PNS | 23 | [8,20-23] |
| Neuroblastoma | 29 | |
| Bone | 62 | [8,19,24,25,27] |
| Osteosarcoma | 62 | |
| Breast | 3 | [8,20,28] |
| CNS | 15 | [19,20,29-32] |
| GBM | 17 | |
| Colorectal | 6 | [20,33] |
| Hematopoietic | 0 | [20] |
| Kidney | 3 | [20] |
| Liver | 7 | [20] |
| Lung | 1 | [8,20,34] |
| Neuroendocrine | 40 | [20,35,36] |
| PanNET | 53 | |
| Ovary | 1 | [8,20] |
| Pancreas | 0 | [20] |
| Prostate | 0 | [20] |
| Skin | 4 | [8,20] |
| Soft tissue | 32 | [19,20,25,27,39–46] |
| Leiomyosarcoma | 56 | |
| MFH | 58 | |
| Liposarcoma | 26 | |
| Stomach | 8 | [20,47] |
| Testis | 7 | [20] |
| Thyroid | 8 | [19,20,48] |
| Urinary bladder | 2 | [20] |
| Uterus | 8 | [20,49] |
| Total | 11 | |
PNS=peripheral nervous system, CNS=central nervous system, GBM=glioblastoma multiforme, PanNET=pancreatic neuroendocrine tumor, MFH=malignant fibrous histiocytoma.
It is clear that certain tumor types commonly use ALT to maintain their telomeres (Table 1). Specifically, ALT activity is most prevalent in cancers arising from mesenchymal tissues [50], including bone (62%), soft tissues (32%), neuroendocrine systems (40%), peripheral nervous system (PNS; 23%), and central nervous system (CNS; 15%). ALT has also been documented in a small percentage of epithelial cancers (Table 1) [50]. The biological underpinnings and significance of this mesenchymal enrichment remain unknown. It is speculated that tighter regulation of telomerase expression in mesenchymal tissues may force these cells to choose ALT when telomere lengthening is needed [51]. It is also possible that altered control of senescence and/or crisis in mesenchymal cells could favor the emergence of ALT through excessive telomere damage and replication stress. Additional possibilities include negative consequences of ALT activity in epithelial cells, differential regulation of recombination pathways in mesenchymal cells, and specific mutations that drive both mesenchymal cancer development and ALT activity. The first possibility is less likely given evidence that ALT can occur in epithelial tumors. Although most recombination factors appear to be ubiquitous, their direct regulation or the broader recombination potential of telomeric chromatin may be divergent between tissue types. Indeed, ALT can be suppressed in hybrids of ALT cells with normal cells and telomerase-positive cells, suggesting the existence of an ALT repressor [52]. Whether loss of such a repressor is more prevalent in mesenchymal cells is unknown. Interestingly, biallelic loss of function mutations of chromatin remodeler ATRX and the histone chaperone DAXX are strongly associated with ALT and thus could represent candidates for the putative ALT suppressor. However, in some contexts telomerase can be suppressed by hybrids with ALT cells, indicating a more complex genetic relationship [53].
Early studies revealed a preponderance of p53 inactivation in ALT cell lines and tumors [8,54]. It has been postulated that elimination of p53 is important for the preservation of high levels of telomeric DNA damage signals and chromosomal instability commonly seen in ALT-positive cells [55,56]. Although relevant, p53 alterations are prevalent across a wide range of human cancers and not specific to ALT [57]. By contrast, recently identified somatic alterations in ATRX and DAXX appear to be common in, and specific to ALT-positive tumors, as first shown in pancreatic neuroendocrine tumors [35,56]. In another study, inactivating mutations in ATRX, DAXX, and neomorphic, gain of function H3.3 missense mutations that globally disrupt histone methylation were identified in 44% of pediatric glioblastoma and shown to correlate with ALT activity [58]. ATRX, DAXX, and H3.3 mutations were often mutually exclusive, suggesting epistasis. Mutations in the canonical histone H3.1 were also identified in pediatric brain tumors, albeit at a lower frequency (12%) [59]. Interestingly, ATRX and DAXX have been reported to participate in depositing H3.3 onto telomeric chromatin, providing a biological rationale for these genetic observations. Although alterations in chromatin modifications appear to be a characteristic of ALT cells, the exact role of ATRX, DAXX, and H3.3 in the pathogenesis of ALT remains a topic of intense study, particularly since ATRX mutations are common in primary cells that achieve immortalization in culture via ALT [56]. Alterations in such chromatin factors may create an environment permissive for HR at telomeres. For example, ATRX depletion alters cell cycle regulation of the long-noncoding RNA telomeric repeat-containing RNA (TERRA) and RPA at ALT telomeres, but does not enhance all ALT characteristics [56,60–62]. Regardless, the enrichment of certain mutations in ALT-positive tumors may provide a useful diagnostic or prognostic tool.
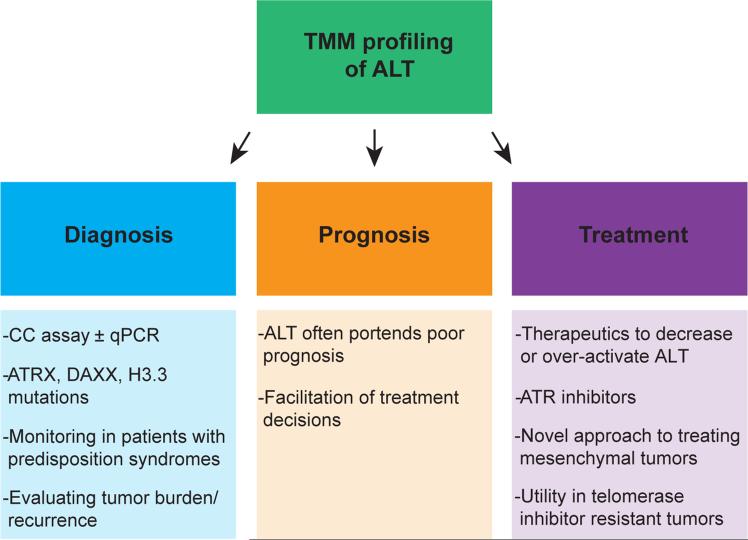
The clinical rationale for determining the ALT status of tumors lies partly in its ability to convey prognostic information (Figure 2). Indeed, utilization of ALT confers a poor prognosis in most settings, such as neuroblastoma and certain soft tissue sarcomas [21,23,42,43,45,63]. It can be speculated that this phenomenon is due to the chromosomal instability of these tumors, which may confer resistance to therapies [21]. Importantly, the poor clinical course of patients with ALT-positive tumors highlights the need to define targeted therapies against ALT. In contrast, ALT portends a better survival in glioblastoma [19,29–32]. Thus, the consequences of ALT activity in human cancers are complex. Future studies using novel animal models of ALT, such as the recently described glioma model [64], should begin to address such consequences.
Figure 2.
Telomere maintenance mechanism (TMM) profiling of alternative lengthening of telomeres (ALT) may have clinical utility for diagnosis, prognosis, and treatment of ALT-positive tumors. Diagnosis could be determined through the C-circle (CC) assay with or without quantitative PCR (qPCR) techniques to increase sensitivity and specificity. Additionally, mutational profiling of ATRX/DAXX/H3.3 may predict ALT activity in some tumor types. These tests may facilitate monitoring for tumor formation in patients with syndromes predisposing to ALT-positive tumors or for tumor dynamics or recurrence of established ALT-positive tumors. Since ALT has prognostic value in many settings, identification of ALT could enable more informed treatment decisions. Finally, TMM profiling may allow for personalized telomere-targeted therapies. In particular, treatments to decrease or increase ALT activity could exploit vulnerabilities present in ALT-positive tumors and lead to selective cell killing. ATR inhibitors represent a promising therapeutic strategy. By targeting ALT, we may in principle benefit patients with ALT-positive tumors, including those that have escaped telomerase inhibitors through ALT activation.
Accurate determination of ALT status remains a hurdle for further clinical development. The C-circle assay (CC assay) has become the forerunner as a clinically useful test for ALT (Figure 2) [65]. Developed to specifically amplify C-circles, a partially single-stranded species of ECTR, the CC assay was shown to be specific and responsive to changes in ALT activity, and detected C-circles in the blood of ALT-positive osteosarcoma patients. When combined with quantitative PCR (qPCR) methods and telomere length, the CC assay may allow for rapid determination of ALT status of tumors, with high sensitivity and specificity [66]. The rapidity and ease of these tests make them well suited for clinical application. However, the question of utility remains unanswered. At the moment, it is possible that a non-invasive test, such as the CC assay of blood, may facilitate monitoring of tumor development in patients with syndromes predisposing to ALT-positive tumors (e.g. multiple endocrine neoplasia syndromes [67]) or for monitoring tumor burden or recurrence of established ALT-positive tumors. However, the sensitivity to changes in tumor cell number in patients is not defined. It is the hope that, in addition to monitoring, physicians will be able to utilize the TMM status of a patient's tumor to direct treatment choices (Figure 2). For example, patients with ALT-positive tumors could receive targeted therapies against ALT, were they to become available.
New insights into the mechanism of ALT
Despite progress in characterizing ALT, we are only beginning to uncover the permissive events and mechanistic details that lead to productive telomere elongation in this TMM. Several recent lines of evidence have converged on the importance of the telomeric chromatin status in ALT [68]. In general, somatic mutations in the chromatin factors ATRX, DAXX, and H3.3 are enriched in ALT-positive tumors [35,58], disruption of the histone chaperone ASF1a/b paralogs can lead to induction of ALT activity [69], and decreased nucleosomal density and altered histone marks are coupled with increased expression of TERRA at ALT telomeres [61]. It is likely that a permissive environment for HR emerges from these chromatin alterations (Figure 1, Key Figure), as such associations are seen to promote HR [70,71]. However, it is also possible that replication stress due to altered histone dynamics and resultant fork stalling may facilitate recombination at ALT telomeres.
A recent study identified telomeric RNA-DNA hybrids as regulators of the recombinogenic potential of ALT telomeres [72]. Despite evidence that telomeric DNA could be transcribed into TERRA and that ALT cells displayed high levels of TERRA species, its role in ALT was unclear [56,61,72–74]. It is now surmised that TERRA has functional consequences for ALT cells through the creation of RNA-DNA hybrids (R-loops) that occur from base pairing of TERRA with complementary regions of telomere DNA (Figure 1, Key Figure) [72]. The RNA-DNA hybrid endonuclease RNaseH1 degrades TERRA-DNA R-loops at telomeres. Depletion of RNaseH1 leads to an increase in recombinogenic potential with rapid telomere excision, whereas its overexpression weakens recombination and leads to gradual telomere shortening. However, whether telomeric R-loops promote recombination due to replication stress or another mechanism, such as priming recombination associated DNA synthesis within APBs, remains to be determined. Additionally, it is unclear whether RNaseH1 is differentially regulated in ALT cells. These results are enticing given the mounting evidence that R-loops can have important regulatory roles as well as be a source of genomic instability [75,76]. In light of these findings, it becomes evident that ALT telomeres must require a balance of pro- and anti-recombinogenic signals to maintain a beneficial cellular state. Tipping the balance in either direction (i.e. through R-loop regulation) may allow for a disruption of ALT activity and may represent a therapeutic vulnerability.
Two studies have shown the role of TERRA in regulating competition between single-stranded telomeric DNA binding proteins [60,77]. Specifically, TERRA inhibits the ribonucleoprotein hnRNPA1 from displacing single stranded DNA binding protein, RPA on telomeres during S-phase [77]. Decreasing TERRA levels in G2 can then facilitate replacement of RPA with POT1, a member of the Shelterin complex that specifically recognizes single stranded telomere DNA. Interestingly, in ALT cells, dysregulated expression leads to persistent TERRA through G2 and RPA retention at telomeres (Figure 1, Key Figure) [60]. Since RPA is important for the initiation of HR, retained RPA is hypothesized to promote recombination and ALT activity. Furthermore, RPA has been identified in a subset of APBs in ALT cells [78], suggesting it is indeed a part of the ALT repertoire. One function of RPA is to activate the ATR kinase [79]. Strikingly, ATR inhibitors were shown to selectively kill ALT-positive cell lines as compared to telomerase-positive lines [60]. Although it may be inferred that increased RPA at ALT telomeres would create an environment more reliant on ATR signaling and consequently allow for selective killing of ALT cells, this link has not been formally proven. It may be that ALT cells have higher levels of replication stress at telomeres, consistent with the presence of APBs, thus creating an increased reliance on ATR.
In addition to being recombinogenic, ALT telomeres are distinguished by the presence of variant telomeric repeats (e.g. TCAGGG, TGAGGG) [80,81]. The TCAGGG repeat was shown to recruit the nuclear receptors COUP-TF2 (NR2F2) and TF4 [81,82], which in turn recruit the zinc-finger nucleosome remodeling complex NuRD-ZNF827 to alter telomeric chromatin [83]. Depletion of nuclear receptors and ZNF827 suppressed ALT phenotypic characteristics, suggesting an important functional role of these factors in ALT [81,83]. In a recent study, the NR2C/F class of nuclear receptors was shown to promote clustering of ALT telomeric foci, thus facilitating telomere recombination (Figure 1, Key Figure) [84]. An unexpected finding was the localization of the telomeric binding factor TRF2 at hundreds of non-telomeric NR2C/F binding sites, with a subset of these sites containing targeted telomere insertions (TTI) in ALT cells. Since telomeric sequences pose a challenge to the replication machinery [85,86], these TTI could act as common fragile sites, leading to chromosomal rearrangements and genomic instability in ALT-positive cancers. In support of this hypothesis, NR2C/F localized to telomeres in ALT-positive tumor specimens and correlated with the extent of karyotype rearrangements [84]. Future research may define the relative contribution of TTI to the genomic instability phenotype in ALT tumors and whether this is a universal phenomenon in patient samples. Although it is clear that the variant repeats in ALT telomeres have functional consequences, the nature of the proximal event(s) creating such variants also remains an outstanding question.
While we are rapidly gaining an appreciation for the cellular context in which ALT is favored and the pathways that regulate recombinogenic potential, relatively few studies have been conducted that detail the precise initiating stimuli and molecular mechanisms of the recombination. ALT telomeres are known to have high levels of DNA damage signals and DNA damaging agents can increase certain ALT characteristics [55,87]. A recent study using a telomere-specific endonuclease (TRF1-FokI) provided definitive proof that DSB responses at telomeres initiate recombination in ALT [88]. Specifically, telomere damage led to a homology search characterized by long-range, directed telomere movement across microns of nucleoplasm to capture another telomere. The damage initiated movement and clustering were dependent on the essential recombination protein Rad51, intimately connecting ALT mechanisms to telomere dynamics. Surprisingly, the meiotic Hop2-Mnd1 heterodimer, which stimulates Rad51 and Dmc1 mediated recombination during meiosis [89,90], was also essential for movement and clustering of ALT telomeres (Figure 1, Key Figure) [88]. Hop2 was broadly expressed in both ALT and telomerase positive cancer cell lines, but uniquely localized to ALT telomeres [88]. Thus, it appears that ALT utilizes a special form of HR for telomere elongation that relies on meiotic factors. Indeed, ALT resembles meiosis in its preference for non-sister HR (i.e. recombination between homologous sequences on different chromosomes). The identification of other meiotic proteins in cancer associated genomic instability (e.g. HORMAD1 [91]) suggests that activation of meiotic pathways may be a common mechanism during tumor pathogenesis. The contribution of other meiotic proteins to the ALT phenotype should be pursued in future studies.
While these exciting new studies provide avenues of further investigation, several major aspects of the mechanism of ALT recombination remain poorly defined. Namely, although telomeric DSB signals initiate recombination in ALT, the source of the endogenous DSB remains to be seen. What is the relative contribution of stalled replication forks, structure-specific endonucleases, or uncapped telomeres to the generation of excessive single-stranded DNA that is thought to initiate ALT telomere recombination? Additionally, what are the substrates used for recombination during ALT? It is assumed that telomeric 3’-overhangs prime Rad51 nucleoprotein filaments for strand invasion, as is the case for classical HR. However, ALT cells contain abundant 5’-C-rich telomeric overhangs [92,93]. The role of these overhangs is still open for debate, but it is possible that they are functional given evidence that 5’-overhangs can be used for strand invasion and priming synthesis [94]. Another area of limited understanding involves mechanisms of DNA synthesis after homology capture during ALT. Based on studies in yeast, ALT telomere synthesis is hypothesized to occur via a homology directed pathway known as break-induced replication (BIR) that is used to repair one-ended DSBs and extend for over 100kb [95,96]. BIR is distinguished by the downstream formation of a replication fork-like structure, extensive DNA synthesis, and high mutagenic potential [97], possibly contributing to the variability of ALT telomere sequences. Recent evidence suggests BIR in yeast proceeds through a non-canonical fork structure and leads to conservative strand inheritance, making it fundamentally different from S-phase synthesis [98,99]. Although many of the same replication factors are important for both BIR and S-phase synthesis [100], the nonessential yeast DNA polymerase δ subunit Pol32 was shown to be important for BIR as well as telomerase-negative survivors [101]. Whether ALT synthesis resembles BIR in yeast is yet to be determined.
Therapeutic outlook for ALT
The requirement for cancers to utilize a TMM begs the question of whether such pathways can be targeted for clinical purposes (Figure 1, Key Figure, 2). Telomerase has been the main focus of strategies to disrupt TMMs in cancer [102,103], and clinical trials of the telomerase inhibitor GRN163L and TERT-based vaccines are ongoing in several types of cancer. However, utilization of ALT occurs in a significant proportion of cancers (~10%), often of mesenchymal lineage, where telomerase inhibitors would be unlikely to have clinical efficacy. Thus, strategies to specifically target ALT could in principle benefit patients with these often poor prognosis tumors. Importantly, the utility of ALT inhibitors could extend beyond ALT-positive tumors given the possibility that targeting telomerase may lead to ALT utilization as a form of resistance (Figure 2). Indeed, this idea is substantiated by a mouse model where telomerase is deleted in established tumors [104] and an in vitro model of cultured human esophageal cell lines [105]. The converse situation, where ALT inhibition leads to reactivation of telomerase, is also plausible. Therefore, combined TMM inhibition using selective agents against telomerase and ALT may be needed to achieve full clinical efficacy.
The phenotypic characteristics of ALT can provide insights into possible ways to disrupt this TMM. For instance, APBs are found in ALT cells and are hypothesized to form structural platforms that mediate telomere recombination [18,78]. Disruption of APBs by overexpression of Sp100, a component of PML bodies, leads to sequestration of the Mre11-Rad50-Nbs1 (MRN) complex and inhibition of ALT activity [65,78,106]. Additionally, depletion of MRN components also inhibits ALT activity [107]. In light of these observations it is worth considering the therapeutic utility of disrupting structural components needed for ALT, such as that seen by disrupting PML by arsenic trioxide in acute promyelocytic leukemia [108]. Furthermore inhibition of integral components that localize to APBs, such as the MRN complex may be a suitable strategy [109].
Central to ALT is the atypical activation of telomere recombination. ALT cells likely depend on a tight balance between pro- and anti-recombination signals. Tipping the balance in either direction could inhibit beneficial ALT activity, causing cell death. In theory, preventing recombination would lead to a gradual loss of telomere sequence with each cell cycle, which could take significant time to fully inhibit proliferation. However, ALT cells have very short telomeres in addition to extremely long ones. Thus, complete ALT inhibition may accentuate damage signals by persistent uncapping of these critically short telomeres in a manner sufficient to induce toxic chromosome fusions, senescence, or apoptosis [110]. Alternatively, tipping the balance towards pro-recombination could lead to further genomic instability and cell lethality, potentially by preventing resolution of toxic recombination intermediates in APBs. Indeed, BLM helicase, which resolves such intermediates, is required for ALT [69,111].
Recent studies have honed in on several emerging features of cells that utilize the ALT mechanism, including: (1) altered telomere chromatin environment, (2) increased expression of telomere encoded TERRA, (3) increased association with nuclear receptors, (4) HR driven telomere movement and recombination between non-sister chromosomes, and (5) elevated activation of ATR. These mechanistic insights provide specific opportunities to rationally develop therapeutics targeting ALT. While each of these processes occurs during ALT, in most cases, it is unknown if they are actually essential for telomere maintenance and long-term survival in ALT cells. Inhibiting the specialized form of HR that predominates in ALT cells may prove fruitful, although it is presently unclear how this could be achieved. Currently, ATR inhibition is a strong candidate to move forward to preclinical and clinical studies in patients with ALT-positive tumors. Indeed ATR inhibitors were reported to confer increased toxicity in ALT dependent cells [60], are already in clinical development [112], and could be tested in cancers with a high prevalence of ALT.
A model of ALT as an aberrant, specialized form of HR provides a framework for conceptualizing treatment strategies. This four step model consists of: (1) Initiating DNA damage signals, (2) homology search and capture, (3) templated DNA synthesis, and (4) resolution (Figure 1, Key Figure). As discussed above, therapies that inhibit or over-activate these processes may selectively kill ALT cells. For instance, ALT synthesis, which may be significantly different from canonical replication, could potentially be manipulated in a manner that does not impede normal, rapidly proliferating cell types. Indeed, the ATR-dependent killing seen in ALT cells may function at the level of DNA synthesis. In this case, ALT can be viewed as a state of perturbed DNA synthesis. Thus, an armamentarium of DNA synthesis inhibitors, including newly developed PCNA and BLM inhibitors [113,114] and G-quadruplex stabilizing agents [115,116], may exploit vulnerabilities present in ALT cells.
Concluding Remarks
Tremendous progress towards understanding ALT has been made since its initial discovery two decades ago. Indeed, ALT has swiftly graduated from a poorly characterized phenomenon to a pivotal TMM enriched in mesenchymal tumors. We have reached a turning point where TMM status, such as ALT activity, may alter diagnosis, prognosis, or therapeutic decision-making in clinical oncology. Furthermore, the ubiquitous nature of telomere maintenance in cancer makes these pathways an attractive therapeutic target. ALT also provides a unique platform of study at the convergence of several fields, including DNA repair, telomere function, and cancer biology. By examining ALT we are sure to make strides towards solving fundamental issues in these fields, at the basic, translational, and clinical levels. As the details of ALT begin to mount, it is important to keep in mind the unanswered questions regarding the ALT mechanism, as these may reveal further aspects of telomere recombination to be therapeutic targets (Outstanding Questions Box). While many avenues of investigation should be pursued to understand ALT in the context of cancer, a more detailed understanding of the specialized form of HR used in ALT is essential for continued progress. Specifically, we must strive to uncover the substrates used for homology search and priming synthesis of telomeric DNA, the mode of DNA synthesis (including the DNA polymerases and helicases), and the steps leading to the resolution of recombination intermediates.
Trends Box.
Alternative lengthening of telomeres (ALT) is a recombination-based telomere maintenance mechanism (TMM) utilized by 10-15% of human cancers. ALT is enriched in tumors of mesenchymal origin and often portends a poor prognosis.
Clinical tests to identify ALT, such as the C-circle assay, may be used for diagnostic, prognostic, or therapeutic purposes. New mechanistic insights are paving the way to clinical translation.
Alterations in the telomeric chromatin environment are increasingly being discovered in ALT. Recent studies have implicated the long-noncoding RNA telomeric repeat-containing RNA (TERRA), nuclear receptors, and RPA in the recombinogenic potential of ALT telomeres.
ALT recombination is initiated by DNA double strand break signals and proceeds by a specialized homology search that relies on Rad51 and the meiotic Hop2-Mnd1 heterodimer.
Outstanding Questions Box.
What is the relative contribution of stalled replication forks, structure-specific endonucleases, uncapped telomeres, and TERRA to the initiation of ALT telomere recombination?
What is the role of TTI in the genomic instability characteristic of ALT cells?
How do alterations in the ATRX/DAXX/H3.3 axis, as well as other chromatin alterations, influence ALT activity?
What are the molecular substrates used for ALT recombination?
What is the role of 5’ overhangs in ALT?
What are the mechanisms of DNA synthesis and resolution of recombination during ALT?
How do ATR inhibitors selectively kill ALT cells in culture?
Can the C-circle assay be used to direct clinical decisions based on the TMM status of a tumor?
Will ATR inhibitors prove useful against ALT-positive tumors in the clinic?
Acknowledgements
This work was supported by NIH grants GM101149, CA138835, and CA17494 to RAG, who is also supported by funds from the Abramson Family Cancer Research Institute and Basser Research Center for BRCA. RLD was supported in part by NIH grants T32GM007170 and T32GM008216.
Glossary
- Alternative lengthening of telomeres (ALT)
A recombination-based mechanism of telomere maintenance used by 10-15% of human cancers. ALT is characterized by heterogeneous, fluctuating telomere lengths, high levels of telomere sister chromatid exchanges (t-SCEs), abundant extrachromosomal telomeric repeat DNA (ECTR), and a specialized telomeric DNA nuclear structure termed ALT-associated promyelocytic leukemia (PML) bodies (APBs).
- ATRX/DAXX/H3.3
A chromatin remodeler while DAXX functions as a histone chaperone. Together, these proteins are involved in depositing the histone H3 variant H3.3 at pericentric heterochromatin and at telomeres. Mutations in the ATRX/DAXX/H3.3 axis have recently been identified in ALT-positive tumors.
- Break-induced replication (BIR)
A recombination-based mechanism used to repair one-ended DNA double strand breaks, such as telomeres. BIR is distinguished from canonical DNA replication by the presence of a migrating D-loop structure, conservative inheritance of nascent DNA, high mutagenicity, and reliance on the accessory polymerase δ subunit Pol32. Eukaryotic BIR has been most extensively studied in yeast and remains speculative in human cells.
- C-circle
Partially single-stranded species of extrachromosomal telomeric repeat DNA (ECTR), the abundance of which closely correlates with ALT activity. A C-circle assay is currently being tested for clinical applications.
- Homologous recombination (HR)
One of two major pathways used to repair DNA double strand breaks. HR makes use of a homologous template to copy DNA sequences from donor to recipient molecules for repair.
- Hop2-Mnd1
A heterodimer that stimulates Rad51 and Dmc1 mediated recombination during meiosis. A recent paper uncovered a new role for these proteins in telomere movement and clustering during ALT recombination.
- Mesenchymal tumor
Tumors derived from connective tissues, including bone and soft tissues. ALT is enriched in tumors of mesenchymal origin.
- NR2C/F
A class of orphan nuclear receptors that bind specifically to ALT telomeres and are important for the ALT phenotype. A recent study implicated these receptors in telomere clustering and targeted telomere insertion (TTI) at non-telomeric sites.
- RPA/ATR
Replication protein A (RPA) is a DNA single stranded binding protein. Ataxia telangiectasia and Rad3-related protein (ATR) is a serine/threonine kinase involved in DNA damage response and checkpoint signaling. ATR is activated by single stranded DNA coated with RPA. A recent study showed that RPA is persistently bound to ALT telomeres and that these cells are hypersensitive to ATR inhibitors.
- Telomerase
A reverse transcriptase capable of adding telomeric repeats directly to ends of chromosomes. Telomerase is used to lengthen telomeres in the majority of human cancers.
- Telomere
Nucleoprotein structures, composed of TTAGGG DNA repeats and the six-member Shelterin protein complex, that protect the ends of linear chromosomes.
- Telomeric repeat-containing RNA (TERRA)
A long-noncoding RNA transcribed from the sub-telomere that contains telomeric repeat sequences. TERRA expression is increased at ALT telomeres. A recent study revealed that TERRA can promote recombination at ALT telomeres through R-loop (RNA-DNA hybrid) formation.
- Targeted telomere insertion (TTI)
Refers to the insertion of telomeric sequences at non-telomeric sites throughout the genome. These TTIs could act as common fragile sites, leading to chromosomal rearrangements and genomic instability in ALT-positive cancers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–81. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 6.Draskovic I, Vallejo AL. Telomere recombination and alternative telomere lengthening mechanisms. Front. Biosci. 2013 doi: 10.2741/4084. [DOI] [PubMed] [Google Scholar]
- 7.Bryan TM, et al. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–8. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan TM, et al. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–4. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 9.Dunham MA, et al. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–50. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 10.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 1993;73:347–60. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 11.McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–34. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 12.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–93. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng SC, et al. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000;6:947–52. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 14.Murnane JP, et al. Telomere dynamics in an immortal human cell line. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Londoño-Vallejo JA, et al. Alternative Lengthening of Telomeres Is Characterized by High Rates of Telomeric Exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 16.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 2004;24:9948–57. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 2009;29:703–13. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeager TR, et al. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 19.Henson JD, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 2005;11:217–25. [PubMed] [Google Scholar]
- 20.Heaphy CM, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011;179:1608–15. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onitake Y, et al. Telomere biology in neuroblastoma: telomere binding proteins and alternative strengthening of telomeres. J. Pediatr. Surg. 2009;44:2258–66. doi: 10.1016/j.jpedsurg.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 22.Else T, et al. Evaluation of telomere length maintenance mechanisms in adrenocortical carcinoma. J. Clin. Endocrinol. Metab. 2008;93:1442–9. doi: 10.1210/jc.2007-1840. [DOI] [PubMed] [Google Scholar]
- 23.Pezzolo A, et al. Intratumoral diversity of telomere length in individual neuroblastoma tumors. Oncotarget. 2015;6:7493–7503. doi: 10.18632/oncotarget.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulaner GA, et al. Absence of a telomere maintenance mechanism as a favorable prognostic factor in patients with osteosarcoma. Cancer Res. 2003;63:1759–63. [PubMed] [Google Scholar]
- 25.Ulaner GA, et al. Divergent patterns of telomere maintenance mechanisms among human sarcomas: sharply contrasting prevalence of the alternative lengthening of telomeres mechanism in Ewing's sarcomas and osteosarcomas. Genes. Chromosomes Cancer. 2004;41:155–62. doi: 10.1002/gcc.20074. [DOI] [PubMed] [Google Scholar]
- 26.Sanders RP, et al. Telomerase expression predicts unfavorable outcome in osteosarcoma. J. Clin. Oncol. 2004;22:3790–7. doi: 10.1200/JCO.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 27.Gocha ARS, et al. Human sarcomas are mosaic for telomerase-dependent and telomerase-independent telomere maintenance mechanisms: implications for telomere-based therapies. Am. J. Pathol. 2013;182:41–8. doi: 10.1016/j.ajpath.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subhawong AP, et al. The alternative lengthening of telomeres phenotype in breast carcinoma is associated with HER-2 overexpression. Mod. Pathol. 2009;22:1423–31. doi: 10.1038/modpathol.2009.125. [DOI] [PubMed] [Google Scholar]
- 29.Hakin-Smith V, et al. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 30.McDonald KL, et al. Presence of alternative lengthening of telomeres mechanism in patients with glioblastoma identifies a less aggressive tumor type with longer survival. J. Neuropathol. Exp. Neurol. 2010;69:729–36. doi: 10.1097/NEN.0b013e3181e576cf. [DOI] [PubMed] [Google Scholar]
- 31.Dorris K, et al. Prognostic significance of telomere maintenance mechanisms in pediatric high-grade gliomas. J. Neurooncol. 2014;117:67–76. doi: 10.1007/s11060-014-1374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangerel J, et al. Alternative lengthening of telomeres is enriched in, and impacts survival of TP53 mutant pediatric malignant brain tumors. Acta Neuropathol. 2014;128:853–62. doi: 10.1007/s00401-014-1348-1. [DOI] [PubMed] [Google Scholar]
- 33.Boardman LA, et al. Correlation of chromosomal instability, telomere length and telomere maintenance in microsatellite stable rectal cancer: a molecular subclass of rectal cancer. PLoS One. 2013;8:e80015. doi: 10.1371/journal.pone.0080015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirotani Y, et al. Alteration in length of telomeric repeats in lung cancer. Lung Cancer. 1994;11:29–41. doi: 10.1016/0169-5002(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 35.Heaphy C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science (80-. ) 2011;333:2011. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marinoni I, et al. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology. 2014;146:453–60. e5. doi: 10.1053/j.gastro.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Villa R, et al. Multiple mechanisms of telomere maintenance exist and differentially affect clinical outcome in diffuse malignant peritoneal mesothelioma. Clin. Cancer Res. 2008;14:4134–40. doi: 10.1158/1078-0432.CCR-08-0099. [DOI] [PubMed] [Google Scholar]
- 38.Au AYM, et al. Telomerase activity in pleural malignant mesotheliomas. Lung Cancer. 2011;73:283–8. doi: 10.1016/j.lungcan.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montgomery E, et al. Telomere lengths of translocation-associated and nontranslocation-associated sarcomas differ dramatically. Am. J. Pathol. 2004;164:1523–9. doi: 10.1016/S0002-9440(10)63710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venturini L, et al. Telomere maintenance mechanisms in malignant peripheral nerve sheath tumors: expression and prognostic relevance. Neuro. Oncol. 2012;14:736–44. doi: 10.1093/neuonc/nos083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan P, et al. Telomerase activity and hTERT mRNA expression can be heterogeneous and does not correlate with telomere length in soft tissue sarcomas. Int. J. Cancer. 2002;98:851–6. doi: 10.1002/ijc.10285. [DOI] [PubMed] [Google Scholar]
- 42.Matsuo T, et al. Telomere-maintenance mechanisms in soft-tissue malignant fibrous histiocytomas. J. Bone Joint Surg. Am. 2009;91:928–37. doi: 10.2106/JBJS.G.01390. [DOI] [PubMed] [Google Scholar]
- 43.Matsuo T, et al. Alternative lengthening of telomeres as a prognostic factor in malignant fibrous histiocytomas of bone. Anticancer Res. 2010;30:4959–62. [PubMed] [Google Scholar]
- 44.Liau J-Y, et al. Leiomyosarcoma with alternative lengthening of telomeres is associated with aggressive histologic features, loss of ATRX expression, and poor clinical outcome. Am. J. Surg. Pathol. 2015;39:236–44. doi: 10.1097/PAS.0000000000000324. [DOI] [PubMed] [Google Scholar]
- 45.Costa A, et al. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res. 2006;66:8918–24. doi: 10.1158/0008-5472.CAN-06-0273. [DOI] [PubMed] [Google Scholar]
- 46.Johnson JE, et al. Multiple mechanisms of telomere maintenance exist in liposarcomas. Clin. Cancer Res. 2005;11:5347–55. doi: 10.1158/1078-0432.CCR-05-0684. [DOI] [PubMed] [Google Scholar]
- 47.Omori Y, et al. Alternative lengthening of telomeres frequently occurs in mismatch repair system-deficient gastric carcinoma. Cancer Sci. 2009;100:413–8. doi: 10.1111/j.1349-7006.2008.01063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang N, et al. Telomerase-dependent and independent telomere maintenance and its clinical implications in medullary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2014;99:E1571–9. doi: 10.1210/jc.2014-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee Y-K, et al. Prognostic value of alternative lengthening of telomeres-associated biomarkers in uterine sarcoma and uterine carcinosarcoma. Int. J. Gynecol. Cancer. 2012;22:434–41. doi: 10.1097/IGC.0b013e31823ca017. [DOI] [PubMed] [Google Scholar]
- 50.Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–11. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Henson JD, et al. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 52.Perrem K, et al. Repression of an alternative mechanism for lengthening of telomeres in somatic cell hybrids. Oncogene. 1999;18:3383–90. doi: 10.1038/sj.onc.1202752. [DOI] [PubMed] [Google Scholar]
- 53.Katoh M, et al. A repressor function for telomerase activity in telomerase-negative immortal cells. Mol. Carcinog. 1998;21:17–25. doi: 10.1002/(sici)1098-2744(199801)21:1<17::aid-mc4>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y-J, et al. Association of mutant TP53 with alternative lengthening of telomeres and favorable prognosis in glioma. Cancer Res. 2006;66:6473–6. doi: 10.1158/0008-5472.CAN-06-0910. [DOI] [PubMed] [Google Scholar]
- 55.Cesare AJ, et al. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 2009;16:1244–51. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 56.Lovejoy C. a, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olivier M, et al. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 59.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–3. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flynn RL, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science (80−. ) 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Episkopou H, et al. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014;42:4391–405. doi: 10.1093/nar/gku114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eid R, et al. Genetic inactivation of ATRX leads to a decrease in the amount of telomeric cohesin and of telomere transcription in human glioma cells. Mol. Cell. Biol. 2015 doi: 10.1128/MCB.01317-14. DOI: 10.1128/MCB.01317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venturini L, et al. Prognostic relevance of ALT-associated markers in liposarcoma: a comparative analysis. BMC Cancer. 2010;10:254. doi: 10.1186/1471-2407-10-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeitany M, et al. A preclinical mouse model of glioma with an alternative mechanism of telomere maintenance (ALT). Int. J. Cancer. 2015;136:1546–58. doi: 10.1002/ijc.29171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henson JD, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009;27:1181–5. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 66.Lau LMS, et al. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. 2013;41:e34. doi: 10.1093/nar/gks781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Wilde RF, et al. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Mod. Pathol. 2012;25:1033–9. doi: 10.1038/modpathol.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Sullivan RJ, Almouzni G. Assembly of telomeric chromatin to create ALTernative endings. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.07.007. DOI: 10.1016/j.tcb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 69.O'Sullivan RJ, et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014;21:167–74. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aymard F, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:366–74. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang J, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013;20:317–25. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arora R, et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Azzalin CM, et al. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 74.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–36. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 75.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 76.Bhatia V, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–5. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 77.Flynn RL, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–6. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Draskovic I, et al. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15726–31. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oakley GG, Patrick SM. Landmark, editor. Replication protein A: directing traffic at the intersection of replication and repair. Front. Biosci. 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Varley H, et al. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat. Genet. 2002;30:301–5. doi: 10.1038/ng834. [DOI] [PubMed] [Google Scholar]
- 81.Conomos D, et al. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012;199:893–906. doi: 10.1083/jcb.201207189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Déjardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–86. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conomos D, et al. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 2014;21:760–770. doi: 10.1038/nsmb.2877. [DOI] [PubMed] [Google Scholar]
- 84.Marzec P, et al. Nuclear-Receptor-Mediated Telomere Insertion Leads to Genome Instability in ALT Cancers. Cell. 2015;160:913–927. doi: 10.1016/j.cell.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 85.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bosco N, de Lange T. A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma. 2012;121:465–74. doi: 10.1007/s00412-012-0377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fasching CL, et al. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 2007;67:7072–7. doi: 10.1158/0008-5472.CAN-07-1556. [DOI] [PubMed] [Google Scholar]
- 88.Cho NW, et al. Interchromosomal Homology Searches Drive Directional ALT Telomere Movement and Synapsis. Cell. 2014;159:108–121. doi: 10.1016/j.cell.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petukhova GV, et al. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell. 2003;5:927–36. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- 90.Petukhova GV, et al. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–53. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- 91.Watkins J, et al. Genomic complexity profiling reveals that HORMAD1 overexpression contributes to homologous recombination deficiency in triple-negative breast cancers. Cancer Discov. 2015;5:488–505. doi: 10.1158/2159-8290.CD-14-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oganesian L, Karlseder J. Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol. Cell. 2011;42:224–36. doi: 10.1016/j.molcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oganesian L, Karlseder J. 5′ C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair (Amst) 2013;12:238–45. doi: 10.1016/j.dnarep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mazloum N, Holloman WK. Brh2 promotes a template-switching reaction enabling recombinational bypass of lesions during DNA synthesis. Mol. Cell. 2009;36:620–30. doi: 10.1016/j.molcel.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anand RP, et al. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 2013;23:271–9. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saini N, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–92. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson MA, et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–6. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lydeard JR, et al. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–44. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lydeard JR, et al. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–3. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 102.Harley CB. Telomerase and cancer therapeutics. Nat. Rev. Cancer. 2008;8:167–79. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 103.Mocellin S, et al. Telomerase and the search for the end of cancer. Trends Mol. Med. 2013;19:125–33. doi: 10.1016/j.molmed.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 104.Hu J, et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148:651–63. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Queisser A, et al. Inhibition of telomerase induces alternative lengthening of telomeres during human esophageal carcinogenesis. Cancer Genet. 2013;206:374–86. doi: 10.1016/j.cancergen.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 106.Jiang W-Q, et al. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 2005;25:2708–21. doi: 10.1128/MCB.25.7.2708-2721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhong Z-H, et al. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 2007;282:29314–22. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 108.Zhang X-W, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–3. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 109.Dupré A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 2008;4:119–25. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaul Z, et al. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 2012;13:52–9. doi: 10.1038/embor.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stavropoulos DJ, et al. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002;11:3135–44. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 112.Fokas E, et al. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat. Rev. 2014;40:109–17. doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 113.Punchihewa C, et al. Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J. Biol. Chem. 2012;287:14289–300. doi: 10.1074/jbc.M112.353201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nguyen GH, et al. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem. Biol. 2013;20:55–62. doi: 10.1016/j.chembiol.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bochman ML, et al. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–80. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang F-C, et al. Induction of senescence in cancer cells by the G-quadruplex stabilizer, BMVC4, is independent of its telomerase inhibitory activity. Br. J. Pharmacol. 2012;167:393–406. doi: 10.1111/j.1476-5381.2012.01997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]