Abstract
The counter-regulatory hormone glucagon inhibits lipogenesis via downregulation of sterol regulatory element binding protein 1 (SREBP-1). The effect of glucagon is mediated via protein kinase A (PKA). To determine if SREBP-1 is a direct phosphorylation target of PKA, we conducted mass spectrometry analysis of recombinant n-terminal SREBP-1a following PKA treatment in vitro. This analysis identified serines 331/332 as bona-fide phosphorylation targets of PKA. To determine the functional consequences of phosphorylation at these sites, we constructed mammalian expression vector for both nSREBP-1a and 1c isoforms in which the candidate PKA phosphorylation sites were mutated to active phosphomimetic or non-phosphorylatable amino acids. The transcriptional activity of WT SREBP and mutant forms was reduced by the phosphomimetic mutation of S332 of nSREBP-1a and the corresponding serine (S308) of nSREBP-1c. This site is a strong candidate for mediating the negative regulatory effect of glucagon on SREBP-1 and lipogenesis.
Keywords: Sterol Regulatory Element Binding Protein (SREBP), Protein Kinase A (PKA), Immobilized Metal Affinity Chromatography (IMAC), Phosphorylation, Site-Directed Mutagenesis, Mass Spectrometry (MS)
Introduction
Obesity is a major public health issue in the developed world. The prevalence of obesity is closely correlated with the increased incidence of major diseases such as type 2 diabetes (T2DM), nonalcoholic fatty liver disease, atherosclerotic heart disease, etc. [1,2]. Although the molecular mechanisms by which obesity causes its associated diseases are still unknown, one common feature is dysregulation of lipid homeostasis [1,3]. Among known lipogenic regulators, the sterol regulatory element-binding proteins (SREBPs) are pivotal activators of enzymes responsible for biosynthesis of fatty acids and cholesterol [4,5]. Therefore, understanding the underlying molecular mechanisms that inhibit the SREBPs is important for development of effective treatments of the dyslipidemia that accompanies obesity and T2DM.
In mammals, there are three SREBPs including SREBP-1a, -1c and -2, which are encoded by two genes, srebf1 and srebf2 [6]. SREBPs are basic helix loop helix leucine zipper (b/HLH/LZ) transcription factors [7,8]. SREBP-2 controls cholesterol homeostasis by activating genes mediating cholesterol synthesis and lipoprotein uptake [7]. The SREBP-1 isoforms (1a and 1c) are the predominant inducers of hepatic fatty acid and triglyceride synthesis. The target genes of SREBP-1 include the rate-limiting lipogenic enzymes fatty acid synthase (FASN), stearoyl-CoA desaturase (SCD) and acetyl Co-A carboxylase-1 (ACC-1) [9,10]. Aberrant regulation of SREBP-1c has been implicated in the pathogenesis of dyslipidemia and hepatic steatosis [8,11]. In this regard, understanding the molecular mechanisms by which hormones which suppress lipogenesis such as glucagon inhibit SREBP-1 is critically important.
In the fasting state, glucagon reduces hepatic lipid synthesis [12]. The metabolic effects of glucagon are mediated through the cAMP-dependent protein kinase A (PKA) [13,14]. In animal models, PKA activation by adrenergic stimulation results in lean phenotypes and improves insulin sensitivity [15,16]. PKA activation attenuates SREBP-1 activity and SREBP-1-mediated lipogenesis [16,17]. We have previously shown that glucagon and dibutyryl-cAMP effectively block activation of SREBP-1c transcription by insulin [18,19] and further that db-cAMP inhibits the ER to Golgi transport and proteolytic activation of nascent SREBP-1c [20,21]. However the mechanism(s) underlying the ability of glucagon and its surrogate cAMP to inhibit SREBP-1c remain unclear.
A growing body of evidence suggests that posttranslational modification of SREBPs regulate their stability and transcriptional activity [21,22]. Phosphorylation of SREBP-1 by PKA has been proposed as mediating inhibition of its transcriptional activity by cAMP [17]. To determine the effect of PKA mediated phosphorylation of SREBP-1c, we identified serines S331/332 of rat SREBP-1a as phosphorylation targets of PKA. We determined that phosphorylation of S332 of rat SREBP-1a and the corresponding S308 of SREBP-1c inhibits transcriptional activity of SREBP-1. This site corresponds to S338 of human SREBP-1a previously implicated by Lu, et al. [17] as a potential phosphorylation site for PKA. Our direct identification of the corresponding site on rat SREBP-1a by mass spectrometry analysis further strengthens the biologic relevance of this site. Based on these findings, it is likely that phosphorylation of this serine by PKA mediates the negative regulatory effect of cAMP on SREBP-1.
1. Materials and methods
1.1. Bacterial expression plasmid construction
Bacterial expression plasmid pET28a-nSREBP-1a (1–403aa), driven by T7 promoter, was constructed by inserting the coding sequence of amino acids (aa) 1–403 of rat SREBP-1a into pET28a vector on the basis of pSREBP-1a (NCBI Reference Sequence: NP_001263636.1). The coding sequence was amplified by PCR from rat hepatocyte cDNA using HotStar HiFidelity Taq Polymerase kit (Qiagen). The forward primer (EcoRI sited added) was CCGGAATTCATGGACGAGCTACCCTTCGGT, and the reverse primer (XhoI sited added) was CCGCTCGAGTGTGCCTCCTCCACTGCCACAA. The PCR conditions were 95°C for 5 min, 35 cycles: 94°C 15 sec, 50°C 1 min, 68°C 2 min, last extension at 72°C for 10 min..
1.2. Mammalian expression plasmid construction
The coding sequence of rat nSREBP-1a was amplified by PCR from rat hepatocyte cDNA using HotStar HiFidelity Taq Polymerase kit. The PCR conditions were as follows. SREBP-1a-F: CCGGAATTCATGGACGAGCTACCCTTCGGT (EcoRI site added) and SREBP-1a-R: CCGCTCGAGCAGGGCCAGGCGGGAGCG (XhoI site added). The insert of nSREBP-1a was cloned into pcDNA3.1/Zeo+ between EcoRI and XhoI sites. The coding sequence of rat nSREBP-1c was amplified by PCR from rat hepatocyte cDNA. The PCR primers were SREBP-1c-EcoRI-F: CCGGAATTCCCACCATGGATTGCACATTTGAAGACATG (EcoRI site added, Kozak sequence added) and SREBP-1c- XhoI-R: CCGCTCGAGCAGGGCCAGGCGGGAGCG (XhoI site added). Thermal Cycle conditions were described above. The insert of nSREBP-1c was cloned into pcDNA3.1/Zeo+.
1.3. Site-directed mutagenesis
Site-directed mutagenesis was performed according to the manual of QuikChange Site-Directed Mutagenesis Kit from Stratagene. For S308A or S332A, the following primers were used. Mut S to A-F: GAAGCGCTACCGTTCCGCTATCAATGACAAGATTG and Mut S to A-R: CAATCTTGTCATTGATAGCGGAACGGTAGCGCTTC. For S308D or S332D, the following primers were used. Mut S to D-F: GAAGCGCTACCGTTCCGATATCAATGACAAGATTG and Mut S to D-R: CAATCTTGTCATTGATATCGGAACGGTAGCGCTTC.
1.4. Expression of pET28a-nSREBP-1a (1–403aa)
200ml of LB-broth was inoculated with 5 ml of a stationary phase culture of BL21(DE3) transformed with pET28a-nSREBP-1a (1–403aa) and grown at 37°C until an absorbance of 0.5 at 590 nm was reached. After adding IPTG to a final concentration of 1 mM, the cells were grown for an additional 4 h at 37°C.
1.5. Purification of the Recombinant nSREBP-1a (1–403aa)
The buffers used in the purification process contained 50mM NaH2PO4, 10mM Tris, 300mM NaCl and varying concentrations of imidazole. The lysis buffer contained 50 mm imidazole, the wash buffer 150 mM imidazole and the elution buffer 250 imidazole. The transfected BL21(DE3) cells were resuspended in 5 ml of lysis buffer and sonicated with a sonic dismembrator (Fisher Scientific) for 30 sec with a 30 sec interval 10 times. After centrifugation at 12,000 × g for 15 min at 4°C, 5ml of the supernatant was loaded on the 1m HiPur Ni-NTA Spin column (Thermo Scientific). Following incubation for 1h at 4°C, the resin was washed three times with 5ml wash buffer. The His-tagged nSREBP-1a was eluted from the resin by adding 0.5ml elution buffer for four times. The samples were loaded on a SDS-PAGE gel and the gels were stained with GelCode Blue Stain Reagent (Thermo Scientific).
1.6. In vitro PKA phosphorylation
5µg of purified nSREBP-1a was incubated at 30°C for 2hrs with 2500U PKA under 1× PKA Reaction Buffer (pH 7.5): 50 mM Tris-HCl, 10 mM MgCl2 and supplemented with/without 200 µM ATP.
1.7. Pro-Q Diamond and GelCode Blue staining
Gels were stained with either regular GelCode Blue or the Pro-Q Diamond (Invitrogen). SDS-polyacrylamide gel electrophoresis was performed by standard methods utilizing 5% stacking gels, pH 6.8 and 10% separating gels, pH 8.8. Fluorescent staining of SDS-polyacrylamide gels using Pro-Q Diamond phosphoprotein gel stain was performed by fixing the gels in 45% methanol, 5% acetic acid for 1h, washing with three changes of deionized water for 10–20 min per wash, followed by incubation in Pro-Q Diamond phosphoprotein gel stain for 90 min, and destaining with 3 washes in 20% acetonitrile in 50 mM sodium acetate, pH 4.0, for 30–60 min each. Before imaging, the gels were rehydrated in deionized water for 30 min. Images were acquired on the stained gels on a 300 nm UV transilluminator from an AlphaImager EP System (Alpha Innotech). GelCode Blue staining was performed by according to the manufacture manual and the images were taken using the same system.
1.8. LC-MS/MS
Following SDS-PAGE fractionation, the band corresponding to phosphorylated nSREBP-1a was excised from the gel, the gel pieces placed in a polypropylene tube, de-stained twice with 50% acetonitrile in 50 mM NH4HCO3, and dehydrated in a vacuum centrifuge. To digest the phosphoprotein, the gel pieces were re-hydrated for 45 min with a solution containing 100 ng trypsin or Lys-C in 50 mM ammonium bicarbonate. Excess trypsin or Lys-C digestion solution was removed and replaced with 50mM ammonium bicarbonate; the digestion proceeded for 12 h at 37°C. The peptide digest solution was collected and residual peptides were extracted from the gel pieces with a solution of 60% acetonitrile/35% water/5% trifluoroacetic acid; the extract was combined with the original digest solution and dried in a vacuum centrifuge. To enrich for phosphopeptides, the digests were subjected to Immobilized Metal Ion Affinity Chromatography (IMAC) as previously described [23]. The enriched digests were analyzed by LC-MS/MS performed with a high-sensitivity system consisting of a nanoflow LC (FAMOS/ULTIMATE from Dionex) coupled to a linear ion trap mass spectrometer (LTQ from ThermoElectron). The LC separation of the enriched phosphopeptide mixtures was performed on-line with a C18 reversed-phase column, using a fused silica capillary column/spray needle packed in-house with reversed-phase particles (MAGIC C18 from Michrom Bioresources). The peptides eluted from LC were ionized by nano-electrospray and analyzed with the linear ion trap mass spectrometer. The LC was performed at a flow rate of 200 nL/min using a 90-min linear gradient from 0% to 90% mobile phase B. Mobile phase B was 10% water/90% methanol/0.05% formic acid; mobile phase A was 98% water/2% methanol/0.05% formic acid [24]. For each peptide, MS and MS/MS data were acquired in the data-dependent mode. The LC-MS/MS data were used to query the UniProt protein sequence database with the program TurboSEQUEST, part of the Bioworks 3.2 LTQ software. The search parameters included phosphorylation of S, T, Y, and oxidation of M as dynamic modifications, and carbamidomethylation of C as a static modification. The structure-diagnostic fragmentation patterns generated by MS/MS were used to: (a) obtain the primary sequence of the peptide, and (b) locate the phosphorylated amino acid(s) within the peptide.
1.9. Sequence alignment
Human and rat SREBP-1a and 1c protein sequence alignment was performed by Vector NTI 9 and GeneDoc 2.6.
1.10. Immunoblot Analysis
Western analysis was conducted as previously described [25]. The immunoblots were scanned and quantified using Quantity One software from Bio-Rad.
1.11. Luciferase Assay
HEK293 cells were plated in 48-well plate (2×104 cells/well) in DMEM containing 5 mM glucose. On the second day, 250ng pcDNA3.1 or 220ng pcDNA3.1-nSREBP-1c (WT, 308A and 308D)/pcDNA3.1-nSREBP-1a (WT, 332A and 332D) plus 30ng pGL4-FASN-Luciferase (or pGL3-5xSRE-Luciferase) were transfected into the cells. On the fourth day, cells were harvested in 200µl passive lysis buffer and luciferase activity was quantified with a fluorometer (Turner Designs) using the Dual-Luciferase Reporter Assay System (Promega)..
2. Results
2.1. Expression, purification and in vitro PKA phosphorylation of nSREBP-1a
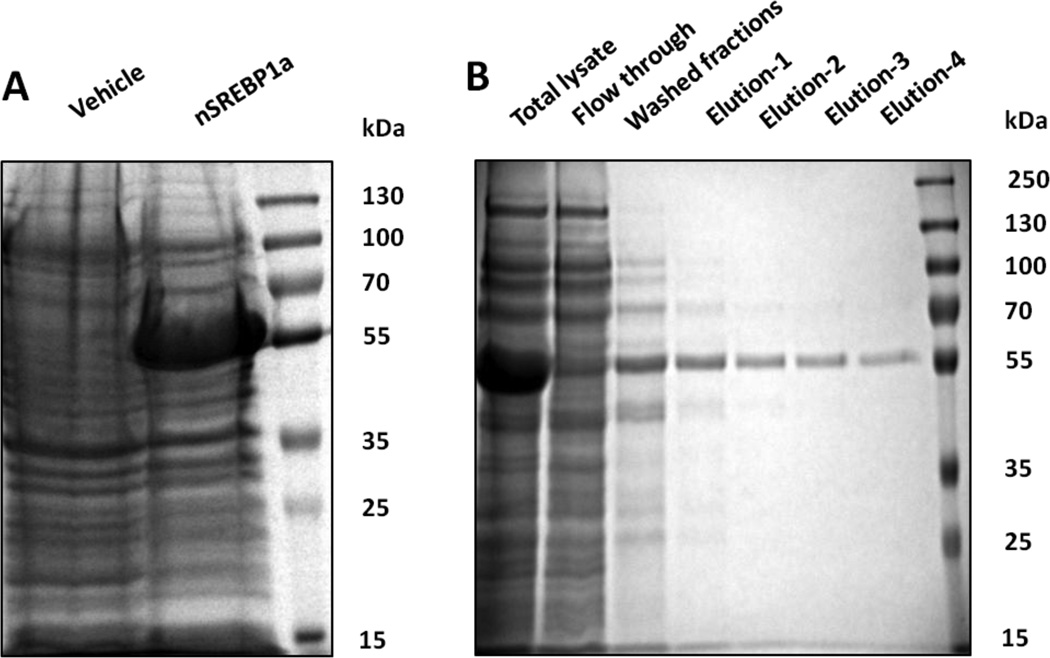
Expression of nSREBP-1a in BL21(DE3) E. coli yielded a high level of the recombinant protein shown as a 46 kDa band in total cell extract of induced cells transformed with pET28a-nSREBP-1a (Fig. 1A). Recombinant His-tagged nSREBP-1a was purified with an IMAC column. As shown in Figure 1B, this purification method yielded highly pure protein fractions.
Fig. 1.
Expression and purification of nSREBP-1a (1–403aa) in E. coli. (A) pET28a and pET28anSREBP-1a were transformed into E. coli BL21(DE3) and expressed as described in the methods. The cell extract was loaded on SDS-PAGE gel and analyzed by staining with GelCode Blue Stain Reagent. The two his tagged nSREBP-1a (1–403aa) proteins are about 46kDa. (B) Eluted fractions from the Ni-NTA resin are shown.
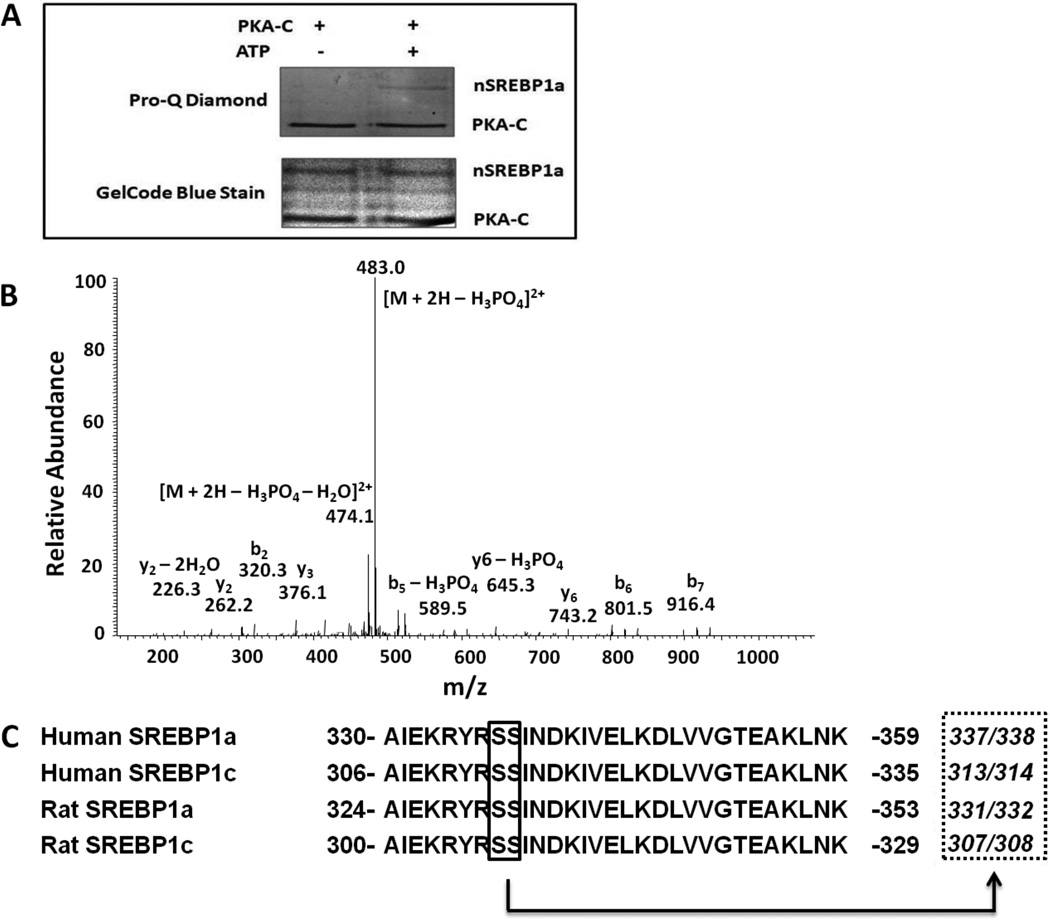
To confirm that nSREBP-1a was phosphorylated by PKA, we assessed phosphorylation of the protein following incubation with the catalytic subunit PKA-C in the presence and absence of a phosphate donor (ATP) (Fig. 2A). In the presence of ATP, this reaction generated phosphorylated nSREBP-1a, as confirmed with phospho-specific gel staining (Pro-Q Diamond; Fig. 2A). An additional phosphorylated protein band was detected in both ATP-treated and control sample. This band, which appeared at ca. 38 kDa, corresponds to the constitutively phosphorylated catalytic subunit of PKA.
Fig. 2.
Mass spectrometry analysis of SREBP-1a. (A) In vitro phosphorylation of bacterial expressed nSREBP-1a (1–403aa). Purified nSREBP-1a was treated with PKA-C. The protein was loaded on SDS-PAGE gel and stained with Pro-Q Diamond Phosphoprotein gel stain (upper panel) or with GelCode Blue Stain Reagent (lower panel). The upper band of the GelCode Blue Stain reagent stained bands was cut for LC-MS/MS. (B) Mass spectrometry (LC-MS/MS) analysis of PKA-treated SREBP-1a. The MS/MS spectrum of a phosphorylated peptide that matched the amino acid sequence YRSSINDK of rat SREBP -1a. The fragment ions in the spectrum localize the phosphorylation site at either S331 or S332. (C) Sequence alignment of human and rat SREBP-1a and 1c around the PKA phosphorylation site. The conserved amino acid sequence between human and rat SREBP-1a and 1c is shown.
2.2. Mass spectrometry
To identify the phosphorylation sites in nSREBP-1a that are target of PKA, the phosphorylated protein was subjected to digestion followed by LC-MS/MS and database searches. To maximize protein coverage, in silico digestion of the protein sequence with different proteases was performed. This in silico analysis indicated that Lys-C and trypsin provided complete coverage of potential phosphorylation sites in nSREBP-1a. Thus, these two proteases were selected for the digestion of recombinant phosphorylated nSREBP-1a. To enrich the mixtures for phosphopeptides, the digests were subjected to IMAC purification. The enriched fractions were analyzed by LC-MS/MS, and the datasets used for searches of protein sequence database. A single phosphorylated peptide was revealed. The MS/MS spectrum, shown in Fig. 2B, displays a phosphate-diagnostic product ion corresponding to loss of phosphoric acid from the activated molecular ion. Furthermore, the spectrum contains a number of sequence-determining product ions that match the amino acid sequence of YRSSINDK of rat SREBP-1a. This peptide contains three potential phosphorylation sites. The MS/MS data (Fig. 2B) exclude Y329 as a phosphorylation candidate site, and localize the phosphorylation to either S331 or S332.
2.3. Sequence alignment
The sequence alignment of human and rat SREBP-1a and 1c in the sequence region around the PKA phosphorylation sites is shown in Fig. 2C. This region is highly conserved between human and rat SREBP-1a and 1c suggesting that PKA phosphorylates human and rat SREBP-1a and 1c in vivo.
2.4. Stability and Transcriptional activity
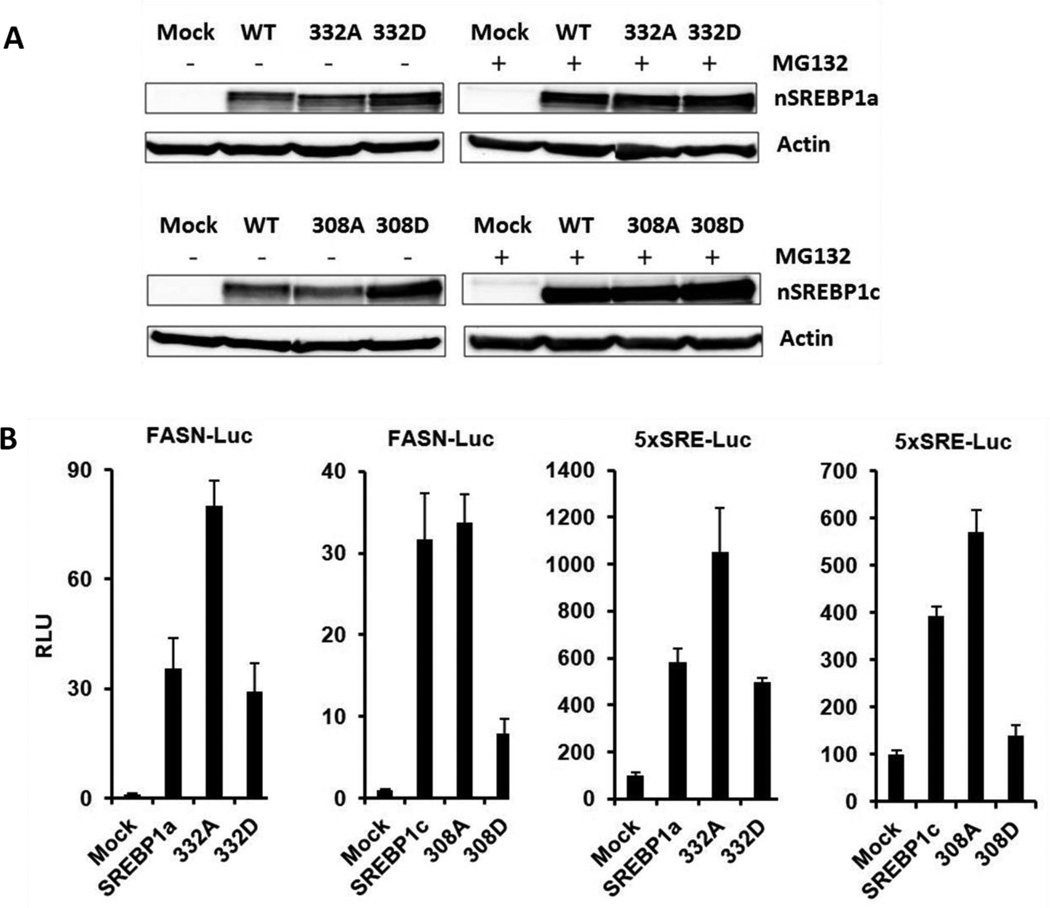
We assessed the effect of loss of function (Ser to Ala, S to A) and gain of function (Ser to Asp, S to D) mutations on stability of SREBP1c and 1a. The results showed that the stability of both nSREBP1a and nSREBP1c containing aspartic acid substitution at S332 or S 308 (D mutant) was not decreased but rather was slightly increased (Fig. 3A). On the contrary, alanine substitution at S332 or S308 (A mutant) had no effect on stability of nSREBP1a and 1c (Fig. 3A). When the transfected cells were treated with 10µM protease inhibitor MG132, the protein levels of WT and both mutants were the same (Fig. 3A). These results showed that phosphomimetic mutation of the putative PKA sites S332D of nSREBP1a and S308D of nSREBP1c does not decrease SREBP-1a and 1c stability, respectively.
Fig. 3.
Stability and transcriptional activity of SREBP mutants. (A) HEK293 cells were transfected with expression plasmids encoding nSREBP-1a (WT, 332A and 332D) or nSREBP-1c (WT, 308A and 308D). After incubation for 18h, the cells were treated with/without proteasomal inhibitor MG132 (10 µM) for 6h. Total cell lysates were extracted and Western blots were performed. Actin was used as the loading control. (B) HEK293 cells were cotransfected with a luciferase reporter (Luc) containing the promoter region of the FASN gene (FASN-Luc) or 5xSRE-Luc along with expression plasmids encoding either the WT or the mutant forms of nSREBP-1a or nSREBP-1c. Luciferase assays were conducted. Data represent the means ± SEMs of three independent experiments. During reporter assays, the level of mutant proteins was normalized to that of WT nSREBP-1a or 1c.
The ability of WT and mutant forms of nSREBP-1 to activate transcription of the known target gene FASN and a synthetic Sterol Response Element (5xSRE) was assessed using luciferase (Luc) assays. In these reporter assays, the level of PKA mutant proteins was normalized to that of WT nSREBP-1a or 1c. WT nSREBP-1a increased relative luciferase activity of pGL4-FAS-Luc and 5xSRE about 35 and 5.8 fold, respectively, as compared to mock vector transfected cells (Fig. 3B). Similarly WT nSREBP-1c increased relative luciferase activity of pGL4-FAS-Luc and 5xSRE about 31 and 3.9 fold, respectively (Fig. 3B). Mutation of S332 and S308 to the non-phosphorylatable amino acid alanine (332A, 308A) of n-terminal SREBP-1a and 1c enhanced transactivation of the luciferase report genes FASN and 5xSRE (Fig. 3B). Conversely, mutation of these serines to the phosphomimetic amino acid aspartic acid (332D, 308D) strongly inhibited the transactivating capacity of nSREBP-1a and 1c (Fig. 3B).
Discussion
During fasting or acute stress, the biosynthesis of lipids in the liver is inhibited in part by cAMP [26]. Glucagon, adrenaline and other hormones that raise cAMP levels reduce the activity and abundance of lipogenic enzymes, which are critical for the synthesis of fatty acids and triglycerides [27,28]. Glucagon represses SREBP-1c activity through PKA signaling cascades [17,20]. In addition, the expression of SREBP-1c is suppressed by fasting or nutritional deprivation [29,30]. Although the activity of SREBP-1c is regulated by PKA or AMP-activated protein kinases (AMPKs), the exact underlying mechanisms have yet to be determined in vivo [17,31].
As the key regulatory factor in lipogenesis, SREBPs are targets of hormones such as insulin, glucagon, and growth factors [19,32,33]. Activation of SREBPs is tightly regulated at multiple levels, including transcription, precursor maturation, nuclear protein stability and transcriptional activity. The abundance of the nuclear form of SREBPs is controlled by transcriptional upregulation followed by proteolytic cleavage. However, increasing evidence supports the hypothesis that posttranslational modifications of SREBPs modulate their transcriptional activity and stability [34,35,36]. Lu, et al. [17] identified two PKA consensus sites in the NH2 terminus of human SREBP-1a, S337 and S338 and postulated that S338 might mediate the inhibitory effect of PKA activation on SREBP-1a. Using unbiased mass spectrometry, we have directly confirmed phosphorylation of the corresponding serines (S331/S332) of rat SREBP-1a by PKA. Furthermore, we determined that phosphorylation of S332 and the corresponding site (S308) of rat SREBP-1c reduces transcriptional activity. Thus it is likely that phosphorylation of SREBP-1 isoforms at this site mediates the inhibitory effect of cAMP on SREBP-1. Recently, Lee, et al. [37] linked phosphorylation of SREBP-1c to sumoylation at Lys98 leading to enhanced ubiquitination and degradation. They observed that the extent of sumoylation and ubquitination was reduced following mutation of S308 to alanine in nSREBP-1c and postulated that sumoylation may be regulated by phosphorylation of this serine by PKA [37]. Interestingly, in our studies we observed that mutation of this site, either in the context of full-length nascent SREBP-1c or its n-terminal fragment nSREBP-1c did not appreciably alter steady state levels of SREBP-1c protein. This does not eliminate a role of PKA mediated phosphorylation in stability of SREBP-1c protein but does not support the hypothesis that phosphorylation of this serine mediates the effect of PKA on stability. Rather, our data indicates that the predominant effect of PKA mediated phosphorylation SREBP1a at S332 and the corresponding site on SREBP-1c (S308) is to decrease its transcriptional activity.
In conclusion, we used a discovery based mass spectrometry approach to find the phosphorylation target sites of PKA in SREBP-1a. With this unbiased methodology, we have identified S331/S332 of SREBP-1a as a phosphorylation target of PKA. Mutational analysis of this site confirmed that phosphorylation of SREBP-1a at S332 and SREBP-1c at S308 by PKA decreases transcriptional activity of nSREBP-1a and 1c. This site is a strong candidate for mediating the negative regulatory effect of glucagon on SREBP-1c and lipogenesis.
Highlights.
Phosphorylation of SREBP-1 regulates its processing and transcriptional activity.
PKA mediates the effects of Glucagon and cAMP by downregulating SREBP-1 activity.
Using MS we identified S331/S332 of SREBP-1a as phosphorylation targets of PKA.
S332D of SREBP-1a and S308D of SREBP-1c reduce their transcriptional activity.
S331/S332 of SREBP-1 are candidates for PKA activators to reduce lipid synthesis.
Acknowledgements
This project was funded by VA Research and Development Program Merit Review Grants to M.B.E. and R.R. Funds for the LTQ mass spectrometer were provided by the NIH Shared Instrumentation Grant S10RR16679, and by the UTHSC College of Pharmacy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reaven GM. Insulin resistance: the link between obesity and cardiovascular disease. Med Clin North Am. 2011;95:875–892. doi: 10.1016/j.mcna.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Kaidar-Person O, Bar-Sela G, Person B. The two major epidemics of the twenty-first century: obesity and cancer. Obes Surg. 2011;21:1792–1797. doi: 10.1007/s11695-011-0490-2. [DOI] [PubMed] [Google Scholar]
- 3.Cheung O, Sanyal AJ. Abnormalities of lipid metabolism in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28:351–359. doi: 10.1055/s-0028-1091979. [DOI] [PubMed] [Google Scholar]
- 4.Ferre P, Foufelle F. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm Res. 2007;68:72–82. doi: 10.1159/000100426. [DOI] [PubMed] [Google Scholar]
- 5.Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiaoping Z, Fajun Y. Regulation of SREBP-Mediated Gene Expression. Sheng Wu Wu Li Hsueh Bao. 2012;28:287–294. doi: 10.3724/SP.J.1260.2012.20034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghow R, Yellaturu C, Deng X, Park EA, Elam MB. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J. Clin. Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan TO, Pascal JM, Armen RS, Rodeck U. Autoregulation of kinase dephosphorylation by ATP binding in AGC protein kinases. Cell Cycle. 2012;11:475–478. doi: 10.4161/cc.11.3.19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKnight GS, Cummings DE, Amieux PS, Sikorski MA, Brandon EP, Planas JV, Motamed K, Idzerda RL. Cyclic AMP, PKA, and the physiological regulation of adiposity. Recent Prog Horm Res. 1998;53:139, 159. discussion 160–131. [PubMed] [Google Scholar]
- 15.Cummings DE, Brandon EP, Planas JV, Motamed K, Idzerda RL, McKnight GS. Genetically lean mice result from targeted disruption of the RII beta subunit of protein kinase A. Nature. 1996;382:622–626. doi: 10.1038/382622a0. [DOI] [PubMed] [Google Scholar]
- 16.Schreyer SA, Cummings DE, McKnight GS, LeBoeuf RC. Mutation of the RIIbeta subunit of protein kinase A prevents diet-induced insulin resistance and dyslipidemia in mice. Diabetes. 2001;50:2555–2562. doi: 10.2337/diabetes.50.11.2555. [DOI] [PubMed] [Google Scholar]
- 17.Lu M, Shyy JY. Sterol regulatory element-binding protein 1 is negatively modulated by PKA phosphorylation. Am J Physiol Cell Physiol. 2006;290:C1477–C1486. doi: 10.1152/ajpcell.00374.2005. [DOI] [PubMed] [Google Scholar]
- 18.Deng X, Cagen LM, Wilcox HG, Park EA, Raghow R, Elam MB. Regulation of the rat SREBP-1c promoter in primary rat hepatocytes. Biochem Biophys Res Commun. 2002;290:256–262. doi: 10.1006/bbrc.2001.6148. [DOI] [PubMed] [Google Scholar]
- 19.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Liepvre X, Berthelier-Lubrano C, Spiegelman B, Kim JB, Ferre P, Foufelle F. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yellaturu CR, Deng X, Cagen LM, Wilcox HG, Park EA, Raghow R, Elam MB. Posttranslational processing of SREBP-1 in rat hepatocytes is regulated by insulin and cAMP. Biochem Biophys Res Commun. 2005;332:174–180. doi: 10.1016/j.bbrc.2005.04.112. [DOI] [PubMed] [Google Scholar]
- 21.Yellaturu CR, Deng X, Park EA, Raghow R, Elam MB. Insulin enhances the biogenesis of nuclear sterol regulatory element-binding protein (SREBP)-1c by posttranscriptional down-regulation of Insig-2A and its dissociation from SREBP cleavage-activating protein (SCAP).SREBP-1c complex. J Biol Chem. 2009;284:31726–31734. doi: 10.1074/jbc.M109.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotzka J, Knebel B, Haas J, Kremer L, Jacob S, Hartwig S, Nitzgen U, Muller-Wieland D. Preventing phosphorylation of sterol regulatory element-binding protein 1a by MAP-kinases protects mice from fatty liver and visceral obesity. PLoS One. 2012;7:e32609. doi: 10.1371/journal.pone.0032609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Fang B, Giorgianni F, Gingrich JR, Beranova-Giorgianni S. Investigation of phosphoprotein signatures of archived prostate cancer tissue specimens via proteomic analysis. Electrophoresis. 2011;32:1984–1991. doi: 10.1002/elps.201100101. [DOI] [PubMed] [Google Scholar]
- 24.Giorgianni F, Cappiello A, Beranova-Giorgianni S, Palma P, Trufelli H, Desiderio DM. LC-MS/MS analysis of peptides with methanol as organic modifier: improved limits of detection. Anal Chem. 2004;76:7028–7038. doi: 10.1021/ac0493368. [DOI] [PubMed] [Google Scholar]
- 25.Yellaturu CR, Deng X, Cagen LM, Wilcox HG, Mansbach CM, 2nd, Siddiqi SA, Park EA, Raghow R, Elam MB. Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J Biol Chem. 2009;284:7518–7532. doi: 10.1074/jbc.M805746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarlow DM, Watkins PA, Reed RE, Miller RS, Zwergel EE, Lane MD. Lipogenesis and the synthesis and secretion of very low density lipoprotein by avian liver cells in nonproliferating monolayer culture. Hormonal effects. J Cell Biol. 1977;73:332–353. doi: 10.1083/jcb.73.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girard J, Perdereau D, Foufelle F, Prip-Buus C, Ferre P. Regulation of lipogenic enzyme gene expression by nutrients and hormones. FASEB J. 1994;8:36–42. doi: 10.1096/fasebj.8.1.7905448. [DOI] [PubMed] [Google Scholar]
- 28.Lefevre P, Diot C, Legrand P, Douaire M. Hormonal regulation of stearoyl coenzyme-A desaturase 1 activity and gene expression in primary cultures of chicken hepatocytes. Arch Biochem Biophys. 1999;368:329–337. doi: 10.1006/abbi.1999.1315. [DOI] [PubMed] [Google Scholar]
- 29.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, Musi N, Hirshman MF, Goodyear LJ, Moller DE. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason RJ, Pan T, Edeen KE, Nielsen LD, Zhang F, Longphre M, Eckart MR, Neben S. Keratinocyte growth factor and the transcription factors C/EBP alpha, C/EBP delta, and SREBP-1c regulate fatty acid synthesis in alveolar type II cells. J Clin Invest. 2003;112:244–255. doi: 10.1172/JCI16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth G, Kotzka J, Kremer L, Lehr S, Lohaus C, Meyer HE, Krone W, Muller-Wieland D. MAP kinases Erk1/2 phosphorylate sterol regulatory element-binding protein (SREBP)-1a at serine 117 in vitro. J Biol Chem. 2000;275:33302–33307. doi: 10.1074/jbc.M005425200. [DOI] [PubMed] [Google Scholar]
- 36.Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Lee GY, Jang H, Lee JH, Huh JY, Choi S, Chung J, Kim JB. PIASy-mediated sumoylation of SREBP1c regulates hepatic lipid metabolism upon fasting signaling. Mol Cell Biol. 2014;34:926–938. doi: 10.1128/MCB.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]