Summary
Somatic stem cells (SCs) maintain tissue homeostasis by dynamically adjusting proliferation and differentiation in response to stress and metabolic cues. Here, we identify Ca2+ signaling as a central regulator of intestinal SC (ISC) activity in Drosophila. We find that dietary L-glutamate stimulates ISC division and gut growth. The metabotropic glutamate receptor (mGluR) is required in ISCs for this response and for an associated modulation of cytosolic Ca2+ oscillations that results in sustained high cytosolic Ca2+ concentrations. High cytosolic Ca2+ induces ISC proliferation by regulating Calcineurin and CREB - regulated transcriptional co-activator (CRTC). In response to a wide range of dietary and stress stimuli, ISCs reversibly transition between Ca2+ oscillation states that represent poised or activated modes of proliferation, respectively. We propose that the dynamic regulation of intracellular Ca2+ levels allows effective integration of diverse mitogenic signals in ISCs to tailor their proliferative activity to the needs of the tissue.
Introduction
Somatic SCs can shift between quiescent and active states and between asymmetric and symmetric division modes according to the regenerative need of a tissue 1–4. How SCs decode and integrate various environmental inputs to adjust their proliferative behaviors remains an unresolved question with implications for our understanding of degenerative and neoplastic diseases. Intestinal SCs (ISCs) represent the majority of mitosis-competent cells in the Drosophila intestinal epithelium, and can undergo symmetric or asymmetric divisions in response to dietary or stress stimuli, respectively 1–3,5. Complex cell-autonomous and non-autonomous interactions between conserved signaling pathways (including JAK/STAT, EGFR, InR, and JNK signaling) govern these responses 1–3,5. Through asymmetric divisions, ISCs form lineage-restricted diploid progenitor cells called EnteroBlasts (EBs), which differentiate into large polyploid EnteroCytes (ECs) or small diploid EnteroEndocrine cells (EE) 1,3,4. Symmetric divisions can be induced by insulin signaling and allow adaptive resizing of the gut upon feeding 1. Growth and physiology of flies is influenced by the protein concentration in the food, yet the role of specific nutrients in adaptive resizing has remained unexamined 6,7. L-glutamate (L-Glu) is among the most abundant amino acids in proteins and is a critical energy source for the intestine 8. At the same time, it serves as a signaling molecule, stimulating specific membrane receptors in a wide range of cells 9. L-Glu promotes cell proliferation in the intestinal epithelium of mice, mediated by the metabotropic glutamate receptor mGluR5 10, yet it remains unclear whether and how this signal impacts the proliferative activity of ISCs.
Results
Glutamate regulates gut growth through mGluR
To assess if dietary L-Glu influences adaptive resizing of the intestinal epithelium, we used a modified version of the feeding protocol developed by O’Brien 1 (Fig. 1a, Extended Data Fig. 2a). As early as 4 hours after starting feeding, flies maintained on 0.1% yeast supplemented with 1% L-Glu exhibited a higher mitotic index along the intestinal epithelium (both anterior and posterior gut) than controls, and after 2 days the posterior midgut significantly increased in length, width, and cell density (Fig. 1b and Extended Data Fig. 2a,d; feeding rates were similar between the different food recipes; Extended Data Fig. 2b). L-Glu had to be ingested with the diet for these effects, as injection of 1% L-Glu into the animal did not increase ISC proliferation rates (Extended Data Fig. 2c). Food supplemented with other amino acids, or in which the caloric content was increased using sugar, did not stimulate ISC proliferation (Extended Data Fig. 2e). L-Glu feeding also increased the growth rate of ISC lineages marked by Flp-out or MARCM lineage tracing systems 11,12 (Extended Data Fig. 2f–h).
Figure 1. Glutamate regulates ISC proliferation and gut growth through mGluR.
a, Schematic of starvation / refeeding experiments performed.
b, Mitotic figures (phospho-histone H3 expressing nuclei), gut length, gut width, and number of Delta (Dl) expressing and non-expressing cells / field in the posterior midgut (PM).
Averages and s.e.m. are shown. P values are from Student’s t –test. For mitotic figures, n=14 flies for each genotype, representative of three independent experiments shown. For gut length, width, and cell numbers, n=9 for each control condition, n=12 flies for each mGluRNAi condition, representative of two independent experiments.
We tested whether this L-Glu response is mediated by ionotropic (NMDA and GluRIIA, B, C) or the one metabotropic L-Glu receptor (mGluR) encoded by the fly genome, and found that ISC-specific knock down of mGluR or homozygosity for the mGluR loss of function allele mGluR112b impaired the response (Fig. 1b and Extended Data Fig.2i). Desensitization / internalization of mGluR, or oxidative glutamate toxicity may explain the observation that higher levels (5%, 10%) of L-Glu impaired ISC proliferation rather than activated it (Extended Data Fig. 2a)13,14.
The concentration of freely available L-Glu in the intestinal epithelium is expected to reflect a balance between L-Glu absorption by ECs and input from the diet. In the nervous system, excess L-Glu is recycled by astrocytes through Excitatory Amino Acid Transporters (EAATs) 15. In mammals, EAAT3 (also known as EAAC-1) is expressed in the apical, brush border membrane of ECs throughout the small intestine 16. RNAseq, qRT-PCR, in situ hybridization and an eaat1::Gal4 reporter 17 suggested expression of eaat1 in ECs of the fly gut, with specifically elevated activity in the anterior midgut (Extended Data Fig. 3a,b,c, and not shown). Silencing eaat1 in ECs using the EC-specific driver NP1::GAL4 18 increased mitotic figures and the number of cells expressing the ISC marker Delta (Dl) throughout the gut without triggering the canonical JAK/STAT-mediated EC stress response or inducing apoptosis in ECs (Extended Data Fig. 3d,f,g). Over-expression of eaat1 significantly reduced the ISC response to L-Glu feeding (Extended Data Fig. 3e), suggesting that L-Glu resorption through EAAT1 influences free L-Glu levels in the intestine, modulating ISC proliferation and adaptive resizing. The limited glutamatergic innervation of the midgut epithelium suggests that direct regulation of ISCs by glutamatergic neurons is unlikely (Extended Data Figure 4a–d).
Ca2+ oscillations in ISCs
mGluR, a G-Protein Coupled Receptor (GPCR), influences cytosolic Ca2+ levels via Phospholipase C and inositol-trisphosphate (IP3) 19. Cytosolic [Ca2+] is low due to active transport that sequesters Ca2+ into the ER, the mitochondria, or the extracellular space. Stimulation by growth factors, hormones, or neurotransmitters triggers Ca2+ release from these stores, leading to acute spikes in cytosolic [Ca2+] as well as [Ca2+] oscillations that influence a wide range of cellular functions 20–22 23. We used UAS::GCaMP3, a genetically encoded Ca2+ reporter 24,25, to test whether intracellular [Ca2+] in ISCs responds to dietary L-Glu and to mGluR activity. Variable activity of this reporter in ISCs (Extended Data Fig. 4e–g) suggested fluctuations of [Ca2+] that we confirmed by live imaging using two-photon microscopy (Fig. 2a–c). Quantification of [Ca2+] in individual ISCs (as fluorescence emission ratio between GCaMP and co-expressed mCherry) revealed frequent and robust [Ca2+] oscillations in ISCs of wild-type intestines (Suppl. Discussion, Figures 2b, c, Extended Data Fig. 5a, e and Supplemental Video 1, 2). Experiments using Ca2+ channel inhibitors and Ca2+ chelators, as well as genetic knockdown of IP3 receptors, suggest that these [Ca2+] oscillations depend on both influx through plasma membrane Ca2+ channels as well as on Ca2+ release from intracellular stores through IP3 receptors (Suppl. Discussion, Fig. 3a, and Extended Data Fig. 5b–d). We confirmed reliability of GCaMP3 by recording Ca2+ oscillations using a bi-cistronic tdTomato-GCaMP5 construct 26,27 and a new, ultra-sensitive (higher affinity, dynamic range and brightness) Ca2+ reporter, IVS-GCaMP6s 28 (Extended Data Fig. 5e and Supplemental Video 2).
Figure 2. Dietary L-Glu influences ISC [Ca2+] oscillations and proliferation through the mGluR/Gαq/Plcβ pathway.
a, Experimental setup for live recordings of Ca2+ oscillations in ISCs.
b, Representative traces of GCaMP3/mCherry ratios in individual wild-type ISCs (pooled from 4 guts; 50 frames, 15 second intervals).
c, Representative frames from live recording (frame and time in minutes indicated). Arrowheads mark typical ISCs.
d, [Ca2+] oscillation frequency and average GCaMP3/mCherry ratio in ISCs of flies of the indicated genotypes re-fed for 4hrs.
e, L-Glu – induced proliferation analyzed after Gαq and Plcβ were knocked down in ISCs for 7 days.
Averages and s.e.m.; P values from Student’s t-test (N.S.: not significant). In d, individual ISCs pooled from 4–5 guts (n = 12, 14, 18, 30, 12, 13, 17, 15, 9, 10, 13, 11 cells from left to right). In e, n = 13, 16, 10, 11, 11, 12 guts from left to right. Representative of 6 experiments in b and c, of three experiments in d and e.
f, Regulation of Ca2+ homeostasis in non-excitable cells. GPCR/IP3R cascade causes release of Ca2+ (red dots) from the ER. A decline of [Ca2+] in the ER is sensed by the Stim-Orai complex and induces extracellular Ca2+ influx into the cytoplasm (Store-operated Ca2+ entry, SOCE). Excessive cytoplasmic Ca2+ is pumped into the ER by SERCA or out of the cell by PMCA.
Figure 3. CaN/CRTC regulates ISC proliferation in response to elevated cytosolic Ca2+.
a, Ca2+ oscillation patterns in ISCs perturbed as indicated.
b, Mitotic figures in guts of 2–3 day old flies maintained at 29°C for 4 days.
c, Cross section of posterior midguts of the indicated genotypes. Visceral muscle identified by Phalloidin (red), and ISCs/EBs by GFP (green). DAPI blue.
d, Mitotic figures in control or crtc null mutants (crtc25-3) combined with SERCA knockdown.
e, Mitotic figures in control or crtc25-3 homozygotes after re-feeding.
f, Mitotic figures in indicated genotypes after re-feeding.
Averages and s.e.m. in all cases. a, individual ISCs (pooled from 4–5 different guts). P values from ANOVA. n = 13, 12, 13, 10, 16, 12, 14, 8, 17, 11, 21, 12, 11, 10, 16, 12, 8, 6, 20, 12 cells from left to right. Representative of three experiments. b, one-way ANOVA (left and middle) and Student’s t-test (right). n = 22, 24, 15, 13, 16, 15, 22, 18, 12, 16 guts. Representative of two experiments. d, one-way ANOVA. n = 12, 22, 11, 19 guts. Representative of two experiments. e, Student’s t-test. n = 12, 8, 10, 8 guts. Representative of three experiments. f, ANOVA. n = 10, 12, 11, 11, 10, 12 guts. Representative of two experiments. N.S.: not significant; ***: P<0.001; ****: P<0.0001.
Re-feeding with 1% L-Glu supplemented food reduced [Ca2+] oscillation frequency and increased the average GCaMP3 signal intensity in wild-type ISCs (Fig. 2d, Extended Data Fig.6a, Supplemental Videos 3–4). Knockdown of mGluR resulted in reduced oscillation frequency while maintaining low signal intensity under mock conditions, and prevented the L-Glu - induced increase in signal intensity (Fig. 2d, Extended Data Fig.6a, c, Supplemental Videos 5–6). These data suggested that increased ISC proliferation is associated with elevated cytosolic [Ca2+]. Accordingly, both oscillations and signal intensity were reduced in feeding conditions in which ISC proliferation is inhibited (Extended Data Fig. 6b; compare Extended Data Fig. 2a).
The Drosophila genome encodes six different Gα subunits, three different Gβ subunits and two Gγ subunits. PLCβ, Gαq, Gβ13F and Gγ1 (but not GαO, Gαi and GαS) are required for [Ca2+] oscillations in ISCs (Extended Data Fig. 6d, e and Supplemental Video 7), and knockdown of Gαq or PLCβ inhibited L-Glu-induced changes in [Ca2+] oscillations and in ISC proliferation (Fig. 2d, e). A canonical GPCR signaling pathway through heterotrimeric G proteins and PLCβ thus regulates cytosolic [Ca2+] and proliferation of ISCs in response to dietary L-Glu (Fig. 2f).
Prolonged increase of cytosolic Ca2+ concentration is sufficient to trigger ISC proliferation
Ca2+ concentrations in the ER are dynamically controlled by store-operated Calcium entry (SOCE) 20–22. Cytosolic Ca2+ is pumped into the ER by Sarco-Endoplasmic Reticulum Calcium ATPase (SERCA) or out of the cell by Sodium Calcium Exchanger (NCX) or Plasma Membrane Calcium ATPase (PMCA) 20–22. A decrease in ER [Ca2+] is sensed by Stromal Interaction Molecule (STIM), an ER membrane protein that opens the plasma membrane Ca2+ channel Orai, allowing influx of extracellular Ca2+ into the cytosol 20–22. This coupled influx/efflux process is essential for Ca2+ oscillations in non-excitable cells 20–22 and is conserved in Drosophila (Fig. 2f, Extended Data Figure 1b). Genetic or pharmacological perturbation of conserved SOCE components revealed that [Ca2+] oscillations in ISCs are regulated by the canonical GPCR / IP3 / SOCE pathway, while receptors that influence membrane polarization do not contribute to Ca2+ signaling in these cells (Supplemental Discussion; Fig. 3a, Extended Data Fig. 6f, 7a, Supplemental Videos 8, 9). Perturbations in which oscillations were impaired while cytosolic [Ca2+] increased (e.g. knocking down SERCA or PMCA) strongly stimulated ISC proliferation (Fig. 3b, c, Extended Data Fig. 7b–d; see supplemental discussion for detailed description of genetic perturbations). This could be prevented by inhibiting the increase in cytosolic [Ca2+] (by also knocking down STIM; Fig. 3b, c, Extended Data Fig. 7e, f). Sustained elevation of cytosolic [Ca2+] is thus sufficient and required to induce ISC proliferation. ISC proliferation is also induced by over-expression of STIM and Orai or IP3R, conditions that elevate cytosolic [Ca2+] independently of SERCA (Fig. 3a, b, Extended Data Fig.7f).
SERCA deficiency can cause mis-folding and mis-processing of Notch (N) receptors, resulting in ISC tumors 29,30. We therefore assessed a possible role for N modulation in the proliferative response of ISCs to elevated [Ca2+]. Studies of N reporter expression, of N-dependent differentiation phenotypes, and of N gain-of-function conditions suggest that the acute mitogenic effect of sustained elevated cytosolic [Ca2+] can be decoupled from longer-term effects of SERCA deficiency on N signaling, and that elevating [Ca2+] through SERCA independent means or inducing ISC proliferation by over-expressing the Ca2+ effector CRTC (see below) does not influence N activity (Suppl. Discussion, Extended Data Fig. 8). SERCA deficiency further does not induce ISC proliferation by triggering an ER stress response (Supplemental Discussion, Extended Data Fig. 9a, b), and knockdown of SERCA or PMCA, or over-expression of CRTC do not result in elevated diphospho-ERK levels in ISCs (Extended Data Fig. 9c). Our experiments thus fail to detect deregulation of N, ERK, and ER stress signaling as associated with the acute proliferative response of ISCs to elevated cytosolic [Ca2+]. Although an as yet undetected perturbation of these pathways by short-term knockdown of SERCA or PMCA cannot be ruled out, we therefore sought to identify additional Ca2+ - sensitive regulators that may mediate the proliferative response of ISCs.
CaN / CRTC regulates ISC proliferation in response to elevated cytosolic [Ca2+]
Silencing the regulatory subunit CanB2 or the catalytic subunit CanA1 of the Ca2+-dependent phosphatase Calcineurin (CaN), but not the Ca2+-dependent kinases CaMKI and CaMKII, significantly abrogated ISC proliferation induced by either SercaRNAi, PmcaRNAi, or by co-overexpression of Stim and Orai (Extended Data Fig.9d, e). Knockdown of other CaN subunits encoded by the Drosophila genome (CanA-14F, Pp2B-14D and CanB) did not limit ISC proliferation (not shown), but over-expression of any of the three constitutively activated catalytic subunits 31 was sufficient to promote ISC proliferation (extended Data Fig. 9f). MARCM clones generated by ISCs homozygous for the CanB2KO null allele 32 or expressing CanB2RNAi grow slower than wild-type clones, while over-expressing constitutively active CanA-14F promotes clone growth (Extended Data Fig.9g).
The transcription factor CRTC (CREB regulated transcription co-activator, also called transducer of regulated CREB activity, TORC; not to be confused with Target of Rapamycin Complex I, TorC1) 33 is a conserved substrate of CaN, which promotes its nuclear translocation. Over-proliferation of SercaRNAi, Pp2B-14Dact, or PmcaRNAi expressing ISCs is suppressed in flies homozygous for the null allele crtc25–334, (Fig. 3d, Extended Data Fig. 9h, j), while over-expression of wild type CRTC or constitutively active CRTC 35 alone is sufficient to induce ISC proliferation (Extended Data Fig. 9i, j). Finally, L-Glu-induced ISC proliferation is not observed in crtc25–3 homozygotes (Fig. 3e), while CRTC is sufficient to induce proliferation independently of mGluR, Gaq, and IP3R (Fig. 3f, Extended Data Fig. 9k). Overexpressing of the conserved CRTC effector CREB (CrebB-17A) also causes ISC over-proliferation (Extended Data Fig. 9l; note that knockdown of CREB and its partner CBP leads to ISC loss, suggesting a broader role of these factors in ISC maintenance).
Ca2+ signaling integrates mitogenic stress and growth factor signals to stimulate ISC proliferation
ISCs sample a variety of environmental, systemic and local cues to adjust proliferative activity to the needs of the tissue, and we asked whether [Ca2+] modulation is a general feature of conditions that promote ISC proliferation. We monitored Ca2+ oscillations in the following mitogenic conditions 1–3,5: over-expression of Insulin Receptor (InR), RasV12 (a constitutively active allele of Ras), or of Upd2 (a ligand of the JAK/Stat pathway that stimulates ISC proliferation after tissue damage), infection with Ecc15 (Erwinia carotovora carotovora 15; oral infection with Ecc15 transiently stimulates ISC proliferation), loss of Notch (resulting in frequent symmetric divisions of ISCs), and Bleomycin treatment (which causes DNA damage in ECs, resulting in compensatory proliferation of ISCs). In all cases, oscillation frequency decreased, while average signal intensity increased (Extended Data Fig. 10a–c). Changes in Ca2+ oscillations are reversible: concomitant with the transient induction of proliferation after Ecc15 infection 36, signal intensity increases and frequency decreases at 4 hours after infection, returning to basal states at 24 hours (when ISC proliferation subsides 36; Extended Data Fig. 10c). Similar results were obtained with the bi-cistronic dTomato-V2-GCaMP5 construct (Extended Data Fig. 10d), and changes in oscillation pattern were also found in ISCs from old guts (30 days old; Extended Data Fig. 10a, b), which exhibit general hyperproliferation of ISCs 2,5. Conditions in which ISC proliferation was inhibited (over-expression of CncC 37 or of dominant-negative Insulin Receptor, InRDN 38) increase oscillation frequency without changing average signal intensity (Extended Data Fig. 10a, b, e, Supplemental Video10). Expression of InRDN (InRK1409A) in ISCs inhibits Bleomycin or feeding-induced proliferation (Extended Data Fig. 10f).
To test if Ca2+ signaling is involved in the integration of different upstream stimuli that control ISC proliferation, we examined if it is required for proliferation induced by the over-expression of InR and RasV12 (representing nutrient- and growth factor- induced ISC proliferation) or the Jun-N-terminal Kinase Kinase Hemipterous (Hep, representing stress-induced ISC proliferation), by knockdown of N, by infection by Ecc15, or by exposure to Bleomycin. While these perturbations strongly activated wild-type ISCs, activation of ISCs in Orai, IP3R, CanB2, Stim, or crtc loss of function conditions was significantly reduced (Fig. 4a, b; Extended Data Fig. 8i, 10g, i, j). Accordingly, loss of IP3R or Orai prevented the increase in cytosolic [Ca2+] by these perturbations (Fig. 4b; Extended Data Fig. 10h). mGluR deficiency did not impair Bleomycin- or Ecc15- induced proliferation and the associated cytosolic [Ca2+] increase (Extended Data Fig. 10i, k).
Figure 4. Ca2+ signaling integrates stress and mitogenic signals to stimulate ISC proliferation.
a, Mitotic figures in indicated genotypes.
b, Frequency of [Ca2+] oscillations and average fluorescence ratio in individual ISCs of indicated genotypes.
c, Quantification of mitotic figures in the midgut, and of [Ca2+] in ISCs of the indicated genotypes.
d, Quantification of mitotic figures in the midgut of the indicated genotypes.
e, ISC [Ca2+] oscillation patterns segregate into three different modes (shaded areas) that correlate with proliferative activity.
ISCs with perturbed Ca2+ signaling machinery are proliferation deficient. Blue arrows indicate ISC activation upon L-Glu feeding or Ecc15 infection. Orange arrow indicates return to quiescence 24 hrs after Ecc15 infection.
Averages and s.e.m. in all cases. a, Student’s t-test. n = 12, 12, 11, 14, 20, 14, 14, 15 guts from left to right. Representative of two experiments. b, ANOVA. n = 18, 27, 15, 15, 10, 11, 25, 20, 15, 22, 22, 17 cells. Representative of three experiments. c, Student’s t-test. For mitotic figures, n = 12, 18, 10, 9 guts. Representative of two experiments. For Ca2+ recordings, individual ISCs pooled from 4–5 guts. n = 11, 12, 15, 19, 8, 14, 10, 16 cells. Representative of three experiments. d, Student’s t-test. n = 9, 12, 11, 11, 12, 12, 13, 12 guts from left to right. Representative of two experiments. e, Averages and s.e.m. of oscillation frequency and average fluorescence ratio are plotted for each condition.
Elevating cytosolic [Ca2+] by knocking down SERCA or PMCA, or over-expressing CRTC is sufficient to bypass the requirement for InR in ISC proliferation (Fig. 4c, d), as well as the requirement for JAK/STAT signaling in Ecc15-induced ISC proliferation (Extended Data Fig. 10l). Knockdown of SERCA was not sufficient to induce proliferation in loss of function conditions for the ERK-regulated transcription factor Fos39, however, demonstrating a parallel requirement for CRTC and RTK/ERK-regulated transcription factors in the proliferative response to mitogens (Extended Data Fig.10m).
When [Ca2+] oscillation frequency is plotted relative to GCaMP3 fluorescence across a range of conditions, proliferative and resting states of ISCs segregate into two distinct modes (Fig. 4e, Extended Data Fig. 10n): quiescent ISCs poised for division (as in young guts under homeostatic conditions) show frequent and robust Ca2+ oscillations with a low level of cytosolic Ca2+, while highly proliferative ISCs have reduced Ca2+ oscillations with high levels of cytosolic Ca2+. ISCs in which Ca2+ signaling is impaired by knocking down components of the mGluR/PLC/IP3 pathway, on the other hand, show a third pattern: reduced oscillation frequency with low [Ca2+], and are proliferation deficient. ISC modes segregate regardless of whether average fluorescence intensity (calculated based on raw fluorescence values, Fig. 4e) or average baseline (calculated based on Gaussian fits, Extended Data Fig. 10n) are plotted against oscillation frequencies, but not when mean local amplitudes are plotted (Extended Data Fig. 10n), indicating that the primary driver of ISC proliferation is not the intensity of individual Ca2+ spikes, but the increase in average [Ca2+] in the ISC cytosol.
Discussion
We propose that the dynamic control of cytosolic [Ca2+] is critical for the ability of ISCs to undergo rapid and reversible activation and to dynamically control their proliferative activity in response to a wide range of nutrient and stress signals. IP3R/Ca2+/CaN/CRTC signaling coordinates and integrates cellular responses to these signals (Extended Data Fig. 1a). Exploring this integration further by assessing the role of Ca2+ signaling in mediating proliferative responses to ER stress40 and hemocyte-derived growth factors41, as well as in the shift from asymmetric to symmetric division modes in response to insulin-like peptides 1 will be of interest.
Our data suggest that cytosolic [Ca2+] changes in a wide range of conditions, and a detailed characterization of the kinetics of these changes, of the relative requirement for CRTC in the mitogenic response to EGF- and insulin-like ligands, as well as of the contribution of N signaling deregulation in [Ca2+] – induced ISC proliferation will shed further light onto the integration of the complex array of mitogenic signals sensed by ISCs.
In vertebrates, high plasma L-Glu levels and expression and activity of mGluR homologues are associated with malignancy 42,43 and our results indicate that chronic mGluR stimulation in stem and progenitor-like cells may be a driver of proliferative deregulation. This may be particularly relevant in the intestine, where L-Glu is not only introduced by the diet, but can also be a product of the commensal microbiome 44,45. Understanding mGluR signaling in ISCs is thus likely to provide significant new leads for possible therapies of intestinal cancers and inflammatory diseases.
Methods
Drosophila Stocks and Culture
The following strains were obtained from the Bloomington Drosophila Stock Center: w1118, Tub-Gal80ts, UASmCherry, UASmCD8GFP, UASnlsGFP, Act5C:::GAL4, UAS-InRDN, UAS-InRWT, UAS::RasV12, 20XUAS-GCaMP3 (BL32235), 20XUAS-IVS-GCaMP6s (BL42746), VGlutGal4oK371 (BL26160), Eaat1RNAi (BL43287), Eaat1Gal4 (BL8894), SercaRNAi (BL44581), Eaat1OE (BL8202), SercaKum170 (BL26700), StimA (BL52395), StimRNAi (BL27263), CamKIRNAi (BL 26726), CamKIIRNAi (BL 29401), IP3RRNAi (BL25937), RyRRNAi (BL28919), PmcaRNAi (BL31572), UAS-IP3R (BL30742), Pp2B-14DRNAi (BL40872), CanB2RNAi (BL27270 and BL38971), mGluRRNAi (BL41668), GαqRNAi (BL30735), Gβ13FRNAi (BL35041, UAS::CREB-b (BL9232), CREB-bRNAi (BL29332). The following lines were obtained from the Vienna Drosophila RNAi Center: SercaRNAi (transformant ID 107446), DomeRNAi (transformant ID 106071), HopRNAi (transformant ID 102830), OraiRNAi (transformant ID 12221), mGluRRNAi (transformant ID 103736), EGFRRNAi (transformant ID 43267), CanA1RNAi (transformant ID 32283). mGluR112b was a gift from Marie-Laure Parmentier, VGlutGal4CNS from A. DiAntonio, Crtc25–3by M. Montminy, esg-gal4 from S. Hayashi, Su(H)-GBE-Gal4 and Gbe-Gal80 are from S. X. Hou, UAS-HepWT by M. Mlodzik, hsFlp;FRT40,tub-Gal80;tub-Gal4,UASGFP from B. Ohlstein. NP1Gal4ts from D. Ferrandon, UAS-Stim and UAS-Orai from G. Hasan, GαqRNAi and Plcβ RNAi from W. Lee, UAS::FosRNAi from Dirk Bohmann, UAS::xbp1spliced by Dr. P. Domingos, UAS-NotchRNAi by N. Perrimon, Su(H)-GBE-lacZ by S. Bray, UAS-CRTC-HA and UAS-CRTC-SA-HA from Yukinori Hirano, CanB2KO, Pp2B-14DACT, CanA-14FACT from Aigaki Toshiro and UAS-tdTomato-P2A–GCaMP5G from R.W. Daniels.
See Supplemental Information for complete list of genotypes for each Figure.
Flies were cultured at 25°C with a 12 hour light/dark cycle on standard yeast/molasses-based food unless otherwise indicated (recipe for 1L food: 750ml H2O, 90ml Molasses, 25g Dry yeast, 6.5 g Agar and 69.6g Cornmeal).
For Ca2+ recording in aged flies, flies were aged at 25°C for 30 days in cages containing populations of 20–30 flies. Flies were shifted to 29°C one day before dissection and recording. Food was changed every 2 days. SercaKum170, FRT42D and CanB2KO, FRT42D chromosomes were recombined by neomycin selection. For MARCM clone induction, 2–3 days old flies were heat-shocked at 37°C for one hour. Midguts from female flies were dissected for analysis at the time indicated.
ISC-specific knockdown was achieved by expressing dsRNA against mGluR using the ISC/EB specific driver esg::GAL4 combined with EB-specific expression of Gal80 using Su(H)GBE::Gal80 40; this driver was rendered temperature sensitive by combining it with tub::Gal80ts (Fig. 1b).
The efficiency of the mGluRRNAi construct used, VDRC #103736, was tested by qPCR, Extended Data Fig. 8, and was further enhanced by co-expressing Dicer2; no off-targets are predicted for this construct).
Bleomycin feeding and Ecc15 infection
2~3 days old flies were dry starved in empty vials for 4hrs before treatment. Whatman paper (Fisherbrand; Thermo Fisher Scientific) was saturated with 25 µg/ml bleomycin (Sigma-Aldrich) dissolved in 5% sucrose. Flies were dissected for pH3 staining or for Ca2+ imaging. Bacteria ECC15 were cultured overnight at 30°C in LB medium. A concentrated bacterial pellet (OD of ≈200), which was centrifuged from 1-ml overnight culture media, was dissolved in 1ml 5% sucrose in Whatman paper. Flies were infected for 6hrs or 24hrs before dissection for pH3 staining or for Ca2+ imaging.
Calcium imaging
Cytosolic [Ca2+] was monitored in ISCs by expressing UAS::GCaMP3, UAS::GCaMP5 or UAS::IVS-GCaMP6s under the control of esg::GAL4 combined with Su(H)Gbe::GAL80. UAS::mCherry (or tdTomato from the bi-cistronic construct with GCaMP5) was introduced as an internal control and an indicator of stem cells, and a temperature-sensitive Gal80 transgene was included to allow temporal control of transgene expression. Real time recordings were performed 4 days after transgenes were induced at 29°C. Similar Ca2+ oscillations were observed in ISCs of age matched animals reared at 25°C and without the temperature-sensitive Gal80 transgene (not shown). No oscillations were observed in ECs when GCaMP3 was expressed in these cells using NP1::Gal4 (not shown).
To visualize [Ca2+] in live ISCs, fresh intact midguts were dissected in adult-hemolymph like (AHL) media (108 mM NaCl, 5 mM KCl, 2 mM CaCl2, 8.2 mM MgCl2, 4 mM NaHCO3, 1 mM NaH2PO4, 5 mM trehalose (Sigma), 10 mM sucrose, 5 mM HEPES, pH 7.5. Alternatively, Shields and Sang M3 insect medium (Sigma) can be used) and imaged using two-photon microscopy. The guts were slightly stretched and pinned down in a 35 mm Petri dish previously one-third filled with Sylgard (Dow Corning). Peristaltic movements were minimized by paralyzing the visceral muscle with Isradipine (10µg/ml, Ca2+ oscillations in ISCs are unaffected by Isradipine; Extended Data Fig. 5f). Ca2+ signals were imaged using a Zeiss LSM 7MP two-photon microscope equipped with a Chameleon XR laser (Coherent, Santa Clara, CA) running at 950nm and using a W Plan-Apochromat 20× 1.0NA lens. The posterior midgut area was recorded as time lapses of z-stacks comprising 17 4-µm-spaced planes every ~15 s. GCaMP3/5/6s emission was captured at 500–550 nm and mCherry/tdTomato emission at 575–610 nm.
Using automatic and manual image stabilization in Image Analyst MKII (Image Analyst Software, Novato, CA) the movement of the gut was compensated for and mean intensities were determined in regions of interests in mean intensity projection images of the recorded z-stacks. Oscillation patterns were either calculated ‘manually’ or in an unsupervised, automated manner in Mathematica 8.0 (Wolfram Research, Champaign, IL). For the ‘manual’ calculation, oscillation frequency was determined by counting individual peaks of GCaMP3:mCherry fluorescence emission ratio observed during 12min recordings, and average Ca2+ levels were calculated for each cell as the average fluorescence ratio for the entire timeline of the experiment using Excel. Automated analysis was performed by first detecting peaks based on the second temporal derivative of the GCaMP:mCherry fluorescence emission ratio followed by a Gaussian fit of each detected oscillation spike. The local amplitude of oscillation spikes and the baseline Ca2+ level between oscillations were calculated from the fit parameters. Statistical analysis using unpaired Student’s t-tests or ANOVA was performed in GraphPad Prism5 as indicated in the figures. Videos were generated using NIH Image J.
For pharmacological manipulations, guts were incubated in AHL media containing the respective compounds for 5–10 min and then imaged immediately. Isradipine, CdCl2, LaCl3, Thapsigargin and EGTA were purchased from Sigma and 2-APB was purchased from Tocris.
Immunofluorescence imaging
Intact guts were dissected, fixed, and stained by immunohistochemistry following standard protocols 18,40,46. Guts were dissected in phosphate-buffered saline (PBS) and fixed for 45 min at room temperature in 4% formaldehyde. All subsequent washes (1 hour) and antibody incubations (4°C overnight) were performed in PBS, 0.5% bovine serum albumin and 0.1% Triton X-100. Delta staining was performed following the methanol-heptane fixation method described in 47.
Primary antibodies and dilution: Rabbit anti-β-galactosidase (Cappel), Rabbit anti-dpERK (Cell Signaling, 1:200)1:5000; Rabbit anti-peIF2α antibody (Cell Signaling: 3597, 1∶150); Rabbit anti-VGlut (Dr. A. DiAntonio), galactosidase rat anti-Delta (gift from Dr. MD Rand, University of Rochester, 1:1000), anti-phospho-Histone H3 Ser10 (Upstate), 1:1000; and anti-Prospero, anti-Armadillo, anti-β-galactosidase, and anti-Delta (Developmental Studies Hybridoma Bank), 1:300, 1:250, 1:100, 1:500, and 1:100, respectively. Alexa Fluor 647-anti HRP and all fluorescent secondary antibodies were from Jackson Immunoresearch. Phalloidin (Invitrogen), 1:400, was used to label the visceral muscle in the midgut in Figure 2C. DAPI was used to stain DNA. All images were taken on a Zeiss LSM 710 confocal microscope and processed using Adobe Photoshop, Illustrator and Image J.
For gut cross sections, guts were fixed using methanol / heptane and briefly washed twice in PBS. The posterior midgut was cut into sections of 0.2–1mm in thickness by a fine steel blade. The gut pieces were flipped and mounted sagittally.
In situ hybridization
The protocol was adapted from48 using Digoxigenin (DIG) labeled RNA probes detected by NBT/BCIP assay (DIG nucleic acid detection kit, Roche). The following primers were used to generate RNA probes for Eaat1: F 5’- AGATTGCTACCAGAGCCATTAC-3’ and R 5’- GCACCAACAGGAATGACAAATC-3’.
Gut growth measurement
The width of the “distal hairpin” was measured as an indicator of gut growth. The distal hairpin region is adjacent to the copper cells in the posterior midgut. The width of this region was measured using ZEN software and reported as arbitrary pixel [px] values. Gut length (pixel) was measured by ImageJ. For cell density measurement, a 70µm × 70µm area in the posterior midgut was chose and cell number was counted based on DAPI staining. At least 10 guts of each genotype were counted.
CAFÉ assay and food intake experiment
Food naïve control flies were water starved for 2 days and re-fed with the CAFÉ assay based on 49 with minor modification. In brief, capillaries filled with liquid food (5% sucrose or 5% sucrose+1% glutamate) were marked with lines before and after 4hr feeding. The length in between (in cm) was calculated and compared in control food and glutamate supplemented food. At least 3 sets of biological replicates were compared for each food condition.
For colored food ingestion experiments, Groups of ddH2O starved food naïve control flies were transferred onto fresh food medium as indicated containing 2.5% (w/v) blue food dye (F D & C Blue Dye no. 1). Images were taken by Zeiss dissection microscope after 4hr ingestion.
Glutamate feeding
The feeding regimen is based on 1 with minor modification. In brief, black pupae raised on regular cornmeal/molasses food were collected and transferred to new empty vials with ddH2O soaked Whatman paper. To allow for transgene induction, flies were shifted to 29°C 2 days before hatching. After eclosion, food naïve flies were water starved (soaked Whatman paper) for another two days at 29 °C. Flies were then re-fed protein restricted food supplemented with various amounts of L-glutamate for the periods indicated in Figures. Restricted food recipe: 5% sucrose, 1% Agar and 0.1% yeast. L-glutamate (G5889), L-Glycine (G2879), L-Aspartate (A9256), L-Serine (S4500), L-Cysteine (168149), L-Glutamine (90114C) were purchased from Sigma. Amino acids were dissolved in water and mixed with the boiled restricted food at different concentrations. Food was changed daily. For pH3 staining and gut growth, flies were dissected, fixed and stained by standard protocols. For Ca2+ imaging after L-Glu treatment, food naïve flies were ddH2O starved for two days at 29°C and recorded in regular AHL media or AHL plus 1% glutamate.
Glutamate Microinjection
The injection protocol was as described in41. In brief, thin sterile glass capillaries were filled with the indicated solutions and 50nl were injected into the thorax of female adults using a NanojectII apparatus (Drummond). Fluid dispersed throughout the animal as indicated by blue food dye (F D & C Blue Dye no.1) mixed into the solutions. To study whether circulating glutamate can influence ISC proliferation in the starvation/refeeding paradigm, food naïve flies were starved on water for two days and injected with 1 × PBS or 1% glutamate +1 × PBS solutions. The injected flies were recovered and raised in restricted food condition (0.1% yeast + 5% sucrose + 1% agar) for the indicated time periods before dissection.
Statistical Methods
Statistical Analysis was performed using GraphPad Prism5 and Microsoft Excel. Statistical methods used and sample sizes are listed in Figure Legends. One-way ANOVA with post-hoc Dunnett test performed for Fig.3a, 3b, Fig.4b, exFig.2a (left chart), exFig.2e, exFig.5c, exFig.5e, exFig.6d, exFig.6f, exFig.7c, exFig.8b, 8f, 8h, 8i, exFig.9a, 9c, 9d, 9f, 9i, 9k, and exFig. 10b, 10i, 10j, 10l. One-way ANOVA with post-hoc Tukey test for Fig.3f, Fig.4b (right chart), exFig.2a (right chart), exFig.3f, exFig.3d (middle chart), exFig.9e, and exFig.10h. P values for other Figures determined by Student’s t-test. Sample sizes were chosen empirically based on observed effect sizes.
qRT-PCR Analysis of Gene Expression
Total RNA was extracted from dissected guts (10 per sample) or heads (4 per sample) using Trizol, and cDNA was synthesized using Superscript III (Invitrogen). Real time PCR was performed in triplicate for each sample from two independent (ExFig.9m) or three independent (ExFig.3c and ExFig.3f) experiments using SYBR Green on a Biorad IQ5 system. Expression was normalized with actin5C.
Primer Sequences for RT-PCR
The following primers pairs were used:
Actin5C (F): 5’- CTCGCCACTTGCGTTTACAGT-3’
Actin5C (R): 5’- TCCATATCGTCCCAGTTGGTC-3’
CAMKI (F): 5’- CGCTACAGTGATCCGACAAA-3’
CAMKI (R): 5’-CTGTTGGAGTGGTAGAGTCTTG-3’
CAMKII (F): 5’-GTGGATTTGTCAACGCGAAC-3’
CAMKII (R): 5’-GTCGTAAGTATGGCTCCCTTTAG-3’
Stim (F): 5’-CCAGAGAGGCTTGTGAGAAAT-3’
Stim (R): 5’-CGACCGATCCACATCATCAATA-3’
CanB2 (F): 5’-GTTCGATGAGTTCTGCTCTGT-3’
CanB2 (R): 5’-GAAGGATGCCGACGTGTATAA-3’
IP3R (F): 5’-TACCTCGGGAGTCTGGAATTAG-3’
IP3R (R): 5’-CTCCCAGTAAGCAGAGAGTAGA-3’
EAAT1 (F): 5’-CCAGACGGAGAGCATTGATAAG-3’
EAAT1(R): 5’-GGAACATGGTCGACTGAATGA-3’
mGluR (F): 5’-CGTCAGTGGCTCTTGTATGT-3’
mGluR (R): 5’-GTAGACGGTGCTGTTCATAGTC-3’
Upd3 (F): 5’-CCCAGCCAACGATTTTTATG-3’
Upd3 (R): 5’-TGTTACCGCTCCGGCTAC-3’
Extended Data
Extended Data Figure 1. Integration of stress and dietary signals by Ca2+ signaling.
a, We propose that Ca2+ signaling integrates and transduces nutritional and stress signals from the environment and from systemic and local paracrine sources to respond appropriately to tissue needs. In homeostatic conditions, ISCs are largely quiescent. Glutamate derived from protein in the food is absorbed by ECs through EAAT1, which is expressed throughout the gut, but at relatively higher levels in the anterior midgut. We propose that excessive dietary L-Glu is not efficiently resorbed by ECs and activates mGluR in ISCs. The L-Glu signal is integrated with local and systemic stress and growth factor signals to promote ISC proliferation in a CaN/CRTC dependent manner. CaN/CRTC signaling is required and sufficient downstream of InR and JAK/STAT signaling to induce ISC proliferation. We propose that CRTC acts in parallel to other transcription factors regulated by InR and JAK/STAT signaling.
b, Ca2+ signaling components in Drosophila (Extended Data Table 1).
Extended Data Figure 2. Dietary L-Glu stimulates mGluR-dependent gut growth.
a, Left: Mitotic figures in intestines re-fed with a range of L-Glu (wt/vol %) in food with varying yeast concentrations for 4 hours. Flies were starved upon eclosing for 48 hours, and then re-fed with food containing varying concentrations of yeast as only amino acid source, but supplemented with varying concentrations of L-Glu (between 0.1 and 10% L-Glu in food containing between 0.1% and 5% yeast; Note that the L-Glu concentration in standard fly food is about 0.1–0.3% 50,51). Middle: 10% L-Glu refeeding inhibits ISC proliferation. Mitotic figures were quantified 24hrs after flies were refed with yeast enriched (5%) or yeast restricted food (0.1%) supplemented with 10% glutamate. Blue food dye was included in the food to monitor food intake. Right: Distribution of mitotic ISCs along the GI tract after L-Glu refeeding. Number of proliferating ISCs (pH3+) in anterior and posterior midguts (AM and PM) is quantified.
b, Food intake is not affected by changing the L-Glu concentration in the food. CAFÉ assay 49 and ingestion of colored food are shown after 4hrs of re-feeding.
c, Injection of L-Glu fails to promote ISC proliferation. Food naïve flies were starved for 2 days before injection with the indicated solutions. Injected or non-injected flies were examined after recovery for the indicated time points. A typical example of injected fly was shown on the top. Blue food dye was mixed into the injected solutions to monitor its distribution throughout the body
d, Representative images of guts after refeeding. The red lines indicate the length and width measurements used to quantify relative gut size.
e, Left: ISC proliferation in animals re-fed sucrose solution supplemented with indicated amino acids (1% w/v final). The number of pH3+ ISCs was determined 6hrs after refeeding. Right: ISC proliferation in animals re-fed isocaloric sucrose-only solution (5.2% sucrose, the calorie content of 0.2% sucrose is equivalent to 1% glutamate) or re-fed excess sucrose (10%).
f, Feeding L-Glu for 7 days promotes growth of EsgtsF/O clones. Note large clones in L-Glu fed intestines (box). Genotype of EsgtsF/O: esgGal4, UAS::FLP, tub::Gal80ts; UAS::nlsGFP; tub::>FRT-CD2-FRT>Gal4.
g, Increased cell density, lineage growth and intestinal diameter in L-Glu treated guts. Cell density in the posterior midgut (PM) was determined in esgts F/O flies exposed to mock or L-Glu supplemented food for 7 days (esgts F/O allows lineage tracing from ISCs. Genotype is esg::Gal4, UAS::Flp, UAS::GFP, Act>FRT-stop-FRT>Gal4; tub::Gal80ts). Cross sections through the posterior midgut and overview images are shown highlighting the increase in intestinal diameter and in clonal growth upon L-Glu supplementation. Clones were analyzed 7 days after clone induction at 29°C.
h, L-Glu supplementation promotes growth of ISC lineages. ISC lineages were marked by MARCM (genotype: hsFlp, UAS::GFP;;tub::Gal4,FRT82B,tub::Gal80/FRT82B). Cell numbers in GFP+ ISC clones were counted 7 days after heat shock induction and re-feeding.
i, Related to Fig.1b. Mitotic figures and gut width measured after L-Glu refeeding. mGluR112b is a null allele of mGluR. For pH3 number, n=12 per condition, for gut width measurement, n=8 per condition.
Averages and s.e.m. are shown. P values from Student’s t-test for a (middle), b, c, g, h and i, from one-way ANOVA for a (left and right) and e. The sample size is as follows: n=8 per condition for a (left), n=7 per condition for a (middle), n=11 per condition for a (right), n=12 per condition for b and c , n=9 per condition for d, n= 11 per condition for e (left), n=6 per condition for e (right), n=6 per condition for g, n=14 per condition for i (left) and n=9 for i (right)). For h, clones (n=32 for control and n= 42 for glutamate food) from 5 guts were analyzed. Data shown in a, e, and g are representative of 3 independently performed experiments, and those shown in b, c, h and i are representative from 2 separate experiments. n.s.: not significant.
Extended Data Figure 3. ISC regulation by EAAT1.
a, Eaat1 transcript is enriched in the anterior midgut. In situ hybridization with antisense RNA probes against eaat1 (detected using NBT/BCIP; sense probes shown in bottom panels). Guts over-expressing Eaat1 in ECs (Eaat1OE), or depleted of eaat1 in ECs (Eaat1RNAi) used as positive/negative controls.
b, Expression pattern of Eaat1::Gal4 in midgut. Expression of eaat1::GAL4>UAS::GFP (green) in anterior midgut (AM) and posterior midgut (PM) is shown in higher magnification in the lower panels. Cell membranes highlighted by anti-armadillo staining (red).
c, Knockdown efficiency of Eaat1 RNAi line determined by qRT-PCR. Eaat1RNAi (BL43287) was used to knock down Eaat1 in the gut using NP1::Gal4, tub::Gal80ts (29°C for 10 days). NP1Gal4ts (NP1::Gal4; tub::Gal80ts) drives expression in ECs throughout the gut when flies are shifted to 29°C. 10 guts were pooled for RNA extraction and three independent groups were repeated for evaluation.
d, Knocking down eaat1 in ECs promotes ISC proliferation. Distribution of mitotic ISCs and percentage of Delta positive cells in intestines of flies in which eaat1 was knocked down in ECs shown in the middle panels. Representative image from confocal microscopy is shown on the right. Arrowheads point to select ISCs identified by anti-Dl staining (Dl, white).
e, Overexpressing eaat1 in EC suppresses L-Glu-mediated ISC proliferation. Eaat1 was over-expressed in ECs using NP1Gal4ts (NP1::Gal4; tub::Gal80ts) at 29°C. Flies were shifted to 29°C before hatching, then maintained on normal food (01.-0.3% L-Glu) for 4 days. Flies were then starved (with water) for another 2days. Mitotic index was determined in intestines of flies refed with 1% glutamate for 6hrs as shown in a.
f, Upd3 transcript in whole guts quantified by qRT-PCR (transcript levels normalized to actin5C).
g, Knocking down eaat1 in ECs by NP1::Gal4ts does not increase JAK/STAT signaling activity in the posterior midgut. 2xSTAT::GFP is an activity reporter for JAK/STAT signaling (green). Apoptosis in ECs is indicated by TUNEL staining (bottom panel, apoptotic nuclei in red).
As a positive control, Bleomycin (25ug/ml) treatment strongly induces 2xSTAT::GFP expression in visceral muscle and epithelium, results in widespread TUNEL positive nuclei in the intestinal epithelium, and substantially increases Upd3 transcripts in the posterior midgut.
Averages and s.e.m. are shown, P values are from ANOVA for d (middle) and f; from Student’s t-test for c, d (left and right) and e. The sample size is n=3 independent samples for each condition in c, n=12 guts for each condition (mitotic figures), and n=14 guts for Control and n=24 guts for Eaat1RNAi (percentage of Delta positive cells) in d. n=15 for Control and n=21 guts for Eaat1OE in e. n=3 independent samples for each condition in f.
For d and e, a representative experiment is shown (three biological replicates). For a, b, g, representative images from a representative of two independent experiments are shown (a: n=8 for each condition; b: n=12; g (left): n=12 per condition, and g (right): n=9 per condition).
Extended Data Figure 4. Neuronal projections and GCaMP3 fluorescence pattern in the intestinal epithelium.
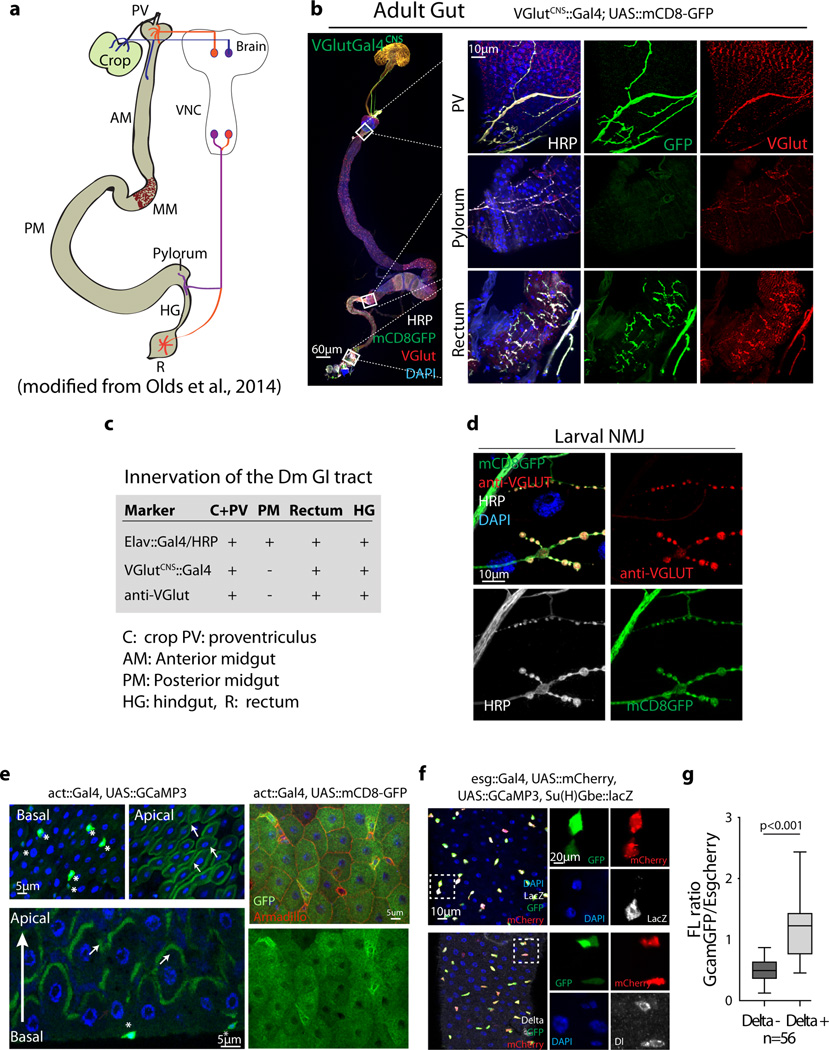
a, Schematic showing neuronal projections in adult Drosophila gastrointestinal tract (modified from Olds et al, 2014). Only three segments are innervated by neurons from the brain: Crop, Proventriculus (PV) and Foregut, Hindgut (HG) Pylorus, and the Rectum (R).
b, No glutamatergic neurites are observed in the posterior midgut (52,53). All neurites are stained by anti-HRP staining in white. mCD8GFP driven by VGlutGal4CNS (a glutamatergic neuron driver readily expressed in adults) labels all glutamatergic neurons. Glutamatergic neurons are also detected by VGlut (vesicular glutamate transporter) immunostaining. Higher magnification is shown on the right.
c, Summary of neurites innervating the Drosophila GI tract (see also Cognini et al, 2011). While L-Glu in the diet may stimulate glutamatergic neurons innervating the intestine, and a role for these neurons in stimulating ISC activity under certain circumstances cannot be ruled out, the widespread induction of ISC proliferation throughout the gut, as well as the ISC-specific requirement for mGluR suggests that the locally restricted glutamatergic innervation is not directly involved in stimulating ISC proliferation.
d, VGlutGAL4CNS>mCD8GFP and anti-VGlut stain the presynaptic neurons of 3rd instar larval NMJs. HRP (white) stains all neurites.
e, Detection of Ca2+ levels in the Drosophila posterior midgut (fixed tissue). Ubiquitous expression of UAS::GCaMP3 (green) in the posterior midgut using actin5C::GAL4 reveals high Ca2+ levels in subsets of small, basally located cells (likely to be ISCs and EBs; asterisks), and at the brushed border of ECs (arrows). Superficial view in top panels, sagittal view in bottom panels. (Right) Actin5C::Gal4 drives expression of mCD8GFP homogeneously in the posterior midgut. Armadillo (red) marks cell boundaries. In ECs, Ca2+ is enriched in the microvilli (marked with arrows), potentially to facilitate Ca2+ dependent absorption and innate immune processes 4.
f, Ca2+ concentration is higher in ISCs than in EBs. ISCs and EBs are labeled by expression of mCherry (red) under the control of esgGAL4. Ca2+ in these cells is detected using GCaMP3 (green). Su(H)Gbe::lacZ marks EBs (white in top panels) and Delta marks ISCs (white in lower panels).
g, Quantification of relative Ca2+ levels in ISCs versus EBs measured as fluorescence ratio between GCaMP3 and mCherry. Note the high variability of [Ca2+] in ISCs. Averages and s.e.m. are indicated. P values from Student’s t-test. n=56 cell pairs from 5 different guts. Four independent experiments were performed.
One representative image from 5 flies in a single experiment (two independent duplicates) is shown in b, e and f. One representative image from 6 larvae in a single experiment (two independent duplicates) is shown in d.
Extended Data Figure 5. Ca2+ oscillations in ISCs.
a, Typical ex vivo recording of Ca2+ oscillations in ISCs from young flies. GCaMP::GFP (green) and mCherry (red) are expressed specifically in ISCs. 56 frames with 15 second intervals are shown (see materials and methods for details). Frames were exported from ZEN software. Genotype: UAS::GCaMP3; esg::GAL4, UAS::mCherry; Su(H)Gbe::Gal80, tub::GAL80ts.
b, Traces of Ca2+ oscillations in ISCs of intestines incubated with EGTA(5mM) or 2-APB (10µM) for 5–10 minutes and then recorded immediately.
c, Ca2+ oscillation frequency of ISCs incubated in AHL with or without EGTA, CdCl2 (5 µM) or LaCl3 (5mM). Values for individual ISCs (collected from 3–4 different guts) are plotted,
d, Ca2+ oscillation in ISCs is inhibited by LaCl3. Scheme of oscillation parameters is shown on the left. The local amplitude of oscillation spikes and the baseline Ca2+ level between oscillations were derived from Gaussian fits on detected oscillation spikes. Average oscillation frequencies and average baselines are shown.
e, Observed Ca2+ oscillation pattern in ISCs is independent of the genetic reporter system used. Oscillation frequency and average fluorescence ratio are compared between different genetic Calcium reporters: UASGCaMP3, UAS::tdTomato-2A–GCaMP5 and UAS-IVS-GCaMP6s. Genetic reporters are expressed specifically in ISCs using esgGal4ts; Su(H)Gbe::Gal80.
f, Ca2+ oscillation pattern in ISCs of guts incubated with or without Isradipine (10µg/ml incubation in AHL medium). Isradipine inhibits L-type VGCCs to paralyze visceral muscles.
Averages and s.e.m. are shown. P value from ANOVA for c and e. Other P values from Student’s t-test. Individual ISCs pooled from 3–4 guts were plotted in c, e and f. The sample size for c is n = 12, 8, 9, 14; for e, n = 12, 16, 13, 14, 20, 24; and for f, n = 22, 14, 25, 22 (from left to right for each panel). Two independent experiments were performed for b-d, and three independent experiments for e and f.
Extended Data Figure 6. Glutamate feeding modulates Ca2+ oscillation pattern through mGluR/Gαq/PLCβ pathway.
a, Automated quantification of oscillation parameters in recordings from wild-type and mGluRRNAi expressing intestines refed L-Glu containing or control food (compare Fig. 3A). Average oscillation frequencies and average baselines are shown.
b, 10% L-Glu feeding inhibits Ca2+ oscillations. Ca2+ oscillations recorded in animals after 6–8h refeeding with 10% L-Glu and incubation of the intestine in AHL supplemented with 10% L-Glu. Prolonged incubation (20 min) in 10% L-Glu causes further decrease of average GCaMP3 fluorescence.
c, Manual and automated quantification of oscillation parameters on recordings from individual guts of the indicated genotypes. Values for individual cells from 3–4 guts are plotted on the left. Values for individual cells from single guts are shown in the third and fourth panels. Typical traces of recordings for individual cells are shown in the middle and the corresponding oscillation parameters calculated automatically by Gaussian fits in Mathematica 8.0 are shown on the right. Average oscillation frequencies and average baselines are shown.
d, Gαq, but not other Gα subunits (GαO, GαS or Gαi) is required for Ca2+ oscillations in ISCs. While knockdown of Gαq, G β 13F and Plc β impairs Ca2+ oscillations, oscillation frequency was not affected when GαO, GαS or Gαi were knocked down in ISCs for 8 days at 29°C (driver genotype is UAS::GCaMP3; esg::Gal4, UAS::mCherry, Su(H)Gbe::Gal80, tub::Gal80ts. Pertussis Toxin (UAS::PTX) was used to specifically inhibit Gαq. Typical traces are shown on the right.
e, Average oscillation frequencies and average baselines from Gaussian fits are shown.
f, Voltage Gated Calcium Channel (VGCC) components Cavβ or Cac are not required for Ca2+ oscillations in ISCs. Oscillations are also not affected in ISCs expressing molecules expected to affect plasma membrane potential (ΔOrk1 (hyperpolarizing) and UAS-NaChBac (hypo-polarizing)). Driver Genotype: UAS::GCaMP3; esg::GAL4, UAS::mCherry; Su(H)Gbe::Gal80, tub::GAL80ts.
Averages and s.e.m. are shown. P values in d, f and the right charts of panel c are from ANOVA; P values for a, b and e and the left charts of c are from Student t-test. Individual ISCs pooled from 3–4 guts are plotted in b-d and f. The sample size for a and e is n=3; for b n =10, 8, 12, 17, 15, 20, 10, 11; for c, n = 16, 20, 17, 12; for d, n =15, 18, 13, 22, 22, 10, 8, 9; and for f, n = 13, 15, 15, 14, 16 (from left to right for all panels). For c, values for individual cells from single guts are shown in the third and fourth panels. Data are representative of two independent experiments for d-f, and for three independent experiments for a-c.
Extended Data Figure 7. Prolonged increase of [Ca2+]cyto promotes ISC proliferation.
a, Typical traces of Ca2+ recordings of indicated genotypes. Transgenes were induced at 29°C for 4 days and 3–4 guts of each genotype were recorded. Thapsigargin concentration was 2 µM in the medium while recording. Genotypes: UAS::GCaMP3; esg::GAL4, UAS::mCherry; Su(H)Gbe::Gal80, tub::GAL80ts combined with w1118 for control or with the indicated transgenes.
b, Knockdown of SERCA in ISCs, but not EBs promotes ISC proliferation. SERCA was knocked down using esg::Gal4 (targeting ISCs+EBs), Su(H)Gbe::Gal4 (EBs), or esg::GAL4; Su(H)Gbe::GAL80 (ISCs) combined with tub::Gal80ts by incubating at 29°C for 4 days. ISCs and/or EBs are labeled by nlsGFP in green and mitotic ISCs are stained by pH3 (red). Number of dividing ISCs (pH3+) per gut were quantified and analyzed.
c, Knocking down SERCA or Pmca promotes growth of ISC-derived clones. Typical MARCM clones expressing SercaRNAi, PmcaRNAi and StimRNAi 7 days after induction are shown. ISC clones are marked in green and pH3 staining was used to detect dividing ISCs. Note: SercaRNAi clones contain many more, but smaller cells, resulting in similar clonal area compared to wild-type. Quantification is shown on the right.
d, (Left) Mitotic figures quantified in intestines of flies homozygous for SERCAkum170, a temperature sensitive SERCA loss of function allele. Flies were exposed to heat shock (42°C, 5 min for two consecutive days, this permanently inactivates SERCA, 54 and proliferation was assessed 7days after heat shock. (Right) Size of MARCM clones of ISCs homozygous for Sercakum170 analyzed 7 days after clone induction.
e, MARCM clones of ISCs homozygous for the Stim null allele, StimA ,analyzed 7 days after clone induction. Dashed lines delineate individual clones (GFP green, DAPI blue, Armadillo (red membrane), Prospero (red nuclei), Dl (white). Dl channel is shown separately in grayscale. Quantification of clone sizes (cells/clone) is shown on the right.
f, Guts of the indicated genotypes visualized in basal views (6 panels) and in cross section (2 panels). In basal views, ISCs are identified by Dl immuno-staining (red) and GFP (green). In cross sections, visceral muscle is identified by Phalloidin staining (red), and ISCs/EBs by GFP (green). DAPI is blue in all panels.
Averages and S.E.M. are shown, P value from ANOVA for c, the other P values from Student’s t-test. For mitotic figures in b, n=11 for each genotype. For mitotic figures in d (left), n=10 for each condition. For clonal analysis in c, clones (FRT40A, n=50; SercaRNAi, n=42; StimRNAi, n=40; PmcaRNAi, n=56) from 8–10 guts were quantified. For clonal analysis in d (right), clones (FRT42D, n=80; Sercakum170, n=60) from 10 guts were quantified. For clonal analysis in e (FRT19A, n=120; stimA, n=102) from 7 guts were assessed. For panel a, traces from individual ISCs from a representative gut of each indicated genotype are shown, 3–4 guts were recorded for each experiment and two independent experiments were performed. For panel b-f, one of the three independent experiments is presented.
Extended Data Figure 8. Time course analysis of Notch activity in ISC lineage after manipulation of Ca2+ signaling.
a, ISC lineage. After an asymmetric division, ISCs (expressing Delta) activate Notch in EBs (thus activating Su(H)Gbe::LacZ). EBs with high Notch activity differentiate into polyploid ECs (expressing Pdm1), EBs with low Notch activity differentiate into EEs (expressing Prospero).
b, ISC proliferation rates (mitotic nuclei / gut) at different time points after perturbation of N or Ca2+ signaling. Number of days after shift to 29°C is listed.
c-e, Representative images of guts perturbed as in b, immuno-stained for Su(H)Gbe::lacZ (bGal, red, reporter for Notch activity) and for Dl (white). GFP green, DAPI blue. For 7d timepoint, higher mag images (boxed area) of bGal channel are shown in lower row in grayscale.
Note that knockdown of Notch results in rapid loss of Su(H)Gbe::lacZ+ cells and accumulation of Dl+ cells. SERCA knockdown results in loss of Su(H)Gbe::LacZ only after prolonged expression (14 days), even though proliferation is induced more strongly than with NRNAi already at 4 days. Su(H)Gbe::lacZ expression is not lost in PMCARNAi or CRTC over-expressing (OE) guts, although proliferation is induced as strongly as in NRNAi expressing guts at 4 days. Quantification of ratio of Delta positive cells and GbeLacZ positive cells in the gut after at the indicated timepoints after shift to 29°C is shown in panel e.
f, Quantification of Prospero+ cells of posterior midguts (PM) in which cytosolic Ca2+ was increased by knocking down SERCA or PMCA, or by over-expressing Stim and Orai using esgts. Increased numbers of EEs (an indication of impaired N signaling) were only observed in animals in which SERCA was knocked down for a prolonged period of time (14 days 29°C). Anti-Prospero stains EE cell nuclei.
g, Clonal analysis of ISC differentiation process after manipulation of Ca2+ signaling or N activity. ISC MARCM clones of Control, NotchRNAi, SercaRNAi, or PmcaRNAi were analyzed at 4 days or 7 days after heat shock induction. Clones (marked in GFP) are circled in dashed line. The nuclei of differentiated cells are stained by Pdm1 in red. Although PmcaRNAi clones are significantly larger, differentiation process is largely normal based on Pdm1 staining. While differentiation of SercaRNAi clones at 7d AHS is significantly perturbed based on the Pdm1 staining, comparing with surrounding WT EC cells. As expected, no Pdm1 positive cells were observed in NRNAi clones.
h, Related to panel g, clone size and percentage of Pdm1 positive cells per clone was quantified 4days after heat shock (AHS) induction.
i, NotchRNAi induced proliferation can be partially rescued by knocking down Ca2+ signaling components, such as STIM or CanB2. Mitotic index was shown on right.
j, Inhibition of Serca stimulates ISC proliferation even when N signaling is induced by over expression of NICD.
Averages and s.e.m. are shown. P values (*, P<0.05; **, P<0.01; ***, P<0.001; n.s, not significant) are from ANOVA (between control and first four perturbations) in panel b, e-f (between control and PmcaRNAi, and stimOE;OraiOE) and h-i, and from Student’s t- test (between SERCARNAi and SERCARNAi; STIMRNAi) in panel b, f (between control and PmcaRNAi), and j. For b, mitotic cells are calculated at indicated timepoints for each genotype: 4d, Control (n=16), PmcaRNAi (8), CRTCOE (9), NotchRNAi (12), SercaRNAi (14) , SercaRNAi;StimRNAi (9); 7d, Control (12), PmcaRNAi (10), CRTCOE (9), NotchRNAi (11), SercaRNAi (10) , SercaRNAi;StimRNAi (8); 14d , Control (6), PmcaRNAi (18), CRTCOE (9), NotchRNAi (12), SercaRNAi (8) , SercaRNAi;StimRNAi (11); For c-e, cells in posterior midgut from 4–7 guts of each genotype and time point were analyzed.
For e, fraction of bGal+ or Dl+ cells in 100–200 total cells counted in a field of the posterior midgut for each condition was quantified. Average and s.e.m. for the following number of guts (left to right) n= 4, 4, 5, 4, 4, 4, 4, 7, 4, 6, 4, 4, 4, 5, 4, 4, 4, 5, 4, 4, 5, 4, 6, 5, 4, 4, 5, 4, 5, 5, 6, 4, 5, 7, 5, 6, 4, 5. For f, Prospero positive cells of indicated genotype at 4 days (n=5 guts) and 14 days (n=7 guts) were analyzed. For clonal analysis in g-h, clones (FRT40A, n=50; SERCARNAi , n=45; NRNAi, n=30; and PMCARNAi, n=43) were analyzed. For mitotic analysis in panel i and j, n=12 for each genotype. Data shown in b-f and i are representative of two independent experiments and those shown in g-h and j represent one of the triplicate experiments.
Extended Data Figure 9. Cytosolic Ca2+ regulates ISC proliferation through CaN/CRTC pathway.
a, Acute knock-down of Serca in ISCs does not induce ER stress. Phospho-eIF2alpha (red, a PERK-mediated phosphorylation and marker of ER stress; see also 40), is only weakly detected in control ISCs. No significant increase of phospho-eIF2alpha staining is observed in ISCs expressing SERCARNAi, while Tunicamycin treatment (a bona fide inducer of ER stress; 50µM, 24hrs) increases phospho-eIF2a strongly in ISCs (asterisk). Quantification of average fluorescent intensity of p-eIF2alpha was shown on right.
b, Elevating ER folding capacity by over-expressing spliced Xbp1 (Xbp1s) does not limit ISC proliferation in SERCA loss of function conditions. Representative images shown on the left. Mitotic ISCs stained by anti-pH3 staining (red). Mitotic index quantified on the right‥
c, Elevated cytosolic [Ca2+] induces proliferation without perturbing EGFR pathway activity. Indicated Ca2+ signaling components were knocked down or over-expressed using esgts;UAS::nlsGFP. dpERK staining as a readout of EGFR pathway activity was quantified after 4 day induction at 29°C. InR was overexpressed using esgts;UAS::mCD8GFP as a positive control.
d, Proliferation of Serca deficient ISCs is suppressed when CaN subunits are silenced simultaneously, but not when CaMKI and CaMKII are knocked down.
e, Proliferation of ISCs in which cytosolic [Ca2+] is increased by over-expression of STIM and Orai, or by knock down of PMCA, is rescued when CaNB2 is silenced simultaneously‥
f, Overexpression of constitutive active forms of CaN catalytic subunits (CanA14F, Pp2B14D, CanA1) promotes ISC proliferation. Mitotic figures were quantified after 4 days of transgene expression at 29°C‥
g, CaN is required for growth of ISC lineages. Quantification of clone sizes of MARCM clones homozygous for the null allele CanB2KO, or expressing dsRNA against CanB2 (canB2RNAi), or constitutively active CanA14Fact.
h, Loss of CRTC (homozygosity for null allele crtc25-3) rescues increased ISC proliferation when Pmca is knocked down or when Pp2B-14Dact is over-expressed using esg::Gal4, tub::Gal80ts (esgts).
i, Over-expression of CRTC promotes ISC proliferation. Mitotic index of intestines over-expressing HA-tagged forms of wild type (UAS::CRTC-HA) or constitutively nuclear forms of CRTC (UAS::CRTC-SA-HA) using esg::GAL4ts was analyzed after 4 days of incubation at 29°C.
j, Increasing cytosolic Ca2+ promotes ISC proliferation via Ca2+/Calcineurin/CRTC pathway. Guts of indicated genotypes were dissected and stained with anti-pH3 (indicating mitotic ISCs), anti-Armadillo (arm, labeling cell boundaries) and anti-Prospero (Pro, labeling entero-endocrine cells).
k, CRTC overexpression is sufficient to promote ISC proliferation when Gaq or IP3R are silenced.
l, CREB and its partner CBP are required for ISC survival, while over-expressing CREB promotes proliferation. Representative images of intestines in which CREB and CBP were genetically perturbed in ISC/EBs using esg::GAL4. Increased numbers of GFP+ ISCs/EBs are observed when CREB is over-expressed, while a significant loss of GFP+ cells is observed when CREB or CBP are knocked down. Guts are stained for Armadillo (membrane, white) and Prospero (nuclear, white) to identify EEs and ECs; DAPI is blue. Genotype: esg::GAL4; UAS::nlsGFP; tub::Gal80ts / UAS::X
m, Knockdown efficiency of RNAi lines determined by qRT-PCR. PmcaRNAi (BL31572) and CanB2RNAi(BL27270) were used to knock down respective genes in the gut using NP1::Gal4, tub::Gal80ts (29°C for 10 days). mGluRRNAi (BL41668) was used to knock down mGluR in the brain using elav::GAL4. Expression levels were normalized using actin5C and to un-induced controls (NP1::Gal4/+; tubGal80ts/+ or elav::Gal4/+). Effectiveness of other constructs used has been reported in the literature: UAS::GaqRNAi and UAS::PLCβRNAi were obtained from and verified by Ha et. al. 55, UAS::CamKIRNAi (BL26726), UAS::CamKIIRNAi (BL 29401), UAS::IP3RRNAi (BL25937), and UAS::RyRRNAi (BL28919) were verified by Shim et al. 23, and UAS::SercaRNAi (BL 44581) by Roti et al. 29.
Average and s.e.m. are shown throughout. P values from Student’s t-test in b, and g-h, and from ANOVA for a, c, d-f, i and k. For a and c, fluorescence intensities for peIF2alpha (a) or dpERK (c) in 30–50 ISCs/EBs doublets in single fields of several independent posterior midguts were averaged for each condition. Averages and s.e.m. shown represent the following sample sizes: a, Control, n=5 guts, SercaRNAi, n=5, Tunicamycin, n=6; c, Control, n=6, SercaRNAi, n=4, CRTCOE, n=9, and PmcaRNAi, n=8. For mitotic analysis in b, d-f, h-i and k: b, n=13 guts for control, n=10 for the rest; d, n=17 for each genotype; e, n=18 for control, n=12 for the rest; f and k, n=11 each genotype; and i, n=12 for each condition. For clonal analysis in g, clones (FRT42D, n=56 clones; CanB2KO, n=60; FRT40A, n=50; CanB2RNAi, n=70; FRT82B, n=62; CanA14FACT, n=58) from 10 guts each were assessed. One representative image from 10 flies in a single experiment (two independent experiments) is shown in j and l. Data shown in a-i, and k are representative of three independent experiments. Data in m is average and s.e.m. from n=3 technical replicates of samples pooled from 10 guts each for Pmca and CanB2, 4 heads each for mGluR. Representative of two independent experiments.
Extended Data Figure 10. Ca2+ oscillation pattern as an indicator of ISC proliferation status.
a, Typical traces of live recordings of indicated genotypes. Genotype for GCaMP3 control: w1118 X UAS::GCaMP3; esg::GAL4, UAS::mCherry; Su(H)Gbe::Gal80, tub::GAL80ts. Genotype for bicistronic control: w1118 X; esg::GAL4, UAS::tdTomato-2A–GCaMP5;Su(H)Gbe::Gal80, tub::GAL80ts
b, Ca2+ oscillation patterns of ISCs in which proliferation was stimulated or inhibited by genetic or environmental perturbations: knockdown of Notch, over-expression of InR, Unpaired2 or RasV12, infection with Ecc15, or aging results in high proliferative activity. Over-expression of InRDN and CncC inhibits proliferation of ISCs.
c, Acute Ecc15 infection transiently increases cytosolic [Ca2+] while decreasing oscillations in ISCs.
d, Acute Ecc15 infection increases cytosolic Ca2+ while decreasing oscillations in ISCs as determined using the bi-cistronic Calcium reporter UAS::tdTomato-P2A–GCaMP5G.
e, ISCs in which proliferation is impaired exhibit more frequent oscillations than controls. Oscillation frequency of ISCs from indicated genotypes is plotted individually (3 guts for each genotype and each dots represents one ISC).
f, Expressing InRDN in ISCs is sufficient to inhibit stress- or diet- induced proliferation. Quantification of mitotic figures of indicated genotype is shown. For Bleomycin treatment, flies were dry starved for 4hrs before feeding on 25ug/ml (final) Bleomycin for 24hrs. For refeeding, Flies were maintained on normal food for 4 days at 29°C, then starved for 2 days, and re-fed with yeast supplemented food. Genotype: esg::Gal4, UAS::GFP; Su(H)Gbe::Gal80, tub::Gal80ts / UAS::InRDN.
g, ISC proliferation induced by oral infection with Ecc15 or by Bleomycin treatment is suppressed by silencing IP3R or Orai. Mitotic figures were analyzed 6 hrs after oral infection with Ecc15 or 24 hrs after feeding with Bleomycin .
h, Elevated cytosolic [Ca2+] in ISCs activated by oral infection with Ecc15 or by Bleomycin treatment. This elevation is suppressed by silencing IP3R or Orai.
i, ISC proliferation induced by Bleomycin treatment or Ecc15 infection is suppressed by silencing CRTC or IP3R or CanB2, or in Crtc25-3 homozygous mutants. mGluR, in turn, is not required for Bleomycin-induced proliferation. Mitotic figures were analyzed 24 hrs after feeding with Bleomycin or 6 hrs after oral infection with Ecc15.
j, Quantification of mitotic figures in animals of the indicated genotypes. ISC proliferation induced by over-expression of InR or Hep can be suppressed by knockdown of IP3R, Crtc, Stim, or CanB2 (see also Fig. 4a).
k, mGluR is not required for Ecc15 - induced proliferation and changes in cytosolic Ca2+. Mitotic figures and Ca2+ oscillation patterns analyzed 6hrs after oral infection.
l, Increasing cytosolic [Ca2+] promotes ISCs proliferation in JAK/STAT loss of function conditions. JAK/STAT pathway (Dome and Hop) is required for ISC proliferation induced by Ecc15 infection. Increasing cytosolic [Ca2+] by knocking down Pmca (left) or Serca (right) is sufficient to rescue ISC proliferation.
m, Knocking down Fos can substantially suppress CRTC over-expression induced ISC proliferation.
n, Left, segregation of active and resting ISCs into Ca2+ oscillation modes as calculated by automatic peak detection and Gaussian fits. As shown for ‘manual’ calculations in Figure 4, Ca2+ oscillation patterns segregate into two modes associated with the proliferative status of the ISCs. ISCs in which the core components of Ca2+ homeostatic machinery are perturbed exhibit both low oscillation frequency and low average signal intensity (lower left corner). Transition from quiescence to active proliferation upon L-Glu feeding is indicated by the blue arrow.
Right: ISC activity does not segregate when local oscillation amplitudes are plotted against oscillation frequency, suggesting that the primary driver of ISC proliferation is not the amplitude of individual Ca2+ spikes, but the increase in basal or average cytosolic Ca2+ concentration within ISC.
Average and s.e.m. are shown. P values from ANOVA in b, h, i j (left), and l (right); P values from Student’s t-test in c-d, f-g, k, m, j (right) and l (left). For Ca2+ recordings in b-e, h and k, individual ISCs pooled from 3–4 guts were plotted.
The sample size for b is n = 10, 14, 14, 12, 16, 18, 9, 12, 11, 20, 22, 38, 29, 12, 19, 8, 12; for c, n = 7, 8, 9, 7, 13, 18, 10, 7; for d, n =14, 16, 15, 18; for e, n = 5, 4, 5, 9, 4, 6, 8, 6, 5; for h, n =12, 11, 12, 11, 12, 14, 12, 11, 17, 12, 14, 11, 19, 9, 10, 8; for k, n=11, 12, 11, 11, 13, 10, 11, 11 (from left to right for all panels).
For mitotic analysis in f-g, n = 12, 10, 10, 10, 12, 11, 10, 12, 12, 14, 10, 9, 9, 10, 9, 9, 10, 13; and in i-m, n = 13, 12, 11, 18, 11, 18, 12, 13, 13, 12, 10, 10, 13, 9, 12, 12, 16, 12, 9, 10, 10, 8, 9, 9, 10, 9, 12, 13, 13, 14, 14, 15, 12, 10, 19, 10, 12, 10, 9, 8, 8, 9, 10 (from left to right for all panels). Data in a-h, j and m are representative of three independently performed experiments, and those shown in i, k and l are a composite from two separate experiments.
Supplementary Material
Acknowledgments
This work was supported by the National Institute on Aging (R01 AG028127), the National Institute on General Medical Sciences (R01 GM100196), and by a Glenn Medical foundation postdoctoral fellowship to H.D. The Zeiss 7MP was purchased using NIH grant S10OD010414. We would like to thank Drs. G. Hasan, Y. Hirano, A. Toshiro, M. Parmentier, W. Lee, S.X. Hou, M. Montminy, N. Perrimon, B. Ohlstein A. DiAntonio, H. Hayashi, R.W. Daniels, K. Venkatachalam, the Vienna Drosophila RNAi Center, and the Bloomington Stock Center for flies, and Developmental Studies Hybridoma Bank for antibodies. We also thank Jasper lab members for discussion.
Footnotes
Author Contributions H.D. and H.J. designed and conceived the study. H.D. and A. G. performed Ca2+ recording and analysis, H.D. performed all other experiments in the study. H.J. and H.D. analyzed data and wrote the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. A.A.G. declares financial interest in Image Analyst Software.
References
- 1.O’Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchon N, Broderick NA, Lemaitre B. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat Rev Microbiol. 2013;11:615–626. doi: 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- 3.Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaitre B, Miguel-Aliaga I. The Digestive Tract of Drosophila melanogaster. Annu Rev Genet. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- 5.Ayyaz A, Jasper H. Intestinal inflammation and stem cell homeostasis in aging Drosophila melanogaster. Front Cell Infect Microbiol. 2013;3:98. doi: 10.3389/fcimb.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 2008;7:478–490. doi: 10.1111/j.1474-9726.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KP, et al. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc Natl Acad Sci U S A. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeds PJ, Burrin DG, Stoll B, Jahoor F. Intestinal glutamate metabolism. J Nutr. 2000;130:978S–982S. doi: 10.1093/jn/130.4.978S. [DOI] [PubMed] [Google Scholar]
- 9.Newsholme P, Procopio J, Lima MM, Pithon-Curi TC, Curi R. Glutamine and glutamate--their central role in cell metabolism and function. Cell Biochem Funct. 2003;21:1–9. doi: 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- 10.Xiao W, et al. Glutamate prevents intestinal atrophy via luminal nutrient sensing in a mouse model of total parenteral nutrition. FASEB J. 2014;28:2073–2087. doi: 10.1096/fj.13-238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H, et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 13.Lewerenz J, et al. The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxidants & redox signaling. 2013;18:522–555. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman NJ, Lefkowitz RJ. Desensitization of G protein-coupled receptors. Recent progress in hormone research. 1996;51:319–351. discussion 352-313. [PubMed] [Google Scholar]
- 15.Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 16.Burrin DG, Stoll B. Metabolic fate and function of dietary glutamate in the gut. Am J Clin Nutr. 2009;90:850S–856S. doi: 10.3945/ajcn.2009.27462Y. [DOI] [PubMed] [Google Scholar]
- 17.Rival T, et al. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol. 2004;14:599–605. doi: 10.1016/j.cub.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Qi Y, Jasper H. Dpp signaling determines regional stem cell identity in the regenerating adult Drosophila gastrointestinal tract. Cell Rep. 2013;4:10–18. doi: 10.1016/j.celrep.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride SM, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile × syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 20.Brini M, Carafoli E. Calcium pumps in health and disease. Physiol Rev. 2009;89:1341–1378. doi: 10.1152/physrev.00032.2008. [DOI] [PubMed] [Google Scholar]
- 21.Dupont G, Combettes L, Bird GS, Putney JW. Calcium oscillations. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Shim J, et al. Olfactory control of blood progenitor maintenance. Cell. 2013;155:1141–1153. doi: 10.1016/j.cell.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong CO, et al. A TRPV channel in Drosophila motor neurons regulates presynaptic resting Ca2+ levels, synapse growth, and synaptic transmission. Neuron. 2014;84:764–777. doi: 10.1016/j.neuron.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniels RW, Rossano AJ, Macleod GT, Ganetzky B. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS One. 2014;9:e100637. doi: 10.1371/journal.pone.0100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roti G, et al. Complementary genomic screens identify SERCA as a therapeutic target in NOTCH1 mutated cancer. Cancer Cell. 2013;23:390–405. doi: 10.1016/j.ccr.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Periz G, Fortini ME. Ca(2+)-ATPase function is required for intracellular trafficking of the Notch receptor in Drosophila. EMBO J. 1999;18:5983–5993. doi: 10.1093/emboj/18.21.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakai Y, et al. Calcineurin and its regulator sra/DSCR1 are essential for sleep in Drosophila. J Neurosci. 2011;31:12759–12766. doi: 10.1523/JNEUROSCI.1337-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeo S, Tsuda M, Akahori S, Matsuo T, Aigaki T. The calcineurin regulator sra plays an essential role in female meiosis in Drosophila. Curr Biol. 2006;16:1435–1440. doi: 10.1016/j.cub.2006.05.058. [DOI] [PubMed] [Google Scholar]
- 33.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 2008;7:434–444. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirano Y, et al. Fasting launches CRTC to facilitate long-term memory formation in Drosophila. Science. 2013;339:443–446. doi: 10.1126/science.1227170. [DOI] [PubMed] [Google Scholar]
- 36.Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8:152. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochmuth CE, Biteau B, Bohmann D, Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell. 2011;8:188–199. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci U S A. 2005;102:13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138:1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Zeng X, Ryoo HD, Jasper H. Integration of UPRER and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation. PLoS Genet. 2014;10:e1004568. doi: 10.1371/journal.pgen.1004568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayyaz A, Li H, Jasper H. Haemocytes control stem cell activity in the Drosophila intestine. Nat Cell Biol. 2015;17:736–748. doi: 10.1038/ncb3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Julio-Pieper M, Flor PJ, Dinan TG, Cryan JF. Exciting times beyond the brain: metabotropic glutamate receptors in peripheral and non-neural tissues. Pharmacol Rev. 2011;63:35–58. doi: 10.1124/pr.110.004036. [DOI] [PubMed] [Google Scholar]
- 43.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan D. Protection of the glutamate pool concentration in enteric bacteria. Proc Natl Acad Sci U S A. 2007;104:9475–9480. doi: 10.1073/pnas.0703360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
References (Methods)
- 46.Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee WC, Micchelli CA. Development and characterization of a chemically defined food for Drosophila. PLoS One. 2013;8:e67308. doi: 10.1371/journal.pone.0067308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ja WW, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olds WH, Xu T. Regulation of food intake by mechanosensory ion channels in enteric neurons. Elife. 2014;3 doi: 10.7554/eLife.04402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cognigni P, Bailey AP, Miguel-Aliaga I. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 2011;13:92–104. doi: 10.1016/j.cmet.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanyal S, Jennings T, Dowse H, Ramaswami M. Conditional mutations in SERCA, the Sarco-endoplasmic reticulum Ca2+-ATPase, alter heart rate and rhythmicity in Drosophila. J Comp Physiol B. 2006;176:253–263. doi: 10.1007/s00360-005-0046-7. [DOI] [PubMed] [Google Scholar]
- 52.Ha EM, et al. Regulation of DUOX by the Galphaq-phospholipase Cbeta-Ca2+ pathway in Drosophila gut immunity. Dev Cell. 2009;16:386–397. doi: 10.1016/j.devcel.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 53.Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- 55.Takashima S, et al. Development of the Drosophila entero-endocrine lineage and its specification by the Notch signaling pathway. Dev Biol. 2011;353:161–172. doi: 10.1016/j.ydbio.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.