Abstract
Background
A central problem in neuroscience is elucidating synaptic connections, the connectome. Because mammalian forebrains contain many neurons, labeling specific neurons with unique tags is desirable. A novel technology, Brainbow, creates hundreds of hues by combinatorial expression of multiple fluorescent proteins (FPs).
New method
We labeled small numbers of neurons, and their axons, with unique hues, by expressing Brainbow from a helper virus-free Herpes Simplex Virus (HSV-1) vector.
Results
The vector expresses a Brainbow cassette containing four FPs from a glutamatergic-specific promoter. Packaging HSV-brainbow produced arrays of seven to eight Brainbow cassettes, and using Cre, each FP gene was in a position to be expressed, in different cassettes. Delivery into rat postrhinal (POR) cortex or hippocampus labeled small numbers of neurons with different, often unique, hues. An area innervated by POR cortex, perirhinal (PER) cortex, contained axons with different hues. Specific axons in PER cortex were matched to specific cell bodies in POR cortex, using hue.
Comparison with existing methods
HSV-Brainbow is the only technology for labeling small numbers of neurons with unique hues. In Brainbow mice, many neurons contain the same hue. Brainbow-adeno-associated virus vectors require transduction of the same neuron with multiple vector particles, confounding neuroanatomical studies. Replication-competent Brainbow-pseudorabies virus vectors label multiple neurons with the same hue.
Conclusions
Attractive properties of HSV-Brainbow include each vector particle contains multiple cassettes, representing numerous hues, recombination products are stabile, and experimental control of the number of labeled neurons. Labeling neurons with unique hues will benefit mapping forebrain circuits.
Keywords: Brainbow, fluorescent protein, Cre-mediated recombination, hue, axon projection, herpes simplex virus vector
1. Introduction
Nervous system functions, ranging from physiological homeostasis to advanced cognitive tasks, are encoded in specific neuronal circuits, composed of specific neurons and synapses. Thus, elucidating all the synaptic connections in a specific nervous system, the connectome, is a central problem in neuroscience, and a challenging task, due to the large numbers of neurons and synapses in a nervous system. Nonetheless, some remarkable progress has been made. The entire connectome for the nematode C. elegans was determined by electron microscopy (White et al., 1986). C. elegans contains a relatively small number, ~300, neurons; in contrast, mammalian nervous systems, and especially the mammalian forebrain, are considerably more complex. The mouse forebrain contains 103 to 104 types of neurons (Sugino et al., 2006), a single rat neocortical column contains ~7,500 neurons (Peters and Jones, 1984), and human neocortex contains ~109 synapses per cubic mm (Alonso-Nanclares et al., 2008). Of note, specific projections in mammalian nervous systems have been mapped using a wide range of techniques (Luo et al., 2008; Zaborszky et al., 2006), including electron microscopy, classical anterograde and retrograde tracers, genetic tracers (Lo and Anderson, 2011), genetic synaptic markers (Feinberg et al., 2008), and viruses (Ekstrand et al., 2008). Specific circuits have been mapped, such as the connections between the different primate neocortical areas that process visual information (Felleman and Van Essen, 1991). However, efficient, high-resolution mapping of forebrain circuits, at the level of individual neurons and axons, remains a challenge.
One challenge for track tracing technologies is to uniquely label individual neurons. Many track tracing technologies label multiple neurons with the same tag, and obtain resolution from serial section reconstruction and process tracing. These technologies range from Golgi staining, as pioneered by Cajal, to specific genetic track tracers, such as expression of a single fluorescent protein (FP). Specific modifications to these techniques support labeling neurons with several different tags, such as several fluorescent dyes or two FPs; these approaches are advantageous for many studies, but, nonetheless, label multiple neurons with the same tag. Recently, a powerful new technology, Brainbow, was developed that creates hundreds of hues by combinatorial expression of different FPs (Livet et al., 2007); a Brainbow cassette contains two to four different FPs, Cre-mediated recombination probabilistically determines which FP is expressed, and an array of Brainbow cassettes results in expression of one specific combination of FPs, from hundreds of potential combinations, or hues. Nonetheless, in Brainbow transgenic mice (Livet et al., 2007), multiple neurons are labeled with the same hue, because the number of labeled neurons far exceeds the number of hues produced by Brainbow; this issue exists even with advantageous Brainbow array integration site, Cre-driver mouse line, and Brainbow promoter. Thus, in Brainbow mice, serial section reconstruction and individual axon tracing would be required to map the projections of individual neurons. To address these issues, mice with Brainbow recombination limited to a specific neuron type might be advantageous; Brainbow mice with serotonergic-specific Cre-expression have been reported (Weber et al., 2009), but analogous approaches in the forebrain will likely label large numbers of neurons, resulting in a challenging axon mapping problem. Further, a number of second-generation Brainbow constructs in transgenic mice have been explored to increase expression levels, and better control Cre-mediated recombination; nonetheless, the use of Brainbow mice for mapping forebrain circuits remains problematic (Cai et al., 2013).
Neurons might be labeled with unique hues by expressing Brainbow in a limited number of neurons, using a virus vector. However, as a single Brainbow cassette is several kb, an array of five to ten Brainbow cassettes will challenge, or exceed, the capacity of a number of virus vectors. Moreover, a procedure is required to obtain Cre-mediated recombination of Brainbow in the chosen virus vector. Adeno-associated virus (AAV) vectors that express Brainbow require multiple infections of the same cell to generate a modest number of colors, color spectrum varies with distance from the injection site, and this system appears difficult to use for neuroanatomical applications (Cai et al., 2013). Another approach obtained multiple hues by coinfecting cells with multiple viruses that each express a single, but different, FP: A replication-competent pseudorabies virus (PRV) vector containing a single Brainbow cassette, propagated in the presence of Cre, supports production of progeny virus that express different FPs, and coinfection of the same cell by multiple progeny viruses produces different hues (Kobiler et al., 2010). Further, this approach has been used in transneuronal mapping studies (Card et al., 2011). Nonetheless, this approach labels multiple neurons with the same hue, due to use of a replication-competent vector that labels many neurons. Further, PRV vectors that coexpress membrane-bound and soluble FPs have been used to determine the sequence of connections in a circuit (Boldogkoi et al., 2009). This approach is useful for transneuronal mapping studies, but expression of the FPs is predetermined rather than probabilistic, so the number of hues is limited, and these replication-competent vectors label many neurons with the same hue.
Helper virus-free Herpes Simplex Virus (HSV-1) plasmid (amplicon) vectors (Fraefel et al., 1996; Geller and Breakefield, 1988) have attractive properties for supporting Brainbow expression in small numbers of neurons, thus labeling specific neurons with unique hues. As these vectors are replication-defective, the number of transduced neurons can be experimentally controlled, via the titer of the inoculum. Importantly, the molecular biology of HSV-1 DNA replication and packaging provides a convenient method to produce an array of Brainbow cassettes: HSV-1 DNA replication is biphasic; first, the input DNA molecules are replicated, and, second, rolling circle replication produces concatamers (Roizman and Sears, 1993). With helper virus-free packaging of HSV-1 vectors, an HSV-1 genome-sized (~152 kb) array of concatamers is packaged into HSV-1 particles (Fraefel et al., 1996). For example, using an ~20 kb vector, seven or eight concatamers are packaged into a single HSV-1 particle. Thus, a single infection event could label neurons with a wide range of hues. Moreover, Brainbow recombination might be supported during vector packaging; Cre-mediated recombination can occur during HSV-1 vector replication and packaging (Logvinoff and Epstein, 2001). Thus, as Cre can be present only during packaging, and can be absent from the mammalian brain, the specific Brainbow configuration produced during packaging will be stable thereafter.
Here we describe labeling small numbers of forebrain neurons, and their distant axons, with unique hues, by expressing a Brainbow array from a HSV-1 vector. A Brainbow cassette, containing four axon-targeted FPs, was placed under the control of a glutamatergic neuron subtype-specific promoter (Rasmussen et al., 2007; Zhang and Geller, 2010), and inserted into a HSV-1 vector. Packaging this vector produced arrays of Brainbow, and, following packaging in the presence of Cre, each of the four FP genes was in a position to be expressed, in different Brainbow cassettes. Injection of a vector stock into rat postrhinal (POR) cortex or hippocampus labeled small numbers of neurons in each area with different, and often unique, hues. Further, in an area that receives a large projection from POR cortex, perirhinal (PER) cortex (Agster and Burwell, 2009; Burwell and Amaral, 1998), specific axons were labeled with different hues. Specific axons in PER cortex could be matched to specific neuronal cell bodies in POR cortex, based on hue. As detailed in the discussion, the primary advantage of HSV-Brainbow is the unique capability to label individual neurons with unique hues; in contrast, Brainbow transgenic mice or replication-competent Brainbow-PRV vectors label multiple neurons with the same hue.
2. Methods
2.1. Materials
Restriction endonucleases and DNA modifying enzymes were from New England Biolabs. DNAs, including PCR primers, were synthesized by Genscript. CMV-Cre, pBS185, was obtained from Addgene. OptiMEM, penicillin/streptomycin, Dulbecco’s modified minimal essential medium (DMEM), and fetal bovine serum (FBS) were obtained from Invitrogen. G418 was obtained from RPI, and Lipofectamine® was from Life Technologies. Vectashield was from Vector Laboratories.
2.2. Plasmids and HSV-1 vectors
Each FP gene/transcription unit contained a Kozak consensus translation start sequence (Kozak, 1987) followed by an axon-targeting sequence (the 22 N-terminal amino acids from GAP-43 (Liu et al., 1994)) fused to a specific FP gene, and a specific polyadenylation site. The Brainbow cassette (Fig. 1A) contained Asc I, a 6 bp spacer, Pme I, LoxP (inverted orientation, no ATG), emerald green FP (EmGFP)/mouse α-globin polyadenylation sequence, Cla I, Sac II, mOrange2/bovine growth hormone polyadenylation sequence (non-coding complementary strand, inverted), Nhe I, LoxP (standard orientation), 10 bp spacer, LoxP (inverted orientation), Spe I, LSSmKate2/human β-globin polyadenylation sequence, Xho I, Bgl II, enhanced blue FP-2 (EBFP2)/SV40 early region polyadenylation sequence (non-coding complementary strand, inverted), LoxP (standard orientation), SnaB I, a 6 bp spacer, and Pac I. The Brainbow cassette was synthesized (Genscript) as four fragments, each in pUC57, and then assembled. The first fragment contained the Brainbow backbone, the EmGFP/mouse α-globin polyadenylation sequence, and an initial red FP gene (TagRFP-T/human β-globin polyadenylation sequence); the second fragment contained mOrange2/bovine growth hormone polyadenylation sequence; the third fragment contained EBFP2/SV40 early region polyadenylation sequence; and the fourth fragment contained LSSmKate2/human β-globin polyadenylation sequence. The mOrange2/bovine growth hormone polyadenylation sequence fragment and the EBFP2/SV40 early region polyadenylation sequence fragment were sequentially inserted into the Brainbow backbone/EmGFP/TagRFP-T construct using the indicated flanking sites (listed above); the initial RFP (TagRFP-T) was excised and the LSSmKate2/human β-globin polyadenylation sequence was inserted to yield the Brainbow cassette. The DNA sequence of this Brainbow cassette is in Supporting Fig. S1.
Fig. 1.
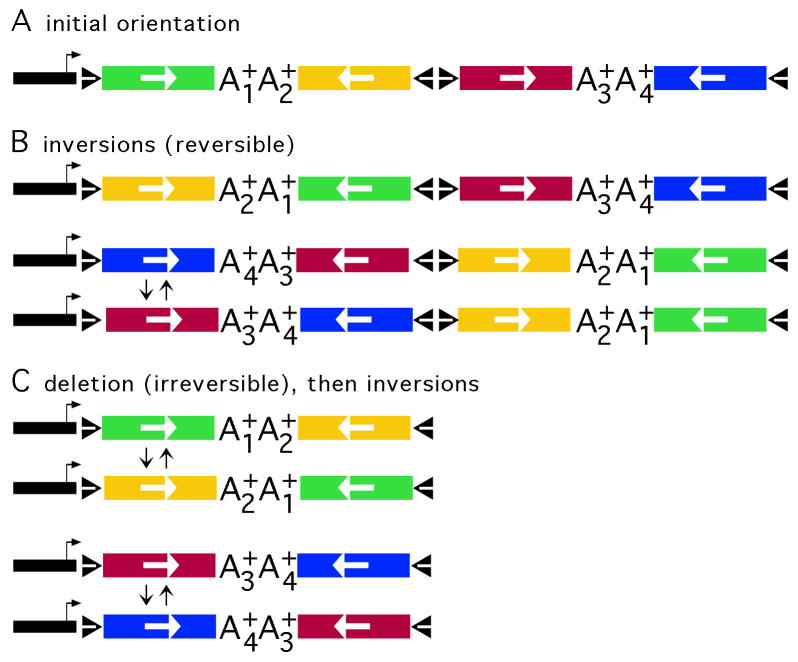
The Brainbow design, and recombination events that can support expression of each FP. (A) The Brainbow transcription unit, as constructed in E. coli, before Cre-mediated recombination; the initial order of the FP genes. The Brainbow transcription unit contains the VGLUT1 promoter (black segment with arrow), loxP sites (black triangles), and four FP genes (EmGFP (Tsien, 1998), green; mOrange2 (Shaner et al., 2008), yellow; LSSmKate2 (Piatkevich et al., 2010), red; or EBFP2 (Ai et al., 2007), blue); the orientation of each gene is indicated by a white arrow; and each gene is followed by a unique polyadenylation site (A1+ to A4+). Cre-mediated recombination between loxP sites in an inverted orientation leads to inversion, which is reversible; Cre-mediated recombination between loxP sites in the same orientation leads to deletion, which is irreversible. (B) Inversions can support expression of each FP gene. The first line shows the product of an inversion between the first and second loxP sites. The second line shows the product of an inversion between the first and fourth loxP sites; and the third line shows the product of a subsequent inversion between the first and second loxP sites. (C) A deletion followed by an inversion can support expression of each FP gene. The first line shows the product of a deletion between the second and fourth loxP sites in the initial orientation; the second line shows the product of a subsequent inversion. The third line shows the product of a deletion between the first and third loxP sites in the initial orientation; the fourth line shows the product of a subsequent inversion.
pVGLUT1linker contains the mouse VGLUT1 promoter and first intron (7 kb and 4.6 kb fragments, respectively) (Rasmussen et al., 2007; Zhang et al., 2012). pVGLUT1linker was digested with Asc I and Pac I, the Brainbow cassette was excised from pUC57 using the same enzymes, and the Brainbow cassette was inserted into pVGLUT1linker to yield pVGLUT1brainbow.
2.3. Brainbow recombination assay
Recombination was assayed by PCR; the 5′ primer was 5′ GAGTCTTTGCTTCTCGCTTCCCTGC 3′ (proximal to the 3′ end of the VGLUT1 promoter, sense); and the 3′ primers were 5′ CAGAAGGCAGCTTAACGGTACTTGG 3′ (from the mouse α-globin polyadenylation sequence, antisense; amplifies the EmGFP gene); 5′ ATGGCTGGCAACTAGAAGGCACAGC 3′ (from the bovine growth hormone polyadenylation sequence, antisense; amplifies the mOrange2 gene); 5′ TAGGCAGAATCCAGATGCTCAAGGC 3′ (from the human β-globin polyadenylation sequence, antisense; amplifies the LSSmKate2 gene); and 5′ AGAAAGCAGACCAAACAGCGGTTGG 3′ (from the SV40 early region polyadenylation sequence, antisense; amplifies the EBFP2 gene).
2.4. Cells and vector packaging
Vectors were packaged into HSV-1 particles using a modified form of the helper-virus free packaging protocol described previously (Fraefel et al., 1996; Sun et al., 1999). A CMV-Cre plasmid was added to specific packaging procedures (70 ng CMV-Cre (pBS185, Addgene) per 0.4 μg pVGLUT1brainbow; excess CMV-Cre inhibits packaging). Vector stocks were purified and concentrated as described (Fraefel et al., 1996; Sun et al., 1999). Purified vectors were titered on BHK cells by counting GFP positive cells at 24 hours post-transduction; for three vector stocks prepared in the presence of Cre, and used for experiments in the rat brain, the titers were 1.0 to 1.5 × 106 infectious vector particles (IVP)/ml, as measured in the EmGFP channel (see below). To optimize the packaging and recombination procedures, over ten vector stocks were examined, and exhibited similar titers. Wild-type HSV-1 was not detected (<10 plaque forming units/ml) in any of these vector stocks.
2.5. Stereotactic injections of vector stocks into rat POR cortex
The Seton Hall University Institutional Animal Care and Use Committee (IACUC) and the VA Boston Healthcare System IACUC approved all the animal procedures. Adult male Sprague-Dawley rats (250–300 g) were anesthetized by ip injection of a Ketamine (20 mg/ml) Xylazine (2 mg/ml) mixture with a final dose of 60 mg/kg and 6 mg/kg, respectively. Additional anesthesia was administered as needed. Each rat received one injection of a pVGLUT1brainbow vector stock into either POR cortex or hippocampus. The injection coordinates in POR cortex were anterior-posterior (AP) −8.0, medial-lateral (ML) +6.0, dorsal-ventral (DV) −5.2 (Paxinos and Watson, 1986). The injection coordinates in the dentate area of the hippocampus were AP −3.3, ML +2.0, DV −3.5. AP is measured relative to bregma, ML is relative to the sagittal suture, and DV is relative to the bregma-lambda plane. A micropump (model 100, KD Scientific) was used for the injections; 3 μl of vector stock was injected at each site over 5 minutes, and after 5 additional minutes, the needle was slowly retracted.
2.6. Section preparation
Brains were perfused with 50 ml phosphate buffered saline (PBS) followed by 200 ml of 4 % paraformaldehyde in PBS, postfixed in 4 % paraformaldehyde in PBS (4 hr, 4 °C), cryoprotected in 25 % sucrose in PBS (2 days, 4 °C) (Zhang et al., 2000), and 25 μm coronal sections were prepared using a freezing microtome. In POR cortex or hippocampus, sections were prepared proximal to the injection site. In PER cortex, sections were prepared from regions that receive projections from neurons proximal to the injection site, as previously determined (Zhang et al., 2010b). Sections were mounted in Vectashield and stored at −20 °C before imaging (Livet et al., 2007).
2.7. Microscopy
The excitation excitation/absorption and emission spectra have been published for LSSmKate2 (Piatkevich et al., 2010), mOrange2 (Shaner et al., 2008), EmGFP (Tsien, 1998), and EBFP2 (Ai et al., 2007). The maximum efficiency wavelengths for excitation and emission for each FP are in Table 1.
Table 1.
The excitation/absorption and emission maximum wavelengths for the four FPs, and descriptive statistics for the relative intensities of these FPs in 167 neuronal cell bodies in POR cortex.
| FP | Absorption λmax (nm) | Emission λmax (nm) | Channel intensities in POR cortex (%)
|
||
|---|---|---|---|---|---|
| Mean ± s.e.m. | Range | Outliers | |||
| LSSmKate2 | 460 | 605 | 29.3±0.2 | 22–35 | 18, 18, 19 |
| mOrange2 | 549 | 565 | 24.0±0.3 | 14–31 | 33, 36 |
| EmGFP | 487 | 509 | 26.9±0.2 | 20–32 | none |
| EBFP2 | 383 | 448 | 20.2±0.5 | 11–36 | 8, 8, 40, 47 |
The excitation/absorption and emission spectra have been reported for LSSmKate2 (Piatkevich et al., 2010), mOrange2 (Shaner et al., 2008), EmGFP (Tsien, 1998), and EBFP2 (Ai et al., 2007).
Using a macro in ImageJ, each cell body was outlined, and the signal in each channel (color) was determined. For each cell body, the percentage of the signal in each channel was calculated. The means and s.e.m.s were calculated based on all 167 cell bodies. Within each channel, outliers were separated by ≥2 % from the range.
Imaging was performed on two different microscopes, with similar results (one microscope was located at Harvard Medical School, and as the laboratory relocated during this study, the second microscope was located at Rutgers Medical School). First, a Zeiss LSM 510 confocal microscope: All images were obtained using a Plan-Apochromat 63x/1.4 oil objective (Zeiss) and with the same settings. Four sets of excitation and emission conditions were used; each condition is specific for one of the four FPs in pVGLUT1brainbow. The 458 nm Argon laser line was used for imaging LSSmKate2 with emission bandpath set as LongPass 560 nm; the 543 nm HeNe laser line was used for imaging mOrange2 with emission bandpath set as 565–615 nm; the 488 nm Argon laser line was used for imaging EmGFP with emission bandpath set as 500–550 nm; a pulse laser line set at 740 nm (Coherent Chameleon) was used for imaging EBFP2 with emission bandpath set as 390–465 nm. Second, imaging was performed on a Nikon A1r spectral confocal microscope: All images were obtained with the same settings, and all high power images were obtained using a Nikon Plan-Apochromat 60x/1.4 oil objective. Again, four sets of excitation and emission conditions were used, and each condition is specific for one of the four FPs in pVGLUT1brainbow. The 488 nm Argon ion laser line was used to excite both LSSmKate2 and EmGFP, the 561 laser line was used to excite mOrange2, and the 405 laser was used to excite EBFP2. To detect the fluorescence emission, the following band pass filters were prepared using the spectral detector of the A1r confocal microscope: 626–696 nm for LSSmKate2, 576–626 nm for mOrange2, 506–556 nm for EmGFP, and 426–476 nm for EBFP2. The settings used with each microscope were designed to match each FP’s excitation and emission spectra and minimize cross-talk effects.
2.8. Image processing
Images were obtained in imageJ, and converted to .tif files for each channel, for a specific image. These .tif files were imported into Photoshop, converted to RGB 8 bit format, uniformly adjusted for levels, brightness, contrast, dust and scratches, and gamma, assigned colors, and then merged.
2.9. Relative color levels and matching axons to cell bodies
To quantify the levels of each channel in a cell body or axon, we wrote a macro in ImageJ in which the user draws a contour around a cell body or axon, and the program then calculates the average intensity within the contour for each channel. These data were used to calculate the relative fraction of each channel compared to sum of all four channels, for each cell body and axon.
We developed a metric to calculate the distance between a specific axon and a specific cell body. For each color (channel), the differences between the axon and cell body were calculated and squared (for example, [Baxon 5 − Bcell body 36]2), and these four values were summed. This sum is the metric for the color distance.
This is an empirical method for quantifying the hue and matching cell bodies to axons based on hue. We did not attempt to correlate the observed hues to the predicted hues based on vector structure, as hue distribution is likely determined by a number of parameters, in addition to the genetically encoded hues. Other variables that affect the hue distribution include the relative stabilities of the different FPs in different neuron types, and the penetrance of different light frequencies through the section.
3. Results
3.1. Construction of a HSV-1 vector that expresses brainbow
We designed a new Brainbow cassette that followed the Brainbow-2.1 model (Livet et al., 2007), but contained current FP genes (Fig. 1A). We chose four monomeric FPs with readily separable excitation and emission spectra, and favorable protein stabilities and fluorescent-quantum properties. For red, orange, green, or blue FPs, we used LSSmKate2 (Piatkevich et al., 2010), mOrange2 (Shaner et al., 2008), EmGFP (Tsien, 1998), or EBFP2 (Ai et al., 2007), respectively. To target each FP to axons, we fused the axon-targeting domain from GAP-43 (Liu et al., 1994) to the N-terminus of each FP. Each FP gene was followed by an efficient polyadenylation sequence; to facilitate selective detection of each FP gene by PCR, a different polyadenylation sequence followed each FP gene. In this Brainbow design, each of the four FPs can be placed in a position to be expressed by either inversions alone (Fig. 1B) or a deletion followed by an inversion (Fig. 1C). If Cre-mediated recombination occurs at a significant level, then the probabilities for expression of each FP are likely to be similar.
This Brainbow cassette was inserted into a HSV-1 vector that contains the vesicular glutamate transporter-1 (VGLUT1) promoter and a standard vector backbone, yielding pVGLUT1brainbow (Fig. 2A). The VGLUT1 promoter, in HSV-1 vectors, restricts expression to VGLUT1-containing glutamatergic neurons in the rat forebrain (Rasmussen et al., 2007; Zhang and Geller, 2010); VGLUT1-containing glutamatergic neurons are the predominate type of glutamatergic neuron in the neocortex and hippocampus (Fremeau et al., 2004).
Fig. 2.
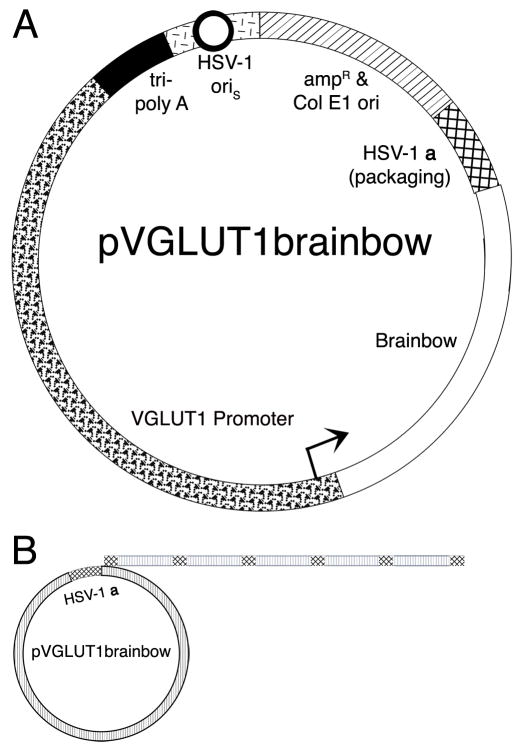
The structure of pVGLUT1brainbow and the production of Brainbow arrays by rolling circle DNA replication. (A) A schematic diagram of pVGLUT1brainbow. The VGLUT1 promoter (wavy segment with arrow) supports expression of Brainbow (clear segment). The vector backbone contains an HSV-1 origin of DNA replication (oriS, black circle with white interior in the short line segment) and an HSV-1 a sequence (cross hatched segment), which contains the packaging site; these elements support replication and packaging of the vector, respectively. A cassette of three polyadenylation sites (tri-poly A, black segment) was placed 5′ to the VGLUT1 promoter to reduce any effects on expression from the HSV-1 immediate early 4/5 promoter (contained in short line segment). Sequences from pBR322 (diagonal line segment) support growth in E. coli. (B) Concatamers of the vector are produced during the second, rolling circle phase, of HSV-1 DNA replication (Roizman and Sears, 1993). A HSV-1 genome-sized concatamer of the vector is packaged into a HSV-1 particle (Fraefel et al., 1996; Roizman and Sears, 1993); the HSV-1 genome is ~152 kb, and pVGLUT1brainbow is ~20 kb; thus, an array of 7 or 8 pVGLUT1brainbow is packaged into a HSV-1 particle.
3.2. Packaging pVGLUT1brainbow into HSV-1 particles, with Cre-mediated recombination
pVGLUT1brainbow was packaged into HSV-1 particles, using our standard helper-virus free packaging system (Fraefel et al., 1996; Sun et al., 1999), without or with the addition of a plasmid that expresses Cre. The rolling circle phase of HSV-1 DNA replication produces concatamers (Roizman and Sears, 1993) (Fig. 2B), and with HSV-1 plasmid vectors, a HSV-1 genome-sized array of concatamers is packaged into HSV-1 particles, as shown by pulse field gel electrophoresis and Southern analysis (Fraefel et al., 1996). The HSV-1 genome is ~152 kb, and pVGLUT1brainbow is ~20 kb; thus, 7 or 8 concatamers of pVGLUT1brainbow should be packaged into each HSV-1 vector particle. With vector packaging in the absence of Cre, we expected that only the initial configuration of Brainbow (Fig. 1A) would be produced and packaged into HSV-1 particles; only EmGFP would be in a position to be expressed from the VGLUT1 promoter. Importantly, Cre-mediated recombination can occur during HSV-1 vector replication and packaging (Logvinoff and Epstein, 2001). Thus, with vector packaging in the presence of Cre, we expected that Cre-mediated recombination would position each of the four FP genes in pVGLUT1brainbow in a position to be expressed from the VGLUT1 promoter (Fig. 1B and C), in different Brainbow cassettes.
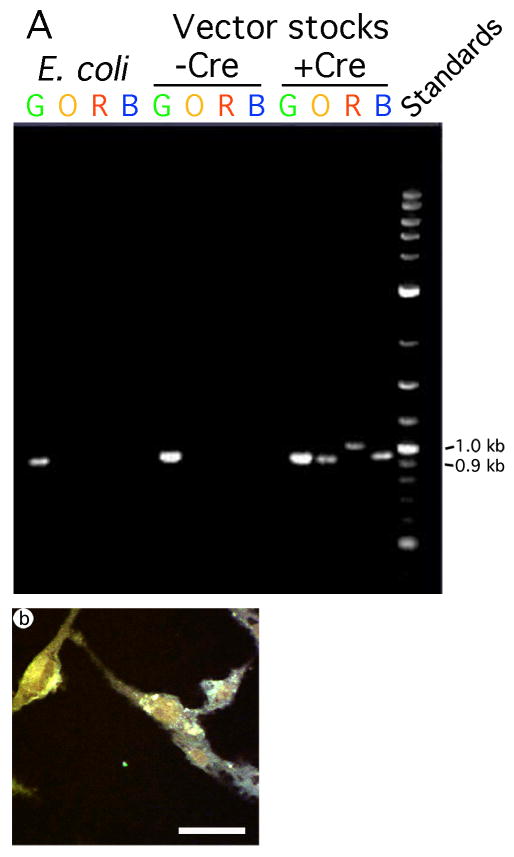
We identified the FP gene or genes proximal to the VGLUT1 promoter in different Brainbow cassettes, either without or with Cre-mediated recombination. We performed PCR on DNA isolated from either E. coli or from vector stocks prepared without or with Cre expression. The 5′ PCR primer was located at the 3′ end of the VGLUT1 promoter, and each of four 3′ primers were located in each of the four polyadenylation sequences at the 3′ end of each FP gene (Fig. 1A). Thus, in a specific Brainbow cassette, PCR will amplify only the FP gene proximal to the VGLUT1 promoter, and in a position to be expressed from this promoter. The results showed that using DNA isolated from either E. coli or from a vector stock prepared in the absence of Cre, only the EmGFP gene was detected by PCR (Fig. 3A); only this FP gene was in a position to be expressed from the VGLUT1 promoter. In contrast, using a vector stock prepared in the presence of Cre, each of the four FP genes was detected by PCR (Fig. 3A); thus, in different Brainbow cassettes, each FP gene was in a position to be expressed from the VGLUT1 promoter. This is a semi-quantitative assay, and the different intensities of the four bands should be interpreted cautiously (see discussion).
Fig. 3.
Packaging pVGLUT1brainbow in the presence of Cre resulted in recombination that placed each FP in a position to be expressed from the VGLUT1 promoter, and multiple FPs were coexpressed in the same fibroblast cells. (A) PCR analysis of pVGLUT1brainbow DNA prepared from E. coli, and after packaging in the absence or presence of Cre. The 5′ primer was located proximal to the 3′ end of the VGLUT1 promoter, and the four 3′ primers were located in each of the four polyadenylation sequences following each FP gene. Before recombination, the predicted size of the PCR products is 910 bp (EmGFP gene (G)). Following recombination to place each FP gene proximal to the VGLUT1 promoter, the predicted sizes of the PCR products are 910 bp (EmGFP gene, size unchanged by recombination), 903 bp (mOrange2 gene (O)), 958 bp (LSSmKate2 gene (R)), and 915 bp (EBFP2 gene (B)). DNA was isolated from E. coli, or from vector stocks prepared in either the absence of presence of Cre. For each sample DNA, four PCR reactions were performed; each reaction contained the 5′ primer and one of the 3′ primers. The analysis showed that only a vector stock prepared in the presence of Cre supported recombination that placed each FP gene proximal to the VGLUT1 promoter. (B) A confocal image showing that BHK fibroblast cells, fixed at 1 day after transduction with pVGLUT1brainbow packaged in the presence of Cre, contained different hues. Scale bar: 50 μm.
pVGLUT1brainbow is likely to support expression of 100 to 200 hues, similar to that observed in Brainbow mice (Livet et al., 2007). The theoretical maximum hue number for a Brainbow array is given by the formula for sampling with replacement independent of order (DeGroot and Schervish, 2002), the binomial (b+f−1, b); b=sampling number, or the number of Brainbow cassettes in the array, f=number of sample choices, or the number of FP genes in a Brainbow cassette; or (b+3, b) for pVGLUT1brainbow. pVGLUT1brainbow is likely to be packaged as arrays of 7 or 8 Brainbow, resulting in theoretical maxima of 120 or 165 hues, respectively. Similarly, a well-studied Brainbow mouse contained 8 arrays and supported expression of 89 to 166 hues (Livet et al., 2007).
3.3. pVGLUT1brainbow labels cultured fibroblast cells with different hues
The four FPs contained in pVGLUT1brainbow; LSSmKate2 (Piatkevich et al., 2010), mOrange2 (Shaner et al., 2008); EmGFP (Tsien, 1998), and EBFP2 (Ai et al., 2007); were chosen because they have well-separated excitation/absorption and emission maxima (Table 1), enabling each FP to be selectively detected in the presence of the other three FPs, by using four different excitation and emission conditions, or channels, as detailed in the methods. In particular, LSSmKate2 is advantageous for spectral separation from mOrange2 because it is excited by green light and emits red light (Table 1 and cited references). Further, each of the other three FPs have excitation and emission spectra that are non-overlapping.
Cultured fibroblast cells were transduced with a pVGLUT1brainbow vector stock prepared in the presence of Cre, and one day later fixed. Expression from the VGLUT1 promoter in fibroblast cells represents ectopic expression that declines rapidly at longer times after gene transfer (Rasmussen et al., 2007; Zhang and Geller, 2010). HSV-1 DNA in a virus (or vector) particle is in a condensed state and lacks nucleosomes (Roizman and Sears, 1993). Thus, immediately following infection, HSV-1 virus or vector DNA is not coated with nucleosomes (Muggeridge and Fraser, 1986), which may facilitate ectopic expression from the VGLUT1 promoter (or other promoters). Over time, HSV-1 DNA is coated with nucleosomes, and latent HSV-1 virus DNA contains nucleosomes (Deshmane and Fraser, 1989), enabling regulated expression from specific promoters. Expression of each FP was analyzed using confocal microscopy. The results revealed that different fibroblast cells contained different hues, and these hues usually represented expression of multiple FPs (Fig. 3B). Specifically, in this photomicrograph, one cell is yellow (left side), and another cell is greenish-blue (right bottom). Over ten vector stocks supported similar hue distributions in this assay.
3.4. pVGLUT1brainbow labels neurons in the rat forebrain, and their axons, with different hues
pVGLUT1brainbow vector stocks prepared in the presence of Cre were injected into POR cortex (one injection site) in the left hemisphere of six rats, the rats were sacrificed at 4 or 8 days after gene transfer, and expression was analyzed. Using these gene transfer conditions and a vector containing a neuron-specific promoter, POR cortex contains the vast majority of the transduced cells, which are spatially grouped together, located mostly in the deeper layers of neocortex, and are neurons, as established by costaining for recombinant gene products and a neuronal marker (Zhang et al., 2005). Further, cell counts showed that POR cortex contained ~300 transduced cells, but only low numbers of transduced cells (1–4 β-galactosidase (β-gal)-immunoreactivity (IR) cells) were observed in specific neocortical areas with large projections to POR cortex, including PER cortex (Zhang et al., 2005). Further, using these gene transfer conditions, the VGLUT1 promoter in a HSV-1 vector supports >90 % specific expression in glutaminergic neurons (costaining for β-gal-IR and phosphate-activated glutaminase (PAG)-IR, a glutamatergic neuron marker), and the VGLUT1 promoter supports >90 specific expression in VGLUT1-containing neurons (costaining for β-gal-IR and VGLUT1-IR) (Rasmussen et al., 2007; Zhang and Geller, 2010).
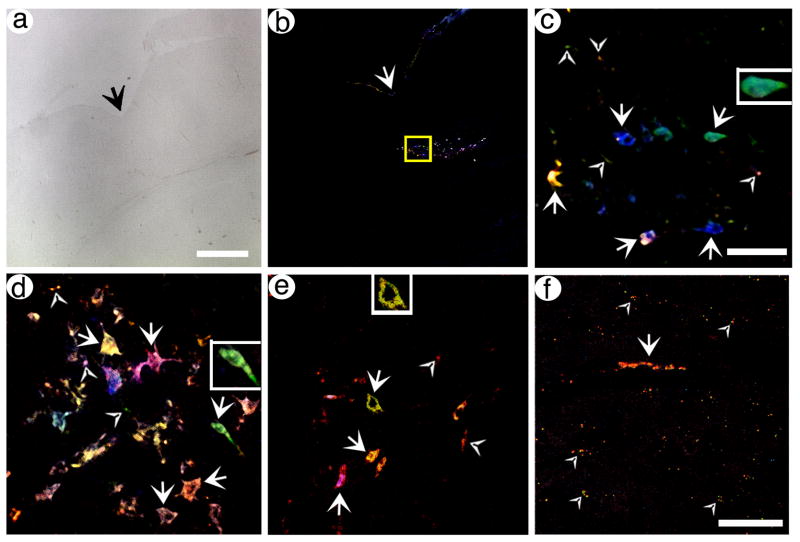
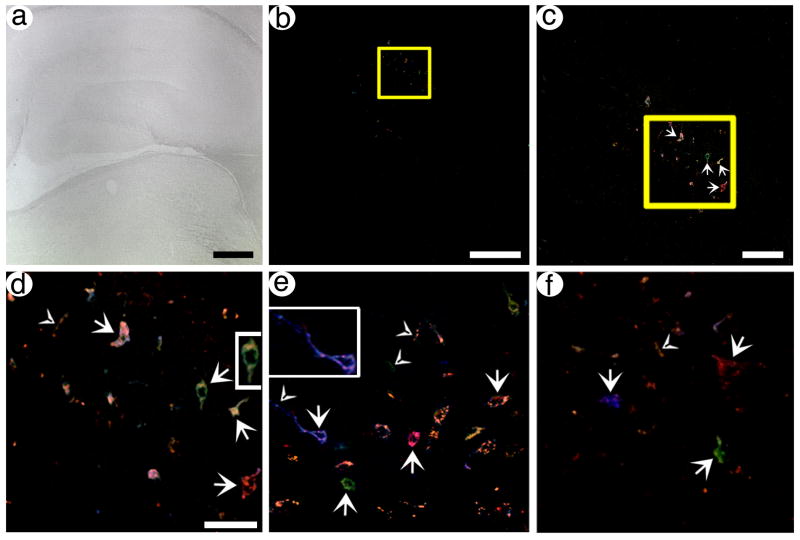
The results showed that specific neurons in POR cortex were labeled with different hues, and these hues usually represented expression of multiple FPs. A low power view using brightfield illumination shows a section containing POR cortex (Fig. 4A), and a low power view of this section using the four channels designed to detect expression of each FP revealed several tens of labeled neurons in POR cortex (Fig. 4B). Under high power, a confocal stack of some of these neurons revealed cell bodies labeled with different hues, and short segments of axons that were also labeled with different hues (Fig. 4C). Further, two other sections contained cell bodies labeled with different hues, and some of these cell bodies also contained labeling in proximal processes (Fig. 4D and E); also, short segments of axons not connected to a cell body were labeled with different hues. The fluorescent signal in these sections is restricted to small numbers of well-separated neurons, and thus is not background auto-fluorescence from most or all of the cells in the sections. Specific cell bodies or axons were labeled blue, green, orange, yellow, pink, or red, or different shades of each hue. Most of these hues represent expression of multiple FPs. The signal exhibits a range in intensity; some neurons are entirely filled, and some neurons exhibit signal primarily proximal to the membrane, due to the axon/membrane-targeting domain (Liu et al., 1994) fused to each FP. This range in signal intensity is similar to that observed using a HSV-1 vector that expresses a single axon-targeted gene, an axon-targeted β-gal, from the VGLUT1 promoter (Zhang et al., 2010b). Further, the inserts in Fig. 4C–E show that specific labeled neurons exhibit normal morphology, consistent with the known properties of this vector system to produce minimal cytopathic effects and inflammatory response (Fraefel et al., 1996; Olschowka et al., 2003).
Fig. 4.
pVGLUT1brainbow supports labeling of POR cortex neurons and axons, and distant axons in PER cortex, with different hues. Rats were sacrificed at 4 days (A–E) or 8 days (F) after gene transfer, brains were sectioned, and confocal stacks were analyzed. (A) A brightfield, low power view of POR cortex in a section proximal to the injection site. The arrow indicated the rhinal sulcus. (B) A low power view of Brainbow labeled neurons in the same section as in (A). (C) A high power view of the boxed area from (B). Neurons and proximal axons that contain different hues are shown. Arrows, cell bodies; arrowheads, axons. (D and E) High power views of neurons and proximal axons, in POR cortex, that contain different hues, from different sections. Some of the labeled cell bodies contain proximal processes, particularly in (D). Also, some axons that are not connected to a cell body are visible. (F) A high power view of PER cortex that shows one axon for ~50 um in length (arrow) and other axons in cross section (arrowheads). Scale bars: (A and B) 250 μm, (C–E) 50 μm, (F) 50 μm.
Next, we analyzed expression in axons in PER cortex, as PER cortex receives a large projection from POR cortex (Agster and Burwell, 2009; Burwell and Amaral, 1998). Of note, using these gene transfer conditions, many of the transduced neurons in POR cortex project to PER cortex, as shown by expressing an axon-targeted β-gal from the VGLUT1 promoter, and the 8 day survival time used for this rat is sufficient to label axons in distant neocortical areas (Zhang et al., 2010b). Importantly, using pVGLUT1brainbow, a confocal stack from a region of PER cortex that is innervated by the transduced neurons revealed axons were labeled with different hues (Fig. 4F), including one extended axon and a number of other axons in cross section. As detailed above, using these gene transfer conditions, the vast majority of the transduced neurons are in POR cortex; PER cortex contains ≥1 % of the number of transduced cell bodies as POR cortex (Zhang et al., 2005). Thus, after injecting pVGLUT1brainbow into POR cortex, the Brainbow-expressing axons in PER cortex likely represent the axons of transduced neurons in POR cortex.
Next, to show that HSV-Brainbow can label neurons in another forebrain area, pVGLUT1brainbow vector stocks prepared in the presence of Cre were injected into the dentate gyrus of the hippocampus (one injection site) in the left hemisphere of three rats, the rats were sacrificed at 4 days after gene transfer, and expression was analyzed. Using these gene transfer conditions and a vector containing a neuron-specific promoter, neurons in the granule cell layer and the hilar region are transduced (Zhang et al., 2009). A low power view under brightfield illumination shows a section containing the hippocampus and proximal areas (Fig. 5A), and a low power view of this section using four channels to detect each FP revealed labeled neurons in the dentate gyrus (Fig. 5B). Under medium power (Fig. 5C), or high power (Fig. 5D), confocal stacks of some of these neurons revealed cell bodies labeled with different hues, and some of these cell bodies also contained proximal processes; also, short segments of axons that were not attached to a cell body were labeled with different hues. These cell bodies were located in either the granule cell layer or the hilar region. Further, two other sections contained cell bodies labeled with different hues, and some of these cell bodies also contained labeling in proximal processes (Fig. 5E and F); again, short segments of axons not connected to a cell body were labeled with different hues. One neuron in the granule cell layer, labeled with a blue hue, contains a proximal axon that extends for ~50 μm (Fig. 5E, left side). As in POR cortex, the labeling is restricted to small numbers of well-separated neurons, and is not background auto-fluorescence. Again, specific cell bodies or axons were predominately blue, green, orange, yellow, pink, or red, or different shades of each hue, representing expression of multiple FPs. Some neurons were entirely filled, and other neurons were labeled primarily proximal to the membrane, due to the axon/membrane-targeting domain (Liu et al., 1994) fused to each FP; this range in signal intensity is similar to that supported by a HSV-1 vector that expresses a single axon-targeted gene (Zhang et al., 2010b). Further, the inserts in Fig. 5D and E show that specific labeled neurons exhibit normal morphology; this vector system to produces minimal cytopathic effects and inflammatory response (Fraefel et al., 1996; Olschowka et al., 2003), consistent with the normal neuronal morphology.
Fig. 5.
pVGLUT1brainbow can label neurons and proximal axons in the hippocampal dentate gyrus with different hues. Rats were sacrificed at 4 days after gene transfer, and confocal stacks were collected. (A) A brightfield, low power view of the hippocampus and adjacent areas in a section proximal to the injection site. (B) A low power view of Brainbow labeled neurons in the same section as in (A). (C) A medium power view of the boxed area from (B) showing Brainbow labeled cells. Arrows, cell bodies. (D) A high power view of the boxed area from (C) showing Brainbow labeled neurons and proximal axons. Arrows, cell bodies; arrowheads, axons. (E and F) High power views of Brainbow labeled neurons and proximal axons from different sections that contain the dentate gyrus. Some of the labeled cell bodies contain proximal processes, particularly a blue neuron on the left side of (E) contains an axon that extends for ~50 μm. Also, some axons that are not connected to a cell body are visible. Scale bars: (A) 500 μm, (B) 250 μm, (C) 100 μm, (D–F) 50 μm.
3.5. Specific axons can be matched to specific neuronal cell bodies, based on hue
We quantified the relative hue distributions in specific neuronal cell bodies in POR cortex and specific axons in PER cortex. Using a macro in imageJ, we drew a contour around a specific cell body or axon, and the macro determined the average density within the contour, in each of the four channels.
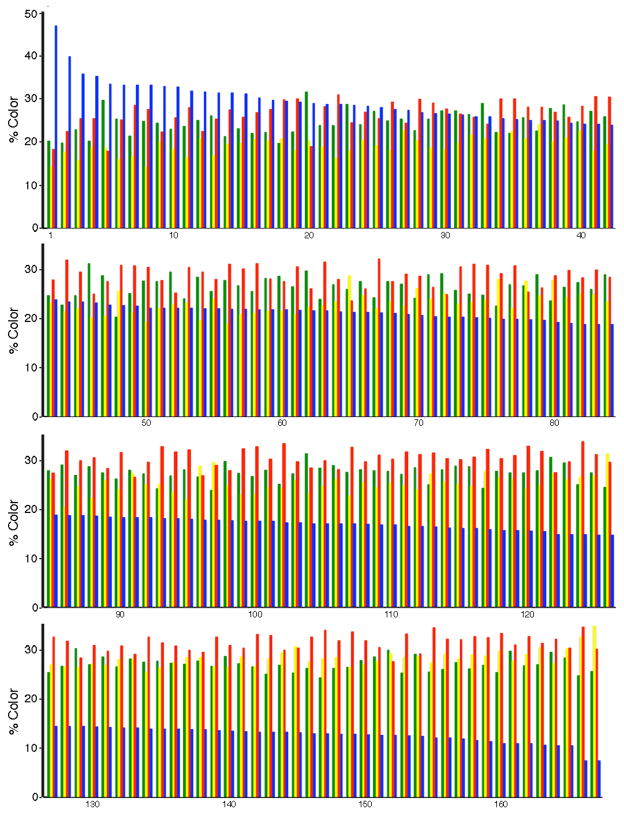
We quantified the hue, or channel, distributions for 167 neuronal cell bodies in serial coronal sections that contained the injection site and proximal regions from a rat sacrificed at 8 days after gene transfer. This analysis captured most of the transduced neurons, as these sections contained most of the transduced neurons. The results showed that the chosen laser intensities excited the four FPs to approximately equal levels, as shown by the similar average intensities (Table 1). More importantly, for the different cell bodies, each channel exhibited a range in the intensity relative to the other channels, indicating that different cell bodies were labeled with different hues. The four channels exhibited a 1.6- to 3.3-fold range, and up to a 5.9-fold range with inclusion of a small number of outlier cell bodies. The blue channel exhibited a modestly larger range than the other three channels. This larger range may reflect a larger range in stability of this FP compared to the other three FPs in different neuron types, and/or a larger range in blue light transmission through the section, or other issues. Thus, a graph of the four channels for the 167 cell bodies was arranged based on the signal in the blue channel (Fig. 6). Examination of the relative intensities of the four channels for these cell bodies showed that many cell bodies had unique channel distributions, or unique hues, and some cell bodies exhibited similar distributions to other cell bodies.
Fig. 6.
The distribution of hues for 167 neuronal cell bodies in POR cortex. Proximal to the injection site in POR cortex, specific transduced cell bodies were outlined. Within each contour, the percentages of the four colors (channels) were determined using a computer program. The cell bodies were ordered based on the size of the signal in the EBFP2 channel (EmGFP, green; mOrange2, yellow; LSSmKate2, red; or EBFP2, blue). The graph shows the color profiles for these 167 cell bodies.
Next, we determined the hue, or channel, distributions for 17 axons in PER cortex (Fig. 7), found in four proximal 25 μm sections. These 17 axons showed a range of hue distributions. Of note, specific subsets of these 17 axons showed similar hue distributions, and may represent axon collaterals from the same neuron.
Fig. 7.
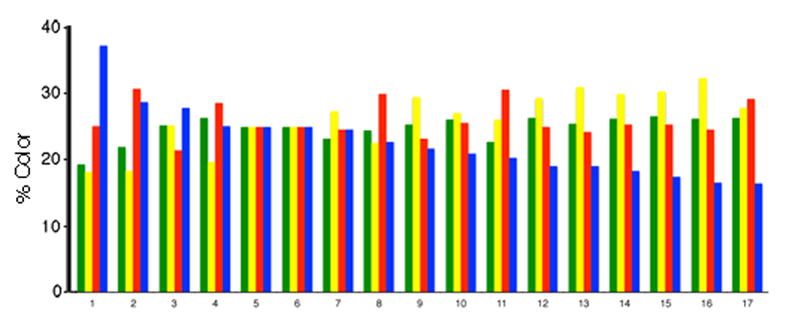
The distribution of hues for 17 axons in PER cortex. In several proximal sections in PER cortex, axons were outlined. Within each contour, the percentages of the four colors (channels) were determined using a computer program. The axons were ordered based on the size of the signal in the EBFP2 channel. The graph shows the color profiles for these 17 axons. Some of these axons displayed similar hue profiles, and were likely axons collaterals from the same neuron.
We used the color, or channel, distributions to match specific axons in PER cortex to specific neuronal cell bodies in POR cortex. For each axon and each cell body, for each color (channel), the differences between a specific axon and a specific cell body were calculated and squared (for example, [Baxon 5 − Bcell body 36]2), and these four values were summed. This sum was used as the metric for the color distance between a specific axon and a specific cell body. For each of the 17 axons, the color distance to each of the 167 cell bodies was calculated. The results showed that specific axons could be matched to a specific cell body; there was a several fold difference between the best match and the second-best match (Table 2). However, some specific axons showed similar color matches to more than one cell body, suggesting that transducing 2–3 fold fewer neurons than the 167 transduced neurons in this rat might benefit this analysis.
Table 2.
Matching of axons in PER cortex to neuronal cell bodies in POR cortex based on hue.
| Axon | Shortest distance (cell body number) | Fold difference, shortest and next distance (cell body number) |
|---|---|---|
| 1 | 0.00054 (4) | 2.9 (2) |
| 2 | 0.00011 (19) | 6.3 (22) |
| 3 | 0.0012 (27) | 2.7 (32) |
| 4 | 0.00015 (36) | 2.5 (42) |
| 5 | 0.0011 (27) | 1.1 (43) |
| 6 | 0.0011 (27) | 1.1 (43) |
| 7 | 0.0019 (37) | 1.5 (43) |
| 8 | 0.000099 (53) | 1.8 (45) |
| 9 | 0.00034 (65) | 5.6 (78) |
| 10 | 0.00026 (78) | 3.1 (66) |
| 11 | 0.00073 (70) | 1.0 (76) |
| 12 | 0.00032 (78) | 1.6 (96) |
| 13 | 0.0010 (65) | 1.4 (78) |
| 14 | 0.00051 (96) | 1.9 (78) |
| 15 | 0.00049 (96) | 1.8 (78) |
| 16 | 0.0022 (96) | 1.4 (126) |
| 17 | 0.00072 (118) | 1.0 (132) |
For each axon and each cell body, for each color (channel), the differences between the axon and cell body were calculated and squared (for example, [Baxon 5 − Bcell body 36]2), and these four values were summed. This sum is the metric for the color distance between a specific axon and a specific cell body. For each of the 17 axons, the color distance to each of the 167 cell bodies was calculated.
4. Discussion
In this study, we establish that mammalian forebrain neurons, and their axons, can be labeled with unique hues, using a HSV-1 vector that expresses Brainbow. The two critical properties of Brainbow, arrays and recombination to probabilistically choose the expressed FP from each Brainbow cassette, were achieved during vector packaging. Following delivery of HSV-Brainbow vector stock into two forebrain areas, small numbers of neurons, and their axons, were labeled with differed hues. Some neurons were labeled with unique hues. Axons in a specific neocortical area could be matched to cell bodies in a distant neocortical area, based on hue.
Within a specific Brainbow array that was packaged into a specific vector particle, Cre-mediated recombination likely supported the expression of different FPs from different Brainbow cassettes. Using this packaging system, we previously showed that concatamers of a vector, produced during the rolling circle phase of HSV-1 DNA replication (Roizman and Sears, 1993), are packaged into HSV-1 particles (Fraefel et al., 1996). Here, we used PCR to show that packaging a HSV-Brainbow vector, in the presence of Cre, supported recombination; after packaging, each FP was in a position to be expressed, in different Brainbow cassettes. However, we did not directly show that within one specific Brainbow array, packaged into a specific HSV-1 particle, different Brainbow cassettes express different FPs. Of note, if replication occurred after recombination, then adjacent Brainbow cassettes would express the same FP, and, potentially, all the Brainbow cassettes in a specific array might express the same FP. Inversely, if recombination occurred after replication, then adjacent Brainbow cassettes will express different FPs, except for the probability that recombination results in expression of the same FP from adjacent cassettes. Previous and current results favor the latter possibility. Cre supports efficient recombination during HSV-1 replication and packaging; using a HSV-1 helper virus that contained a packaging site flanked by loxP sites, packaging in the presence of Cre resulted in vector stocks that contained <0.1 % HSV-1 (Logvinoff and Epstein, 2001): Thus, Cre excised the packaging site from >99.9 % of the HSV-1 genomes, suggesting that Cre can support recombination in most Brainbow cassettes. Further, using pVGLUT1brainbow, the vast majority of the transduced neurons were labeled with hues that require expression of multiple FPs, indicating that recombination occurred within specific Brainbow arrays. Different HSV-Brainbow vector stocks likely encode similar distributions of hues; as recombination during packaging is efficient, reaching >99.9 % in one study (Logvinoff and Epstein, 2001), recombination likely approaches equilibrium for the different potential recombination products, resulting in different vector stocks encoding similar hue distributions. Moreover, HSV-Brainbow vector stocks likely encode most of the potential hues, as recombination likely approaches equilibrium during packaging, and over ten vector stocks supported similar hue distributions after transduction of fibroblast cells.
HSV-Brainbow produced a large number of hues, as determined by quantifying the hues in 167 neurons in POR cortex, in one rat. A number of variables likely determine hue distribution, including the hues encoded in a specific HSV-Brainbow after recombination, the relative stabilities of the different FPs in different neuron types, the penetrance of different light frequencies through the section, and other variables. Thus, we empirically quantified and compared hues for different neurons, and we did not attempt to assign specific observed hues to specific predicted hues based on vector structure. Individual HSV-Brainbow particles encode a large number of potential hues, and single transduction events will label neurons with a wide range of hues; a single HSV-Brainbow particle contains seven or eight arrays of Brainbow, encoding theoretical maxima of 120 or 165 hues, respectively. In contrast, as AAV-Brainbow or PRV-Brainbow encode small number of hues (one to four hues), these approaches depend upon transducing the same neurons with multiple vector particles to generate a larger hue distribution, and hue distribution varies with distance from injection site (Boldogkoi et al., 2009; Cai et al., 2013; Card et al., 2011; Kobiler et al., 2010). With HSV-Brainbow, neurons that are proximal to the injection site likely receive multiple HSV-Brainbow particles, but we did observe a change in hue distribution with distance from the injection site; this is likely because single HSV-Brainbow particles encode large numbers of hues, and the hues produced by multiple transduction of the same neurons are likely similar to at least one of the hues encoded by single HSV-Brainbow particles. Thus, using HSV-Brainbow, the hue distribution is likely to display minimal variation with the amount of injected vector, and the percentage of transduced neurons that contain unique hues can likely be improved by varying the titer of injected HSV-Brainbow. Of note, in the experiments reported here, specific neurons in POR cortex were labeled with unique hues, although we did not systematically optimize the percentage of transduced neurons that contain unique hues by varying the amount of injected HSV-Brainbow vector stock.
The primary advantage of HSV-Brainbow is the unique capability to label individual neurons with unique hues. Additional attractive properties of HSV-Brainbow include that a single vector particle contains a sufficient number of Brainbow cassettes to produce a large number of hues, the recombination products are stabile, and control of the number of labeled neurons. In contrast, AAV-Brainbow vectors express only one fluorescent protein from each vector genome, consequently require multiple infections of the same cell to produce different hues, and display different classes of hue profiles at different distances from the injection site (Cai et al., 2013). Thus, the number of neurons expressing unique, or useful, hues is difficult to control. Similarly, Brainbow expressed from a replication-competent PRV vector requires coinfection of the same cell by multiple viruses that each express a single FP (Kobiler et al., 2010); PRV-Brainbow can support transneuronal mapping studies (Card et al., 2011), but this approach labels multiple neurons with the same hue, due to use of a replication-competent vector that transduces many neurons. Brainbow mice label many more neurons than there are hues, thus requiring serial section reconstructions to trace axons, and such analyses have been problematic to date, given the large numbers and high densities of labeled axons (Cai et al., 2013; Livet et al., 2007). While the use of both a cell type-specific promoter and a regulated Cre to mediate Brainbow recombination may produce analyzable numbers of labeled neurons in specific midbrain nuclei (Weber et al., 2009), such approaches are likely to be problematic in the more complex forebrain. Further, Brainbow mice require stabile Brainbow recombination products, thereby constraining potential Brainbow designs by requiring irreversible recombination, or temporal regulation of Cre expression, or other complex solutions (Cai et al., 2013). In contrast, HSV-Brainbow addresses many of these limitations: HSV-Brainbow produces a useful number of hues from single HSV-1 vector particles, which contain arrays of seven or eight Brainbow cassettes; multiple infections of the same neuron by multiple vector particles is not required. Further, the Brainbow recombination products are stable; HSV-Brainbow recombination occurs during vector packaging, and Cre is absent from the vector stocks and the transduced animals. Additionally, specific neurons can be labeled with unique hues by transducing a predetermined number of neurons; these HSV-1 vectors are replication defective, and the number of transduced neurons can be controlled by choosing an appropriate amount of vector to inject into the brain.
The major limitation of the current HSV-Brainbow is that although expression levels are sufficient to clearly label neurons, different neurons were labeled to different intensities. Some neurons were filled, whereas other neurons exhibited fluorescence predominately near the plasma membrane. These levels of labeling are similar to those observed with HSV-1 vectors expressing a single gene from the VGLUT1 promoter (Zhang et al., 2010b). Of note, higher levels of expression might be obtained by modifying the vector. In particular, expression levels from the VGLUT1 promoter might be improved. An initial analysis of the VGLUT1 promoter identified a basal promoter that is not glutamatergic-specific, and glutamatergic-specific elements, likely enhancers, in both the upstream promoter and first intron (Zhang et al., 2011). Thus, substitution of the VGLUT1 basal promoter with a strong basal promoter, and/or use of cancatamers of the glutamatergic-specific upstream elements might increase expression levels. Also, the four polyadenylation sites used here were all from non-neuronal genes, and use of polyadenylation site(s) from highly expressed neuronal genes might be beneficial. Further, a woodchuck hepatitis virus post-transcriptional regulatory element often increases mRNA expression levels from specific virus vectors, and could be explored.
This HSV-Brainbow vector labeled VGLUT1-containing glutamatergic neuron cell bodies in POR cortex or hippocampus with different hues, and many cell bodies were labeled with unique hues. Because these HSV-1 vectors are replication defective, the number of transduced neurons can be selected by adjusting the amount of vector injected into the brain. Here, we labeled 167 neurons in POR cortex, and for an array of seven or eight of these Brainbow cassettes (4 FPs per cassette), the theoretical maximum number of hues is 120 or 165, respectively. Thus, some specific sets of neurons were labeled with the same hue, and specific hues that are proximal in color space may be difficult to separate using present methodology. A higher percentage of the transduced neurons might be labeled with unique hues by reducing the amount of injected vector, thereby transducing fewer neurons.
We matched distant axons in PER cortex to transduced neuronal cell bodies in POR cortex based on hue. Brainbow hues were previously shown to be stable over considerable distances in axons (Livet et al., 2007). The four FPs used in the present Brainbow were efficiently transported into axons; each of these FPs is a monomer (Ai et al., 2007; Piatkevich et al., 2010; Shaner et al., 2008; Tsien, 1998), facilitating intracellular transport, and each FP was fused to the axon-targeting domain from GAP-43 (Liu et al., 1994). Consistent with both previous results and the present Brainbow design, the hue was relatively uniform in axons we imaged for over 50 μm. To determine the hue for a cell body, we outlined the entire cell body and determined the average hue within the cell body. Specific axons could be matched to specific neuronal cell bodies, as reflected by the small color distance between the axon and cell body, and with confidence, reflected by a ≥2.5 fold difference between the best color match and the next-best match. However, some axons matched well to more than one cell body, reflected by a relatively small fold difference between the best and the next-best match. As noted above, transduction of fewer neurons might result in a higher percentage of transduced neurons that contain unique hues, and a higher percentage of unambiguous matches of specific axons to specific cell bodies.
Mapping forebrain circuits will benefit from the capabilities to label small numbers of neurons with unique hues, and to match specific axons to distant cell bodies, based on hue. For example, glutamatergic neurons in POR cortex project to more than ten other neocortical areas, but the specific areas that receive axon collaterals from the same neurons remains to be determined (Agster and Burwell, 2009). Of note, the capability to match specific axons to specific cell bodies based on hue could be used to map multiple axon collaterals, in multiple neocortical areas, that arise from a single neuron. Additional specificity for the subtype of neuron that is labeled could be obtained from using specific neuron-subtype specific promoters to express Brainbow and/or from targeting gene transfer to specific types of neurons (Cao et al., 2011). Further, postsynaptic neurons could be identified by either coexpressing a transneuronal marker, such as wheat germ agglutinin (Zhang et al., 2010b), with Brainbow, or by targeted gene transfer of HSV-1 vectors across specific synapses (Zhang et al., 2012). Moreover, local circuits might be mapped by developing HSV-Brainbow that express dendrite-targeted FPs; a dendrite-targeted GFP (Kameda et al., 2008) has been established in HSV-1 vectors (Zhang et al., 2012). Of note, the neurons that were studied here are part of circuit that can encode essential information for specific visual object discriminations (Zhang et al., 2010a); thus, mapping the projections of these neurons may illuminate visual information processing. In summary, the capabilities to label neurons with unique hues and determine the projections of these neurons may benefit elucidating the mammalian connectome, and the physiological functions of specific circuits.
Supplementary Material
Highlights.
Brainbow creates hundreds of hues by combinatorial expression of fluorescent proteins
In Brainbow mice, multiple neurons contain the same hue, as many neurons are labeled
HSV-brainbow labels small numbers of neurons, and their axons, with unique hues
Specific axons can be matched to specific neuronal cell bodies, based on hue
Acknowledgments
We thank Drs. N. Brose and C. Rosenmund for the VGLUT1 promoter; Dr. A. Davison for HSV-1 cosmid set C; Lai Ding and Daniel Tom at the Harvard NeuroDiscovery Center, Enhanced Neuroimaging Core for assistance with microscopy; and Luke Fritzky at the Digital Imaging and Histology Core of Rutgers-NJMS Cancer Center for assistance with microscopy. This work was supported by NIH Grants AG025894 (G.Z.), NS045855, NS057558, and NS086960 (A.I.G.).
Abbreviations
- AAV
adeno-associated virus
- AP
anterior-posterior
- β-gal
β-galactosidase
- DV
dorsal-ventral
- DMEM
Dulbecco’s modified minimal essential medium
- EmGFP
emerald green FP
- EBFP2
enhanced blue FP-2
- FBS
fetal bovine serum
- FPs
fluorescent proteins
- HSV-1
Herpes Simplex Virus
- IR
immunoreactivity
- IVP
infectious vector particles
- ML
medial-lateral
- PER
perirhinal
- PAG
phosphate-activated glutaminase
- POR
postrhinal
- PRV
pseudorabies virus
- VGLUT1
vesicular glutamate transporter-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agster KL, Burwell RD. Cortical efferents of the perirhinal, postrhinal, and entorhinal cortices of the rat. Hippocampus. 2009;19:1159–86. doi: 10.1002/hipo.20578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46:5904–10. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- Alonso-Nanclares L, Gonzalez-Soriano J, Rodriguez JR, DeFelipe J. Gender differences in human cortical synaptic density. Proc Natl Acad Sci U S A. 2008;105:14615–9. doi: 10.1073/pnas.0803652105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogkoi Z, Balint K, Awatramani GB, Balya D, Busskamp V, Viney TJ, Lagali PS, Duebel J, Pasti E, Tombacz D, Toth JS, Takacs IF, Scherf BG, Roska B. Genetically timed, activity-sensor and rainbow transsynaptic viral tools. Nat Methods. 2009;6:127–30. doi: 10.1038/nmeth.1292. [DOI] [PubMed] [Google Scholar]
- Burwell RD, Amaral DG. Perirhinal and postrhinal cortices of the rat: interconnectivity and connections with the entorhinal cortex. J Comp Neurol. 1998;391:293–321. doi: 10.1002/(sici)1096-9861(19980216)391:3<293::aid-cne2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–7. [PubMed] [Google Scholar]
- Cao H, Zhang GR, Geller AI. Antibody-mediated targeted gene transfer of helper virus-free HSV-1 vectors to rat neocortical neurons that contain either NMDA receptor 2A or 2B subunits. Brain Research. 2011;1415:127–35. doi: 10.1016/j.brainres.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Kobiler O, McCambridge J, Ebdlahad S, Shan Z, Raizada MK, Sved AF, Enquist LW. Microdissection of neural networks by conditional reporter expression from a Brainbow herpesvirus. Proc Natl Acad Sci U S A. 2011;108:3377–82. doi: 10.1073/pnas.1015033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGroot MH, Schervish MJ. Probability and statistics. 3. Addison Wesley; Boston: 2002. [Google Scholar]
- Deshmane SL, Fraser NW. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J Virol. 1989;63:943–7. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Enquist LW, Pomeranz LE. The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends in molecular medicine. 2008;14:134–40. doi: 10.1016/j.molmed.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) Defines Cell Contacts and Synapses in Living Nervous Systems. Neuron. 2008;57:353–63. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Fraefel C, Song S, Lim F, Lang P, Yu L, Wang Y, Wild P, Geller AI. Helper virus-free transfer of herpes simplex virus type 1 plasmid vectors into neural cells. J Virol. 1996;70:7190–7. doi: 10.1128/jvi.70.10.7190-7197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremeau RT, Jr, Voglmaier S, Seal RP, Edwards RH. VGLUTs define subsets of excitatory neurons and suggest novel roles for glutamate. Trends Neurosci. 2004;27:98–103. doi: 10.1016/j.tins.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Geller AI, Breakefield XO. A defective HSV-1 vector expresses Escherichia coli beta-galactosidase in cultured peripheral neurons. Science. 1988;241:1667–9. doi: 10.1126/science.241.4873.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda H, Furuta T, Matsuda W, Ohira K, Nakamura K, Hioki H, Kaneko T. Targeting green fluorescent protein to dendritic membrane in central neurons. Neurosci Res. 2008;61:79–91. doi: 10.1016/j.neures.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Kobiler O, Lipman Y, Therkelsen K, Daubechies I, Enquist LW. Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat Commun. 2010;1:146. doi: 10.1038/ncomms1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–48. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fisher DA, Storm DR. Intracellular sorting of neuromodulin (GAP-43) mutants modified in the membrane targeting domain. J Neurosci. 1994;14:5807–17. doi: 10.1523/JNEUROSCI.14-10-05807.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lo L, Anderson DJ. A cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–50. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logvinoff C, Epstein AL. A novel approach for herpes simplex virus type 1 amplicon vector production, using the Cre-loxP recombination system to remove helper virus. Hum Gene Ther. 2001;12:161–7. doi: 10.1089/104303401750061221. [DOI] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–60. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggeridge MI, Fraser NW. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J Virol. 1986;59:764–7. doi: 10.1128/jvi.59.3.764-767.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olschowka JA, Bowers WJ, Hurley SD, Mastrangelo MA, Federoff HJ. Helper-free HSV-1 amplicons elicit a markedly less robust innate immune response in the CNS. Mol Ther. 2003;7:218–27. doi: 10.1016/s1525-0016(02)00036-9. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic Press; Sidney: 1986. [DOI] [PubMed] [Google Scholar]
- Peters A, Jones E, editors. Cellular components of the cerebral cortex. Plenum Press; New York: 1984. [Google Scholar]
- Piatkevich KD, Hulit J, Subach OM, Wu B, Abdulla A, Segall JE, Verkhusha VV. Monomeric red fluorescent proteins with a large Stokes shift. Proc Natl Acad Sci U S A. 2010;107:5369–74. doi: 10.1073/pnas.0914365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Kong L, Zhang G, Liu M, Wang X, Szabo G, Curthoys NP, Geller AI. Glutamatergic or GABAergic neuron-specific, long-term expression in neocortical neurons from helper virus-free HSV-1 vectors containing the phosphate-activated glutaminase, vesicular glutamate transporter-1, or glutamic acid decarboxylase promoter. Brain Res. 2007;1144:19–32. doi: 10.1016/j.brainres.2007.01.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Sears AE. Herpes simplex viruses and their replication. In: Roizman B, Whitley RJ, Lopez C, editors. The human herpesviruses. Raven Press; New York: 1993. pp. 11–68. [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–51. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Sun M, Kong L, Wang X, Holmes C, Gao Q, Zhang W, Pfeilschifter J, Goldstein DS, Geller AI. Coexpression of tyrosine hydroxylase, GTP cyclohydrolase I, aromatic amino acid decarboxylase, and vesicular monoamine transporter 2 from a helper virus-free HSV-1 vector supports high-evel, long-term biochemical and behavioral correction of a rat model of Parkinson’s disease. Hum Gene Ther. 2004;15:1177–96. doi: 10.1089/hum.2004.15.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Zhang GR, Yang T, Yu L, Geller AI. Improved titers for helper virus-free herpes simplex virus type 1 plasmid vectors by optimization of the packaging protocol and addition of noninfectious herpes simplex virus-related particles (previral DNA replication enveloped particles) to the packaging procedure. Hum Gene Ther. 1999;10:2005–11. doi: 10.1089/10430349950017365. [DOI] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–44. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Weber T, Bohm G, Hermann E, Schutz G, Schonig K, Bartsch D. Inducible gene manipulations in serotonergic neurons. Frontiers in molecular neuroscience. 2009;2:24. doi: 10.3389/neuro.02.024.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Zaborszky L, Wouterlood FG, Lanciego JL, editors. Neuroanatomical Tract-Tracing 3: Molecules, Neurons, and Systems. Springer; New York: 2006. [Google Scholar]
- Zhang G, Cao H, Kong L, O’Brien J, Baughns A, Jan M, Zhao H, Wang X, Lu X, Cook RG, Geller AI. Identified circuit in rat postrhinal cortex encodes essential information for performing specific visual shape discriminations. Proc Natl Acad Sci USA. 2010a;107:14478–83. doi: 10.1073/pnas.0912950107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cao H, Li X, Zhao H, Geller AI. Genetic labeling of both the axons of transduced, glutamatergic neurons in rat postrhinal cortex and their postsynaptic neurons in other neocortical areas by Herpes Simplex Virus vectors that coexpress an axon-targeted β-galactosidase and wheat germ agglutinin from a vesicular glutamate transporter-1 promoter. Brain Res. 2010b;1361:1–11. doi: 10.1016/j.brainres.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Geller AI. A helper virus-free HSV-1 vector containing the vesicular glutamate transporter-1 promoter supports expression preferentially in VGLUT1-containing glutamatergic neurons. Brain Res. 2010;1331:12–9. doi: 10.1016/j.brainres.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li X, Cao H, Zhao H, Geller AI. The vesicular glutamate transporter-1 upstream promoter and first intron each support glutamatergic-specific expression in rat postrhinal cortex. Brain Research. 2011;1377:1–12. doi: 10.1016/j.brainres.2010.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu M, Cao H, Kong L, Wang X, Cook RG, Geller AI. Improved spatial learning in aged rats by genetic activation of protein kinase C in small groups of rat hippocampal neurons. Hippocampus. 2009;19:413–23. doi: 10.1002/hipo.20506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wang X, Kong L, Lu X, Lee B, Liu M, Sun M, Franklin C, Cook RG, Geller AI. Genetic enhancement of visual learning by activation of protein kinase C pathways in small groups of rat cortical neurons. J Neurosci. 2005;25:8468–81. doi: 10.1523/JNEUROSCI.2271-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wang X, Yang T, Sun M, Zhang W, Wang Y, Geller AI. A tyrosine hydroxylase--neurofilament chimeric promoter enhances long-term expression in rat forebrain neurons from helper virus-free HSV-1 vectors. Molec Brain Res. 2000;84:17–31. doi: 10.1016/s0169-328x(00)00197-2. [DOI] [PubMed] [Google Scholar]
- Zhang G, Zhao H, Cao H, Li X, Geller AI. Targeted gene transfer of different genes to presynaptic and postsynaptic neocortical neurons connected by a glutamatergic synapse. Brain Res. 2012;1473:173–84. doi: 10.1016/j.brainres.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.