Abstract
Background
Age-related macular degeneration (AMD), left untreated, is the leading cause of vision loss in people older than 55. Severe central vision loss occurs in the advanced stage of the disease, characterized by either the in growth of choroidal neovascularization (CNV), termed the “wet” form, or by geographic atrophy (GA) of the retinal pigment epithelium (RPE) involving the center of the macula, termed the “dry” form. Tracking the change in GA area over time is important since it allows for the characterization of the effectiveness of GA treatments. Tracking GA evolution can be achieved by physicians performing manual delineation of GA area on retinal fundus images. However, manual GA delineation is time-consuming and subject to inter-and intra-observer variability.
Methods
We have developed a fully automated GA segmentation algorithm in color fundus images that uses a supervised machine learning approach employing a random forest classifier. This algorithm is developed and tested using a dataset of images from the NIH-sponsored Age Related Eye Disease Study (AREDS). GA segmentation output was compared against a manual delineation by a retina specialist.
Results
Using 143 color fundus images from 55 different patient eyes, our algorithm achieved PPV of 0.82±0.19, and NPV of 0:95±0.07.
Discussion
This is the first study, to our knowledge, applying machine learning methods to GA segmentation on color fundus images and using AREDS imagery for testing. These preliminary results show promising evidence that machine learning methods may have utility in automated characterization of GA from color fundus images.
Keywords: Automated delineation, Segmentation, Geographic atrophy of the retinal pigment epithelium, Age-related macular degeneration, AREDS color fundus imagery, Machine learning
1. Introduction
1.1. Age-related macular degeneration and geographic atrophy
Age-related macular degeneration (AMD), left untreated, is a leading cause of irreversible vision loss in older Americans, in which the intermediate stage affects over 8 million persons of at least 55 years of age in the United States [2]. AMD is caused by retinal degeneration, with the intermediate stage characterized by the accumulation of drusen, i.e., long-spacing collagen and phospholipid vesicles between the basement membrane of the retinal pigment epithelium and the basement membrane of the choriocapillaris (Bruch’s membrane). It is estimated that there are at least 1.75 million cases of advanced stage AMD in the US [9].
The advanced stage of AMD can be associated with vision loss. The advanced stage is characterized by damage to the macula through either the “wet” form or “dry” form of AMD. Wet AMD is characterized by the ingrowth of choroidal neovascularization (CNV) due to the production of vascular endothelial growth factor (VEGF) in eyes with drusen. Advanced dry AMD is characterized by geographic atrophy (GA) of the retinal pigment epithelium (RPE) involving the center of the macula. Either the neovascular or geographic atrophic forms of advanced AMD can result in rapid or gradual loss of visual acuity due to a loss of photoreceptors that can be replaced by scar tissue with CNV or degenerate with GA. GA is characterized by any sharply delineated roughly round or oval region of hypopigmentation, depigmentation, or apparent absence of the retinal pigment epithelium in which choroidal vessels are more visible than in surrounding regions. In many classification schemes, the diameter of this region must be at least 175 μm in order to be classified as GA [6]. Geographic atrophy (GA) is present in nearly 20% of legal blindness cases of AMD in North America [22].
Although there is no definite cure for AMD, worsening of vision due to CNV can be slowed substantially through intraocular injections of anti-VEGF agents. This reduces the chance of vision loss compared with no treatment [10], photodynamic therapy with verteporfin [7], or laser photocoagulation [1]. No comparable treatment is currently available for GA. As a result, numerous studies are being conducted [31,42] with the goal of slowing GA growth rate. As noted in a recent review article, the reduction in the worsening of atrophy is an important biomarker for assessing the effectiveness of a given GA treatment [42]. Thus, it is of value to reliably monitor GA evolution and measure the growth of GA area to study the effectiveness of treatment therapies. It is also important to understand GA worsening that occurs as a part of standard care. GA area can be tracked in retinal fundus images, but this requires accurate segmentation.
1.2. Retinal imaging modalities
The most widely available and simplest modality for GA assessment by ophthalmologists is color fundus imagery [3]. In color fundus images, GA is characterized by an often strongly demarcated area, apparent RPE absence, and choroidal vessel visibility. Examples of fundus images are shown in Fig. 1.
Fig. 1.
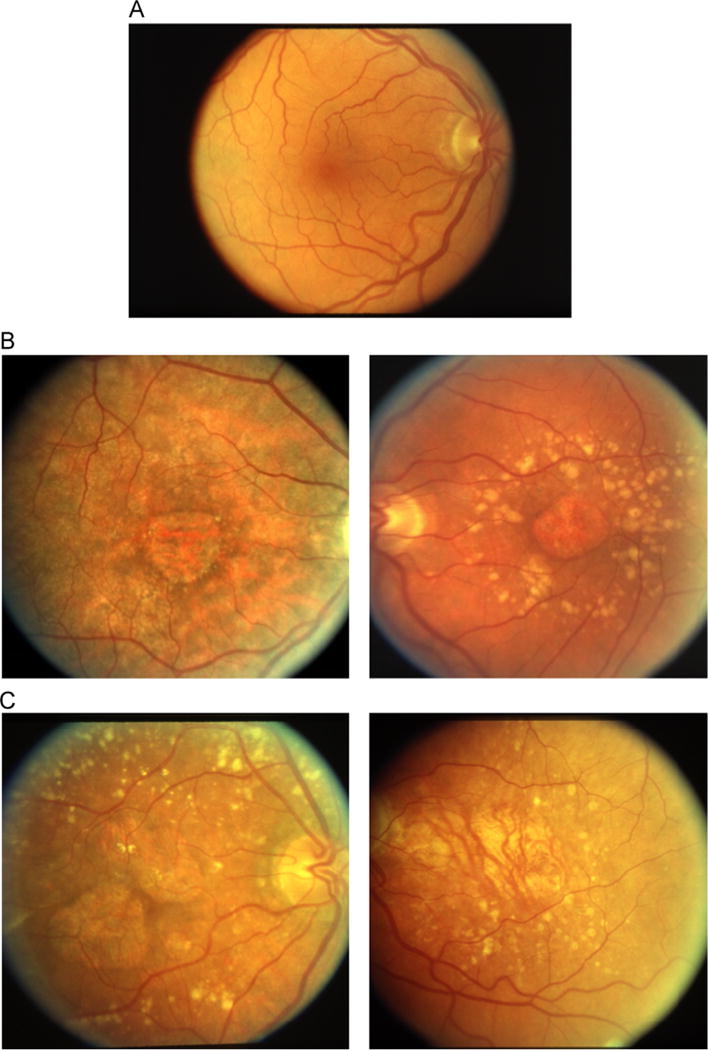
Row A shows a color fundus image of a healthy eye, with an absence of AMD and geographic atrophy. Row B shows two examples of GA that were labeled as unambiguous by our retina specialist. Row C shows two examples of GA that were labeled as ambiguous by our retina specialist. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
While fundus imaging is widely available and deemed a reliable means to measure GA growth [41], some fundus images can be challenging to interpret with regard to actual GA extent and may lead to ambiguous GA delineation. Alternative retinal imaging modalities include fundus autofluorescence (FAF) and optical coherence tomography (OCT). While FAF often is not as available as color fundus imaging, these imaging modalities can result in clearer and more informative GA depiction [13,20,35].
FAF creates an image using the fluorescent emission from lipofuscin, or the accumulation of granules marking cellular aging and oxidative damage in the RPE [14]. Therefore, due to RPE loss, GA regions in FAF can be represented distinctly by very low image intensity, with clear contrast to the background. While cases of GA may appear more prominently in FAF, the foveal area also presents with low image intensity and may be confused with GA [14,37].
In spectral-domain optical coherence tomography (SD-OCT), a 3-D cross-sectional image of the eye is obtained, providing 3-D structural information of the retina [17,25]. It is also possible to visualize GA in a planar manner, similar to what is done with fundus images, by creating a 2-D projection image from a 3-D OCT volume [12,23]. Accurate GA information can be obtained by using solely the signal reflected from beneath the RPE [45]. However, GA segmentation on SD-OCT may be complicated by the difficulty in differentiating the RPE from Bruch’s membrane or the photoreceptor layer in areas of atrophy. A recent development in OCT, polarization-sensitive OCT (PS-OCT), enables the reliable segmentation of the RPE in atrophied areas [34]. PS-OCT images GA lesions using tissue-specific polarizing properties from the RPE [5,34,39].
For all imaging modalities, manual delineation of geographic atrophy is a time-consuming process [40], which motivates the need for automated segmentation methods.
1.3. Prior automated segmentation work
Although there is a substantial body of work in the area of automated retinal image analysis (ARIA) [43], most studies deal with the automated characterization of diabetic retinopathy [3]. Fewer ARIA investigations have been devoted to the automated detection and classification of images of age-related macular degeneration [3,11,18,28,32], and there is a relative paucity of image analysis studies dedicated specifically to automated GA characterization.
Of those studies looking at automated image analysis of GA, nearly all are applied to FAF or OCT. Automated segmentation methods applied to these modalities have produced useful results, and GA segmentation using FAF and OCT has been found to agree in the resulting delineated tissues [39,45].
For GA segmentation from FAF, methods include both automated and interactive approaches including supervised classification [16,24], level sets [23], watershed [30], fuzzy c-means clustering [35], and region growing [15]. Semi-automated GA segmentation of FAF images has also been pursued by using commercial packages such as RegionFinder software (Heidelberg Engineering, Heidelberg, Germany) [33,36].
For GA segmentation from OCT, several studies convert 3-D volumetric image information to 2-D image slices [12,23]. Segmentation methods in the projected 2D images then use techniques similar to those used in FAF, such as geometric active contours [12] or level sets [23]. Commercial segmentation software is also becoming available for OCT modalities. The Cirrus HD-OCT (Carl Zeiss Meditec, Dublin, CA) platform uses advanced RPE analysis to segment GA areas [45]. Interactive automated segmentation on PS-OCT images have also shown promising results for GA segmentation [39].
To the best of our knowledge, there is only one prior study focusing on the automated segmentation of GA using color fundus images. This color fundus segmentation was compared to segmentation on FAF [35]. This study uses a fuzzy c-means method on a cohort of 10 patients. The study found that their automated segmentation method worked very well for fundus autofluorescence (94% sensitivity and 98% specificity) but less well on color fundus images (a sensitivity of 47% and a specificity of 98%). This speaks to the challenges of GA segmentation on color fundus images.
The relevance of automated GA segmentation in color fundus images is predicated on the fact that fundus imagery is the most widely available and simplest retinal imaging modality and has been regarded as the standard for assessing dry AMD [3,27]. This work constitutes one of the first and few studies that considers this problem. It is also unique among ARIA studies for its use of AREDS images taken from an NIH-sponsored clinical study looking at the effect of supplements for mitigating the evolution of AMD. As such, this study establishes an initial benchmark for the performance of fully automated GA segmentation in a moderately sized dataset of color fundus images. Our study also demonstrates a promising – while still preliminary – proof of concept pointing to the potential utility of automated segmentation methods for clinical GA characterization in color fundus images.
2. Methods
In this section, we describe our image processing and supervised machine learning algorithm for fully automated segmentation of GA. The salient steps of this pixel based algorithm are summarized in a diagram in Fig. 2 and discussed in more detail below.
Fig. 2.
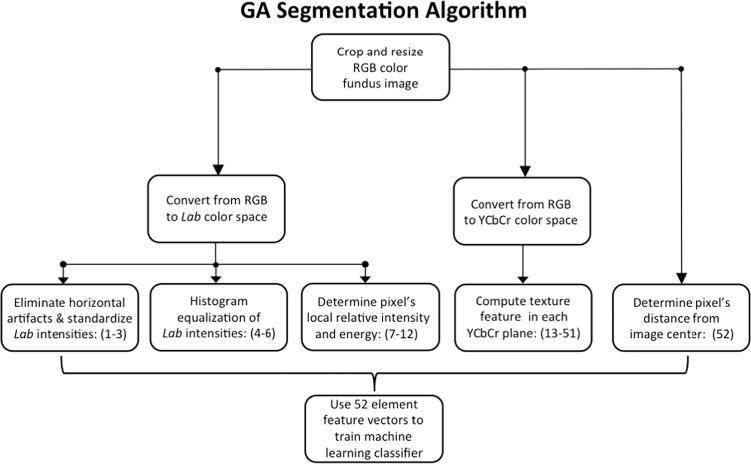
An overview of the segmentation algorithm.
2.1. Image preprocessing
All processes described below are fully automated and implemented in MATLAB (MathWorks, Natick, MA). As shown in Fig. 1, raw color fundus images typically appear as colored circular objects over a black background. We first crop the image to the largest inscribed square in the color region. To reduce processing time, this square region of interest is resized to a canonical size of 256 × 256 pixels. This resolution still allows for clear GA visualization with reasonable processing time. For our fundus images, this 256 × 256 pixel region corresponds to a physical area of approximately 9.85 mm × 9.85 mm. An example of the cropped image is shown in Fig. 3.
Fig. 3.
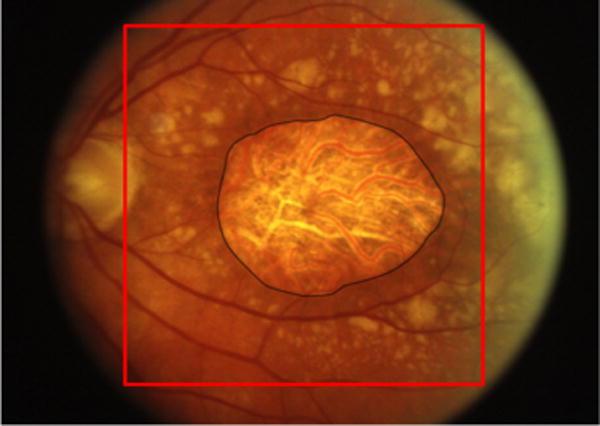
A color fundus image of GA. The black line indicates the expert-assigned segmentation of GA. The red square indicates the region of interest selected by the algorithm for segmentation. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
2.2. Features selection and computation
The algorithm computes a set of 52 features for each image pixel. We use standard image processing features that characterize basic aspects of an image such as color, texture, and intensities. For this purpose, the first step of the algorithm converts the fundus image from RGB to Lab color space. For completeness, we note here that Lab color space has several technical advantages over RGB space. Specifically, metrics for measuring distance between colors are essentially Euclidian. In addition, intensity (L for lightness) and colors (the a channel is green or magenta hue and the b channel is blue or yellow hue) are held separately; thus, one can vary one without varying the others [26].
After converting from RGB to Lab, we compute a histogram-equalized version of each dimension in the Lab color space to improve contrast. Next, we mitigate the effect of artifactual horizontal intensity gradients that are often present in retinal fundus images. To do this, we find the difference between the average intensity of each column of the image and the average intensity of the entire image. This difference is added (column by column) to the original Lab image. This procedure is carried out for each dimension of the Lab image as well as the histogram-equalized Lab images. Next, the Lab images are normalized. The intensities of each pixel in L, a, and b are divided by the mean image intensity of their representative dimension. These normalized Lab intensities constitute features 1–3.
The histogram-equalized Lab intensities make up features 4–6. We note here that histogram equalization ensures that all images span the same intensity range. This creates a more standardized intensity metric across all the images in the study. It also helps mitigate adverse effects of normalization in features 1–3 that may occur in images with large GA regions versus images with very small GA regions.
Next, we incorporate features that quantify information about the pixels’ neighborhood. First, the Lab intensity features 1–3 are each divided by the respective median intensity values in the surrounding 35 × 35 pixel (approximately 1.35 by 1.35 mm) neighborhood. This window size was designed to capture information from smaller-sized GA and its local contrasting surroundings. We selected a window small enough to capture information from relatively small GA, while not being so small as to be the same size as drusen. This helps to prevent isolated drusen or outlier pixels from being misclassified as GA. These relative intensity features constitute features 7–9.
Features 10–12 consist of each Lab pixel’s local energy [16]
where I(x, y) is the image intensity at row y and column x, and is the average intensity in the pixel’s 35 × 35 neighborhood.
Texture is an important characteristic to describe and categorize images, and it is important in discerning GA regions from the rest of the fundus image. Consequently, texture is quantified for each pixel, in each dimension of YCbCr color space. Similar to Lab color space, YCbCr consists of lightness and opponent color dimensions. YCbCr consists of luminance (Y) and blue-difference (Cb) and red-difference (Cr) color components [26]. Thirteen texture features are computed in each plane, as described by Haralick [21]. These make up features 13–51.
The last entry in the feature vector consists of the distance from the pixel to the image center, taken as a proxy of the center of the macula, as GA typically presents near the center of the macula.
In summary, the 52 features computed for each pixel are as follows: 1–3: L, a, and b intensities divided by image mean L, a, and b intensities, respectively 4–6: Histogram equalized L, a, and b intensities 7–9: L, a, and b intensities divided by the 35 × 35 neighborhood median L, a, and b intensities, respectively 10–12: L, a, and b local energies 13–51: 13 Haralick texture features in each Y, Cb, and Cr color planes, resulting in a total of 39 texture features 52: distance from the image center.
2.3. Machine learning
We use a supervised machine learning approach [29] to segment the images into GA regions, and not-GA regions (i.e. we solve a two class classification problem). The general principle of supervised machine learning is to build a model that uses a set of specified features to predict labels (GA or not-GA, in our case). First, broadly summarizing, a training set is used to “teach” the machine. Next, a machine learning method is used to build a model based on this training set. Finally, given a new image, the model will then predict a label for each pixel in the image.
In our case, the training set consists of a sample set of color fundus images in which each pixel is assigned a value for each of the 52 features and then labeled either GA or not-GA. Thus, we did not use healthy control images. However, since the algorithm is pixel-based and uses binary classification, every GA image has an abundance of not-GA pixels which were used as “control” pixels.
In our study, we use an ensemble learning method called a random forest classifier [8]. The random forest method creates a single strong learner resistant to data overfitting and relatively robust to noise [8]. In particular, a random forest is an ensemble approach to classification that employs a group of weak learners (decision trees) with a random selection of features at each node split. Each weak learner will use this random selection of features to decide how a piece of data should be classified. At the end, each of the outputs of the weak decision tree is used in a majority voting scheme to decide the individual pixel label. In our algorithm, for every piece of data, each weak learner “votes” whether a pixel is labeled as GA or not-GA. The threshold for classification is 50%. If a piece of data receives over 50% of the votes from the weak learners, it is classified as GA. Otherwise, it is classified as not-GA, outputting a binary classification map. In this study we use a random forest classifier with 50 decision trees.
3. Validation experiments
3.1. The AREDS data used for validation
Color fundus images from the Age-Related Eye Disease Study (AREDS) database were used for characterizing the performance of our automated GA delineation algorithm. The AREDS was a longitudinal study in which a large number of patients were followed for up to 12 years (median enrollment time 6.5 years) including control patients, neovascular AMD cases, and geographic atrophy cases [4]. As part of the study, the patients were examined by an ophthalmologist on a regular basis, at which time fundus photographs were taken of the left and right eyes and subsequently graded by experts at grading centers for AMD severity. Specifically, each image was assigned a category from 1 to 4 with category 1 corresponding to no evidence of AMD, category 2 corresponding to early stage AMD, category 3 corresponding to intermediate stage AMD, and category 4 corresponding to one of the advanced forms of AMD. During the study, some patients (who were initially diagnosed in category 1, 2, or 3) progressed to one of the advanced forms of AMD including GA.
NIH made available for research purposes a set of anonymized ancillary information on these patients, including fundus photographs and health data. Only a subset of all participants agreed to allow their AMD severity categories available for research purposes. This dataset is known as the AREDS dbGAP and a set of these AREDS dbGAP fundus photographs were digitized and made available by NIH. Although the images are graded, when AMD or GA were present, explicit delineation information was not constructed by the grading centers. Thus, for the purpose of our study, GA delineation had to be performed by a retina specialist. In our study, we used a subset of 143 color fundus images from 55 unique eyes in the AREDS dbGAP (herein shortened to AREDS) database representing varying levels of segmentation challenge.
Our team used Inkscape, an open source graphics editor, to manually delineate regions of GA. Manual delineations were performed and then carefully vetted and corrected by ophthalmologists at the Wilmer Eye Institute of Johns Hopkins University School of Medicine who previously had completed their retina subspecialty training. We then converted the delineations to binary masks that separate the pixels showing GA lesions from the rest of the fundus image so as to compute segmentation metrics.
3.2. Validation method and results
The 143 AREDS images used in this study represented a clear GA diagnostic but a broad spectrum of difficulty and ambiguity with regard to exact delineation of the GA lesion region. In some images, GA lesions were easy to discern and delineate with high contrast to background. GA in other images was very dull, or only faintly distinct as compared to the background. In many ambiguous images, the exact extent of the region where choroidal vessels were visible was not clear, making exact delineation difficult even to a trained grader. Furthermore, many images presented with brighter surrounding drusen.
Of these 143 images, we categorized images as either ambiguous or unambiguous to provide an approximate indication of the extent of the challenge and ambiguity of segmentation for a qualified clinician. 120 images were deemed by our retina specialist to be relatively unambiguous, meaning the GA area was quickly and confidently discerned. For these images, the expected inter-observer variability in GA delineation would be negligible, with no room for argument. The remaining 23 images were categorized as exhibiting some degree of delineation ambiguity. Our retina specialist was confident that GA was present, but its exact extent and borders could be debated.
During the training process, the aggregation of training data was downsampled by a factor of 8. This dramatically reduced training time, with no significant reduction of classification performance.
We used a leave-one-out validation approach to test our machine learning algorithm [38,44]. Given a set of n images, training was performed on n − 1 images, and then the nth image was used for testing. This process was repeated on all n images, therefore all images were tested, with training performed on a non-repetitive data set. Thus, when each image was tested, it was an entirely new image that had never been seen during training.
We used five metrics to assess how well the machine predictions compared to the ground truth segmentations. The Dice coefficient measures the agreement between the segmentation gold standard and the result of the machine-predicted segmentation. The Dice coefficient is quantified as 2(A ∩ B)/(A + B), where A is the area of GA in the ground truth, and B is the area of GA in the machine prediction. Thus, A ∩ B represents the total GA area belonging to both the ground truth and the machine prediction. In the machine-predicted segmentation, every pixel will either be a true positive (TP), false positive (FP), true negative (TN), or false negative (FN). Sensitivity is quantified as TP/(TP + FN), and measures the capability to detect areas of GA. Specificity is defined as TN/(TN + FP), and measures the capability to detect the background. Positive predictive value (PPV) is measured as TP/(TP + FP), and measures the correctly identified proportion the machine labeled as GA. Negative predictive value (NPV) is measured as TN/(TN + FN), and quantifies the correctly identified proportion of pixels that the machine classified as background. Note that all of the above metrics range from 0 to 1, with 1 corresponding to most accurate.
Validation metrics results are presented in Table 1. As expected, performance was markedly better on images of low ambiguity as compared to images of higher ambiguity. Specificity and NPV values were high, indicating stronger correct prediction of background pixels. This is due to the larger quantity of background pixels than GA pixels in the majority of images. Lastly, Dice coefficient, sensitivity, and PPV values have large standard deviations, indicating variability in classification performance.
Table 1.
Results of the segmentation experiment. Results are separated into three categories: all GA images (143 images), GA images with low ambiguity (120 images), and GA images with ambiguity (23 images). Performance is quantified by dice coefficient, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
| Performance metric | All images (n = 143) |
Low ambiguity images (n = 120) |
Ambiguous images (n = 23) |
|---|---|---|---|
| Dice coefficient | 0.68±0.25 | 0.70±0.21 | 0.55±0.25 |
| Sensitivity | 0.65±0.26 | 0.68±0.24 | 0.49±0.27 |
| Specificity | 0.99±0.02 | 0.99±0.01 | 0.98±0.05 |
| PPV | 0.82±0.19 | 0.82±0.19 | 0.82±0.20 |
| NPV | 0.95±0.07 | 0.96±0.06 | 0.91±0.11 |
3.3. Feature importance
To gain a sense of feature importance, we performed an out-of-bag prediction error analysis. During the random forest learning process, the algorithm will “bag” a set of data points to use for learning. The remaining data points are considered “out of bag”. These out of bag data points can be used to test the quality of the predictive power that the random forest learned from the bagged data points [8,19].
To estimate the importance of each feature, the increase in prediction error was calculated when the values of each feature were scrambled across the out-of-bag observations. If the specified feature were important for correct classification, scrambling its values across the out-of-bag observations would degrade the predictive quality of the random forest model developed from the bagged data points. Likewise, if a specified feature were relatively irrelevant for correct classification, scrambling its values across the out-of-bag observations would have little impact on the classification error.
For each variable in this analysis, the increase in prediction error was computed for every tree and then averaged over the entire ensemble of trees. Finally, this error was divided by the standard deviation of the error increase over all trees, leaving a metric quantifying relative importance of features.
A detailed bar graph depicting feature importance is shown in Fig. 8. The most important features tended to be both absolute and local intensity-based features of the a and b color channels. Haralick information correlation textures were the most influential texture features, while most others were less important. The seven most important features were, in ascending order of importance, b and a intensities (divided by mean), a and L intensities divided by local median, b local energy, distance from image center, and b intensity divided by local median.
Fig. 8.
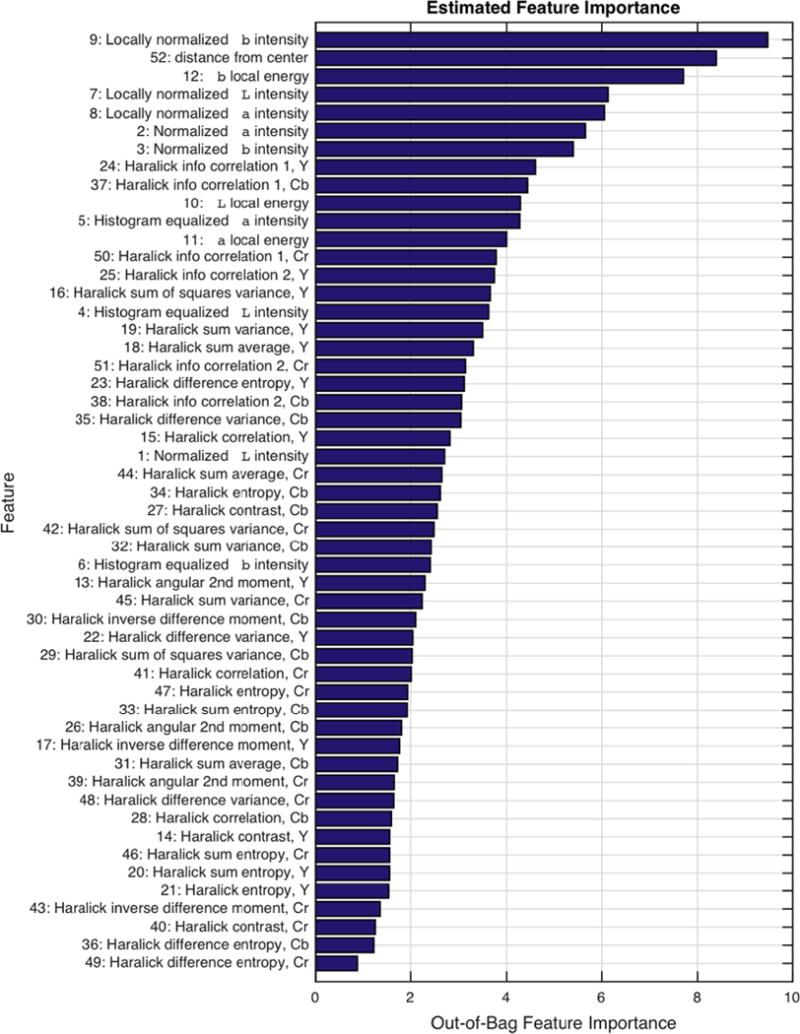
A bar graph of estimated feature importance. Features are labeled on the vertical axis by order of importance.
4. Discussion
4.1. Data set and interpretation of results
Our study proposes a fully automated segmentation method using random forests and applied to a set of 143 AREDS color fundus images. This is a typical test size for GA segmentation studies and ARIA problems in general, in which many studies use of the order of 100 or fewer images [43]. Our segmentation results are promising. When compared to the only other study known to us addressing this specific challenge (GA segmentation on color fundus images) [35], our method produced results that were comparable when we applied our algorithm to only ambiguous GA cases, but improved metrics when we computed our performance on our overall set of images (including ambiguous or unambiguous GA cases). We also performed experiments on a much larger dataset. Overall, this points to a somewhat favorable outcome. However, given the difference in data sets and the remarkable variation of GA presentation, care should be taken not to draw definitive conclusions and interpretations.
4.2. Comparison of segmentation results with prior studies using FAF and OCT
It is important to discuss our results in comparison to the results achieved on FAF and OCT modalities. FAF and OCT segmentation methods undoubtedly result in the high quality segmentation. Segmentation methods on FAF and OCT have achieved high accuracy, reproducibility, and agreement, commonly displaying segmentation accuracy above 0.9 [12,15,23,24,30,33,35,36,39,45]. However, this disparity in segmentation performance between color fundus and FAF and OCT is largely due to differences in imaging modalities rather than faults in segmentation method [35]. Indeed, FAF and OCT have GA presentation with much clearer contrast, making intensity-based segmentation much more straightforward on FAF and OCT images. For the reasons discussed in the subsequent section, the successful segmentation methods applied to FAF and OCT images are difficult to directly translate to the color fundus GA segmentation.
Nevertheless, we believe our method does present some inherent advantages. First, it is a fully automated algorithm. This means the operator does not affect the output of the algorithm, making inter-operator variability a non-factor. Meanwhile, most segmentation methods on FAF and OCT require user initiation. It is likely that by modifying our algorithm to accept human assistance, performance could be substantially improved. Our segmentation method is also pixel-based, meaning at every pixel of the image, the algorithm makes a decision on whether or not to classify the pixel as GA or not-GA. Therefore, it does not matter what shape or configuration the GA assumes, including cases with single lesions, multifocal lesions, or foveal sparing.
4.3. Challenges of color fundus GA segmentation
GA segmentation in color fundus images is a challenging and unsolved problem. As previously recognized [13,40], GA delineation in color fundus images can also be difficult due to poor contrast, variability in choroidal vessel color presentation, or different types of appearances within the same area of atrophy. From our AREDS image set, it is clear that GA presentation is indeed remarkably variable, especially from a machine learning perspective. This is evident from Figs. 1 and 4–7. GA can present as yellow compared to the rest of the fundus. At other times it is red compared to the rest of the fundus. Similarly, it can exhibit rich texture or very little texture. The presence of drusen also complicates the problem. Drusen are labeled as “not-GA” during machine learning, but drusen often have a similar appearance to GA (brighter yellow with respect to the background). Our algorithm recognizes the difference between GA and drusen in some cases. However, as shown in Fig. 6, our algorithm struggled to distinguish drusen from GA in a consistent and reliable manner. A mechanism to reliably separate drusen from GA in an automated approach would improve the capability of automated GA segmentation in color fundus images. Attempting to distinguish GA from not-GA with an automated approach is further complicated by variations in photography lighting conditions.
Fig. 4.
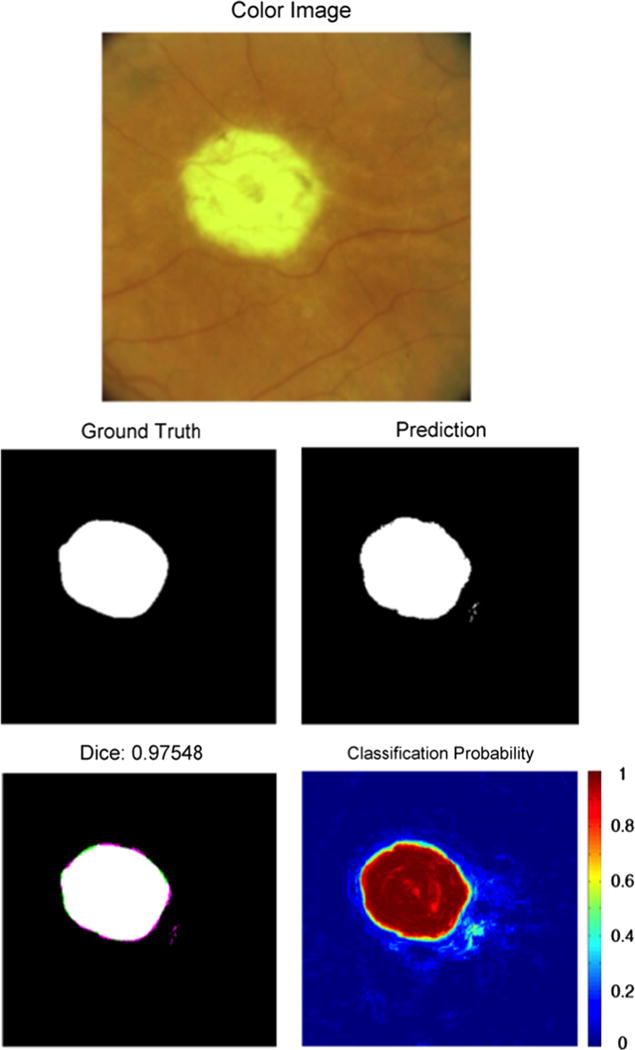
An example of segmentation results of an unambiguous presentation of GA. Note the high classification accuracy in this unambiguous image. The top image is the color fundus image of the eye with GA. The “Ground Truth” image indicates the expert-assigned segmentation, with white pixels corresponding to GA and black pixels corresponding to an absence of GA. The “Prediction” image is the GA prediction from our segmentation algorithm. The “Dice” image provides the Dice coefficient, as well as a comparison image between the ground truth and the segmentation. Pixels that are either white or black were classified the same by both the human expert and the segmentation algorithm. Pink pixels were incorrectly identified as GA by the segmentation algorithm. Green pixels were classified as GA in the ground truth but missed by the segmentation algorithm. The “Classification Probability” image shows the probability that the algorithm would classify a pixel as GA, with red corresponding to probability 1 and blue corresponding to probability 0. 0.5 was used as the classification threshold. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Fig. 7.

An example of segmentation results of GA without clear borders. The GA was labeled as ambiguous. The algorithm correctly identifies a majority of the GA around its center of mass, but fails to identify the GA to the full extent of its soft borders. Refer to Fig. 4 caption for an explanation of images. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Fig. 6.
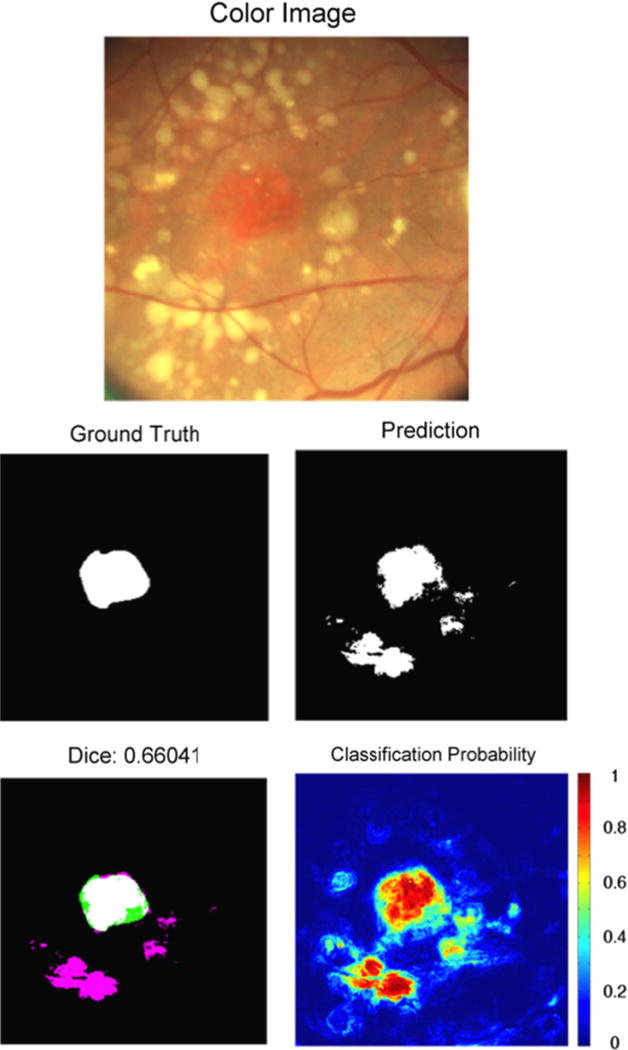
An example of segmentation results of GA with surrounding drusen. The GA was labeled as unambiguous. This GA is reddish in appearance, with surrounding yellow drusen that were misclassified as GA. Note the yellow appearance of the GA in Fig. 4 is similar to the appearance of drusen in this image. Refer to Fig. 4 caption for an explanation of images. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
A variety of GA examples and their machine-predicted segmentation are shown in Figs. 4–7. Only Fig. 7 was labeled as “ambiguous,” Figs. 4–6 were labeled as “unambiguous.” However, despite their lack of ambiguity to a retina expert, the challenges they present for automated segmentation are evident. Furthermore, it is important to note a shortcoming of the supervised machine learning approach. A predictive machine learning approach is developed using training data. So if the training data does not encompass the entire variety of GA appearances, it is likely that performance will be poor on a GA image that presents quite differently from all images in the training set.
4.4. GA feature selection and feature analysis
Because GA presentation on color fundus images is so variable, we selected generic image features in our segmentation algorithm. These features were focused on color intensities and textures, also with respect to local neighborhoods. The local neighborhood features incorporated basic size information of GA versus drusen. Our work relied on the premise that the random forest classifier will – by design – learn a combination of colors or textures that had a strong probability of identifying as GA, and be somewhat immune to incorporating features that may be uninformative as long as informative features are also used. Our results suggest that this is true to a large extent.
In this study we also set out to discover which are the features most important to segmentation performance. Feature analysis showed that color-based intensity features (a and b) were generally more important than texture features. Brightness, L, was also important, but to a lesser extent. This suggests that GA presentation on color fundus images is represented more by color change than brightness or texture change. However, texture is still an important cue, with Haralick “information correlation” textures shown to be the most influential. However, there does not appear to be a single unique texture that defines GA. Instead, certain presentations may have one type of texture, while others may have another. The fact that the distance from the center feature scored high as a feature was not a surprise however, given that GA presents at the peripheral area of the retina much less often than towards the center.
4.5. Future directions and applications
Our results suggest that a fully automated pixel-based machine learning algorithm used to classify all GA color fundus images at a clinically useful level of accuracy is still challenging. However, as a whole, our machine learning approach demonstrates a promising preliminary capability in detecting a large portion of the GA in these color fundus images. In images with very clear GA presentation (Fig. 4), our algorithm obtains very good agreement with the ground truth. This provides numerous pathways for future investigation and potential utility. One is to extend our dataset to more images to train our algorithm to incorporate a greater variety of GA presentation. Furthermore, we could investigate refinement of feature selection and the possibility of segmentation post-processing. The main premise of our method, machine-learning pixel-based classification of GA, could also be applied to GA segmentation of FAF images or OCT projection images.
We believe this allows for possibilities for investigation with potential clinical relevance. First, one could explore coordination of automated GA segmentation on color fundus images with automated GA segmentation on FAF images. It is true that many studies have obtained high-accuracy segmentation results in FAF images. However, in the case of parafoveal GA, it is difficult to differentiate GA from background in FAF images because the center of the macula normally has a decreased AF signal [14,37]. In challenging FAF cases such as this, the automated color segmentation algorithm proposed here could be used for joint segmentation from both FAF and color fundus imagery for a higher confidence segmentation. Most GA segmentation work has not been done using joint FAF and color fundus images, despite the fact that many clinicians use the two types of images in conjunction during examination.
Second, although our current approach is fully automated, one path for future investigation might entail exploring an interactive or semi-automated approach extending from our approach. This could be done by tailoring the machine learning towards each individual patient, while having the classifier still provide predictive information from other images with different GA presentation, drusen presence, and lighting conditions. This approach could be used by incorporating the first visit’s image into training, and then automatically segmenting images from follow-up visits. Or, the clinician could select an initial approximate region containing GA, as well as a region of background [18]. The machine could then incorporate this image-specific information into the training, and then automatically provide a more precise and accurate segmentation of the GA. If approaches to GA segmentation on color fundus images were made to evolve towards reliable segmentation via the aforementioned possibilities, the algorithm could be studied for clinical application of analyzing GA growth rates of patients throughout follow-up visits in longitudinal studies.
In sum, our approach shows a promising step for computer-assisted and automated segmentation of geographic atrophy in color fundus images. This has potential to provide a more robust segmentation, especially in coordination with other retinal imaging modalities such as FAF and OCT. It offers a baseline for further developments in automated segmentation of color fundus images, an area that has been sparsely investigated.
5. Conclusions
We developed a fully automated method using random forest classification for GA segmentation using exclusively color fundus images, an image modality that is widely available. As shown in this study (Table 1), when comparing the results to ground truth obtained from a physician-defined gold standard, we found substantial agreement suggesting that this automated method may offer a good baseline for the future study of such automated methods applied to color fundus images.
Fig. 5.
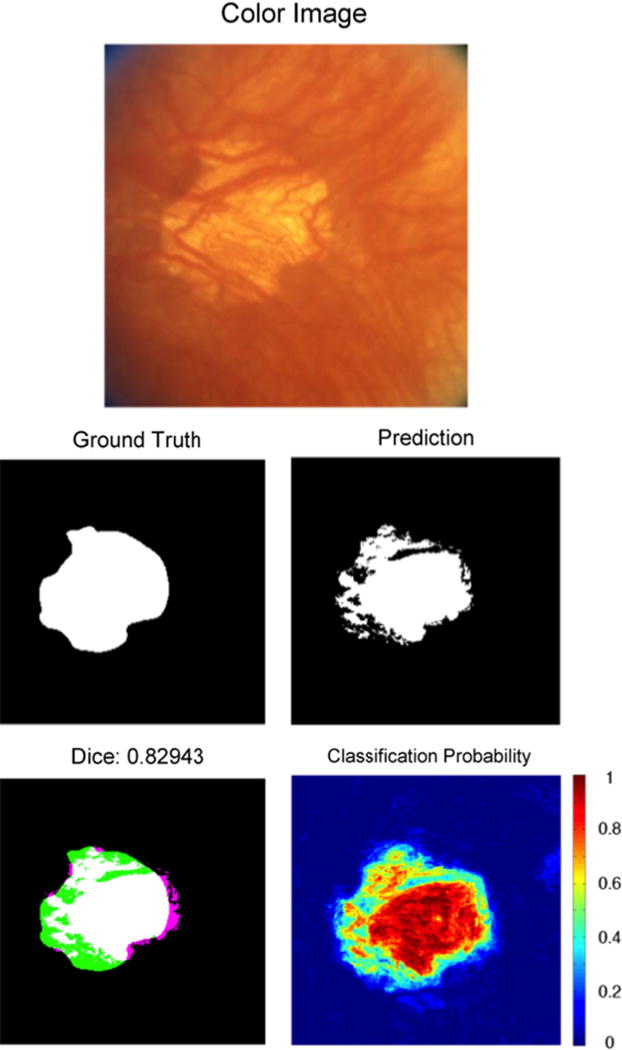
An example of segmentation results of GA with inhomogeneous presentation. The GA was labeled as unambiguous. Dark choroidal vessels appear within the GA, causing the segmentation algorithm to miss some areas of GA. Refer to Fig. 4 caption for an explanation of images. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Footnotes
Research reported in this publication was supported by the National Eye Institute of the National Institutes of Health under award number R21EY024310. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Supported in part by the James P. Gills Professorship and unrestricted research funds to the Retina Division for macular degeneration and related diseases research. Additional support from the JHU Whiting School of Engineering SPUR program is acknowledged.
Conflict of interest statement
None declared.
References
- 1.Subfoveal neovascular lesions in age-related macular degeneration: guidelines for evaluation and treatment in the macular photocoagulation study. Arch Ophthalmol. 1991;1099:1242–1257. URL: http://dx.doi.org/10.1001/archopht.1991.01080090066027. [PubMed] [Google Scholar]
- 2.Potential public health impact of age-related eye disease study results: Areds report no. 11. Arch Ophthalmol. 2003;12111:1621–1624. doi: 10.1001/archopht.121.11.1621. URL: 〈 http://dx.doi.org/10.1001/archopht.121.11.1621〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramoff M, Garvin M, Sonka M. Retinal imaging and image analysis. IEEE Rev Biomed Eng. 2010;3:169–208. doi: 10.1109/RBME.2010.2084567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Age-Related Eye Disease Study Research Group and others. The age-related eye disease study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the age-related eye disease study report number 6. Am J Ophthalmol. 2001;1325:668. doi: 10.1016/s0002-9394(01)01218-1. [DOI] [PubMed] [Google Scholar]
- 5.Baumann B, Gtzinger E, Pircher M, Sattmann H, Schtze C, Schlanitz F, Ahlers C, Schmidt-Erfurth U, Hitzenberger CK. Segmentation and quantification of retinal lesions in age-related macular degeneration using polarization-sensitive optical coherence tomography. J Biomed Opt. 2010;156:061704–061704-9. doi: 10.1117/1.3499420. URL: 〈 http://dx.doi.org/10.1117/1.3499420〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A, Bressler N, Bressler S, Chisholm I, Coscas G, Davis M, de Jong P, Klaver C, Klein B, Klein R, Mitchell P, Sarks J, Sarks S, Soubrane G, Taylor H, Vingerling J. An international classification and grading system for age-related maculopathy and age-related macular degeneration. Surv Ophthalmol. 1995;395:367–374. doi: 10.1016/s0039-6257(05)80092-x. URL: 〈 http://www.sciencedirect.com/science/article/pii/S003962570580092X〉. [DOI] [PubMed] [Google Scholar]
- 7.Blinder KJ, Bradley S, Bressler NM, Bressler SB, Donati G, Hao Y, Ma C, Menchini U, Miller J, Potter MJ, Pournaras C, Reaves A, Rosenfeld PJ, Strong HA, Stur M, Su XY, Virgili G. Treatment of age-related macular degeneration with photodynamic therapy study group, Verteporfin in Photo-dynamic Therapy study group. Effect of lesion size, visual acuity, and lesion composition on visual acuity change with and without verteporfin therapy for choroidal neovascularization secondary to age-related macular degeneration: tap and vip report no. 1. Am J Ophthalmol. 2003;1363:407–418. doi: 10.1016/s0002-9394(03)00223-x. URL: 〈 http://dx.doi.org/10.1016/S0002-9394(03)00223-X〉. [DOI] [PubMed] [Google Scholar]
- 8.Breiman L. Random forests. Mach Learn. 2001;451:5–32. URL: 〈 http://dx.doi.org/10.1023/A%3A1010933404324〉. [Google Scholar]
- 9.Bressler N. Age-related macular degeneration is the leading cause of blindness. J Am Med Assoc. 2004;29115:1900–1901. doi: 10.1001/jama.291.15.1900. URL: 〈 http://dx.doi.org/10.1001/jama.291.15.1900〉. [DOI] [PubMed] [Google Scholar]
- 10.Bressler NM, Chang TS, Suñer IJ, Fine JT, Dolan CM, Ward J, Ianchulev T. Vision-related function after ranibizumab treatment by better- or worse-seeing eye. Ophthalmology. 2010;1174:747–756. e4. doi: 10.1016/j.ophtha.2009.09.002. URL: 〈 http://www.aaojournal.org/article/S0161-6420(09)00981-6/abstract〉. [DOI] [PubMed] [Google Scholar]
- 11.Burlina P, Freund D, Dupas B, Bressler N. Automatic screening of age-related macular degeneration and retinal abnormalities. Engineering in Medicine and Biology Society, EMBC, 2011 Annual International Conference of the IEEE; Boston: IEEE; 2011. pp. 3962–3966. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, de Sisternes L, Leng T, Zheng L, Kutzscher L, Rubin DL. Semiautomatic geographic atrophy segmentation for sd-oct images. Biomed Opt Express. 2013;412:2729–2750. doi: 10.1364/BOE.4.002729. URL: 〈 http://www.opticsinfobase.org/boe/abstract.cfm?URI=boe-4-12-2729〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q, Leng T, Niu S, Shi J, de Sisternes L, Rubin DL. A false color fusion strategy for drusen and geographic atrophy visualization in optical coherence tomography images. Retina. 2014;3412:2346–2358. doi: 10.1097/IAE.0000000000000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choudhry N, Giani A, Miller JW. Fundus autofluorescence in geographic atrophy: a review. Semin Ophthalmol. 2010;255–256:206–213. doi: 10.3109/08820538.2010.518121. URL: 〈 http://dx.doi.org/10.3109/08820538.2010.518121〉. [DOI] [PubMed] [Google Scholar]
- 15.Deckert A, Schmitz-Valckenberg S, Jorzik J, Bindewald A, Holz F, Mansmann U. Automated analysis of digital fundus autofluorescence images of geographic atrophy in advanced age-related macular degeneration using confocal scanning laser ophthalmoscopy (cslo) BMC Ophthalmol. 2005;51:8. doi: 10.1186/1471-2415-5-8. URL: 〈 http://www.biomedcentral.com/1471-2415/5/8〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devisetti K, Karnowski T, Giancardo L, Li Y, Chaum E. Geographic atrophy segmentation in infrared and autofluorescent retina images using supervised learning. Engineering in Medicine and Biology Society, EMBC, 2011 Annual International Conference of the IEEE; Boston. 2011. pp. 3958–3961. [DOI] [PubMed] [Google Scholar]
- 17.Drexler W, Fujimoto JG. State-of-the-art retinal optical coherence tomography. Prog Retin Eye Res. 2008;27(1):45–88. doi: 10.1016/j.preteyeres.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Freund DE, Bressler N, Burlina P. Automated detection of drusen in the macula. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2009. ISBI’09, IEEE; 2009. pp. 61–64. [Google Scholar]
- 19.Genuer R, Poggi JM, Tuleau-Malot C. Variable selection using random forests. Pattern Recognit Lett. 2010;3114:2225–2236. URL: 〈 http://www.sciencedirect.com/science/article/pii/S0167865510000954〉. [Google Scholar]
- 20.Göbel A, Fleckenstein M, Schmitz-Valckenberg S, Brinkmann C, Holz F. Imaging geographic atrophy in age-related macular degeneration. Opthalmologica. 2011;226:182–190. doi: 10.1159/000330420. [DOI] [PubMed] [Google Scholar]
- 21.Haralick RM. Statistical and structural approaches to texture. Proc IEEE. 1979;675:786–804. URL: 〈 http://dx.doi.org/10.1109/proc.1979.11328〉. [Google Scholar]
- 22.Holz FG, Strauss EC, Schmitz-Valckenberg S, van Lookeren Campagne M. Geographic atrophy: clinical features and potential therapeutic approaches. Ophthalmology. 2014;1215:1079–1091. doi: 10.1016/j.ophtha.2013.11.023. URL: 〈 http://www.sciencedirect.com/science/article/pii/S0161642013011020〉. [DOI] [PubMed] [Google Scholar]
- 23.Hu Z, Medioni GG, Hernandez M, Hariri A, Wu X, Sadda SR. Segmentation of the geographic atrophy in spectral-domain optical coherence tomography and fundus autofluorescence images. Invest Ophthalmol Vis Sci. 2013;5413:8375–8383. doi: 10.1167/iovs.13-12552. URL: 〈 http://www.iovs.org/content/54/13/8375.abstract〉. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, Medioni GG, Hernandez M, Sadda SR. Automated segmentation of geographic atrophy in fundus autofluorescence images using supervised pixel classification. J Med Imaging. 2015;21:014501. doi: 10.1117/1.JMI.2.1.014501. URL: 〈 http://dx.doi.org/10.1117/1.JMI.2.1.014501〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang D, Swanson E, Lin C, Schuman J, Stinson W, Chang W, Hee M, Flotte T, Gregory K, Puliafito C, et al. Optical coherence tomography. Science. 1991;254(5035):1178–1181. doi: 10.1126/science.1957169. URL: 〈 http://www.sciencemag.org/content/254/5035/1178.abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain AK. Fundamentals of Digital Image Processing. Prentice-Hall; Upper Saddle, NJ: 1989. [Google Scholar]
- 27.Jain N, Farsiu S, Khanifar AA, Bearelly S, Smith RT, Izatt JA, Toth CA. Quantitative comparison of drusen segmented on sd-oct versus drusen delineated on color fundus photographs. Invest Ophthalmol Vis Sci. 2010;5110:4875–4883. doi: 10.1167/iovs.09-4962. URL: 〈 http://www.iovs.org/content/51/10/4875.abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kankanahalli S, Burlina PM, Wolfson Y, Freund DE, Bressler NM. Automated classification of severity of age-related macular degeneration from fundus photographs. Invest Ophthalmol Vis Sci. 2013;543:1789–1796. doi: 10.1167/iovs.12-10928. [DOI] [PubMed] [Google Scholar]
- 29.Kotsiantis SB. Supervised machine learning: a review of classification techniques. Proceedings of the 2007 Conference on Emerging Artificial Intelligence Applications in Computer Engineering: Real Word AI Systems with Applications in eHealth, HCI, Information Retrieval and Pervasive Technologies; Amsterdam, The Netherlands, The Netherlands: IOS Press; 2007. pp. 3–24. URL: 〈 http://dl.acm.org/citation.cfm?id=1566770.1566773〉. [Google Scholar]
- 30.Lee N, Smith R, Laine A. Interactive segmentation for geographic atrophy in retinal fundus images, in: 42nd Asilomar Conference on Signals, Systems and Computers. 2008:655–658. doi: 10.1109/ACSSC.2008.5074488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;3799827:1728–1738. doi: 10.1016/S0140-6736(12)60282-7. URL: 〈 http://www.sciencedirect.com/science/article/pii/S0140673612602827〉. [DOI] [PubMed] [Google Scholar]
- 32.Mookiah MRK, Acharya UR, Koh JE, Chandran V, Chua CK, Tan JH, Lim CM, Ng E, Noronha K, Tong L, Laude A. Automated diagnosis of age-related macular degeneration using greyscale features from digital fundus images. Comput Biol Med. 2014;530:55–64. doi: 10.1016/j.compbiomed.2014.07.015. URL: 〈 http://www.sciencedirect.com/science/article/pii/S0010482514001802〉. [DOI] [PubMed] [Google Scholar]
- 33.Panthier C, Querques G, Puche N, Le Tien V, Garavito RB, Bechet S, Massamba N, Souied EH. Evaluation of semiautomated measurement of geographic atrophy in age-related macular degeneration by fundus autofluorescence in clinical setting. Retina. 2014;343:576–582. doi: 10.1097/01.iae.0000433986.32991.1e. [DOI] [PubMed] [Google Scholar]
- 34.Pircher M, Gotzinger E, Leitgeb R, Sattmann H, Findl O, Hitzenberger C. Imaging of polarization properties of human retina in vivo with phase resolved transversal PS-OCT. Opt Express. 2004;12(24):5940–5951. doi: 10.1364/opex.12.005940. [DOI] [PubMed] [Google Scholar]
- 35.Ramsey DJ, Sunness JS, Malviya P, Applegate C, Hager GD, Handa JT. Automated image alignment and segmentation to follow progression of geographic atrophy in age-related macular degeneration. Retina. 2014;347:1296–1307. doi: 10.1097/IAE.0000000000000069. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz-Valckenberg S, Brinkmann CK, Alten F, Herrmann P, Stratmann NK, Göbel AP, Fleckenstein M, Diller M, Jaffe GJ, Holz FG. Semiautomated image processing method for identification and quantification of geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;5210:7640–7646. doi: 10.1167/iovs.11-7457. URL: 〈 http://www.iovs.org/content/52/10/7640.abstract〉. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz-Valckenberg S, Fleckenstein M, Göbel AP, Sehmi K, Fitzke FW, Holz FG, Tufail A. Evaluation of autofluorescence imaging with the scanning laser ophthalmoscope and the fundus camera in age-related geographic atrophy. Am J Ophthalmol. 2008;1462:183–192. doi: 10.1016/j.ajo.2008.04.006. URL: 〈 http://www.sciencedirect.com/science/article/pii/S0002939408002754〉. [DOI] [PubMed] [Google Scholar]
- 38.Scholkopf B, Smola AJ. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. MIT Press; Cambridge, MA, USA: 2001. [Google Scholar]
- 39.Schütze C, Bolz M, Sayegh R, Baumann B, Pircher M, Gtzinger E, Hitzenberger CK, Schmidt-Erfurth U. Lesion size detection in geographic atrophy by polarization-sensitive optical coherence tomography and correlation to conventional imaging techniques. Invest Ophthalmol Vis Sci. 2013;541:739–745. doi: 10.1167/iovs.11-7877. URL: 〈 http://www.iovs.org/content/54/1/739.abstract〉. [DOI] [PubMed] [Google Scholar]
- 40.Sunness JS, Bressler NM, Tian Y, Alexander J, Applegate CA. Measuring geographic atrophy in advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 1999;408:1761–1769. URL: 〈 http://www.iovs.org/content/40/8/1761.abstract〉. [PubMed] [Google Scholar]
- 41.The AREDS Research Group. Change in area of geographic atrophy in the age-related eye disease study: Areds report number 26. Arch Ophthalmol. 2009;1279:1168–1174. doi: 10.1001/archophthalmol.2009.198. URL: 〈 http://dx.doi.org/10.1001/archophthalmol.2009.198〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolentino MJ, Dennrick A, John E, Tolentino MS. Drugs in phase ii clinical trials for the treatment of age-related macular degeneration. Expert Opin Invest Drugs. 2015;242:183–199. doi: 10.1517/13543784.2015.961601. URL: http://dx.doi.org/10.1517/13543784.2015.961601. [DOI] [PubMed] [Google Scholar]
- 43.Trucco E, Ruggeri A, Karnowski T, Giancardo L, Chaum E, Hubschman JP, al Diri B, Cheung CY, Wong D, Abràmoff M, et al. Validating retinal fundus image analysis algorithms: issues and a proposal. Invest ophthalmol Vis Sci. 2013;545:3546–3559. doi: 10.1167/iovs.12-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vapnik VN. Statistical Learning Theory. 1st. Wiley; New York: 1998. [Google Scholar]
- 45.Yehoshua Z, Garcia Filho C, Alexandre A, Penha F, Gregori G, Stetson PF, Feuer WJ, Rosenfeld PJ. Comparison of geographic atrophy measurements from the oct fundus image and the sub-rpe slab image. Ophthal Surg Lasers Imaging Retina. 2013;442:127–132. doi: 10.3928/23258160-20130313-05. URL: 〈 http://search.proquest.com/docview/1350204446?accountid=27702〉. [DOI] [PubMed] [Google Scholar]


