Abstract
Background and Objectives
The morphology of red blood cells (RBCs) deteriorates progressively during hypothermic storage. The degree of deterioration varies between individual cells, resulting in a highly heterogeneous population of cells contained within each RBC unit. Current techniques capable of categorizing the morphology of individual stored RBCs are manual, laborious, error-prone procedures that limit the number of cells that can be studied. Our objective was to create a simple, automated system for high-throughput RBC morphology classification.
Materials and Methods
A simple microfluidic device, designed to enable rapid, consistent acquisition of images of optimally oriented RBCs, was fabricated using soft lithography. A custom image analysis algorithm was developed to categorize the morphology of each individual RBC in the acquired images. The system was used to determine morphology of individual RBCs in several RBC units stored hypothermically for 6–8 weeks.
Results
The system was used to automatically determine the distribution of cell diameter within each morphological class for >1,000,000 individual stored RBCs (speed: >10,000 cells/hour; accuracy: 91.9% low-resolution, 75.3% high-resolution). Diameter mean and standard deviation by morphology class: discocyte 7.80±0.49μm, echinocyte 1 7.61±0.63μm, echinocyte 2 7.02±0.61μm, echinocyte 3 6.47±0.42μm, sphero-echinocyte 6.01±0.26μm, spherocyte 6.02±0.27μm, stomatocyte 1 6.95±0.61μm, stomatocyte 2 7.32 ± 0.47μm.
Conclusion
The automated morphology classification procedure described in this study is significantly simpler, faster and less subjective than conventional manual procedures. The ability to evaluate the morphology of individual RBCs automatically, rapidly and in statistically significant numbers enabled us to perform the most extensive study of stored RBC morphology to date.
Keywords: storage, morphology, microfluidic
Introduction
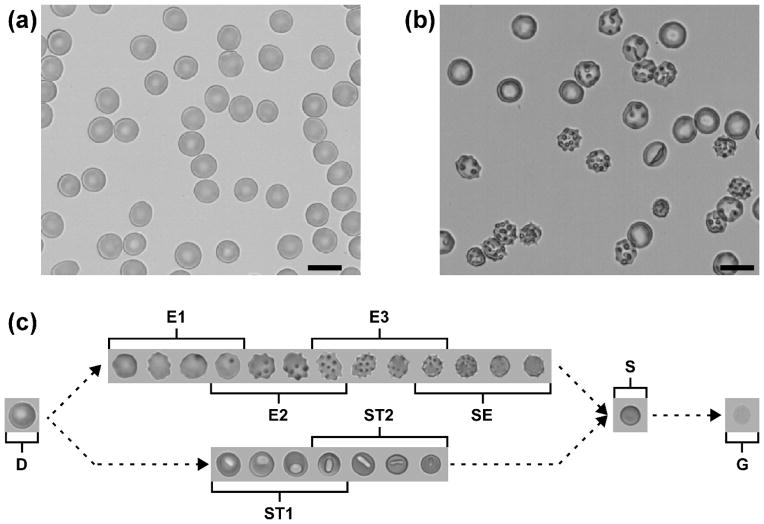
Normal, healthy red blood cells (RBCs) are highly-deformable biconcave discs with a diameter of ~8 μm, thickness of ~2.5 μm and volume of ~90 fL (Fig. 1a).[1–3] High surface membrane area to volume ratio enables RBCs to deform and fit into even the narrowest capillaries, such as those in the spleen.[4, 5] During ex vivo hypothermic storage, RBCs accumulate significant oxidative damage and shed microparticles via vesiculation, progressively losing membrane surface area, cell volume and physiologically relevant deformability.[6–8] As a result of the storage-induced biochemical changes and microvesiculation, stored RBCs undergo a gradual morphological transformation from healthy, flexible discocytes through reversible intermediate forms to irreversibly-damaged, rigid spherocytes with the smallest surface area to volume ratio allowed by geometry, before ultimately undergoing lysis (Fig. 1c).[9, 10]
Figure 1.
Morphological heterogeneity of individual fresh and stored RBCs. (a) Representative image of normal, fresh RBCs. (b) Representative image of RBCs stored for 6 weeks (2–6°C, AS-1 additive solution). Scale bars are 10 μm. (c) Morphological transformation of stored RBCs: discocyte (D), echinocyte 1 (E1), echinocyte 2 (E2), echinocyte 3 (E3), sphero-echinocyte (SE), stomatocyte 1 (ST1), stomatocyte 2 (ST2), spherocyte (S), ghost (G).
Individual RBCs are extraordinarily heterogeneous with respect to this morphological transformation – after the first 2–3 weeks of storage, the population of stored RBCs in a single unit consists of a complex mixture of relatively well-preserved cells, reversibly damaged intermediate cells and an increasing fraction of irreparably damaged cells (Fig. 1b). A manual evaluation of individual RBC morphology in 9 units previously found that after 6 weeks of storage the units consisted of 23.3 ± 4.3 % discocytes, 46.8 ± 6.7 % reversibly damaged RBCs and 29.9 ± 4.0 % irreversibly damaged RBCs.[9] The variation between units is likely the result of intrinsic inter-donor differences, post-collection processing practices and storage conditions. Unlike RBC biochemical properties (e.g. levels of ATP or 2,3-BPG) that are known to recover following infusion, [11] the transformation of stored RBCs into spherocytes is irreversible because the total surface area of the cell membrane lost due to vesiculation is too large for the cell to return to the discoid form.[9, 12] These irreparably damaged RBCs are dysfunctional, non-deformable cells, potentially capable of obstructing capillaries and causing ischemia.[13] They are removed from the circulation by the phagocytic cells of the recipient’s spleen shortly after transfusion, provided they did not lyse in the blood bag or bloodstream before reaching that organ.[5, 14] The infusion and subsequent destruction of a large number of these cells may accelerate iron overload in chronically transfused patients.[7, 15] The non-transferrin-bound iron released due to the extra- or intravascular lysis of these cells may predispose the recipient to inflammation and infection.[16, 17]
In current research and clinical practice, the morphology of individual stored RBCs is evaluated by visually matching and assigning cells to predefined morphological classes based on each cell’s overall appearance (e.g. size, shape, surface texture).[2] The extent of RBC morphological deterioration induced by hypothermic storage for each unit is then quantified by determining the percentage of cells in the RBC population that belong to each of the predefined morphological classes and/or calculating a weighted-average “morphology score”.[18, 19] A conventional approach to performing the measurement is to fix the RBCs in the sample, spread them on a glass slide and acquire several non-overlapping images of the cells in the blood smear via bright-field digital microscopy[20, 21] or using a scanning electron microscope.[9, 19, 22] Individual RBCs captured in the acquired images are then visually classified via manual counting.[23, 24] The highly laborious nature of this manual approach has limited the number of individual cells that are normally counted in a single morphological measurement to a very small number, varying from only 200 to 1500 cells.[9, 19–22] This technical limitation may introduce a significant sampling error, [24, 25] particularly when attempting to quantify the morphological heterogeneity of nearly 2×1012 cells contained in a single RBC unit.
The objective of this study was to develop and validate a simple, automated system for morphological classification of single stored RBCs at high-throughput and to use this system to analyze a statistically meaningful number of individual cells. Once loaded into the microfluidic device, individual RBCs (arranged in a single-cell, non-overlapping layer) were continuously passed through the field-of-view of a microscope equipped with a digital camera. The morphological appearance of individual RBCs was determined via automated segmentation, feature extraction and classification of the acquired images. The use of the microfluidic device alleviated the need for skilled sample preparation and, in combination with the image analysis algorithm, reduced subjectivity and eliminated the need for tedious manual counting. We demonstrated the performance of this automated system by simultaneously classifying the morphology and measuring the diameter of 1,367,852 individual stored RBCs; the largest set of such paired data known to date.
Materials and Methods
Microfabrication
The microfluidic devices used in the study were fabricated using soft lithography.[26] Silicon wafers (University Wafer, South Boston, MA) were spin-coated with a 7 μm layer of SU-8 negative photoresist (MicroChem Corp, Newton, MA). The wafers were then exposed to near-UV light (ETI/6/350/NUV/DCCD/M mask aligner, Evergreen Technology Inc., San Jose, CA) through a transparency photomask (FineLine Imaging, Colorado Springs, CO) and developed. Large domed structures at the inlet and the outlet port were fabricated by depositing small droplets (23 μL) of SU-8 photoresist onto the circular regions outlining the inlet and the outlet, soft-baking at 55°C for 8 hours, exposing to near-UV light (3×100 seconds) and post-baking at 95°C for ~5 hours.[27, 28] Patterned wafers were treated for 8 hours with (tridecafluoro-1,1,2,2-tetrahydrooctyl) trichlorosilane (Gelest Inc., Morrisville, PA) in a vacuum chamber.
The patterned wafers served as templates for replicating the microfluidic devices in polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning Corp, Midland, MI). Inlet and outlet ports were created by punching 4 mm holes through the PDMS replicas using biopsy punches (Acuderm Inc., Fort Lauderdale, FL). A ~1 mm thick slab of PDMS and the patterned surface of the PDMS replica of the device were exposed to air plasma (PDC-3xG, Harrick Plasma, Ithaca, NY) for 100 seconds then sealed together. The resulting devices were filled with 1% (w/v) aqueous solution of mPEG-silane (MW 5000, Laysan Bio Inc., Arab, AL) and let sit for at least 15 minutes before being flushed with GASP buffer (1.3 mM NaH2PO4, 9 mM Na2HPO4, 140 mM NaCl, 5.5 mM glucose, 1% bovine serum albumin; osmolality 290 mmol/kg; pH 7.4) in order to prevent unwanted RBC adhesion to the walls of the microchannels.[29–31] The combined cost of materials needed to fabricate a single PDMS device for our system was less than $0.25. The microfluidic devices were discarded after a single use.
Preparation of stored RBC samples
Six units of packed RBCs were purchased from regional blood centers (The Blood Center, New Orleans, LA; Gulf Coast Regional Blood Center, Houston, TX). RBC units were stored in a blood bank refrigerator (Helmer iB111 i.Series, Helmer Scientific, Noblesville, IN) at 2–6°C for 8 weeks. Prior to sampling, RBC units were removed from the refrigerator and placed on a rocking platform (model 100, VWR, West Chester, PA) for 15 minutes. 5mL RBC samples were withdrawn from each unit after 6, 7 and 8 weeks of storage, using the aseptic technique with a sterile syringe (BD, Franklin Lakes, NJ) through a sampling site coupler (Fenwal Inc., Lake Zurich, IL). Whole blood samples from healthy, consenting volunteers were collected by venipuncture into 4 mL plastic Vacutainer tubes spray-coated with K2EDTA (BD, Franklin Lakes, NJ). A hematology analyzer (Medonic M-Series, Boule Medical AB, Stockholm, Sweden) was used to verify the initial hematocrit and cell count of the blood samples. The samples were diluted to ~1% hematocrit with GASP buffer prior to introduction into the devices. 40μL aliquots of the diluted RBC samples were used for each experiment. Experiments were repeated six times for each unit at each time point.
Imaging and analysis
The imaging system consisted of an inverted microscope (IX71, Olympus America Inc., Center Valley, PA) and a digital camera (MC1362, Mikrotron GmbH, Germany). The microscope was manually focused on the microchannel at a magnification of 64x. The camera was programmed to acquire an image every 5 seconds for 30 minutes (100 fps, 9.617 ms exposure, global shutter). Image sequences were analyzed using a custom algorithm developed in MATLAB (The Math Works Inc., Natick, MA) for automated classification of individual stored RBC morphology in accordance with standard classification.[2, 9] The algorithm first standardized the brightness of captured grayscale images, then created a binary mask separating objects from the background via a combination of contrast stretching, erosion, dilation and thresholding. Partial cells, overlapping cells and image artifacts were removed from the binary mask, and a composite image of the brightness-adjusted grayscale image and the binary mask was then used to measure features of individual RBCs, and a binary decision tree was then used to assign RBCs into morphology categories based on these features.[32]
Results
Design of the microfluidic device
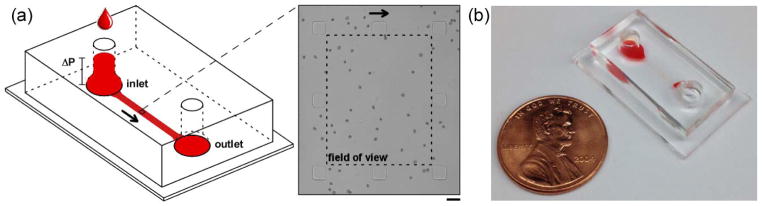
The design of the microfluidic device is illustrated schematically in Figure 2. The device was symmetrical, with two identical cylindrical through-holes serving as the inlet and the outlet ports. To prevent screening of cells based on their shape and size in the inlet, the through-holes lead into a smooth, curved slope allowing for a less abrupt transition into the microchannel.[28] The microfluidic part of the device (Fig. 2a) comprised a relatively wide (1.5 mm) and long (7.5 mm) but shallow (7 μm) channel connecting the inlet and the outlet of the device. The channel contained a sparse array of 30 μm × 30 μm square pillars oriented to conform to the field-of-view of the microscope (280 μm × 225μm) (Fig. 2a, inset). The pillars (i) prevented spontaneous collapse of the shallow microchannel, [33] (ii) simplified alignment of the field-of-view of the microscope with the direction of flow through the device, and (iii) provided a high-definition standard for microscope focusing to ensure consistency of images obtained from different devices.
Figure 2.
Schematic illustration of the microfluidic device used for acquiring non-overlapping images of single RBCs. (a) Loading a 40 μL sample of stored RBCs at 1% hematocrit into the inlet produced a pressure differential (ΔP) between the inlet and outlet sufficient to drive the sample through the device at the desired rate. Inset shows a 280 μm × 225 μm viewing area (dashed line) surrounded by eight 30 μm × 30 μm square pillars. Scale bar is 30 μm. (b) Photograph of the assembled microfluidic device filled with a sample of stored RBCs (one-cent U.S. coin shown for size reference).
To perform the measurement, 40 μL of diluted RBC sample was added to the inlet, the microscope was focused on the microchannel (such that the square pillars were outside the margins of the field-of-view) and image acquisition was initiated. The height of sample in the inlet created a sufficient pressure difference between the inlet and the outlet of the device to drive the flow of sample through the device. Because of the very small rate of flow through the device (~0.5 nL/s), the level of liquid in the inlet did not change significantly during the measurement and, consequently, the driving pressure difference remained relatively constant. The sample flow through the 7 μm microchannel transported thousands of individual RBCs through the field-of-view of the microscope in a single-cell layer, without significant cell flipping, overlap or evidence of morphology-based selection bias at the inlet. The image acquisition was timed with the rate of cells entering and exiting the field-of-view to prevent possible bias due to double-counting. The recorded images were processed off-line to classify the morphology of each individual RBC.
Automated classification of individual RBC morphology
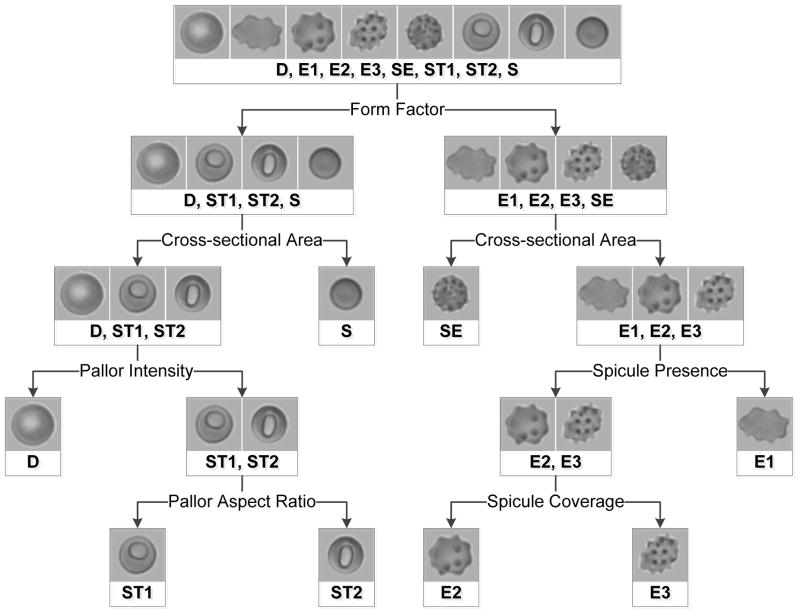
The uniform, high quality images acquired using the microfluidic system enabled consistent extraction of basic cellular features and complex subcellular features. Cell form factor, cross-sectional area, spicule presence, spicule coverage, pallor intensity and pallor aspect ratio were extracted from each isolated RBC image. Individual cells were automatically classified based on these features. The RBCs were categorized at two levels: into 8 specific (“high-resolution”) categories (D, discocyte; E1, echinocyte 1; E2, echinocyte 2; E3, echinocyte 3; SE, sphero-echinocyte; ST1, stomatocyte 1; ST2, stomatocyte 2; S, spherocyte) and into 4 broad (“low-resolution”) categories (D; E [E1, E2, E3, SE]; ST [ST1, ST2]; S) based on Bessis’s general classification.[2] Two levels of precision (high-resolution and low-resolution) were calculated in order to avoid limiting the utility of the system. Some applications (e.g. study of RBC deformability, fragility or morphology recovery) may require high-resolution classification because the number of spicules an echinocyte has is an indicator of the degree of membrane loss due to microvesiculation during storage and therefore of functionally relevant RBC surface area to volume ratio. However, other applications (e.g. microfluidic cell sorting) may only require distinctions between broad categories of RBCs (e.g. D, E and ST vs. S) in order to separate reversibly and irreversibly damaged RBCs.
Figure 3 shows the logic of the simple binary decision tree used to structure the RBC classification algorithm. The layout of the tree was modeled after the decision making process used for manual RBC classification. A multiparameter cluster analysis algorithm employed all six cellular features at each branch point to classify each individual RBC; weighting factors for each feature, and a threshold level with which to classify the result of their combination, at each branch point were determined via previously-described numerical methods.[32] The final categorization was determined by the terminal branch reached by each individual RBC. The RBC classification algorithm was capable of automatically classifying single RBCs at a rate of >10,000 cells per hour (CPU time). After the algorithm is initiated, it does not require user input, so the operator is free to perform other tasks while the algorithm is running – a significant improvement compared to manual classification which requires active user involvement. The cell classification algorithm was optimized using a manually-categorized training set of 1000 randomly-selected individual RBCs, taken from the entire set (n = 1,367,852) of images of single stored RBCs acquired using the system.
Figure 3.
Schematic illustration of the binary decision tree used by the automated RBC morphology classification algorithm. The feature specified at each bifurcation informed the structure of the tree. The multiparameter cluster analysis algorithm uses all six cellular features to sort RBCs at each bifurcation. Final morphology classification is determined by the terminal branch reached by each sorted cell.
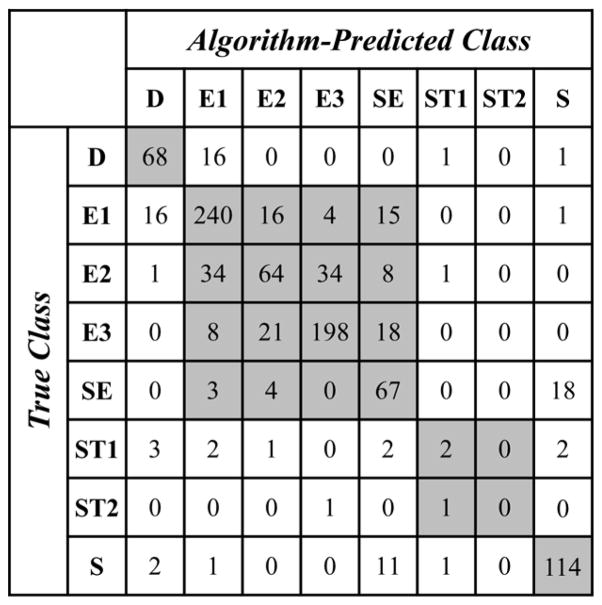
Figure 4 shows a confusion matrix (a method commonly used for evaluating the accuracy of automated classifications)[34, 35] for the morphology classifications based on the multi-parameter cluster analysis algorithm. To produce the confusion matrix, we selected a random set of images of 1000 single stored RBCs (test set) from the set of all images and classified the morphology of each individual RBC via visual examination (“True Class”) and by applying the automated classification (“Algorithm Predicted Class”). The overall accuracy was calculated as the quotient of the number of correctly classified RBCs (i.e. cells in which the true class was the same as algorithm predicted class) and the total number of analyzed RBCs. The overall accuracy of the RBC classification algorithm was 91.9% for low-resolution classification (D, E, ST, S) and 75.3% for high-resolution classification (D, E1, E2, E3, SE, ST1, ST2, S). The overall accuracy for high-resolution sub-classifications (E1, E2, E3, SE) of RBCs classified as echinocytes (E) at low-resolution was 77.5%. The overall accuracy for high-resolution sub-classifications (ST1, ST2) of RBCs classified as stomatocytes (ST) at low-resolution was 66.7%.
Figure 4.
Confusion matrix for the automated stored RBC morphology classification algorithm. Test set consisted of 1000 individual stored RBCs randomly selected from the entire set. Rows correspond to true RBC classes (categorized manually by eye) and columns correspond to algorithm-predicted RBC classes (categorized automatically by algorithm). Cells along the diagonal contain correct high-resolution morphology classifications, and all shaded cells contain correct low-resolution morphology classifications. The overall low-resolution classification accuracy of the algorithm is 91.9%, and the overall high-resolution classification accuracy of the algorithm is 75.3%.
To evaluate the reproducibility of the system, we performed 5 measurements of the same stored blood unit and compared the distributions of morphologies among all individual RBCs analyzed for each measurement. A total of 66,161 RBCs were used to evaluate reproducibility (n = 16,217 – measurement 1, n = 12,262 – measurement 2, n = 12,943 – measurement 3, n = 10,213 – measurement 4 and n = 14,526 – measurement 5). The means and standard deviations of the proportions of the samples composed of each morphology class are as follows: 3.50 ± 0.26 % (CV 7.5%) - D, 25.50 ± 3.40 % (CV 13.3%) - E1, 13.20 ± 1.05 % (CV 8.0%) - E2, 19.16 ± 4.10 % (CV 21.4%) - E3, 16.48 ± 0.65 % (CV 4.0%) - SE, 21.25 ± 1.07 % (CV 5.1%) - S, 0.90 ± 0.19 % (CV 21.2%) - ST1 and 0.01 ± 0.01 % (CV 122.9%) - ST2. The low standard deviation between experiments suggests that the system is highly reproducible. The coefficient of variation for ST2 is very high because very few RBCs with this morphology were present in this unit compared to all other morphologies.
Measurement of the diameter of individual RBCs with different morphologies
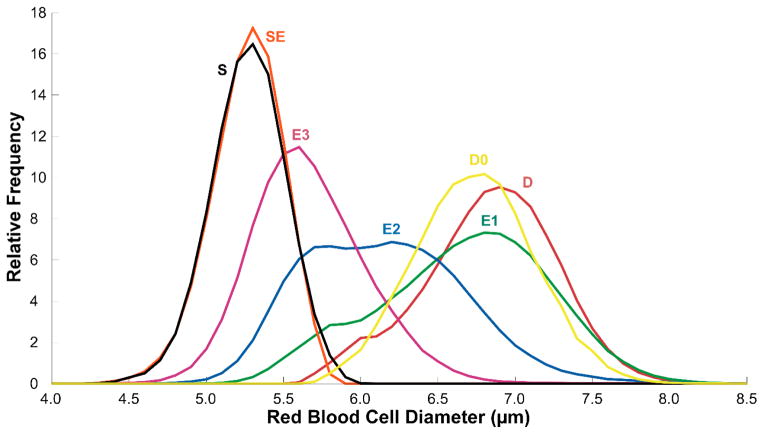
The data from all experiments (6 units × 3 weeks per unit × 6 experiments per week) was combined to form a single representative set of single stored RBCs (n = 1,367,852). We used our system to automatically classify the morphology of these individual stored RBCs and to determine the mean effective diameter (d = (4A/π)0.5, where A is the total cross-sectional area of the cell) of individual RBCs belonging to different stages of storage-induced RBC morphology deterioration. Figure 5 shows the distribution of the diameters of individual RBCs by morphology class for all stages of echinocytosis. A distribution of sizes for fresh RBCs (D0 in Fig. 5) is given for reference. With respect to discocytes from fresh blood samples, stored discocytes were slightly larger (D: 7.80 ± 0.49 μm vs. D0: 7.69 ± 0.43 μm), which is consistent with initial swelling of RBCs during storage, prior to loss of volume by vesiculation. There was significant overlap of the size distributions for individual stored RBCs in various stages of echinocytosis (Fig. 5), which is indicative of the practical difficulty with making a sharp distinction between echinocyte subtypes. Sphero-echinocytes and spherocytes had a virtually identical size distribution (SE: 6.01 ± 0.26 μm vs. S: 6.02 ± 0.27 μm). The mean diameters for all high-resolution and low-resolution morphology categories are summarized in Table 1.
Figure 5.
Distribution of individual stored RBC diameters by morphology class. Data presented as relative frequency polygons (bin size 0.1μm). Cells were categorized automatically by multiparameter cluster analysis. The distribution of a sample (unpaired) of fresh discocytes (D0) is shown for comparison. A total of 1,367,852 individual RBCs were analyzed.
Table 1.
Diameter data by morphology class. (a) Diameter mean and standard deviation data for high-resolution multiparameter cluster analysis classification. (b) Diameter mean and standard deviation data for low-resolution multiparameter cluster analysis classification.
| (a) | D0 (n = 22,985) | D (n = 124,229) | E1 (n =395,472) | E2 (n = 174,836) | E3 (n = 321,762) | SE (n = 162,648) | S (n = 154,570) | ST1 (n = 11,193) | ST2 (n = 157) |
| Diameter mean ± standard deviation (μm) | 7.69 ± 0.43 | 7.80 ± 0.49 | 7.61 ± 0.63 | 7.02 ±0.61 | 6.47 ± 0.42 | 6.01 ±0.26 | 6.02 ± 0.27 | 6.95 ± 0.61 | 7.32 ± 0.47 |
| (b) | D0 (n = 22,985) | D (n = 124,229) | E (n = 1,054,718) | S (n= 154,570) | ST (n= 11,350) | ||||
| Diameter mean ± standard deviation (μm) | 7.69 ± 0.43 | 7.80 ± 0.49 | 6.92 ± 0.80 | 6.02 ± 0.27 | 6.96 ±0.61 | ||||
Discussion
The majority of individual stored RBCs are well-preserved cells (discocytes or early stages of echinocytosis), which largely recover their properties post-transfusion and survive a normal lifespan in circulation [11, 36]. A relatively small fraction of individual stored RBCs, however, is damaged irreversibly, entering into the terminal stages of echinocytosis (sphero-echinocytes, spherocytes) and ultimately undergoing lysis either during storage or soon after transfusion. Conventional approaches to improving the quality of stored blood have focused on optimizing storage conditions (e.g. composition of the anticoagulant-preservative solution, [36] or reduction of oxygen in the storage bag[11]). Given the highly heterogeneous response of individual RBCs to hypothermic storage, however, further fine-tuning of the overall storage conditions may not produce a significant improvement in quality.[36] In this context, a more successful approach could be to simply remove the irreparably damaged cells from the unit before transfusion (e.g. using one of the methods recently developed in the field of high-throughput microfluidics [37–39]). To enable this approach, we need to quantify morphology, size and other properties that could potentially inform the design of microfluidic devices to be used for separating the populations of well-preserved and irreparably damaged cells. Existing manual methods for evaluating morphology of single stored RBCs severely limit the number of cells that can be analyzed and are impractical for making simultaneous measurements of morphology and other properties (e.g. size) for a meaningful number of cells with required precision. The simple, automated system we developed in this study enabled us to accomplish this task for >1,000,000 individual cells.
The most common conventional procedure for measuring the morphology of individual stored RBCs involves preparation of a blood smear, best-effort focusing of the microscope on the cells in the smear, a search for good working areas of correctly oriented cells by repeatedly moving the blood smear preparation in the field-of-view of the microscope (with a possible need for re-focusing), image acquisition, and visual classification of a few hundred individual RBCs (all normally manually performed by a skilled technician). These manual operations are very laborious, highly dependent on the technical skills and experience of the operator performing the measurement and are notoriously difficult to standardize between different laboratories. The primary advantage of our automated approach is its simplicity. To perform the RBC morphology measurement with our approach, an operator only needs to load the diluted RBC sample into the inlet of the microfluidic device, focus the microscope using the support pillars as a high-definition standard, and initiate the automated image acquisition and analysis processes. The microfluidic device completely eliminated the need for preparation of the blood smear and search for good working areas by creating a dynamic blood smear (Fig. 2) which allowed acquisition of numerous unique images of single optimally oriented, non-overlapping RBCs, without moving the blood smear preparation in the field-of-view of the microscope or re-focusing. The automated morphology analysis algorithm eliminated the need for subjective visual classification of individual RBCs. Our approach significantly reduced the time and user-input required to perform the morphological study and simultaneously increased the consistency and quality of the images. A secondary advantage of our approach is it’s relatively low cost, and lack of a need for specialized equipment – the disposable microfluidic devices are compatible with any sufficiently powerful bright field microscope equipped with a digital camera, which may already be in place in many laboratories. Because of these properties, our system could also be useful for studying the impact of different storage conditions, naturally occurring inter-donor variation and various rejuvenation strategies on the morphology of stored RBCs.
The approach to morphometric image analysis described in this paper represents a significant departure from conventional paradigms that emphasize development of ever more complex image analysis algorithms capable of compensating for images with inconsistent quality instead of improving the consistency and quality of images. Recent efforts in developing new analytical methods attempt to counterbalance the physical limitations of poor sample preparation by circumventing the issue of poor image quality through additional layers of complex code. Several clever methods (e.g. the pulse-coupled neural network approach) have been developed that can efficiently de-noise and segment blood cell images, selectively eliminating objects that may confound subsequent analysis steps, and extract specific cells from the image background.[40, 41] However, because these improved methods do not resolve the inherent problem of poor quality images, complex additional codes are required each time new features need to be extracted from the same images, severely limiting the versatility of these systems. The approach implemented in this paper utilizes automated sample preparation and imaging to produce standardized RBC images with consistently-high quality (minimal cell clumping, overlapping and non-standard orientation), thereby eliminating the need for complex image analysis techniques.
By resolving the problem of poor image quality, our system significantly reduced the complexity of the code required for cell feature extraction and measurement, creating a versatile, easily adaptable platform for high-throughput morphometric analysis of single cells. Here, paired stored RBC morphology and diameter data was chosen as a useful demonstration of the capabilities of the system, however the system is capable of extracting other complex cellular and subcellular features (e.g. number of spicules, form factor or central pallor aspect ratio) with single cell resolution. The image analysis algorithm could easily be adapted for use in quality control procedures (e.g. after blood collection and processing) or for the automated detection of RBC pathologies (e.g. sickle cell disease). As such, this simple, versatile, low-cost system has applications in both clinical and research environments.
The overall accuracy of the automated RBC classification algorithm was 91.9% for low-resolution and 75.3% for high-resolution. This compares favorably with the general accuracy of automated image analysis methods throughout other areas, particularly for the low-resolution classification.[34, 42] Interestingly, the majority of incorrectly categorized cells (high-resolution) in the test set were categorized as one of the neighboring morphologies along the transition from discocyte to spherocyte (e.g. E1 identified as D or E2). These ‘off-by-one’ errors likely occurred because the transition from discocyte to spherocyte is a continuous spectrum spanning a range of intermediate forms between these more distinct morphological endpoints. Even experienced technicians have to make subjective judgments regarding how to classify each individual cell, and thus could potentially classify the same cell differently. Manual classification of RBCs, which in itself is not perfectly objective, was used to define ‘true’ classes in the training and test sets. The observed deviation of the diameter distributions from normal distributions for some of the morphological classes (particularly E2) likely reflects these classification inaccuracies.
In summary, we developed an easy-to-use system for automated and high-throughput measurement of the morphology of individual stored RBCs by combining a simple microfluidic device and a morphometric image analysis. Our system completely eliminated the need for laborious manual preparation and visual examination of blood smears, and thus enabled classification of the morphology of more than one million individual cells, the largest dataset of stored RBC morphology data known to date. Using these data, we were able to quantify the distribution of cell diameter across a heterogeneous population of single stored RBCs in various stages of morphological deterioration. This information will be useful for developing technologies aimed at eliminating the irreparably damaged cells (particularly spherocytes and sphero-echinocytes) from stored blood units before transfusion. The ability to evaluate the morphology and other properties of individual RBCs automatically, rapidly and in statistically significant numbers could also improve our ability to assess the quality of stored blood in clinical and research settings.
Acknowledgments
Funding: This work was supported in part by a 2012 NIH Director’s Transformative Research Award (NHLBI R01HL117329, PI: SSS).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data and described custom-written scripts are available from the authors upon reasonable request.
Author contributions: Shevkoplyas, Piety and Yang conceived and designed the study. Piety performed the experiments and collected the data. Shevkoplyas and Piety analyzed and interpreted the data. Piety and Gifford developed the image analysis algorithm. Shevkoplyas obtained funding and supervised the study. Shevkoplyas and Piety wrote the manuscript. All authors critically revised the manuscript and approved the final submitted manuscript.
References
- 1.Evans EA. New membrane concept applied to the analysis of fluid shear- and micropipette-deformed red blood cells. Biophys J. 1973;13:941–54. doi: 10.1016/S0006-3495(73)86036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessis M. Red cell shapes. An illustrated classification and its rationale. Nouvelle revue francaise d’hematologie. 1972;12:721–45. [PubMed] [Google Scholar]
- 3.Fischer TM, Haest CW, Stohr-Liesen M, et al. The stress-free shape of the red blood cell membrane. Biophys J. 1981;34:409–22. doi: 10.1016/S0006-3495(81)84859-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gifford SC, Derganc J, Shevkoplyas SS, et al. A detailed study of time-dependent changes in human red blood cells: from reticulocyte maturation to erythrocyte senescence. Br J Haematol. 2006;135:395–404. doi: 10.1111/j.1365-2141.2006.06279.x. [DOI] [PubMed] [Google Scholar]
- 5.Deplaine G, Safeukui I, Jeddi F, et al. The sensing of poorly deformable red blood cells by the human spleen can be mimicked in vitro. Blood. 2011;117:e88–95. doi: 10.1182/blood-2010-10-312801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenwalt TJ. The how and why of exocytic vesicles. Transfusion. 2006;46:143–52. doi: 10.1111/j.1537-2995.2006.00692.x. [DOI] [PubMed] [Google Scholar]
- 7.Hess JR. Red cell changes during storage. Transfus Apher Sci. 2010;43:51–9. doi: 10.1016/j.transci.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Zimrin AB, Hess JR. Current issues relating to the transfusion of stored red blood cells. Vox Sang. 2009;96:93–103. doi: 10.1111/j.1423-0410.2008.01117.x. [DOI] [PubMed] [Google Scholar]
- 9.Berezina TL, Zaets SB, Morgan C, et al. Influence of storage on red blood cell rheological properties. J Surg Res. 2002;102:6–12. doi: 10.1006/jsre.2001.6306. [DOI] [PubMed] [Google Scholar]
- 10.Park Y, Best CA, Badizadegan K, et al. Measurement of red blood cell mechanics during morphological changes. Proc Natl Acad Sci U S A. 2010;107:6731–6. doi: 10.1073/pnas.0909533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida T, Shevkoplyas SS. Anaerobic storage of red blood cells. Blood Transfus-Italy. 2010;8:220–36. doi: 10.2450/2010.0022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagaprasad V, Singh M. Sequential analysis of the influence of blood storage on aggregation, deformability and shape parameters of erythrocytes. Clin Hemorheol Microcirc. 1998;18:273–84. [PubMed] [Google Scholar]
- 13.Chin-Yee I, Arya N, d’Almeida MS. The red cell storage lesion and its implication for transfusion. Transfus Sci. 1997;18:447–58. doi: 10.1016/S0955-3886(97)00043-X. [DOI] [PubMed] [Google Scholar]
- 14.Cooper RA, Jandl JH. The role of membrane lipids in the survival of red cells in hereditary spherocytosis. J Clin Invest. 1969;48:736–44. doi: 10.1172/JCI106031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosman GJ, Werre JM, Willekens FL, et al. Erythrocyte ageing in vivo and in vitro: structural aspects and implications for transfusion. Transfus Med. 2008;18:335–47. doi: 10.1111/j.1365-3148.2008.00892.x. [DOI] [PubMed] [Google Scholar]
- 16.Hod EA, Spitalnik SL. Harmful effects of transfusion of older stored red blood cells: iron and inflammation. Transfusion. 2011;51:881–5. doi: 10.1111/j.1537-2995.2011.03096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hod EA, Brittenham GM, Billote GB, et al. Transfusion of human volunteers with older, stored red blood cells produces extravascular hemolysis and circulating non-transferrin-bound iron. Blood. 2011;118:6675–82. doi: 10.1182/blood-2011-08-371849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albertini MC, Teodori L, Piatti E, et al. Automated analysis of morphometric parameters for accurate definition of erythrocyte cell shape. Cytometry A. 2003;52:12–8. doi: 10.1002/cyto.a.10019. [DOI] [PubMed] [Google Scholar]
- 19.Berezina TL, Zaets SB, Machiedo GW. Alterations of red blood cell shape in patients with severe trauma. J Trauma. 2004;57:82–7. doi: 10.1097/01.ta.0000135492.24111.24. [DOI] [PubMed] [Google Scholar]
- 20.Hogman CF, Lof H, Meryman HT. Storage of red blood cells with improved maintenance of 2,3-bisphosphoglycerate. Transfusion. 2006;46:1543–52. doi: 10.1111/j.1537-2995.2006.00893.x. [DOI] [PubMed] [Google Scholar]
- 21.de Korte D, Kleine M, Korsten HG, et al. Prolonged maintenance of 2,3-diphosphoglycerate acid and adenosine triphosphate in red blood cells during storage. Transfusion. 2008;48:1081–9. doi: 10.1111/j.1537-2995.2008.01689.x. [DOI] [PubMed] [Google Scholar]
- 22.Blasi B, D’Alessandro A, Ramundo N, et al. Red blood cell storage and cell morphology. Transfus Med. 2012;22:90–6. doi: 10.1111/j.1365-3148.2012.01139.x. [DOI] [PubMed] [Google Scholar]
- 23.Pierre RV. Red cell morphology and the peripheral blood film. Clin Lab Med. 2002;22:25–61. v–vi. doi: 10.1016/s0272-2712(03)00066-0. [DOI] [PubMed] [Google Scholar]
- 24.Xiong W, Ong SH, Lim JH, et al. Automatic area classification in peripheral blood smears. IEEE Trans Biomed Eng. 2010;57:1982–90. doi: 10.1109/TBME.2010.2043841. [DOI] [PubMed] [Google Scholar]
- 25.Anggraini D, Nugroho AS, Pratama C, et al. Automated status identification of microscopic images obtained from malaria thin blood smears. Electrical Engineering and Informatics (ICEEI), 2011 International Conference on; 2011; pp. 1–6. [Google Scholar]
- 26.Whitesides GM, Ostuni E, Takayama S, et al. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–73. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 27.Forouzan O, Burns JM, Robichaux JL, et al. Passive recruitment of circulating leukocytes into capillary sprouts from existing capillaries in a microfluidic system. Lab on a Chip. 2011;11:1924–32. doi: 10.1039/c0lc00547a. [DOI] [PubMed] [Google Scholar]
- 28.Burns JM, Yang X, Forouzan O, et al. Artificial microvascular network: a new tool for measuring rheologic properties of stored red blood cells. Transfusion. 2012;52:1010–23. doi: 10.1111/j.1537-2995.2011.03418.x. [DOI] [PubMed] [Google Scholar]
- 29.Shevkoplyas SS, Gifford SC, Yoshida T, et al. Prototype of an in vitro model of the microcirculation. Microvasc Res. 2003;65:132–6. doi: 10.1016/s0026-2862(02)00034-1. [DOI] [PubMed] [Google Scholar]
- 30.Shevkoplyas SS, Yoshida T, Munn LL, et al. Biomimetic autoseparation of leukocytes from whole blood in a microfluidic device. Anal Chem. 2005;77:933–7. doi: 10.1021/ac049037i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shevkoplyas SS, Yoshida T, Gifford SC, et al. Direct measurement of the impact of impaired erythrocyte deformability on microvascular network perfusion in a microfluidic device. Lab Chip. 2006;6:914–20. doi: 10.1039/b601554a. [DOI] [PubMed] [Google Scholar]
- 32.Gifford S. Methods of multivariate data cluster separation and visualization. 8,423,596 B2. Office USPaT; United States Patent No. US. 2013 Apr 16;
- 33.Decre MM, Timmermans PH, van der Sluis O, et al. Numerical and experimental study of critical roof collapse conditions in soft lithography. Langmuir. 2005;21:7971–8. doi: 10.1021/la050709p. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Osuna-Highley E, Hua J, et al. Automated analysis of protein subcellular location in time series images. Bioinformatics. 2010;26:1630–6. doi: 10.1093/bioinformatics/btq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sammut C, Webb GI. Encyclopedia of Machine Learning. New York: Springer; 2011. [Google Scholar]
- 36.Hess JR. Measures of stored red blood cell quality. Vox Sang. 2014;107:1–9. doi: 10.1111/vox.12130. [DOI] [PubMed] [Google Scholar]
- 37.Gifford SC, Spillane AM, Vignes SM, et al. Controlled incremental filtration: a simplified approach to design and fabrication of high-throughput microfluidic devices for selective enrichment of particles. Lab Chip. 2014;14:4496–505. doi: 10.1039/c4lc00785a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis JA, Inglis DW, Morton KJ, et al. Deterministic hydrodynamics: taking blood apart. Proc Natl Acad Sci U S A. 2006;103:14779–84. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S, Hou HW, Kanias T, et al. Towards microfluidic-based depletion of stiff and fragile human red cells that accumulate during blood storage. Lab on a Chip. 2015;15:448–58. doi: 10.1039/c4lc00768a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Ruberto C, Dempster A, Khan S, et al. Analysis of infected blood cell images using morphological operators. Image Vision Comput. 2002;20:133–46. [Google Scholar]
- 41.Angulo J, Klossa J, Flandrin G. Ontology-based lymphocyte population description using mathematical morphology on colour blood images. Cell Mol Biol (Noisy-le-grand) 2006;52:2–15. [PubMed] [Google Scholar]
- 42.Huh S, Lee D, Murphy RF. Efficient framework for automated classification of subcellular patterns in budding yeast. Cytometry A. 2009;75:934–40. doi: 10.1002/cyto.a.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]