Abstract
In the marine environment, settling particulates have been widely studied for their role as effective vertical transporters of nutrients and metals scavenged from the euphotic zone to the benthos. These particulates are composed of transparent exopolymers, plankton and bacterial cells, detritus and organic matter, and form various size fractions from colloids (<0.2μm) to aggregates, and finally marine snow (>300 μm). As marine snow forms in the water column, anoxic layers form around and within the aggregation potentially creating a prime environment for the methylation of mercury (Hg), which occurs primarily in low oxygen environments. To examine this process, marine aggregates were produced from sieved estuarine seawater (100 μm) in 1-L glass bottles spiked with stable isotope enriched methylmercury (CH3199Hg) and inorganic mercury (200Hg(II)) at 18° C using a roller-table. After the rolling period, different particle-size fractions were collected and analyzed, including: visible marine snow (>300μm), particulates 8 to 300 μm, and particulates 0.2 to 8μm. Particulate analysis indicated higher incorporation of both forms of Hg into marine snow compared to unrolled treatments, with greater incorporation of 200Hg(II) than CH3199Hg. In addition, inorganic Hg was methylated and CH3Hg was demethylated in the larger particulate fractions (>8μm). Methylation and demethylation rates were assessed based on changes in isotopic composition of Hg(II) and CH3Hg, and found to be comparable to methylation rates found in sediments. These results indicate that net Hg methylation can occur in marine snow and smaller aggregates in oxic coastal waters, and that this net formation of CH3Hg may be an important source of CH3Hg in both coastal and open ocean surface environments.
Keywords: Marine snow, Mercury, Methylation Rates, Methylmercury, Organic matter, Partitioning, Fractionation
Introduction
The origin of methylmercury (CH3Hg) in the open ocean has been a topic of deliberation for years (Fitzgerald et al., 2007, Mason et al., 2012; Mason and Fitzgerald, 1990; Ullrich et al., 2001). Many researchers debate whether or not methylation could be occurring in the water column due to abiotic photochemical processes (Ullrich et al., 2001), degradation of detritus in low oxygen or sub-thermocline waters (Mason and Fitzgerald, 1993; Sunderland et al., 2009), or release from phytoplankton cells as they die and settle through the water column (Heimbürger et al., 2010). It has been suggested that Hg(II) is methylated within the mixed layer of the ocean (Hammerschmidt and Bowman, 2012) and methylation has been documented within the upper water column of the Arctic Ocean (Lehnherr et al., 2011). Current consensus is that most of the CH3Hg measured in open ocean surface waters is formed within subsurface waters and vertically mixed from deeper waters (Mason et al., 2012; Mason and Fitzgerald, 1993; Sunderland et al., 2009), given that mostly anaerobic bacteria methylate Hg (Gilmour et al., 2013; Parks et al., 2013). For coastal environments it has been concluded that methylation is mostly occurring in sediments or that CH3Hg is being transported from watersheds (Balcom et al., 2008; Hollweg et al., 2009; Chen et al., 2012; Sunderland et al., 2012; 2010). There is little evidence for in situ water-column production of CH3Hg in estuaries. Large settling particulates in the water column, however, could provide an ideal environment for mercury methylation because of the presence of anoxic “microzones” (Alldredge and Cohen, 1987), and thereby they could be the key to understanding in situ water-column production of CH3Hg. Determining a mechanism for the production of CH3Hg in the water column would assist in understanding the sources of CH3Hg to coastal food chains, and aid in closing the Hg budget in coastal and ocean models.
The current study examined the incorporation and potential methylation and demethylation of Hg and CH3Hg within marine snow and on smaller particles. Particle aggregations in the oceans’ water column have been referred to as flocs or marine aggregates, with the largest of these aggregations designated as marine snow (>300μm; Alldredge et al., 1993; Alldredge and Silver, 1988; Kach and Ward, 2008; Kiørboe et al., 1990). These heterogeneous particulates are composed of transparent exopolymer particles (TEP), living organisms (both prokaryotes and eukaryotes) and their detritus, inorganic particles, fecal pellets, and organic matter (humic and fulvic acids). As particles settle through the water column they coagulate as a result of collisions caused by Brownian motion, physical shear, and differential settling (Alldredge et al., 1993; Alldredge and Silver, 1988; Heinonen et al., 2007; Jackson and Burd, 1998; Kiørboe et al., 1990; Li et al., 2008; de la Rocha, 2006). Settling macroscopic particulates (particles 3–500+ μm) are important not only to the cycling of nutrients from the euphotic to the benthic zones, but also to the transport of trace metals (Alldredge and Silver, 1988; de la Rocha, 2006; Simon et al., 2002).
The chemical environment of marine snow is still unresolved. Some research, however, has demonstrated that particle aggregation can create microzones with chemical characteristics that markedly differ from that of the surrounding water column. Alldredge and Cohen (1987) were the first to present evidence of oxygen and pH gradients that occur within a 1-mm diameter from the outside to the inside of marine snow. Oxygen concentrations measured at the surface of marine snow were higher than the ambient seawater indicating photosynthetic activity. Once inside the marine snow aggregation, oxygen concentrations sharply decreased suggesting that anaerobic conditions can be maintained within the aggregation (Alldredge and Cohen, 1987). Alldredge and Silver (1988) suggested that as marine snow forms, organic-binding substances and biologically labile gasses can become entrained in the growing aggregation. The possible combination of anaerobic conditions with organic-binding substances and biologically available gasses could create suitable oxygen-free conditions for Hg methylation and CH3Hg degradation within marine snow. As the biotic methylation pathways are entirely due to anaerobic bacteria (Gilmour et al., 2013), a necessary condition for methylation within oxic surface waters is reduced oxygen microenvironments, which can be found in marine snow. Additionally, since Hg forms are highly particle reactive, it can be assumed that CH3Hg and Hg(II) will be readily incorporated into marine snow as it forms and settles through the water column, although this possibility has not been examined (Fitzgerald et al., 2007; Kim et al., 2008; Lawrence and Mason, 2001).
Based on the known interactions of Hg and CH3Hg with organic matter and surfaces, we hypothesized that incorporation would be greatest in the marine snow compared to smaller particles, particle aggregations and microorganisms. In addition, we hypothesized that the formation of marine snow would not only enhance the methylation of Hg(II), but also result in enhanced degradation of CH3Hg compared to environments without marine snow. To examine these hypotheses we incubated coastal waters, spiked with stable isotopes of CH3Hg and Hg(II) (CH3199Hg and 200Hg(II)), under conditions conducive to marine snow formation. Rates of incorporation and transformation of the stable isotopes were compared with control samples to determine the extent to which marine snow formation enhanced removal of CH3Hg and Hg from the waters, and whether it resulted in increased methylation or demethylation.
Materials and Methods
Experimental Design
The materials for this experiment consisted of trace-metal clean 1-L jars (Qorpak wide-mouth, borosilicate glass), 15-mL polyethylene Falcon tubes, pre-weighed 0.2- and 8-μm polycarbonate filters, 25-mm frit filter, graduated filter towers, pre-ashed quartz fiber (QF/F) and glass fiber (GF/F) filters,, and 250-mL ICHEM bottles.
Seawater was collected from the intertidal region surrounding the University of Connecticut’sAvery Point campus (41° 18.954″ N, 72° 03.588″ W). Separate water samples were collected on two consecutive days and passed through a 100-μm mesh to remove large particulates, including zooplankton. Water collected on each day was considered a separate experiment and is hereafter referred to as either experiment 1 or experiment 2. The sieved seawater was characterized by dissolved organic carbon (DOC), and total suspended solids (TSS) analysis. The pH, temperature, and salinity were also measured. Then the water was poured into clean 1-L jars and spiked with CH3199Hg and 200Hg(II) aqueous standards to a final concentration of 0.124 nM. Marine snow was generated using the roller-table method described by Shanks and Edmondson (1989). Several control jars were also prepared. First, to control for the production of marine snow, some jars were spiked with Hg and then placed on a stationary surface next to the roller table (i.e., hereafter referred to as unrolled). Second, to control for the addition of Hg, six replicate jars were prepared but not spiked with CH3199Hg or 200Hg(II) For each of the two experiments, six rolled, spiked jars and six unrolled, spiked jars were prepared. All of the jars (rolled and unrolled) were maintained in an environmental chamber at 18° C under a 12h:12h light:dark cycle to represent field conditions at the time of sampling. Preliminary research indicated that the highest incorporation of Hg into marine snow was reached at 4 days (Ortiz, 2014), consequently the experiments were terminated after this period of time.
Sample Collection
Jars were removed from the roller table and allowed to settle for 15 minutes. Given that the 1-L jars were 18cm in height, a 15 minute period would allow marine snow larger than ca. 300 μm to settle to the bottom (Hill et al., 2000; Hill, 1998). After settling, visible marine snow was collected using a clean disposable pipet. Unrolled jars were removed from the stationary table and gently inverted three times and allowed to settle for 15 minutes. After settling, 5 mL of water was randomly sampled from the bottom of the unrolled jars such that the clean pipet did not come in direct contact with the bottom. The remaining liter of water was then filtered through an 8-μm polycarbonate filter. A fraction of the 8-μm filtrate was then passed through a 0.2-μm polycarbonate filter until the filter clogged to ensure an even coating of particles, which typically occurred after filtering 20 to 75 mL of water. The remaining 8-μm filtrate (i.e., water that could not pass through the 0.2-μm filter) was passed through a pre-ashed QF/F filter and weighed for TSS. All filters were rinsed with an ammonium formate (NH4COOH) solution (prepared to the same concentration (w/v) as the experiment seawater). This isotonic, slightly volatile solution was used to remove salts from the marine snow to yield a more accurate dry weight. The same method was used for the material collected from the unrolled jars. The final water filtrate was collected and preserved with hydrochloric acid (0.5% v/v Fisher Trace Metal Grade) in clean 250-mL ICHEM bottles for 200Hg(II) (and other Hg(II) isotope analysis) and 500-mL ICHEM bottles for CH3199Hg (and other CH3Hg isotopes) ICPMS analysis, as discussed further below. All preserved water samples were stored at 4°C in the dark. A sub-sample of the final filtrate was saved for DOC analysis. All polycarbonate filters were frozen then freeze dried for 72 hrs.
Dried filters were weighed, placed in polyethylene falcon tubes containing 7-mL of 4.57M HNO3 (Fisher Trace Metal Grade), and digested in a 60° C water bath for 12 h before analysis. Aliquots of the final digest were used for both CH3199Hg and 200Hg(II) analysis. Both forms of mercury were analyzed using a Perkin-Elmer DRC II ICPMS. Digested filters were analyzed for CH3199Hg and other CH3Hg isotopes using CVAFS purge-trap techniques coupled with the ICPMS, as detailed below. Water CH3199Hg was analyzed using the Tekran 2700 methylmercury autoanalyzer coupled with the ICPMS. Filters and water samples were analyzed for 200Hg(II) using a Perkin Elmer-FIAS coupled with the ICPMS.
The following isotopes were quantified and used in the analysis of the incorporation and in the estimations of methylation and demethylation potential: 1) for methylmercury, CH3199Hg was used to estimate incorporation, CH3202Hg was used to estimate natural levels of all isotopes, and CH3200Hg was used to estimate methylation of the added 200Hg spike; and 2) for inorganic Hg, 200Hg(II) was used to estimate incorporation, 202Hg(II) was used to estimate natural levels of all isotopes, and 199Hg(II) and CH3199Hg were used to estimate demethylation of the added CH3199Hg. Calculations of the excess amounts of the isotopes of CH3Hg and Hg compared to that of natural abundances were estimated using the approach of Hintelmann and Evans (1997), taking into account the relative isotope abundances in the spikes which were respectively 91.95% CH3199Hg and 96.41% 200Hg. The 199CH3Hg standard was certified to contain 91.95% 199CH3Hg, 0.73% 202CH3Hg, and 0.45% 200CH3Hg (Oak Ridge National Laboratory). The 200Hg(II) standard was certified to contain 96.41% 200Hg(II), 1.46% 201Hg(II), 0.99% 199Hg(II), and 0.91% 202Hg(II). The internal 201Hg(II) standard used for isotope dilution analysis of Hg(II) was certified to contain 98.11% 201Hg(II), 1.18% 202Hg(II), 0.45% 200Hg(II), and 0.1% 199Hg(II). From here on, the various isotope that is being quantified will be indicated as CH3xxxHg and yyyHg.
Sample Preparation and Analysis
Digested filters were analyzed for CH3xxxHg by spiking 0.5 to 1mL of digest into 100-mL reagent-grade water (Millipore A10 System) in a glass-purge vessel. The solution was adjusted to a pH of ~7 with 10N KOH. Finally, the solution was buffered to a pH of ~4.9 with 4M acetate buffer and ethylated with sodium tetraethylborate (NaTEB). The final solution was allowed to react for 10 minutes prior to being purged with ultra-high purity argon gas at a flow rate of 100mL min−1 for 12 minutes onto TENAX© traps that were subsequently burned onto a GC column (Bloom, 1989; Hammerschmidt and Fitzgerald, 2006; Hintelmann and Evans, 1997; Tseng et al, 2004; ). The GC gas effluent was connected to the ICPMS. Peaks were integrated and recorded using Perkin-Elmer Chromera software. Water samples were analyzed for CH3xxxHg by digesting 150mL of sample in acid-clean ICHEM bottles with 50% (v/v) H2SO4 at room temperature for at least 12 h. The digested water was adjusted to a pH of ~7 with 10M KOH and then buffered to a pH of 4.9 with 4M acetate buffer. Ascorbic acid (2% w/v) was added to the solution prior to the addition of NaTEB (Munson et al., 2014). The bottle was capped, and the final solution was allowed to react for 30 minutes prior to analyzing on the Tekran Methylmercury Autoanalyzer-ICPMS.
Digests were analyzed for yyyHg(II) by transferring a 3-mL aliquot of the original 7-mL particulate digest, into clean tubes. The aliquots were diluted up to 12 mL with reagent-grade water, oxidized with BrCl, and digested in a 60° C bath for at least 12 h prior to analysis. On the day of analysis the oxidized digest was spiked with a 201Hg(II) internal standard (final concentration of 0.5nM) and hydroxylamine (equivalent in volume to BrCl) and analyzed using the FIAS-ICPMS (Bloom and Crecelius, 1983; Hammerschmidt and Fitzgerald, 2006; Hammerschmidt and Fitzgerald, 2005; US EPA 1630). The water filtrates in the 250-mL ICHEM jars were oxidized with BrCl and digested at room temperature for at least 12 h prior to 200Hg(II) analysis. The oxidized water sample was again spiked with internal 201HgT (final concentration of 0.5nM) standard and hydroxylamine just prior to FIAS analysis.
Calculations
Incorporation
Incorporation percentages (%I) and log Kd values were calculated for each particle fraction, using equations 1 and 2, respectively:
| (1) |
In equation1, [Hg]part represents the concentration of mercury (in nmol) in the particulates and [Hg]water represents the concentration of Hg in the water after filtration. A similar equation was used for CH3Hg.
| (2) |
In equation 2, [Hg]part and [Hg]water represent the same parameters as in equation 1 and “mass” is the suspended mass collected.
Methylation/Demethylation
Concentrations of 200Hg(II) and CH3200Hg isotopes were calculated using the isotope dilution approach of Hintelmann and Evans (1997) in which the natural abundances of all the Hg isotopes (199Hg, 200Hg, and 201Hg) were calculated based on the amounts of 202Hg and CH3202Hg (the most abundant, ambient stable mercury isotope). These calculated abundances were then used to determine the excess concentration of each isotope. Excess CH3200Hg was indicative of methylation of the 200Hg(II) spike. Likewise, demethylation can be calculated using the ratio of excess CH3199Hg to 199Hg(II). Methylation and demethylation rate constants were calculated using equation 3 for methylation (km) and equation 4 for demethylation (kdm), respectively. In equation 3, the methylation rate is based on the formation of the product (i.e. CH3200Hg). Therefore, the ratio of CH3200Hg to 200Hg(II) multiplied by the inverse of the incubation time in days results in the estimated methylation rate. In equation 4, the estimated demethylation rate is based on the decrease in the reactant (CH3199Hg). Equation 4 uses the inverse of the time in days multiplied by the natural log of the ratio of CH3199Hg to 199Hg(II). The different formulas account for the fact that for methylation, the product is being measured (CH3200Hg) while for the demethylation, the change in the reactant is being measured.
| (3) |
| (4) |
QA/QC
Calibration curves of at least r2=0.99 were achieved daily prior to continuing analysis. Given that no true corresponding standard reference material (SRM) for marine snow exists, no SRM could be used to check accuracy for CH3199Hg marine snow analysis. Instrument precision and accuracy was monitored using analytical duplicates of filter digests, laboratory reagent blanks, and standard checks analyzed every 10 samples for CH3Hg analysis. Since the filter digests needed to be analyzed for both forms of Hg, matrix spikes were not an option, but matrix spikes were used in water sample analysis. The precision of the instrument estimated in relative percent difference (RPD) averaged 14 ± 8% (n=12). Duplicates greater than 10% RPD were re-analyzed, but re-analysis was limited by sample volume, therefore replications closest to 10% RPD were accepted for quality assurance. The accuracy of the instrument was estimated from the replication of the standard checks. Standard checks that were >15% different from the expected concentration were re-analyzed. Matrix CH3Hg spikes in the water analysis yielded an average recovery of 84 ± 6%. The recovery value falls within the acceptable 71 to 125% range detailed in the EPA Method 1630.
Similar to CH3xxxHg analysis, yyyHg(II) FIAS analysis quality control parameters consisted of analyzing laboratory reagent blanks, standard checks, and analytical duplicates every 10 samples, and matrix spikes (201Hg internal standard) in each sample. In contrast to CH3xxxHg analysis, MESS-3 marine sediment certified by the National Research Council of Canada (0.45 ± 0.05 nmol g−1) was used as the SRM for FIAS analysis. MESS-3 samples averaged 0.42±0.03 nmol g−1, within the certified range of the SRM. On average, RPD values were <10% for all analytical duplicates. LCS standard checks averaged 83.7± 6% recovery over all analytical days with error propagated over time. Matrix spike recoveries averaged 109.9 ± 22.6% from the digested filters and 105.1 ± 6.4% from the waters. The measured method detection limit (MDL) for CH3199Hg analysis was 1.27nmol g−1 and for 200Hg(II) analysis was 3.11 nmol g−1. This translated into a CH3199Hg incorporation MDL of ~1% and a 200Hg(II) incorporation MDL of ~4%.
Statistical Analysis
All data were log transformed (i.e., log10(x+1)) prior to analyses to improve normality and homoscedasticity. Transformed data were analyzed using a two-way analysis of variance procedure (ANOVA, general linear model) followed by a Tukey’s post-hoc test to compare levels of the independent variables (SYSTAT 13). Single sample t-tests (Bonferroni-adjusted) were used to determine if methylation and demethylation rate data were significantly different than zero. For all statistical tests, an alpha level of 0.05 was used.
Results and Discussion
Particulate Aggregation
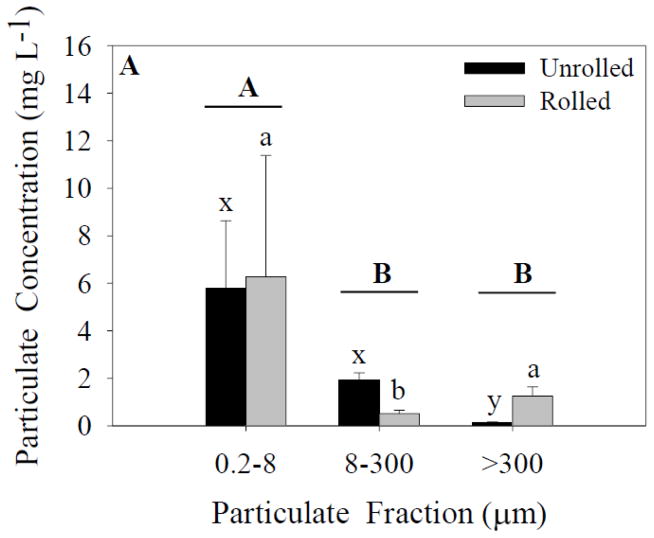
The mass of particulate matter in the water (TSS) collected in experiment 1 was 50% higher than that collected in experiment 2 (Table 1). The DOC concentration in water collected in experiment 2 was about 20% higher than that collected in experiment 1 (Table 2). In experiment 1, there was a significant effect of particle-size fraction, but not treatment on the concentration of particles in the jars after four days of rolling (ANOVA, Figure 1A). There was also a significant interaction effect found between the two independent variables. These results demonstrate that in experiment 1 rolling did not substantially increase the concentration of large aggregations or marine snow. Such an outcome suggests that the concentration of transparent exopolymer particles (TEP) or other organic matter that could enhance aggregation was lower in the water collected on day 1. The difference in particle concentrations with size demonstrates that the highest concentration of particles remained in the smallest size fraction.
Table 1.
Particulate masses filtered from 1-L jars. Values are on a per volume basis.
| Experiment 1 | Experiment 2 | ||||
|---|---|---|---|---|---|
| Fraction (μm) | Treatment | Particulate Mass (mg L-1)% Total Mass |
Particulate Mass (mg L-1)% Total Mass |
||
| TSS | Experiment Start | 14.9 ± 2.97 | NA | 9.90 ± 0.445 | NA |
| 0.2–8 | Unrolled | 5.78 ± 2.86 | 38.9 ± 19.2 | 4.65 ± 2.46 | 46.9 ± 24.9 |
| Rolled | 6.27 ± 5.10 | 42.2 ± 34.3 | 6.91 ± 1.56 | 69.8 ± 15.8 | |
| 8–300 | Unrolled | 1.91 ± 0.308 | 12.9 ± 2.07 | 2.53 ± 0.187 | 25.5 ± 1.89 |
| Rolled | 0.511 ± 0.13 | 93.43 ± 0.937 | 0.625 ± 0.321 | 6.31 ± 3.24 | |
| >300 | Unrolled | 0.115 ± 0.0476 | 0.773 ± 0.320 | 0.157 ± 0.0393 | 1.58 ± 0.397 |
| Rolled | 1.25 ± 0.382 | 8.43 ± 2.57 | 1.74 ± 0.0970 | 17.6 ± 0.979 | |
Values represent the means ± SD; n=6
Table 2.
Summary of measured and calculated parameters for both CH3199Hg and 200Hg(II).
| 199CH3Hg | 200Hg(II) | |||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | Unrolled | Rolled | Unrolled | Rolled | Unrolled | Rolled | Unrolled | Rolled |
| Experiment 1 | Experiment 2 | Experiment 1 | Experiment 2 | |||||
| 0.124 | 0.124 | 0.124 | 0.124 | |||||
| 0.2μm–8μm Incorporation (%) | ND | ND | ND | ND | 9.24 ± 19.3 | 24.4 ± 17.0 | 24.6 ± 3.54 | 18.8 ± 1.38 |
| 8μm–300μm Incorporation (%) | 27.5 ± 3.57 | 12.1 ± 4.21 | 26.5 ± 12.2 | 15.0 ± 18.2 | 33.3 ± 7.31 | 8.43 ± 4.15 | 34.1 ± 1.64 | 11.0 ± 1.57 |
| >300μm Incorporation (%) | 0.773 ± 0.392 | 7.76 ± 1.68 | 5.82 ± 3.82 | 29.3 ± 23.5 | 0.308 ± 0.0803 | 23.0 ± 10.9 | 0.372 ± 0.178 | 30.6 ± 5.50 |
| Dissolved Hg (nM) | 0.0269 ± 0.00649 | 0.0379 ± 0.00992 | 0.0269 ± 0.0107 | 0.0400 ± 0.0189 | 0.0586 ± 0.003 | 010.0708 ± 0.0110 | 0.0915 ± 0.00244 | 0.0928 ± 0.00676 |
| 0.2μm–8μm Log Kd | 5.11c | 4.51c | 4.63c | 4.63c | 4.61 ± 0.367 | 4.99 ± 0.363 | 4.90 ± 0.267 | 4.55 ± 0.115 |
| 8μm–300μm Log Kd | 5.32 ± 0.137 | 5.44 ± 0.167 | 5.15 ± 0.248 | 5.39 ± 0.451 | 5.43 ± 0.135 | 5.23 ± 0.241 | 5.33 ± 0.0517 | 5.39 ± 0.324 |
| >300μm Log Kd | 4.81 ± 0.121 | 4.84 ± 0.147 | 5.51 ± 0.281 | 5.34 ± 0.461 | 4.49 ± 0.111 | 5.35 ± 0.150 | 4.35 ± 0.153 | 5.40 ± 0.115 |
| TSSmg L−1)a | 14.9 ± 2.97 | 9.90 ± 0.445 | 14.9 ± 2.97 | 9.90 ± 0.445 | ||||
| DOC (μM)b | 302 ± 7.60 | 295 ± 30.0 | 364 ± 13.7 | 367 ± 21.7 | 302 ± 7.60 | 295 ± 30.0 | 364 ± 13.7 | 367 ± 21.7 |
Values represent the means ± SD; n=6
ND = not detected
Incorporation Detection Limit (DL) = ~1% CH3Hg incorporation and ~4% Hg(II) incorporation
TSS collected on each experimental day before rolling
Average DOC for each treatment after aggregate collection
Estimated based on the DL since the collected mass was <balance MDL=0.38mg
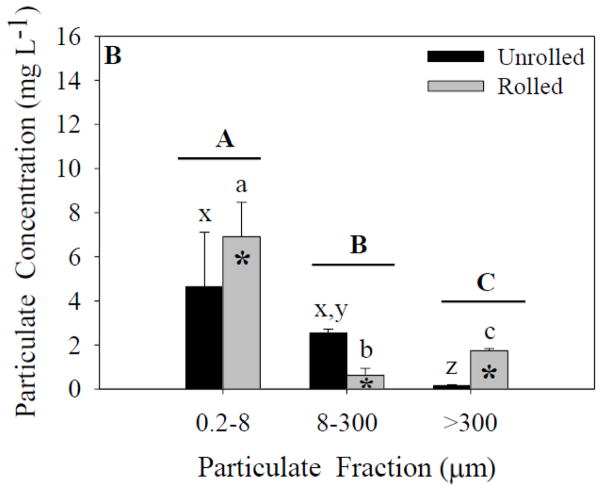
Figure 1.
Particulate concentrations for each size fraction and treatment. Capital letters specify significant differences between particle-size fractions (ANOVA model), whereas lower-case letters specify significant differences between particle-size fractions within a treatment (Tukey’s post-hoc test; x-z for unrolled treatment, a-c for rolled treatment). In experiment 1 (A), a significant effect of particle-size fraction was found (p<0.05). In experiment 2 (B) significant effects of both particle-size fraction and treatment were found (p<0.05). Data represent means ± standard deviation (n=6).
In experiment 2, there was a significant effect of both particle-size fraction and treatment on particle concentration (ANOVA; Figure 1B). A significant interaction effect between these two independent variables was also found. Post-hoc tests indicated a significant difference between the unrolled 0.2- to 8-μm fraction and the >300-μm fraction, and between the 8- to 300-μm fraction and >300-μm fraction. No difference was found between the 0.2- to 8-μm and 8- to 300-μm fractions. There was a significant difference between all three particle-size fractions in the rolled treatments. These results demonstrate that in experiment 2 rolling served to increase the concentration of marine snow (>300 μm), suggesting that conditions in the water collected on day 2 were more conducive to the aggregation of particles. Nonetheless, as in experiment 1, the highest concentration (mg L−1) of particles was measured in the smallest size fraction. Overall, the trends between fractions were similar in the two experiments.
In both experiments the high particulate concentration in the smallest fraction in the rolled treatments indicates that even after four days of rolling a relatively small fraction of the particles were aggregated into large marine snow, and many particles remained in suspension. In further support of this conclusion, Table 1 shows the particle-size fractions as a percent of the total mass. Similar to the particulate concentrations shown in Figures 1A and 1B, the 0.2- to 8-μm fraction comprises the largest percent of the total mass. Furthermore, the >300-μm fraction was a greater percentage of the total mass in the rolled jars than the unrolled, but the marine snow was a lower percentage of the total mass compared to the 8- to 300-μm fraction.
Mercury Partitioning
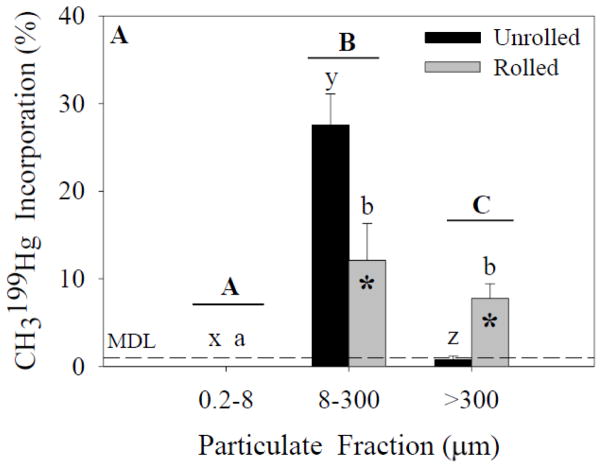
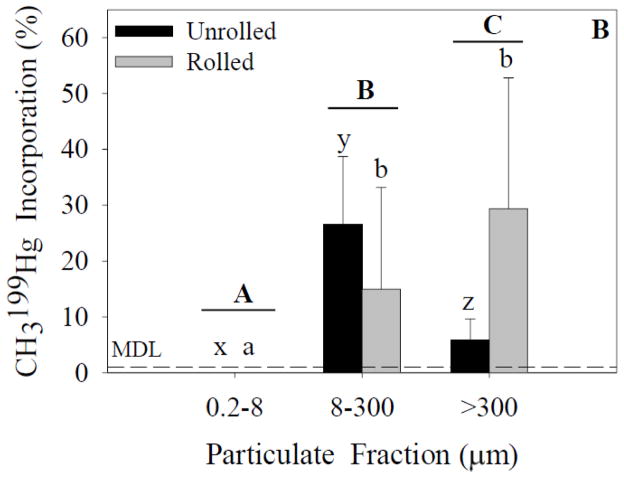
The CH3199Hg and 200Hg(II) incorporation results (see below; Figures 2 and 3, respectively) demonstrate that despite the presence or absence of marine snow, a portion of both forms of Hg remains associated with particulates <300 μm. Analysis of CH3199Hg incorporation (ANOVA; Figure 2A) in experiment 1 indicated a significant effect of particle-size fraction and treatment on incorporation. A significant interaction effect also was found between these independent variables. Post-hoc analysis indicated significant differences between the particle-size fractions within unrolled and rolled treatments. A significant difference was found between the unrolled and rolled treatments within the 8- to 300-μm fraction and within the >300-μm fraction. Analysis of 199CH3Hg incorporation (ANOVA; Figure 2B) in experiment 2 indicated a significant effect of particle-size fraction, but no significant effect of treatment on incorporation. A significant interaction effect also was found. Post-hoc analysis indicated differences between the particle-size fractions within unrolled and rolled treatments.
Figure 2.
CH3199Hg incorporation for each size fraction and treatment. Capital letters specify significant differences between particle-size fractions (ANOVA model), whereas lower-case letters specify significant differences between particle-size fractions within a treatment (Tukey’s post-hoc test; x-z for unrolled treatment, a-c for rolled treatment). Asterisks specify significant differences between treatments within a particle-size fraction (Tukey’s post-hoc test). In experiment 1 (A), significant effects of particle size fraction and treatment were found (p<0.05). In experiment 2 (B), a significant effect of particle size fraction was found (p<0.05). Data represent means ± standard deviation (n=6).
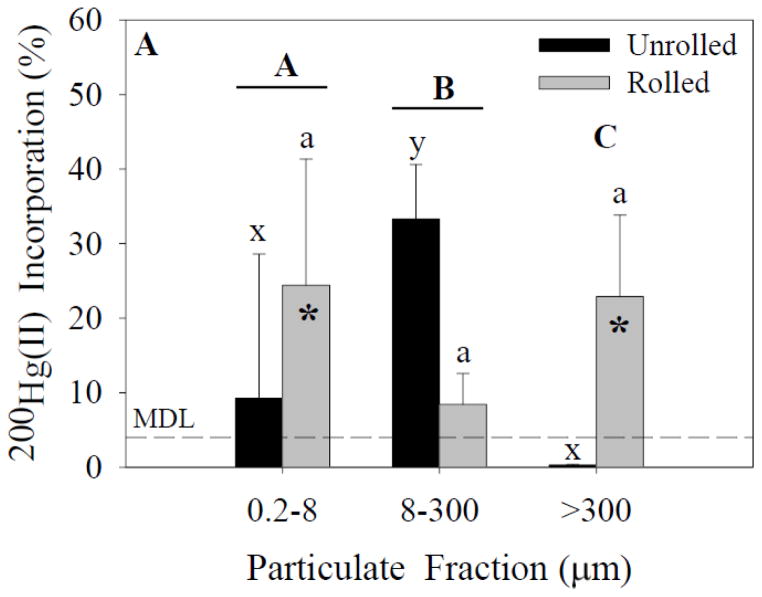
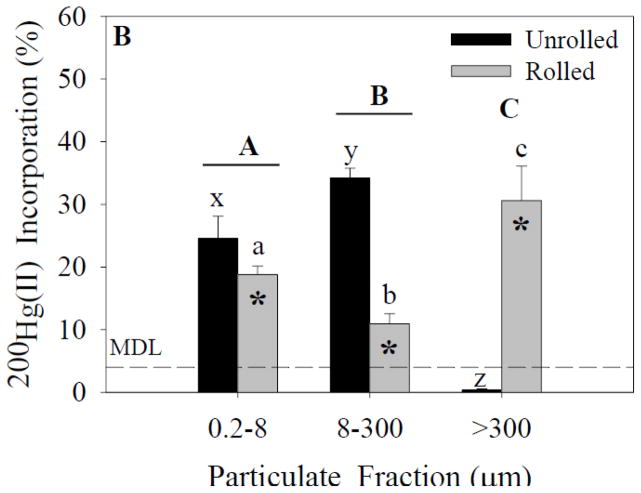
Figure 3.
200Hg(II) incorporation for each size fraction and treatment. Capital letters specify significant differences between particle-size fractions (ANOVA model), whereas lower-case letters specify significant differences between particle-size fractions within a treatment (Tukey’s post-hoc test; x-z for unrolled treatment, a-c for rolled treatment). Asterisks specify significant differences between treatments within a particle-size fraction (Tukey’s post-hoc test). In experiment 1 (A), significant effects of both particle size fraction and treatment were found (p<0.05). In experiment 2 (B), significant effects of both particle size fraction and treatment were found (p<0.05). Data represent means ± standard deviation (n=6).
Analysis of 200Hg(II) incorporation in experiment 1 (ANOVA; Figure 3A) indicated a significant effect of particle-size fraction and treatment on 200Hg(II) incorporation. A significant interaction effect also was found between these independent variables. Post-hoc analysis indicated a significant difference between the 8- to 300-μm fraction and both of the 0.2- to 8-μm and >300-μm fractions within the unrolled treatment. There was no significant difference between particle-size fractions within the rolled treatments. Analysis of 200Hg(II) incorporation in experiment 2 (ANOVA; Figure 3B) indicated significant effects of particle-size fraction and treatment on incorporation. A significant interaction effect also was present. Post-hoc analysis indicated differences between the particle-size fraction within the unrolled and rolled treatments.
The majority of the CH3199Hg was detected in the 8- to 300-μm fraction in both treatments (Figure 2A). A smaller proportion of CH3199Hg was in the marine snow while none was detected in the 0.2- to 8-μm fraction (Table 1). The fact that no CH3199Hg was detected in the smallest size fraction, however, does not mean that incorporation did not occur. The volumes of water filtered to collect the 0.2- to 8-μm fraction were small (20–75 mL of the 1-L sample) and it is likely that CH3Hg would have been detected in this fraction if greater volumes had been filtered. From the data in Table 1, it is apparent that there was complete recovery of the Hg(II) spike based on the measured concentrations in the three particle-size fractions and the water, given the errors associated with the measurements. Similarly, assuming recovery of all the CH3199Hg spike, we can estimate that the percentage of CH3Hg incorporated into the 0.2- to 8-μm fraction was similar to the percentage of Hg(II) incorporated into the same fraction. Unfortunately it is not possible to obtain a more detailed estimate of the extent of incorporation of CH3Hg into the smallest fraction based on the analytical results.
To further analyze the difference in the distribution of CH3Hg and Hg(II) in the three size classes, partition coefficients (Log Kd) were calculated (Table 2). Partition coefficients were of similar magnitude across all experiments, all treatments, and all size fractions, and were similar to those measured in coastal estuaries and in LIS (Coquery et al., 1997; Fitzgerald et al., 2007; Schartup, et al., 2013; Stordal et al., 1996). In experiment 1, the Hg(II) log Kd values were not significantly affected by particle-size fraction or treatment (ANOVA), but in experiment 2 there was a significant effect of both particle-size fraction and treatment on the log Kd. A significant interaction effect was also present. In comparison, the CH3Hg log Kd values were only significantly affected by particle-size fraction. The differences in the 200Hg(II) log Kd values between experiment 1 and 2 may be a result of different concentrations of dissolved organic matter, TEP, and other materials that affect aggregation of particles. As the particles aggregate into larger marine snow, the available binding sites could be protected inside the aggregations preventing further binding of Hg. Additionally, freely-suspended particulates have higher surface to volume ratios that provide more binding sites; thereby resulting in higher partitioning. If these factors were the sole reason for differing log Kd values, then the smallest particulates should have had the highest Kd values. This outcome, however, was not observed; therefore, we conclude this hypothesis of protected binding sites to be proven false.
Counter to the surface to volume hypothesis, the Kd of the smallest fraction was less than the 8- to 300-μm fraction for Hg(II) and for CH3Hg, with the Kd for CH3Hg estimated based on the 1.27 nmol g−1 MDL (Table 2). It is important to note that upon calculating a particle mass balance for these experiments, the total mass of all particle fractions collected at the end of each experiment (7.8 and 8.0 mg L−1 in Exp. 1; 7.3 and 9.3 mg L−1 in Exp. 2; Table 1) did not equal the total TSS at the beginning of each experiment (14.9 mg L−1 in Exp 1; 9.9 mg L−1 in Exp 2). Overall, <10 to 50% of the suspended mass was unaccounted for at the end of the experiments (Table 1). This loss could be a result of microbial degradation of organic matter during the experiments, or could be a result of colloidal formation due to particle disintegration (Honeymann and Santschi, 1988) as colloids (<0.2 μm) would be not be captured on any of the filters. The latter explanation could also account for the slightly lower Kd values compared to field measurements in LIS (Balcom et al., submitted 2015; Chen et al., 2014), as a higher amount of colloids would essentially result in the estimation of a lower Kd. The differences may also be that the TSS and, subsequently, the log Kd values were affected by the presence of plankton that were not removed from the water by the initial sieving (100-μm mesh). Plankton may have accumulated either form of Hg and were likely ingesting particulates and respiring carbon during the rolling period thereby reducing the particle-size fraction. Given these potential effects, future experiments should characterize DOC and the particulate composition more closely to allow for more detailed examination of the factors influencing partitioning.
Mercury methylation and demethylation
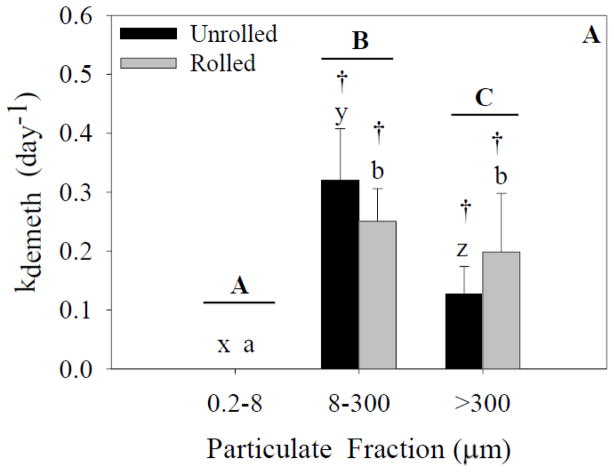
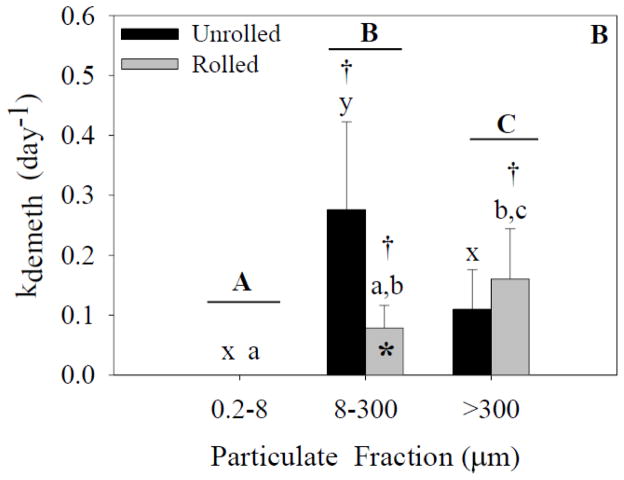
Methylation of Hg and demethylation of CH3Hg were found in both experiments in the larger size fractions, but were not detected in the smallest size fraction (<8 μm) as the CH3Hg concentration was below the MDL (1.27 nmol g−1, Figures 4 and 5). Given the amount of spiked 200Hg isotope in the smallest fraction, however, the CH3Hg MDL, and the mass of particulates in this fraction, we can estimate that the lack of detection is equivalent to a methylation rate of 0.02 day−1. Thus, we cannot conclude that there was no methylation in the smallest fraction even though we could not detect any CH3Hg isotope. In terms of the factors promoting methylation, the ANOVA analysis indicated a significant effect of particle-size fraction, but not treatment on the methylation rates in experiment 1 (Figure 4A). A significant interaction was also present. Post-hoc analysis indicated a significant difference between the unrolled >300-μm fraction and the other fractions. There was a significant difference between the rolled 0.2- to 8-μm and 8–300-μm fractions in both experiments but this entirely reflects the lack of detection of CH3Hg in the smallest fractions. There was no significant difference between the rolled 8- to 300-μm fraction and the other two rolled fractions.
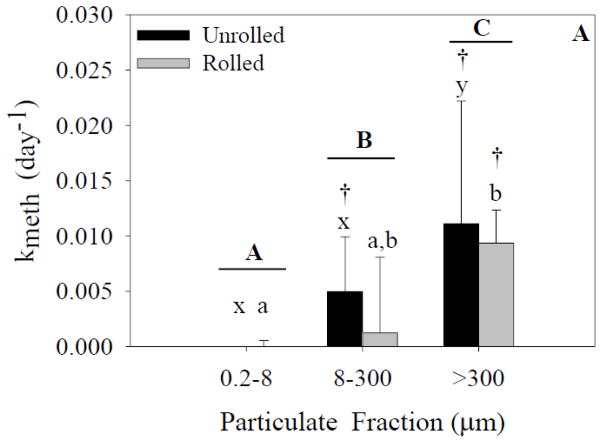
Figure 4.
Methylation rates for each size fraction and treatment. Capital letters specify significant differences between particle-size fractions (ANOVA model), whereas lower-case letters specify significant differences between particle-size fractions within a treatment (Tukey’s post-hoc test; x-z for unrolled treatment, a-c for rolled treatment). Asterisks specify significant differences between treatments within a particle-size fraction (Tukey’s post-hoc test). Crosses specify that means are significantly different than zero (single sample t-test). In experiment 1 (A), a significant effect of particle size fraction was found (p<0.05). In experiment 2 (B), significant effects of both particle size fraction and treatment were found (p<0.05). Data represent means ± standard deviation (n=6).
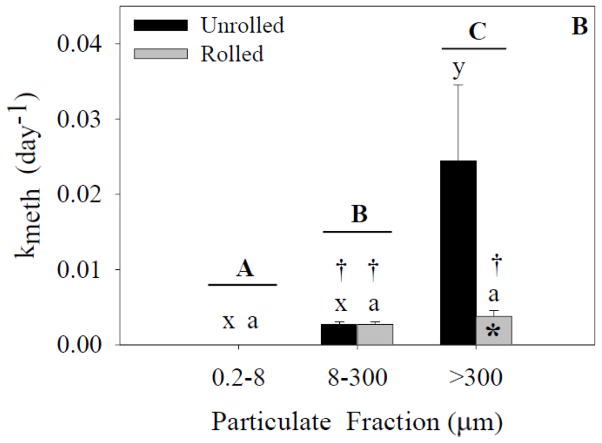
Figure 5.
Demethylation rates for each size fraction and treatment. Capital letters specify significant differences between particle-size fractions (ANOVA model), whereas lower-case letters specify significant differences between particle-size fractions within a treatment (Tukey’s post-hoc test; x-z for unrolled treatment, a-c for rolled treatment). Asterisks specify significant differences between treatments within a particle-size fraction (Tukey’s post-hoc test). Crosses specify that means are significantly different than zero (single sample t-test). In experiment 1(A), a significant effect of particle size fraction was found (p<0.05). In experiment 2 (B), significant effects of both particle size fraction and treatment were found (p<0.05). Data represent means ± standard deviation (n=6).
Analysis of experiment 2 (ANOVA; Figure 4B) indicated significant effects of both particle-size fraction and treatment on methylation rates. Additionally, a significant interaction effect was present. Post-hoc analysis of the unrolled treatment indicated a significant difference between the >300-μm fraction and the other two fractions. There was no significant difference between the unrolled 0.2- to 8-μm and 8- to 300-μm fractions. No significant differences were present between the particle-size fractions in the rolled treatment. There was also a significant difference between the unrolled and rolled treatments in the >300-μm fraction.
In experiment 1 (Figure 4A), one-sample t-tests indicated that methylation rates in the unrolled 8- to 300-μm, rolled 8- to 300-μm, and rolled >300-μm fractions were significantly different from zero. The remaining fractions were not significantly different from zero. In experiment 2 (Figure 4B), one-sample t-tests indicated that the unrolled 8- to 300-μm fraction, the rolled 8- to 300-μm, and >300-μm fractions were significantly different from zero.
In terms of the factors promoting demethylation, ANOVA analysis indicated a significant effect of particle-size fraction, but not treatment on demethylation rates in experiment 1 (Figured 5A). A significant interaction effect was also present. Post-hoc analysis of the unrolled treatment indicated a significant difference between all three particle-size fractions. In the rolled treatment, there was a significant difference between the rolled 0.2- to 8-μm fraction and the other two fractions, again because of the lack of detection of CH3Hg in this fraction. No significant difference was found between the rolled 8- to 300-μm fraction and the >300-μm fraction.
Analysis of experiment 2 (ANOVA; Figure 5B) indicated significant effects of both particle-size fraction and treatment on demethylation rates. A significant interaction effect was also present. Post-hoc analysis of the unrolled treatment indicated a significant difference between the three particle-size fractions. There was no significant difference between the 0.2- to 8-μm fraction and the 8- to 300-μm fraction because of the lack of detection of CH3Hg in the smallest fraction. There was also no significant difference between the 8- to 300-μm fraction and >300-μm fraction, but there was a significant difference between the 0.2- to 8-μm fraction and the >300-μm fraction. There was a significant difference between the treatments within the 8- to 300-μm fraction.
In experiment 1 (Figure 5A), one-sample t-tests indicated that the unrolled 8- to 300-μm and >300-μm fractions and the rolled 8- to 300-μm and >300-μm fractions were significantly different from zero. In experiment 2 (Figure 5B), one-sample t-tests indicated that the unrolled 8- to 300-μm and the rolled 8- to 300-μm and >300-μm fractions were significantly different from zero.
Changes in the isotopic composition of CH3Hg and Hg in the larger particle-size fractions during the experiments are indicative of the net formation and degradation of Hg forms during the experiment. We quantified these rates of change based on changes in the distribution of the isotope spikes. An increase in the concentration of CH3200Hg was found in the particle-size fractions >8 μm in all experiments, with measured amounts significantly above background levels based on natural abundances. The calculated methylation rate constants confirmed the presence of mercury methylation in both the marine snow (>300 μm) and 8-to 300-μm particle fractions (Figures 4A and 4B). Methylation rates (Figure 4A and 4B) ranged from 0 to ~0.02 d−1. Treatment did not have a significant effect on methylation rates in experiment 1 (Figure 4A), but treatment did have a significant effect in experiment 2 in the marine-snow fraction (Figure 4B). It is important to note that methylation rates were significantly higher in the unrolled jars, a result that was not expected. These findings indicate that rolling did not enhance the methylation rates in the larger particulates, but rather that methylation can occur as long as large particulates are present (>8 μm). To further support this hypothesis, the fractions that were significantly different from zero were also independent of treatment. As noted above, we cannot rule out that methylation did not occur in the smallest size fraction even though CH3Hg was not detected, but given that microbial methylation is thought to be mostly due to anaerobic bacteria (Gilmour et al., 2013; Parks et al., 2013), it is unlikely that conditions conducive to methylation would occur in the smallest size fraction, which is likely composed of individual particles, small phytoplankton, and other microbes. The detectable rates in the >300-μm fraction and 8- to 300-μm fraction are more likely given evidence for anoxic “microzones” in such particulates (Alldredge and Cohen, 1987). Overall, our results demonstrate the potential for methylation in oxic environments which would be enhanced if large particulates, such as marine snow, are present. We conclude, based on current knowledge that the methylation by anaerobes in oxic waters must be due to the formation of reduced oxygen microzones within the larger aggregations, a process that has been demonstrated to occur (Alldredge and Cohen, 1987).
Analysis of demethylation rate data indicated no treatment effect, which was different from that found for the methylation rate data. The demethylation rates ranged from non-detectable in the 0.2- to 8-μm fraction to 0.3 d−1 in the unrolled 8- to 300-μm fraction. Again it would appear that the presence of marine snow does not enhance demethylation rates, but rather as long as larger particulates (>8 μm) are present, demethylation is enhanced. In contrast to methylation, demethylation in sediments and the water column has been shown in both aerobic and anaerobic environments (e.g., Heyes et al., 2006; Hollweg et al., 2009; Whalin et al., 2007).
Overall, demethylation rate data indicate a higher rate of demethylation versus methylation. A similar result has been found in most studies on sediments (Heyes et al., 2006; Hollweg et al., 2010; Schartup et al., 2013 ) and the water column (Lehnherr et al., 2011), which is consistent with the relatively low fraction of total Hg as CH3Hg in sediments and suspended particles. The methylation and demethylation rate data indicate that the presence of marine snow does not necessarily guarantee higher rates of net CH3Hg formation, but rather that enhanced rates of both methylation and demethylation would likely occur in settling particulate matter. Overall, a lower or higher net rate of methylation may result depending on the relative rates of aggregate formation and degradation. The observed rates presented here compare to rates published in the literature for sediment methylation. Mitchell and Gilmour (2008) measured methylation rates ranging from 0.01 to 0.05 d−1 within the first 2 centimeters of marsh sediment in the Chesapeake Bay. Hollweg et al. (2009) reported methylation rates ranging from 0.01 to 0.04 d−1 in 12 cm sediment cores in the Chesapeake Bay and adjacent offshore shelf. Similarly, Schartup et al. (2013) measured methylation rates of 0.01 to 0.05 d−1 in Long Island Sound (LIS) sediments. Therefore, the methylation rates in aggregated organic matter in the water column are similar to methylations rates in marine sediments.
There are limited data on the methylation and demethylation rates for the water column of the ocean, especially in oxic waters. Lehnherr et al. (2011) measured methylation rates from 0.5–1 × 10−2 d−1 and demethylation rates of 0.2 to 0.5 d−1 in the chlorophyll maximum region and in low oxygen waters of the Arctic Ocean, rates comparable to those reported here. Monperrus et al. (2007) measured higher methylation rates in coastal and offshore surface waters of the Mediterranean Sea than were found in the present study, but their demethylation rates were comparable to our measurements. Schartup et al. (in review) found methylation rates from the MDL to 0.004 day−1 for the surface waters of Lake Melville, a saline fjord in Labrador, Canada, lower than those reported here. The demethylation rates, however, were also low, with overall net methylation occurring in the surface waters of this system. Whalin et al. (2007), who examined transformations of Hg in the surface and deep waters of the Chesapeake Bay and adjacent offshore shelf and slope, found methylation rates lower than their MDL for methylation (<0.1 d−1). Nonetheless, their measured demethylation rates were comparable to the values found here. In the previous studies, unfiltered water was used for the experiments, and therefore no distinction could be made between methylation rates driven by free-living bacteria and those driven by bacteria inhabiting larger particulates such as marine snow. Considering the results of the present study, it is possible that methylation and demethylation in these field incubations was occurring in larger particle-size fractions, a possibility that may be generally applicable to coastal environments. Overall, it appears that the enhanced microbial communities of marine snow (Balzano et al., 2009) relative to smaller, suspended particles stimulates the production of CH3Hg, but also enhances the reverse reaction of demethylation.
Similarly, there is debate in the freshwater literature over whether biofilms are important in the production of CH3Hg in lakes and wetlands. Lin and Jay (2007), using cultures of sulfate reducing bacteria, showed that methylation in biofilms was enhanced compared to that of free-living bacteria. In a recent study, Hamelin et al. (2015) showed that the rate of net methylation in the biofilms on macroalgae was two orders of magnitude higher than that of the nearby sediment of a temperate wetland. Similarly, Lázaro et al. (2013) showed that methylation within biofilms of floating plants in the Amazon were sites of substantial Hg methylation. They also found that the methylation rate correlated with the algal biomass on which these biofilms grow. The microbial communities of marine snow are likely similar to those of biofilms and, therefore, the hypothesis that formation of aggregates in coastal waters enhances methylation is reasonable. Balzano et al. (2009), for example, found Fe(III) and nitrate reduction in their experiments with marine flocs, and identified bacterial strains capable of Fe reduction. The microbes identified by Balzano et al., including Desulvovibrio species, could also be responsible for CH3Hg production (Gilmour et al., 2013). In the sub-thermocline waters of the ocean, high CH3Hg concentrations have been interpreted as indicative of methylation that occurs during the decomposition of sinking particulate material (Mason et al., 2012; Mason and Fitzgerald, 1993; Sunderland et al., 2009). A similar enhanced decomposition is likely occurring in the marine snow formed even in oxic surface waters.
In studies of methylation and demethylation in sediments there is often a comparison made between the ratio of the rate constants and the fraction of total Hg that is CH3Hg. These two ratios should be the same at steady state, assuming similar bioavailability of both fractions to the microorganisms (Heyes et al., 2006; Liu et al., 2015). The ratio of the measured rate constants for this study ranged from ~0.05 to 0.25, depending on the size fraction and the experiment (Figures 4 and 5). Given the magnitude of the rate constants (the characteristic time to equilibrium = 1/(km + kdm) assuming a reversible reaction), it is likely that chemical processes in the jars did not reach steady state during the experimental period. Therefore, it is more appropriate to compare our data to measurements in suspended particulate matter from LIS (Balcom et al., 2006; 2015; Chen et al., 2014), which have measured fractions of CH3Hg/total Hg that cover a similar range (0.01 to 0.2). These comparisons provide further validation that the rates of reaction measured in the current study are representative of those occurring in the water column of LIS.
There is an ongoing debate about the importance of various sources of CH3Hg to estuarine food chains with much work focused on examining the net methylation of Hg in sediment and the supply from the sediments to the water column (Chen et al., 2014; Hammerschmidt et al., 2004; Hollweg et al., 2010; 2009). There is increasing evidence, however, that sediments may not be the most important source in some coastal locations (Balcom et al., 2008; Chen et al., 2012; Sunderland et al., 2012; 2010). Additionally, Chen et al. (2014) showed through a study of multiple estuaries that there was little correlation between sediment CH3Hg and levels in foraging fish, and that water column particulates were the best predictor of CH3Hg concentrations in fish. These and other studies (Balcom et al., 2015; Conaway et al., 2003) have shown that particulate CH3Hg (on a mass basis) is often much higher than that of surface sediment. Most of these studies have invoked external sources of CH3Hg as important, but the results presented here suggest that further focus should be given to in situ formation of CH3Hg in the water column of estuaries and coastal waters. Our results demonstrate that the formation of larger particulates by aggregation enhances methylation in oxic waters, and is the likely mechanism whereby methylation is occurring in shallow coastal waters. More data is needed to determine the importance of this source of CH3Hg compared to other sources such as sediments, groundwater, and external inputs.
Conclusions
In conclusion, we show that both methylation of Hg and demethylation of CH3Hg can occur in particulates and marine snow in oxic waters of the coastal zone, and demonstrate the possibility of water column net methylation in coastal waters. Furthermore, the stated hypothesis that the highest incorporation of Hg would occur in the marine snow was not confirmed. Whereas there was a portion of both Hg forms incorporated into the >300-μm marine-snow fraction, most of the CH3199Hg and 200Hg(II) was incorporated into the 8- to 300-μm fraction. Particle-size analysis indicated that, despite rolling the seawater for several days, the suspended particle field was mostly composed of small particulates rather than large marine-snow aggregations.
Methylation rates measured in this study demonstrate that settling particulates can form environments conducive to mercury methylation with production rates on the same order of magnitude as sediment methylation. Whether particles form large marine snow aggregations or smaller aggregates (e.g. 8 to 300 μm) it is likely that their organic material is being degraded through bacterial action. This process simultaneously results in net CH3Hg formation, providing a potential source of Hg methylation in the water column. Whereas our results are consistent with the idea of methylation within settling particulates in low oxygen waters of the open ocean, they may also explain the presence of CH3Hg in coastal and open ocean surface waters, even where there is enhanced photodegradation of CH3Hg. This study also stresses the importance of smaller particulates, not just large macroscopic marine snow aggregations, in Hg cycling and methylation.
Highlights.
CH3Hg and Hg(II) were incorporated into laboratory-produced marine snow
Methylation rates in marine snow were of same magnitude as estimated sediment rates
Particulate coagulation enhances net mercury methylation in oxic estuarine waters
Acknowledgments
Veronica Ortiz would first like to thank her father, William Ortiz, for passing on his GI Bill, via the Post-9/11 GI Bill, to her to attend graduate school at the University of Connecticut for her master’s degree. She would also like to thank Mr. Prentiss Balcom, Ms. Bridget Holohan, Dr. Amina Schartup, Dr. Claudia Koerting, and Dr. John J. Doyle for their assistance in training her to use the various instruments and methods that were needed to complete this study. Lastly, she’d like to thank the UConn Department of Marine Sciences for all of their support throughout her graduate studies. This research was also partially supported by prior and continuing grants from the National Institute of Health through collaboration with investigators at Dartmouth College (NIH Grant Number P42 ES007373), and the National Science Foundation (CBET-1336358).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alldredge AL, Cohen CY. Can Microscale Chemical Patches Persist in the Sea? Microelectrode Study of Marine Snow, Fecal Pellets. Science. 1987;235:689–691. doi: 10.1126/science.235.4789.689. [DOI] [PubMed] [Google Scholar]
- 2.Alldredge AL, Passow U, Logan BE. The abundance and significance of a class of large, transparent, organic particles in the ocean. Deep Sea Res I. 1993;40:1131–1140. [Google Scholar]
- 3.Alldredge AL, Silver MW. Characteristics Dynamics and Significance of Marine Snow. Prog Oceanogr. 1988;20:41–82. [Google Scholar]
- 4.Balcom PH, Hammerschmidt CR, Fitzgerald WF, Lamborg CH, O’Connor JS. Seasonal distributions and cycling of mercury and methylmercury in the waters of New York/New Jersey Harbor Estuary. Mar Chem. 2008;109:1–17. [Google Scholar]
- 5.Balcom PH, Schartup AT, Mason RP, Chen CY. 2015 Feb; Submitted to Mar. Chem. Pathways of methylmercury transfer to the water column across multiple estuaries. [Google Scholar]
- 6.Balzano S, Statham PJ, Pancost RD, Lloyd JR. Role of microbial populations in the release of reduced iron to the water column from marine aggregates. Aquat Microb Ecol. 2009;54:291–303. [Google Scholar]
- 7.Bloom N. Determination of pictogram levels of methylmercury by aqueous phase ethylation, followed by cryogenic gas chromatography with cold vapour atomic fluorescence detection. Can J Fish Aquat Sci. 1989;46:1131–1140. [Google Scholar]
- 8.Bloom NS, Crecelius EA. Determination in seawater at sub-nanogram per liter levels. Mar Chem. 1983;14:49–59. [Google Scholar]
- 9.Chen CY, Borsuk ME, Bugge DM, Hollweg T, Balcom PH, Ward DM, Williams J, Mason RP. Benthic and pelagic pathways of methylmercury bioaccumulation in estuarine food webs of the northeast United States. PLOS. 2014:1–11. doi: 10.1371/journal.pone.0089305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CY, Driscoll CT, Lamber KF, Mason RP, Rardin LR, Schmitt CV, Serrell NS, Sunderland EM. Sources to Seafood: Mercury Pollution in the Marine Environment. Hanover, NH: Toxic Metals Superfund Research Program, Dartmouth College; 2012. p. 26. http://www.dartmouth.edu/~toxmetal/assets/pdf/sources_to_seafood_report.pdf. [Google Scholar]
- 11.Conaway CH, Squire S, Mason RP, Flegal AR. Mercury speciation in the San Francisco Bay estuary. Mar Chem. 2003;80:199–225. [Google Scholar]
- 12.Coquery M, Cossa D, Sanjuan J. Speciation and sorption of mercury in two macro-tidal estuaries. Mar Chem. 1997;58:213–227. [Google Scholar]
- 13.Fitzgerald WF, Lamborg CH, Hammerschmidt CR. Marine Biogeochemical Cycling of Mercury. Chem Rev. 2007;107:641–662. doi: 10.1021/cr050353m. [DOI] [PubMed] [Google Scholar]
- 14.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somoenhally AC, Johs A, Hurt RA, Jr, Bailey KL, Elias DA. Mercury methylation by novel microorganisms from new environments. Environ Sci Technol. 2013;47:11810–11820. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 15.Hamelin S, Planas D, Amyot M. Mercury methylation and demethylation by periphyton biofilms and their host in a fluvial wetland of the St. Lawrence River (QC, Canada) Sci Tot Environ. 2015:464–471. doi: 10.1016/j.scitotenv.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 16.Hammerschmidt CR, Bowman KL. Vertical methylmercury distribution in the subtropical North Pacific Ocean. Mar Chem. 2012;132–133:77–82. [Google Scholar]
- 17.Hammerschmidt CR, Fitzgerald WF. Bioaccumulation and trophic transfer of methylmercury in Long Island Sound. Archives of Environ Contamination And Toxicol. 2006;51:416–424. doi: 10.1007/s00244-005-0265-7. [DOI] [PubMed] [Google Scholar]
- 18.Hammerschmidt C, Fitzgerald WF. Bioaccumulation and Trophic Transfer of Methylmercury in Long Island Sound. Arch Environ Contam Toxicol. 2006;51:416–424. doi: 10.1007/s00244-005-0265-7. [DOI] [PubMed] [Google Scholar]
- 19.Hammerschmidt CR, Fitzgerald WF. Methylmercury in mosquitoes related to atmospheric mercury deposition and contamination. Environ Sci Technol. 2005;39:3034–3039. doi: 10.1021/es0485107. [DOI] [PubMed] [Google Scholar]
- 20.Hammerschmidt CR, Fitzgerald WF, Lamborg CH, Balcom PH, Visscher PT. Biogeochemistry of methylmercury in sediments of Long Island Sound. Mar Chem. 2004;90:31–52. [Google Scholar]
- 21.Heimbürger LE, Cossa D, Marty JC, Migon C, Averty B, Dufour A, Ras J. Methyl mercury distributions in relation to the presence of nano- and picophytoplankton in an oceanic water column (Ligurian Sea, North-western Mediterranean) Geochemica et Cosmochimica Acta. 2010;74:5549–5559. [Google Scholar]
- 22.Heinonen KB, Ward JE, Holohan BA. Production of transparent exopolymer particles (TEP) by benthic suspension feeders in coastal systems. J Experim Mar Biol And Ecol. 2007;341:184–195. [Google Scholar]
- 23.Heyes A, Mason RP, Kim EH, Sunderland E. Mercury methylation in estuaries: Insights from using measuring rates using stable mercury isotopes. Mar Chem. 2006;102:134–147. [Google Scholar]
- 24.Hill PS. Controls on floc size in the sea. Ocean. 1998;11:13–18. [Google Scholar]
- 25.Hill PS, Syvitski JP, Cowan EA, Powell RD. In situ observations of floc settling velocities in Glacier Bay, Alaska. Mar Geo. 2000;145:85–94. [Google Scholar]
- 26.Hintelmann H, Evans RD. Application of stable isotopes in environmental tracer studies - Measurement of monomethylmercury (CH3Hg+ ) by isotope dilution ICP-MS and detection of species transformation. Fresenius’ J of Analytical Chem. 1997;358:378–385. [Google Scholar]
- 27.Hollweg TA, Gilmour CC, Mason RP. Mercury and methylmercury cycling in sediments of the mid-Atlantic continental shelf and slope. Limnol Oceanogr. 2010;55:2703–2722. [Google Scholar]
- 28.Hollweg TA, Gilmour CC, Mason RP. Methylmercury production in sediments of Chesapeake Bay and the mid-Atlantic continental margin. Mar Chem. 2009;114:86–101. [Google Scholar]
- 29.Honeymann BD, Santschi PH. Metals in aquatic systems. Environ Sci Technol. 1988;22:862–871. doi: 10.1021/es00173a002. [DOI] [PubMed] [Google Scholar]
- 30.Jackson GA, Burd AB. Aggregation in the Marine Environment. Enviro Sci and Technol. 1998;32:2805–2814. [Google Scholar]
- 31.Kach DJ, Ward JE. The role of marine aggregates in the ingestion of picoplankton-size particles by suspension-feeding molluscs. Mar Biol. 2008;153:797–805. [Google Scholar]
- 32.Kim E, Mason RP, Bergeron CM. A modeling study on methylmercury bioaccumulation and its controlling factors. Ecol Modelling. 2008;218:267–289. [Google Scholar]
- 33.Kiørboe T, Anderson KP, Dam HG. Coagulation efficiency and aggregate formation in marine phytoplankton. Mar Biol. 1990;107:235–245. [Google Scholar]
- 34.Lázaro WL, Guimarães JRD, Ignácio ARA, Da Silva CJ, Díez S. Cyanobacteria enhance methylmercury production: A hypothesis tested in the periphyton of two lakes in the Pantanal floodplain, Brazil. Sci Tot Environ. 2013:231–238. doi: 10.1016/j.scitotenv.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence AL, Mason RP. Factors controlling the bioaccumulation of mercury and methylmercury by the estuarine amphipod Leptocheirus plumulosus. Environ Pollut. 2001;111:217–231. doi: 10.1016/s0269-7491(00)00072-5. [DOI] [PubMed] [Google Scholar]
- 36.Lehnherr I, St Louis VL, Hintelmann H, Kirk JL. Methylation of inorganic mercury in polar marine waters. Nature Geosciences. 2011;4:298–302. [Google Scholar]
- 37.Li B, Ward JE, Holohan BA. Transparent exopolymer particles (TEP) from marine suspension feeders enhance particle aggregation. Mar Ecol Prog Ser. 2008;357:67–77. [Google Scholar]
- 38.Lin CC, Jay JA. Mercury methylation by planktonic and biofilm cultures of Desulfovibrio desulfuricans. Environ Sci Technol. 2007;41:6691–6697. doi: 10.1021/es062304c. [DOI] [PubMed] [Google Scholar]
- 39.Liu B, Schaider LA, Mason RP, Shine JP, Rabalais NN, Senn DB. Controls on methylmercury accumulation in northern Gulf of Mexico sediments. Estuarine Coastal and Shelf Science. 2015 in press. [Google Scholar]
- 40.Mason RP, Choi AL, Fitzgerald WF, Hammerschmidt CR, Lamborg CH, Soerensen AL, Sunderland EM. Mercury biogeochemical cycling in the ocean and policy implications. Environ Res. 2012;119:101–117. doi: 10.1016/j.envres.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mason RP, Fitzgerald WF. The distribution and biogeochemical cycling of mercury in the equatorial Pacific. Deep-Sea Research. 1993;40:1897–1924. [Google Scholar]
- 42.Mason RP, Fitzgerald WF. Alkylmercury species in the equatorial Pacific. Nature. 1990;347:457–459. [Google Scholar]
- 43.Mitchell CPJ, Gilmour CC. Methylmercury production in a Chesapeake Bay salt marsh. J Geophys Res. 2008;113:1–14. [Google Scholar]
- 44.Monperrus M, Tessier E, Amouroux D, Leynaert A, Huonnic P, Donard OFX. Mercury methylation, demethylation and reduction rates in coastal and marine surface waters of the Mediterranean Sea. Mar Chem. 2007;107:49–63. [Google Scholar]
- 45.Munson KM, Babi D, Lamborg C. Determination of monomethylmercury from seawater with ascorbic acid-assisted direct ethylation. Limnol Oceanogr Methods. 2014;12:1–9. [Google Scholar]
- 46.Ortiz VL. Thesis. 2013. An Examination of Marine Snow as a Pathway for Mercury Uptake in the Blue Mussel Mytilus edulis. [Google Scholar]
- 47.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Jr, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L. The genetic basis for bacterial mercury methylation. Science. 2013;339:1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 48.de la Rocha CL. The Biological Pump in: The Oceans and Marine Geochemistry. In: Elderfield H, Holland HD, Turekian KK, editors. Treatise on Geochemistry. Elsevier; Amsterdam, The Netherlands: 2006. pp. 83–111. [Google Scholar]
- 49.Schartup AT, Mason RP, Balcom PH, Hollweg TA, Chen CY. Methylmercury production in estuarine sediments: role of organic matter. Environ Sci Technol. 2013;47:695–700. doi: 10.1021/es302566w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schartup AT, Balcom PH, Soerensen AL, Gosnell KJ, Calder RSD, Mason RP, Sunderland EM. Freshwater discharges drive high levels of methylmercury in Arctic marine biota. PNAS. doi: 10.1073/pnas.1505541112. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shanks AL, Edmondson EW. Laboratory-made artificial marine snow: a biological model of the real thing. Mar Bio. 1989;101:463–470. [Google Scholar]
- 52.Simon M, Grossart H, Schweitzer B, Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquatic Microbial Ecology. 2002;28:175–211. [Google Scholar]
- 53.Stordal MC, Santschi PH, Gill GA. Colloidal pumping: evidence for the coagulation process using natural colloids tagged with 203Hg. Environ Sci Technol. 1996;30:3335–3340. [Google Scholar]
- 54.Sunderland EM, Amirbahman A, Burgess NM, Dalziel J, Harding G, Jones SH, Kamai E, Karagas MR, Shi X, Chen CY. Mercury sources and fate in the Gulf of Maine. Environ Res. 2012;119:27–41. doi: 10.1016/j.envres.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sunderland EM, Dalziel J, Heyes A, Branfireun BA, Krabbenhoft DP, Gobas FAPC. Response of a Macrotidal Estuary to Changes in Anthropogenic Mercury Loading between 1850 and 2000. Environ Sci Technol. 2010;44:1698–1704. doi: 10.1021/es9032524. [DOI] [PubMed] [Google Scholar]
- 56.Sunderland EM, Krabbenhoft DP, Moreau JW, Strode SA, Landing WA. Mercury sources, distribution, and bioavailability in the North Ocean: insights from data and modesl. Global Biogeochemical Cycles. 2009;23:1–14. [Google Scholar]
- 57.Tseng CH, Hammerscmidt CR, Fitzgerald WF. Determination of methylmercury in environmental matrixes by on-line flow injection and atomic fluorescence spectrometry. Anal Chem. 2004;76:7131–7136. doi: 10.1021/ac049118e. [DOI] [PubMed] [Google Scholar]
- 58.Ullrich SA, Tanton TW, Abdrashitova SA. Mercury in the Aquatic Environment: A Review of Factors Affecting Methylation. Crit Rev in ES&T. 2001;31:241–293. [Google Scholar]
- 59.US Environmental Protection Agency. Methyl Mercury in Water by Distillation, Aqueous Ethylation, Purge and Trap, and CVAFS. Jan, 2001. [Google Scholar]
- 60.Whalin L, Kim EH, Mason R. Factors influencing the oxidation, reduction, and methylation and demethylation of mercury species in coastal waters. Mar Chem. 2007;107:278–294. [Google Scholar]