Abstract
Inflammation is a condition stemming from complex host defense and tissue repair mechanisms, often simply characterized by plasma levels of a single acute reactant. We attempted to identify candidate biomarkers of systemic inflammation within the plasma proteome. We applied quantitative proteomics using isobaric mass tags (iTRAQ) tandem mass spectrometry to quantify proteins in plasma of 500 Nepalese children 6–8 years of age. We evaluated those that co-vary with inflammation, indexed by α-1-acid glycoprotein (AGP), a conventional biomarker of inflammation in population studies. Among 982 proteins quantified in >10% of samples, 99 were strongly associated with AGP at a family-wise error rate of 0.1%. Magnitude and significance of association varied more among proteins positively (n = 41) than negatively associated (n = 58) with AGP. The former included known positive acute phase proteins including C-reactive protein, serum amyloid A, complement components, protease inhibitors, transport proteins with anti-oxidative activity, and numerous unexpected intracellular signaling molecules. Negatively associated proteins exhibited distinct differences in abundance between secretory hepatic proteins involved in transporting or binding lipids, micronutrients (vitamin A and calcium), growth factors and sex hormones, and proteins of largely extra-hepatic origin involved in the formation and metabolic regulation of extracellular matrix. With the same analytical approach and the significance threshold, seventy-two out of the 99 proteins were commonly associated with CRP, an established biomarker of inflammation, suggesting the validity of the identified proteins. Our findings have revealed a vast plasma proteome within a free-living population of children that comprise functional biomarkers of homeostatic and induced host defense, nutrient metabolism and tissue repair, representing a set of plasma proteins that may be used to assess dynamics and extent of inflammation for future clinical and public health application.
Introduction
Inflammation is an evolutionarily conserved body response for protecting the host from potentially lethal stresses [1]. It is often understood as a process to isolate and eliminate pathogens or other non-self or intolerant agents by immune cells [2] followed by processes that resolve inflammation, repair damaged tissues and restore homeostasis [3]. However, when the transition to tissue repair fails, inflammatory processes can persist and cause harm [4]. It is well documented that chronic inflammation, often viewed as subclinical, contributes to chronic disease processes leading to obesity, diabetes, atherosclerosis, rheumatoid arthritis and cancer [4]. In addition, inflammation in response to continuous exposure to infectious agents [5–7] and environmental toxins [8, 9] may contribute to childhood undernutrition and developmental deficits in impoverished areas of the world where poor sanitation and frequent infections are common.
An observable characteristic of inflammation is an increase or decrease in the release and concentration of certain proteins in the bloodstream, a process regarded as an acute phase reaction, although it can occur in response to either acute or chronic inflammatory stress [1]. At least 50 acute phase proteins (APP) are known to increase or decrease by at least 25% in humans, a commonly used cut-off, from their baseline concentrations during inflammation [10, 11]. This quantitative alteration is mainly regulated by inflammatory mediators that induce reactions within innate and adaptive immune, neuroendocrine, vascular endothelial, hematopoietic, metabolic, and other defense and repair systems [10]. Due to these systemic effects, APPs are widely used as clinical diagnostic and prognostic indicators of disease processes [12]. Also, APPs are often recommended for use in population studies as correction factors for assessing status with respect to micronutrients, whose indicator concentrations may decrease (e.g., serum retinol for vitamin A) or rise (e.g, serum ferritin for iron) with inflammation [13–15]. Although large in number, only a few APP, primarily α-1-acid glycoprotein (AGP) and C-reactive protein (CRP), are typically assessed in population studies as indicators of inflammation [15]. This is because their responses and prognostic values are different and well-defined, with concentrations that reliably increase during infection, responding to numerous systemic infections and other pathological conditions, and decrease with resolution [10, 14, 16]. Yet this nearly sole reliance on AGP and CRP may also, in part, be due to an incomplete understanding of the many and varied molecular mediators and responders to inflammation, a general inability to measure and interpret their responses to inflammatory stresses, limited resources to quantify multiple APPs, and a general lack of awareness in the public health research community of the highly varied dynamics of inflammation that coexist in populations.
The unbiased approach of plasma proteomics offers a unique opportunity to discover, quantify and explore the utility of a wider array of proteins that respond to, initiate, maintain and resolve inflammation. In a given population, all phases of the inflammatory response are likely to be present, highlighting the potential utility of accessing and interpreting a larger inventory of biomarkers with which to understand homeostatic mechanisms, disease processes, types of inflammation and the range of inflammatory exposures in an environment.
In this population study of 6–8 year old Nepalese children [17], we rely primarily on the circulating concentration of AGP to define inflammation as a continuous measure. AGP, or orosomucoid (ORM), is present in plasma as a mixture of ORM1 and ORM2 each of which is encoded by two tandomly arranged genes, ORM1 and ORM2 [18]. Because the concentration of AGP slowly rises and remains elevated during recovery or convalescence, AGP may be considered more sensitive in detecting chronic and subclinical inflammation in populations than CRP, which tends to react, spike and resolve more quickly, and thus be less often detected in prevalence surveys [14]. We refer to the entire set of plasma proteins that definitively and quantitatively co-vary with AGP, as a “population plasma inflammasome” whose members may include but extend beyond classical acute phase proteins, contain proteins of hepatic origin as well as those secreted or leaked from extra-hepatic tissues [19, 20], and include proteins involved in constitutive and induced homeostatic immune, coagulative and repair processes associated with inflammation. We verify the reliability of this set of inflammatory proteins by repeating the same analysis using CRP, and evaluating overlap. We distinguish the compound term, population plasma inflammasome, from “inflammasome”, used to describe an intracellular multiprotein complex that is responsible for activating the processing of pro-inflammatory cytokines [21]. While limited animal and human subject experimentations have described an endotoxin-induced plasma inflammasome and compilations of inflammatory plasma proteins from databases for clinical use exist in the literature [22–24], definition of a human population plasma inflammasome remains incomplete.
In this population sample [17], 31% and 6% of children exhibited elevated plasma concentrations of AGP and CRP above conventional, clinical disease thresholds of 1g/L and 5 mg/L, respectively, reflecting a population affected by chronic inflammation [13] but not acutely ill, providing an opportunity to explore markers of low-grade (“subclinical”) inflammation that may nonetheless be useful for population assessment [25]. We hypothesized the existence of a quantifiable population plasma inflammasome, defined as a suite of plasma proteins tracking AGP with high confidence, which we verified by also evaluating their concordance of association with plasma CRP concentrations.
Materials and Methods
Field study and assessment of inflammatory biomarkers
In 1999–2001, a community-randomized, placebo-controlled field trial was carried out in the District of Sarlahi, Nepal to assess effects of 4 combinations of antenatal supplemental micronutrients on birth outcomes [26]. Of 3,892 children exiting the trial as infants, 3,524 were followed-up at 6–8 years of age for a nutritional, health and socio-demographic assessment by methods previously described [27, 28].
During home visits, children of consenting parents were asked to fast overnight, and phlebotomists collected venous blood samples the next morning from 94% (n = 3,305) of eligible children. Bio-specimens were brought to a field laboratory for plasma extraction. Plasma samples were stored in liquid nitrogen tanks and shipped to the Center for Human Nutrition, Johns Hopkins Bloomberg School of Public Health in Baltimore, MD, USA. Among the plasma samples, 2,130 samples (64%) were selected based on having multiple plasma aliquots, complete epidemiologic data from both the original trial and follow-up assessment, and valid birth size measures (birth weight measured <72 hours after birth). Of the 2,130, 1,000 child plasma samples were randomly sampled across the five original maternal intervention groups (n = 200 from each) and analyzed for multiple micronutrients and inflammation (AGP and CRP) status by conventional assays [13]. Specifically, concentrations of plasma AGP and CRP were measured by a radial immunodiffusion assay (Kent Laboratories; CV = 10.0%) and a benchtop clinical chemistry analyzer (Immulite 1000; Siemens Diagnostics; CV = 5.8%), respectively. From the 1,000 specimens, specimens were ordered by date of field blood collection within each original maternal supplement allocation stratum of 200 specimens, and following a chance start every other specimen was selected for inclusion into the proteomics archive, yielding a total of 500 samples (n = 100 per maternal group) [17]. Socio-demographic, anthropometric and morbidity status, and dietary intakes among the 500 selected children were comparable to the 500 not in the proteomics study [13].
Ethics statement
The original prenatal micronutrient supplementation trial was approved by the institutional review board (IRB) at Johns Hopkins University, Baltimore, MD, USA and the Nepal Health Research Council (NHRC) in Kathmandu, Nepal and registered with ClinicalTrials.gov: NCT00115271. The follow-up study with biospecimen collection was approved by the IRB and NHRC. During follow-up home visits, oral consent was obtained from mothers of eligible children after being explained the purpose, activities and risks of participation by trained field staff following an approved script. Oral consent was deemed appropriate due to high levels of illiteracy in the study population. Consent was documented by field staff and entered into the study database.
Proteomics and data analysis
Immuno-depletion and proteomics assays have been previously described [17]. Briefly, a master plasma pool was prepared by combining plasma aliquots of 25 μL from each of the 1,000 samples comprising the micronutrient archive. The 500 specimens selected for proteomics analysis each comprised 40 μL of plasma. Individual specimens plus master pool aliquots were depleted of six high abundance proteins (albumin, transferrin, IgG, IgA, anti-trypsin, and haptoglobin) using a Human-6 Multiple Affinity Removal System LC column (Agilent Technologies). Proteomics analysis was performed at the Proteomics and Mass Spectrometry Core within the Johns Hopkins School of Medicine. Immno-depleted samples (100 μg of protein) were digested overnight with trypsin (Promega, sequencing grade). Peptide samples of 7 individuals and one masterpool sample were randomly labeled with 8-plex Isobaric Tag for Relative and Absolute Quantification (iTRAQ) reagents that contained different reporter ions which can be used as measures of peptide relative abundance in the original sample. The combined sample was fractionated into 24 fractions by strong cation exchange chromatography. iTRAQ-labeled peptides were loaded on to a reverse-phase nanobore column. Eluted peptides were sprayed into an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) and interfaced with a NanoAcquity ultra-HPLC (Waters). Full MS scans and fragmented MS/MS scans were acquired and these spectra were searched against Refseq 40 protein database using MASCOT (Matrix Science v2.3) through Proteome Discoverer software (v1.3, Thermo Scientific). Peptides were identified with a confidence threshold of <5% false discovery rate. A total of 72 iTRAQ experiments were performed for this study.
Details of relative abundance estimation have been described previously [29]. Briefly, reporter ion intensities were log2 base-transformed and median normalized for each reporter ion intensity spectrum. The relative abundance of proteins in each channel of each experiment was estimated by calculating the median of all the median-polished log2 ion intensities across all spectra belonging to each protein. Corrections for differences in amounts of material loaded in the channels and sample processing were carried out by subtracting the channel median from the relative abundance estimate, normalizing all channels to have median zero. Because there is no physiologically meaningful cut-off of plasma AGP concentration to dichotomize inflammation status, we employed linear mixed-effects (LME) models to assess the linear association between log2 transformed plasma AGP concentration and relative abundance of individual plasma proteins from multiple iTRAQ experiments. A univariate random intercept model was fit for each protein with AGP as a dependent variable, the protein as a fixed effect, and each iTRAQ experiment as a random effect [28]. Model parameters in these mixed effects models were estimated via Restricted Maximum Likelihood [30]. Estimates of absolute protein abundance were calculated as Best Linear Unbiased Predictors [31].
Observed sample sizes were different among proteins due to missing values from the mass-spectrometry [32]. We report summary statistics for the association between plasma protein abundance and AGP as percent change in AGP per 2-fold (100%) increase in protein relative abundance (derived from the slope of the LME model) and its statistical significance (p-value), and the correlation between estimated absolute protein abundance and plasma AGP concentration. We controlled the family-wise error rate (FWER) at the 0.1% level using a Bonferroni correction, to only select proteins truly associated with AGP (P <1.02e-06). We also present in Supporting Information, proteins passing a false discovery rate threshold of 1% (q<0.01).
Additional analyses were conducted to examine the validity of the identified proteins. Specifically, we applied the same analytical approach and significance threshold to identify, quantify and evaluate the direction and strength of association between plasma protein relative abundance and CRP concentration. In addition, we identified differentially abundant proteins between children who reported at least one day of symptomatic morbidity (fever, diarrhea, productive cough, or rapid breathing) and children without any symptoms in the week prior to blood sampling. All CRP and morbidity-related results are presented in Supporting Information tables.
Since we expected that proteins associated with AGP might be co-regulated or co-vary in the same biological systems, we examined relationships between proteins by constructing a correlation matrix and performed principal component analysis (PCA). Pairwise protein:protein correlation coefficients were calculated within each iTRAQ experiment and averaged coefficients across iTRAQ experiments to construct a correlation matrix and to perform PCA. Bi-plots were constructed to visualize the 1st, 2nd, and 3rd principal components of each protein from PCA [33].
Corresponding gene symbols of protein genInfo identifier (gi) numbers were derived from the Human Genome Organisation (HUGO) gene annotation and used in tables and figures, once linked to protein names in initial descriptive tables, to conserve space [34]. Resources of general description of proteins including cellular compartment, biological/molecular functions, and mRNA expression across tissues were extracted from the NCBI protein database, UniProt beta, Gene ontology Annotation (Uniprot-GOA) database, BioGPS, COMPARTMENTS, and in-depth review of literature [35–39].
All analyses were performed using the R Environment for Statistical Computing (version 3.1.0; R Foundation for Statistical Computing, Vienna, Austria).
Results
Study participant characteristics
Demographic, nutritional, and health characteristics of study children (Table 1) were similar to children in the original, larger follow-up cohort [28]. Children were undernourished, compared to the WHO reference population, reflected by prevalence rates of 40%, 15%, and 50% for stunting, thinness, and underweight. More than half and approximately 30% of children consumed dairy food and dark green leafy vegetables equal to or greater than 3 times in the past week, respectively, but meat, fish, and eggs were less frequently consumed by children. Approximately 8% of children reported at least one episode of fever in the past week, but the prevalence of other symptoms was low (<5%). Fourteen percent of children reported any symptoms of fever, diarrhea, productive cough, or rapid breathing in the past week. Median (interquartile range) of plasma AGP and CRP concentrations were 0.84 (0.70, 1.05) g/L and 0.28 (0.13, 0.78), with 30% and 6% of children having an elevated AGP concentration (>1 g/L) and CRP (>5 mg/L) concentrations, respectively.
Table 1. Demographic, nutritional, and health characteristics of 6–8 year old children in rural Nepal (n = 500). a .
| Characteristics | Values |
|---|---|
| Age, years | 7.5 (0.4) |
| Girls, % | 50.2 |
| Ethnicity (Pahadi), % | 31.8 |
| Anthropometric measurements b | |
| Height, cm | 114.1 (5.8) |
| Weight, kg | 18.2 (2.2) |
| BMI, kg/m2 | 14.0 (1.0) |
| Mid-upper arm circumference, cm | 15.4 (1.1) |
| Height-for-age | -1.77 (0.95) |
| BMI-for-age | -1.20 (0.89) |
| Weight-for-age | -1.99 (0.85) |
| Stunted, % c | 39.1 |
| Thin, % c | 16.4 |
| Underweight, % c | 48.5 |
| Dietary, ≥3 intake in the past week, % | |
| Dairy | 56.6 |
| Meat | 8.6 |
| Fish | 9.0 |
| Eggs | 2.2 |
| Dark green leafy vegetables | 32.4 |
| Morbidity, symptoms reported in the past week, % | |
| Fever | 8.2 |
| Diarrhea | 3.2 |
| Productive cough | 3.8 |
| Rapid breathing | 2.8 |
| Any of above symptoms | 14.0 |
| Plasma Concentration of AGP | |
| AGP, g/L | 0.84 (0.70, 1.05) d |
| AGP > 1.0 g/L e , % | 29.8 |
| CRP, mg/L | 0.28 (0.13, 0.78) d |
| CRP > 5.0 mg/L e , % | 6.0 |
Abbreviations: AGP, α-1-acid glycoprotein; BMI, body mass index; CRP, C-reactive protein.
aValues are means (SD) or percentages (95% CI).
bOne outlier was excluded (n = 499).
cZ-scores were calculated based on World Health Organization reference for 5–19 years. Underweight, weight-for-age Z-score< -2; stunted, height-for-age Z-score< -2; thin, BMI-for-age Z-score< -2 [40].
dMedian (Interquartile range).
eCut-off for inflammation [41].
Plasma proteins associated with AGP
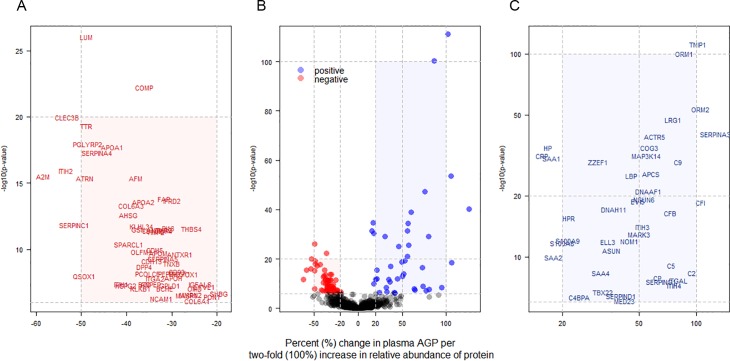
A total of 3,933 proteins were identified and quantified among 72 iTRAQ experiments required to analyze the 500 child plasma samples, of which 982 proteins were quantified in >10% of all samples (n>50). Ninety-nine proteins (~10% of all proteins adequately quantified) significantly co-varied with plasma AGP passing a Bonferroni corrected significance level, of which 41 and 58 proteins were positively and negatively associated with AGP, respectively. Among proteins positively associated with AGP, TNFAIP3 interacting protein 1 (Gene symbol: TNIP1) (P = 7.6x10-112) and orosomucoid 1 (ORM1) (P = 4.6x10-101) showed the strongest associations, followed by orosomucoid 2 (ORM2) (P = 3.1x10-54) (Table 2). Lumican (LUM) (P = 9.1x10-27) and cartilage oligomeric matrix protein (COMP) (P = 5.7x10-23) were most strongly associated among negative correlates (Table 3). A volcano plot shows distinct patterns within the population plasma inflammasome (Fig 1A–1C). Specifically, the percent change in AGP (95% confidence intervals) and strength of significance of association varied more widely within the group of positively than negatively associated proteins. A 86.6 (76.2, 97.6)% and 105.3 (87.4, 124.8)% increase in AGP concentration was associated with a 100% (two-fold) increase in relative abundance of ORM1 and ORM2, respectively (Fig 1C). A comparable 73~105% increase in AGP was also associated with a 100% increase in complement components 2, 5, and 9 (C2/5/9) and complement factors F and I (CFB and CFI). On the other hand, a smaller 15~18% increase in AGP was associated with a 2-fold rise in other acute phase proteins such as CRP, haptoglobin (HP), and serum amyloid A 1 and 2 (SAA1/2) (Fig 1C). Overall, a narrower range in reduction in AGP, 20~40%, was associated with a 2-fold increase in the relative abundance of 46 out of the 58 negatively associated proteins (Fig 1A). A total of 206 plasma proteins passed a false discovery significance threshold of q<0.01 (~20% of all analyzed), representing a larger plasma proteome that appears to covary with plasma AGP, as listed in Supporting Information (S1 and S2 Tables).
Table 2. Plasma proteins positively associated with plasma α-1-acid glycoprotein (AGP) in 6–8 year old children in rural Nepal, ordered by P. a .
| Protein name | Gene symbol | n b | % change in AGP (95% CI) c | P d | q e | r f | Accession g |
|---|---|---|---|---|---|---|---|
| TNFAIP3-interacting protein 1 | TNIP1 | 388 | 101.8 (89.8, 114.6) | 7.6E-112 | 8.4E-109 | 0.82 | 116256481 |
| Orosomucoid 1 | ORM1 | 500 | 86.6 (76.2, 97.6) | 4.6E-101 | 2.5E-98 | 0.77 | 167857790 |
| Orosomucoid 2 | ORM2 | 500 | 105.3 (87.4, 124.8) | 3.1E-54 | 1.1E-51 | 0.67 | 4505529 |
| Leucine-rich alpha-2-glycoprotein 1 | LRG1 | 500 | 75.5 (62.7, 89.4) | 5.6E-48 | 1.5E-45 | 0.65 | 16418467 |
| Serpin peptidase inhibitor, clade A, Member 3 | SERPINA3 | 500 | 125.9 (100.4, 154.5) | 7.0E-41 | 1.5E-38 | 0.63 | 50659080 |
| Actin-related protein 5 | ACTR5 | 255 | 60.4 (49.5, 72.2) | 9.6E-40 | 1.8E-37 | 0.74 | 151301041 |
| Haptoglobin isoform 1 | HP | 354 | 16.8 (14.0, 19.7) | 2.7E-35 | 4.0E-33 | 0.66 | 4826762 |
| Conserved oligomeric golgi complex subunit 3 | COG3 | 215 | 56.8 (46.0, 68.4) | 2.9E-35 | 4.0E-33 | 0.73 | 13899251 |
| C-reactive protein, pentraxin-related | CRP | 438 | 15.6 (12.8, 18.4) | 3.2E-32 | 3.5E-30 | 0.61 | 55770842 |
| Mitogen-activated protein kinase kinase kinase 14 | MAP3K14 | 307 | 55.6 (44.5, 67.5) | 3.2E-32 | 3.5E-30 | 0.66 | 115298645 |
| Serum amyloid A protein | SAA1 | 493 | 17.6 (14.4, 20.9) | 3.1E-31 | 3.1E-29 | 0.59 | 40316912 |
| Complement component 9 | C9 | 500 | 80.2 (62.8, 99.6) | 6.0E-30 | 5.3E-28 | 0.58 | 4502511 |
| Zinc finger ZZ-type and EF-hand domain-containing protein 1 | ZZEF1 | 444 | 30.7 (24.8, 36.9) | 6.2E-30 | 5.3E-28 | 0.60 | 73747881 |
| Serum amyloid P component | APCS | 500 | 57.2 (44.5, 70.9) | 3.0E-26 | 2.2E-24 | 0.56 | 4502133 |
| Lipopolysaccharide-binding protein | LBP | 500 | 45.7 (35.8, 56.4) | 9.9E-26 | 6.7E-24 | 0.56 | 31652249 |
| Leucine-rich repeat-containing protein 50 | DNAAF1 | 207 | 56.2 (42.5, 71.2) | 9.4E-22 | 5.1E-20 | 0.69 | 157674358 |
| Putative methyltransferase NSUN6 | NSUN6 | 249 | 53.4 (39.9, 68.2) | 6.4E-20 | 3.1E-18 | 0.60 | 32698918 |
| Ecotropic viral integration site 5 protein homolog | EVI5 | 271 | 49.5 (37.0, 63.2) | 1.6E-19 | 7.1E-18 | 0.55 | 68299759 |
| Complement factor I | CFI | 500 | 105.7 (75.6, 140.9) | 3.4E-19 | 1.5E-17 | 0.52 | 119392081 |
| Dynein heavy chain 11, axonemal | DNAH11 | 298 | 37.1 (27.5, 47.4) | 8.4E-18 | 3.2E-16 | 0.58 | 51479173 |
| Complement factor B | CFB | 500 | 72.9 (52.2, 96.4) | 3.6E-17 | 1.3E-15 | 0.51 | 67782358 |
| Haptoglobin-related protein | HPR | 431 | 21.6 (16.0, 27.4) | 3.0E-16 | 1.0E-14 | 0.51 | 45580723 |
| Inter-alpha (globulin) inhibitor H3 | ITIH3 | 500 | 51.8 (36.6, 68.7) | 8.0E-15 | 2.4E-13 | 0.49 | 133925809 |
| MAP/Microtubule affinity-regulating kinase 3 isoform E | MARK3 | 231 | 50.3 (34.9, 67.5) | 1.1E-13 | 3.0E-12 | 0.56 | 193083131 |
| Protein S100-A9 | S100A9 | 500 | 21.3 (15.1, 28.0) | 8.9E-13 | 2.2E-11 | 0.48 | 4506773 |
| Nucleolar MIF4G domain-containing protein 1 | NOM1 | 119 | 44.8 (30.6, 60.5) | 1.1E-12 | 2.7E-11 | 0.62 | 61097912 |
| RNA polymerase II elongation factor ELL3 | ELL3 | 214 | 34.5 (23.8, 46.0) | 1.4E-12 | 3.4E-11 | 0.54 | 13376768 |
| Protein S100-A8 | S100A8 | 500 | 20.0 (14.0, 26.3) | 2.2E-12 | 4.9E-11 | 0.47 | 21614544 |
| Cell cycle regulator mat89Bb homolog | ASUN | 235 | 36.2 (24.4, 49.1) | 1.9E-11 | 3.6E-10 | 0.52 | 155030185 |
| Serum amyloid A2 isoform a | SAA2 | 189 | 18.0 (12.2, 24.1) | 1.0E-10 | 1.9E-09 | 0.55 | 188497671 |
| Complement component 5 | C5 | 500 | 73.8 (45.6, 107.5) | 8.7E-10 | 1.4E-08 | 0.45 | 38016947 |
| Complement component 2 isoform 1 | C2 | 423 | 94.8 (55.8, 143.6) | 4.5E-09 | 7.0E-08 | 0.45 | 14550407 |
| Serum amyloid A-4 protein | SAA4 | 493 | 31.8 (20.1, 44.5) | 4.6E-09 | 7.1E-08 | 0.44 | 10835095 |
| Ceruloplasmin | CP | 500 | 63.8 (38.2, 94.3) | 1.3E-08 | 1.8E-07 | 0.44 | 4557485 |
| Integrin alpha L isoform B | ITGAL | 82 | 80.5 (46.3, 122.7) | 2.3E-08 | 3.0E-07 | 0.65 | 167466217 |
| Serpin peptidase inhibitor, clade G, member 1 | SERPING1 | 500 | 64.7 (38.0, 96.5) | 2.9E-08 | 3.8E-07 | 0.44 | 73858570 |
| Inter-alpha (globulin) inhibitor H4 isoform 1 | ITIH4 | 500 | 76.9 (43.9, 117.5) | 5.6E-08 | 6.9E-07 | 0.44 | 31542984 |
| T-box transcription factor TBX22 isoform 1 | TBX22 | 103 | 32.5 (19.0, 47.5) | 2.0E-07 | 2.4E-06 | 0.46 | 18375603 |
| Heparin cofactor II | SERPIND1 | 500 | 40.9 (23.4, 60.9) | 3.6E-07 | 4.1E-06 | 0.43 | 73858566 |
| Complement component 4 binding protein, alpha chain | C4BPA | 500 | 24.4 (14.2, 35.4) | 4.6E-07 | 5.1E-06 | 0.43 | 4502503 |
| Mediator of RNA polymerase II transcription subunit 23 isoform b | MED23 | 196 | 41.7 (23.2, 62.9) | 8.6E-07 | 9.2E-06 | 0.48 | 28558969 |
aPlasma proteins that achieved a Bonferroni corrected significance level (P <0.001/982 = 1.02e-06).
bThe number of child plasma samples of each listed protein (50<n≤500).
cPercent change (%) in plasma AGP per two-fold (100%) increase in relative abundance of protein.
d P value for the hypothesis test of null association between plasma AGP and protein.
eAdjusted P value correcting multiple hypothesis testing (false discovery rate).
fCorrelation between plasma AGP concentration and protein.
gGenInfo Identifier for protein sequence assigned by the National Center for Biotechnology Information.
Table 3. Plasma proteins negatively associated with plasma alpha-1-acid glycoprotein (AGP) in 6–8 year old children in rural Nepal, ordered by P. a .
| Protein name | Gene symbol | n b | % change in AGP (95% CI) c | P d | q e | r f | Accession g |
|---|---|---|---|---|---|---|---|
| Lumican | LUM | 500 | -48.9 (-54.8, -42.2) | 9.1E-27 | 7.1E-25 | -0.56 | 4505047 |
| Cartilage oligomeric matrix protein | COMP | 500 | -36 (-41.4, -30.0) | 5.7E-23 | 3.5E-21 | -0.54 | 40217843 |
| Tetranectin | CLEC3B | 500 | -53 (-60.0, -44.9) | 1.0E-20 | 5.4E-19 | -0.53 | 156627579 |
| Transthyretin | TTR | 500 | -48.8 (-55.7, -40.9) | 4.7E-20 | 2.3E-18 | -0.52 | 4507725 |
| N-acetylmuramoyl-l-alanine amidase | PGLYRP2 | 500 | -48.6 (-55.7, -40.4) | 1.2E-18 | 4.9E-17 | -0.52 | 156616294 |
| Apolipoprotein A-I | APOA1 | 500 | -43.2 (-49.9, -35.5) | 1.9E-18 | 7.6E-17 | -0.52 | 4557321 |
| Serine proteinase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 4 | SERPINA4 | 500 | -46.5 (-53.6, -38.4) | 4.9E-18 | 1.9E-16 | -0.51 | 21361302 |
| Inter-alpha globulin inhibitor h2 polypeptide | ITIH2 | 500 | -53.4 (-61.1, -44.2) | 1.1E-16 | 3.9E-15 | -0.50 | 70778918 |
| Alpha-2-macroglobulin | A2M | 500 | -58.5 (-66.4, -48.7) | 3.1E-16 | 1.0E-14 | -0.50 | 66932947 |
| Afamin | AFM | 500 | -38 (-44.7, -30.4) | 4.1E-16 | 1.3E-14 | -0.50 | 4501987 |
| Attractin isoform 2 | ATRN | 500 | -49.2 (-56.9, -40.2) | 4.3E-16 | 1.3E-14 | -0.50 | 21450863 |
| Fibroblast activation protein, alpha subunit | FAP | 423 | -31.7 (-38.1, -24.7) | 1.5E-14 | 4.6E-13 | -0.51 | 16933540 |
| Interferon-related developmental regulator 2 | IFRD2 | 486 | -29.9 (-36.1, -23.2) | 2.2E-14 | 6.3E-13 | -0.49 | 197333755 |
| Apolipoprotein A-II | APOA2 | 500 | -36.3 (-43.4, -28.5) | 2.7E-14 | 7.7E-13 | -0.49 | 4502149 |
| Alpha 3 type vi collagen isoform 5 | COL6A3 | 472 | -38.9 (-46.2, -30.5) | 4.8E-14 | 1.3E-12 | -0.49 | 55743106 |
| Alpha-2-hs-glycoprotein | AHSG | 500 | -39.3 (-46.9, -30.6) | 2.4E-13 | 6.3E-12 | -0.48 | 156523970 |
| Serpin peptidase inhibitor, clade c, member 1 | SERPINC1 | 500 | -61.6 (-70.6, -49.9) | 1.5E-12 | 3.5E-11 | -0.47 | 4502261 |
| Kelch-like protein 34 | KLHL34 | 403 | -36.7 (-44.2, -28.1) | 1.7E-12 | 4.0E-11 | -0.49 | 23397572 |
| Peptidase inhibitor 16 | PI16 | 500 | -31.0 (-37.8, -23.4) | 2.6E-12 | 5.7E-11 | -0.47 | 70780384 |
| Thrombospondin 4 | THBS4 | 451 | -25.5 (-31.4, -19.1) | 2.7E-12 | 5.8E-11 | -0.47 | 31543806 |
| Gelsolin isoform a | GSN | 493 | -36.9 (-44.6, -28.2) | 3.0E-12 | 6.3E-11 | -0.48 | 4504165 |
| Anthrax toxin receptor 2 isoform 1 | ANTXR2 | 367 | -33.4 (-40.6, -25.3) | 3.3E-12 | 6.9E-11 | -0.53 | 50513243 |
| Retinol-binding protein 4, plasma | RBP4 | 500 | -31.6 (-38.5, -23.8) | 3.7E-12 | 7.4E-11 | -0.47 | 55743122 |
| Cd109 antigen isoform 2 | CD109 | 300 | -34.4 (-41.8, -26.1) | 3.9E-12 | 7.8E-11 | -0.56 | 227430301 |
| Timp metallopeptidase inhibitor 2 | TIMP2 | 368 | -33.5 (-40.8, -25.3) | 4.7E-12 | 9.2E-11 | -0.49 | 4507511 |
| Sparc-like protein 1 | SPARCL1 | 493 | -39.5 (-47.9, -29.7) | 4.0E-11 | 7.5E-10 | -0.46 | 190341024 |
| Cadherin 5, type 2 | CDH5 | 500 | -34.1 (-41.9, -25.1) | 1.1E-10 | 2.0E-09 | -0.46 | 166362713 |
| Olfactomedin related er localized protein isoform 1 | OLFM1 | 465 | -36.8 (-45.1, -27.2) | 1.5E-10 | 2.6E-09 | -0.47 | 17136143 |
| Apolipoprotein M | APOM | 500 | -33.8 (-41.7, -24.8) | 1.9E-10 | 3.4E-09 | -0.46 | 22091452 |
| Anthrax toxin receptor 1 isoform 1 | ANTXR1 | 339 | -28.1 (-35.1, -20.4) | 2.4E-10 | 4.1E-09 | -0.49 | 14149904 |
| Plasma serine protease inhibitor | SERPINA5 | 500 | -31.9 (-39.7, -23.1) | 5.2E-10 | 8.8E-09 | -0.45 | 194018472 |
| Cadherin 13 | CDH13 | 465 | -34.4 (-42.6, -24.9) | 7.6E-10 | 1.3E-08 | -0.46 | 4502719 |
| Tenascin xb isoform 1 | TNXB | 500 | -29.5 (-37.1, -21.1) | 1.2E-09 | 1.9E-08 | -0.45 | 188528648 |
| Dipeptidyl peptidase 4 | DPP4 | 416 | -35.3 (-43.9, -25.4) | 2.0E-09 | 3.1E-08 | -0.43 | 18765694 |
| Cd93 antigen | CD93 | 416 | -28.2 (-35.8, -19.7) | 4.9E-09 | 7.4E-08 | -0.47 | 88758613 |
| Xaa-pro dipeptidase isoform 1 | PEPD | 466 | -31.2 (-39.4, -21.9) | 6.5E-09 | 9.7E-08 | -0.45 | 149589008 |
| Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 isoform a | MACF1 | 430 | -28.2 (-35.8, -19.6) | 6.6E-09 | 9.7E-08 | -0.47 | 33188445 |
| Procollagen c-endopeptidase enhancer 1 | PCOLCE | 500 | -35.3 (-44.2, -25.0) | 6.7E-09 | 9.8E-08 | -0.44 | 157653329 |
| Prenylcysteine oxidase 1 | PCYOX1 | 493 | -27.1 (-34.5, -18.8) | 7.7E-09 | 1.1E-07 | -0.44 | 166795301 |
| Sulfhydryl oxidase 1 isoform a | QSOX1 | 500 | -49.3 (-59.9, -36.0) | 1.0E-08 | 1.5E-07 | -0.44 | 13325075 |
| Apolipoprotein H | APOH | 500 | -29.6 (-37.7, -20.5) | 1.5E-08 | 2.1E-07 | -0.44 | 153266841 |
| Integrin alpha 2 | ITGA2 | 187 | -33.7 (-42.5, -23.5) | 1.6E-08 | 2.1E-07 | -0.49 | 116295258 |
| Membrane alanine aminopeptidase | ANPEP | 500 | -35.2 (-44.5, -24.3) | 4.0E-08 | 5.3E-07 | -0.44 | 157266300 |
| Inter-alpha (globulin) inhibitor h1 isoform a | ITIH1 | 500 | -41.2 (-51.3, -28.9) | 4.0E-08 | 5.3E-07 | -0.44 | 156119625 |
| Biotinidase | BTD | 500 | -35.5 (-44.9, -24.5) | 4.2E-08 | 5.4E-07 | -0.44 | 4557373 |
| Insulin-like growth factor binding protein, acid labile subunit isoform 2 | IGFALS | 500 | -23.7 (-30.7, -15.9) | 4.3E-08 | 5.4E-07 | -0.44 | 4826772 |
| Phosphatidylinositol-glycan-specific phospholipase d isoform 1 | GPLD1 | 500 | -30.1 (-38.6, -20.5) | 5.0E-08 | 6.3E-07 | -0.44 | 29171717 |
| Basement membrane-specific heparan sulfate proteoglycan core protein | HSPG2 | 493 | -40.2 (-50.3, -28.0) | 5.4E-08 | 6.7E-07 | -0.43 | 126012571 |
| Lymphatic vessel endothelial hyaluronic acid receptor 1 | LYVE1 | 479 | -22.9 (-29.8, -15.2) | 6.8E-08 | 8.3E-07 | -0.45 | 40549451 |
| Butyrylcholinesterase | BCHE | 500 | -31.4 (-40.3, -21.2) | 8.9E-08 | 1.1E-06 | -0.44 | 4557351 |
| Osteomodulin | OMD | 465 | -24.9 (-32.4, -16.5) | 9.0E-08 | 1.1E-06 | -0.47 | 4826876 |
| Plasma kallikrein b1 | KLKB1 | 500 | -36.6 (-46.4, -25.0) | 9.4E-08 | 1.1E-06 | -0.44 | 78191798 |
| Sex hormone-binding globulin isoform 1 | SHBG | 486 | -19.6 (-25.9, -12.6) | 2.3E-07 | 2.6E-06 | -0.43 | 7382460 |
| Multimerin-2 | MMRN2 | 444 | -26.3 (-34.5, -17.2) | 2.7E-07 | 3.1E-06 | -0.45 | 221316695 |
| Mannan-binding lectin serine protease 1 isoform 2 | MASP1 | 486 | -26.6 (-34.8, -17.4) | 2.9E-07 | 3.3E-06 | -0.43 | 21264359 |
| Paraoxonase 1 | PON1 | 500 | -21.1 (-28.0, -13.5) | 3.7E-07 | 4.1E-06 | -0.43 | 19923106 |
| Neural cell adhesion molecule 1 isoform 3 | NCAM1 | 453 | -31.9 (-41.4, -20.8) | 5.4E-07 | 5.8E-06 | -0.45 | 115529478 |
| Collagen, type vi, alpha 1 | COL6A1 | 472 | -24.3 (-32.2, -15.4) | 7.9E-07 | 8.5E-06 | -0.45 | 87196339 |
aPlasma proteins that achieved a Bonferroni corrected significance level (P <0.001/982 = 1.02e-06).
bThe number of child plasma samples of each listed protein (50<n≤500).
cPercent change (%) in plasma AGP per two-fold (100%) increase in relative abundance of protein.
d P value for the hypothesis test of null association between plasma AGP and protein.
eAdjusted P value correcting multiple hypothesis testing (false discovery rate).
fCorrelation between plasma AGP concentration and protein.
gGenInfo Identifier for protein sequence assigned by the National Center for Biotechnology Information.
Fig 1. Volcano plot of plasma proteins associated with plasma α-1-acid glycoprotein (AGP) in 6–8 year old children in rural Nepal.
Plot (A) and (C) are enlarged rectangles in plot (B). (A) Plasma proteins negatively associated with AGP, presented by gene symbol (n = 58); (B) Plasma proteins associated with AGP were colored in red and blue (n = 99); (C) Plasma proteins positively associated with AGP, presented by gene symbol (n = 41). x- and y-axes are logarithmic.
Separate verification analyses identified 41 (S3 Table) and 40 (S4 Table) proteins that were positively and negatively associated with CRP, respectively, passing the same Bonferroni corrected significance level as applied to the AGP analysis, of which 72 proteins were associated with both AGP and CRP. The 9 “non-overlapping” CRP-associated proteins (marked in red in S3 and S4 Tables) were still associated with AGP but less significantly (all P < 0.003). In addition, 27 proteins were differentially abundant between children with at least one episode of any morbidity symptom (fever, diarrhea, productive cough or rapid breathing) and children without any symptoms, passing a false discovery significance threshold of q <0.01 (S5 Table). All but one of the 27 proteins associated with morbidity symptoms were strongly correlated with AGP.
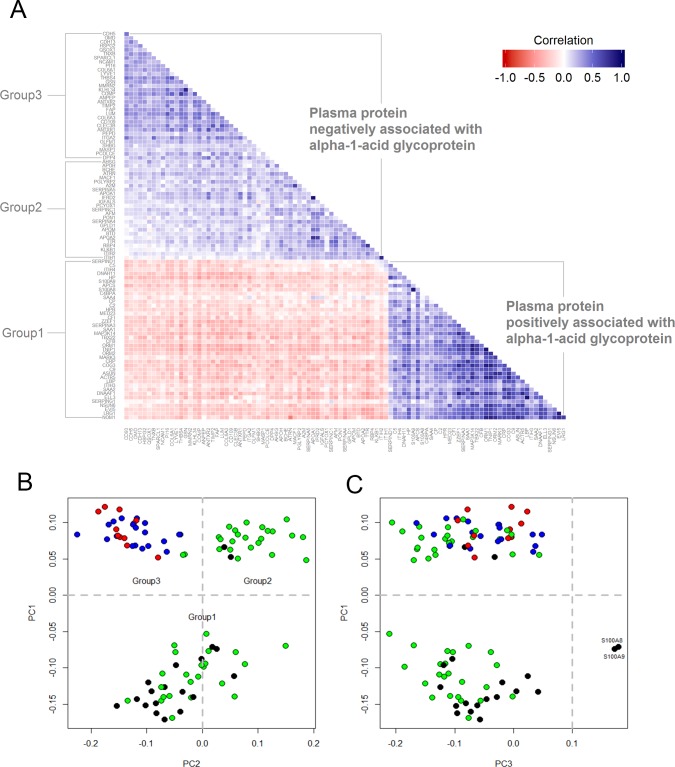
We anticipated that proteins positively and negatively associated with AGP would also be correlated with each other. The correlation matrix, shown in Fig 2A, shows that plasma proteins positively associated with AGP were more highly correlated with each other (i.e., darker blue cells) than plasma proteins negatively associated with AGP (i.e., lighter blue cells). There were 83 and 5 protein-pairs, respectively, whose correlation coefficients (r) were greater than 0.6 within the positive and negative population plasma inflammasomes. Within the former, high correlations were observed among pairs involving ORM1-TNIP1-MAP3K14-COG3-ACTR5-NOM1 (all r >0.80), especially between LBP-ELL3 (r = 0.92), and LRG1-EVI5 (r = 0.86).
Fig 2. Correlation matrix and bi-plots from principal components (PC) analysis using plasma proteins associated with α-1-acid glycoprotein in 6–8 year old children in rural Nepal.
(A) Bottom-triangle is the correlation matrix of plasma proteins positively associated with AGP (Group1). Upper-triangle is the correlation matrix of plasma protein negatively associated with AGP (Group 2 & 3). (B and C) Bi-plot was constructed by the first three principle components. Color depicts representative tissue origins or subcellular localization of proteins: black-intracellular space; green-hepatic origin and secreted into plasma; red-extracellular matrix; blue-extracellular matrix membrane binding. Proteins with PC1 less than 0 were assigned into group 1, proteins with PC1 and PC2 greater than 0 were assigned into group 2, and proteins with PC1 greater than 0 and PC2 less than 0 were assigned into group 3 (4 proteins were not included due to missing values and lack of information about subcellular localization).
As expected, principal components (PC) analysis divided the population plasma inflammasome into proteins that were positively (PC1<0, referred to as Group 1) and negatively associated with AGP (PC1>0). (Fig 2B). However, two subgroups further emerged among proteins negatively associated with AGP. PC2 partitioned negatively associated proteins into two groups (Group 2 vs. Group 3) and PC3 separated two proteins (S100A8 and S100A9) from the rest of the proteins (Fig 2C). To better understand the unexpected partitioning among plasma proteins negatively correlated with AGP, we investigated cellular localization of the proteins. Proteins in PC2 are known to be mainly produced by the liver and secreted into the bloodstream (green) and proteins in PC3 are largely produced by extra-hepatic tissues and localized in extracellular matrix regions (red and blue).
Localization and functions of plasma proteins associated AGP
Proteins associated with AGP are summarized by their most often described cellular localization and biological or molecular functions in Table 4. More than half of the proteins positively associated with AGP were primarily extracellular, secreted into circulation from the liver, and known to promote or regulate innate immune responses and inhibit oxidative activity. These included ORM1/2, CRP, SAA 1/2/4, bacterial lipopolysaccharide binding protein (LBP), LRG1 (a protein involved in granulocyte differentiation), components of the complement cascade, free hemoglobin scavengers, a copper-carrier, and several protease inhibitors. Other positively associated proteins are mainly localized in the membrane or intracellular space and involved in diverse functions, including leukocyte recruitment and trafficking, cell signaling, transcription, translation, DNA repair, protein methylation, modulation of cell cycle, cytokinesis and cytoskeleton, and endoplasmic reticulum-Golgi vesicle transport.
Table 4. Cellular localizati4on and molecular/biological functions of plasma proteins associated with α-1-acid glycoprotein (AGP) in 6–8 year old children in rural Nepal. a .
| Association | Cellular localization | Molecular/biological function | Protein |
|---|---|---|---|
| Positively associated with AGP (n = 40) | Extracellular (plasma) (n = 23) | Immune system | ORM1/2, APCS, LBP, LRG1 |
| Lipoproteins | SAA1/2/4 | ||
| Complement system | C2/5/9, CFB, CFI, C4BPA, CRP | ||
| Transport/scavenger protein | HP/HPR, CP | ||
| Serine proteases inhibitor | SERPINA3, SERPING1, SERPIND1, ITIH3/4 | ||
| Extracellular (plasma membrane) (n = 1) | Leukocyte-endothelial interaction | ITGAL | |
| Extra- & intra-cellular (n = 2) | Leukocyte trafficking | S100A8/9 | |
| Intracellular (n = 14) | Regulation of cell signaling | MAP3K14, TNIP1 | |
| Transcription and translation regulation, DNA/RNA binding | ELL3, NOM1, TBX22, MED23, NSUN6, ACTR5, MARK3 | ||
| Cell cycle, Cell division, Mitosis | EVI5, ASUN | ||
| Cytoskeleton | DNAAF1, DNAH11 | ||
| ER-Golgi vesicle-mediated transport | COG3 | ||
| Negatively associated with AGP (n = 56) | Extracellular (plasma) (n = 25) | Transport | RBP4, TTR, AFM, AHSG |
| Lipoproteins | APOA1/2, APOH, APOM | ||
| Serine proteinase inhibitor | SERPINA5, SERPINC1, A2M, SERPINA4, ITIH1/ ITIH2 | ||
| Serine type endopeptidase | KLKB1, MASP1 | ||
| Other enzymes | BCHE, PON1, BTD, PCYOX1, GPLD1 | ||
| Inflammatory response | ATRN | ||
| Growth factor/Hormone binding | IGFALS, SHBG | ||
| Peptidoglycan recognition | PGLYRP2 | ||
| Extracellular matrix (n = 11) | Collagen | COL6A1/3 | |
| Non-collagenous glycoprotein | COMP, THBS4, TNXB, SPARCL1, MMRN2 | ||
| Proteoglycan | HSPG2, LUM | ||
| Bone matrix, mineralization | CLEC3B, OMD | ||
| Extracellular (plasma membrane) (n = 20) | Aminoprotease or peptidase | PCOLCE, PEPD, DPP4, FAP, ANPEP | |
| Cell-cell/cell-ECM adhesion | CDH13, CDH5, NCAM1, OLFM1, CD93, ANTRX1, ANTRX2, QSOX1, ITGA2 | ||
| Cytoskeleton modulation | GSN, MACF1 | ||
| Peptidase inhibitor | TIMP2, PI16, CD109 | ||
| Hyaluronan receptor | LYVE1 |
aProteins of unknown function were not presented (KLHL34, IFRD2, and ZZEF1).
About half of the proteins negatively correlated with AGP (i.e., that decline in relative abundance with inflammation) are also considered hepatic proteins released into the bloodstream, but more involved in transport and metabolism of nutrients and small molecules (e.g., RBP4 and TTR for vitamin A; apolipoproteins A1/A2/H/M for lipid or cholesterol transport and metabolism; and AHSG for calcium and phosphate metabolism), sex hormone and growth factor binding (e.g., SHBG and IGFALS) or serine endopeptidase or proteinase inhibition in regulating blood coagulation and complement cascades, among other roles. Other negative correlates are physical constituents of the extracellular matrix (ECM), including collagens [e.g, collagen types VI α1 and 3 (COL6A1, COL6A3)], glycoproteins, glucosaminoglycans and bone matrix proteins. Proteins known to facilitate interaction between cells and ECM are aminoproteinases/peptidases (e.g. PCOLCE and PEPD), protease inhibitors (e.g. TIMP2) and numerous cell-cell or cell-matrix adhesion molecules (e.g. CDH5 and ANTRX1).
Discussion
This study provides evidence of a population plasma inflammasome, defined in relation to the continuous distribution of an established biomarker of inflammation, α-1-acid glycoprotein. We virtually ensured identification of nearly 100 plasma proteins associated with AGP by applying a stringent family-wise error rate threshold of 0.1%. Approximately three-quarters of the same proteins were similarly correlated with a second acute phase reactant in plasma, CRP. Study children in rural Nepal were undernourished, similar to many child populations in rural Asia, but were active and not acutely ill, with only 14% reporting any illness symptom in the previous week. As such, the set of proteins observed to covary with AGP can be inferred to reflect homeostatic control of inflammation within the environment of this typical South Asian rural setting.
Positive plasma inflammasome proteins exhibited stronger associations with AGP and greater variation in their degree of change per unit difference in AGP concentration than negatively associated proteins, possibly reflecting higher metabolic priority and functional specificity. In addition to established acute phase proteins, our quantitative proteomics approach identified numerous intracellular signaling, membrane-bound, and extracellular matrix molecules not widely regarded as acute phase reactants, appearing to reflect a systemic repertoire of proteins that respond to inflammation.
Acute phase proteins, complement components, protease inhibitors and transport proteins with anti-oxidant activity positively covaried with plasma AGP, in accordance with their expected roles in responding to stress [1]. Many of these biomarkers are produced in the liver and secreted into plasma [10]. As AGP abundance in plasma is attributable to expression of homologous ORM1 (AGP1) and ORM2 (AGP2) genes [42], ORM1 and ORM2 were expected, and observed, to be among the strongest correlates of AGP (measured by radial immunodiffusion), offering evidence that mass spectrometry is a valid method to detect and quantify relative protein abundance. LRG1, HP, SERPINA3, CRP, SAA1, C9, and LBP were positively associated with AGP (all P <1.0x10-25), suggesting these proteins increase in circulation during inflammation [10]. Substantial variation in the strength of association with AGP likely reflects wide differences in expression across acute and chronic phase proteins [43–45]. It is likely that rises in complement components, LRG1 and SERPINA3 reflect a persistent inflammation that accompanies a modest elevation in AGP where exposure to parasites, bacteria [46–48] and environmental toxins, such as aflatoxin and arsenic, are common [49, 50]. Positively associated proteins we observed are known to largely be involved in immune activation (e.g., CRP, SAA, LBP, and complement components), proteolytic attack processes (SERPINA3, SERPING1, and SEPRIND1), and transport of pro-oxidative metabolites (CP and HP) [44]. ORM1/2 and LRG1 are involved in immunomodulation and granulocyte differentiation, respectively, although their molecular roles have not been fully elucidated [51–53]. Collectively, our results suggest that hepatic-driven proteins that positively covary with inflammation are involved in host defense.
Fourteen of 41 positive correlates of AGP are intracellular proteins whose larger numbers of missing values (Table 2) support the notion that these proteins may be low in abundance, and leaked or secreted from tissues as part of normal metabolism and tissue maintenance. The strongest positive correlate (P = 7.6x10-112), sharing a nearly 1:1 association with AGP, was TNIP1, suggesting their co-regulation during inflammation, although no study to our knowledge has drawn this direct metabolic linkage. TNIP1 regulates inflammation by inhibiting cell signal transduction such as in the NF-kappa-B activation pathway [54]. However, unknown extracellular functions of TNIP1 leave its positive association with AGP in plasma unexplained. A 30~60% increase in AGP concentration was associated with a 100% increase in intracellular proteins involved in signal transduction, and protein transcription, translation, maturation and secretion. High correlations between intracellular proteins and circulating ORM1, LBP, and LRG1 suggest their abundance in plasma could reflect biosynthesis of inflammatory mediators [55], or act themselves as acute phase proteins.
Among proteins negatively correlated with plasma AGP, about half are known to transport and regulate bioavailability of nutrients and hormones. Correlations among these proteins were negligible, suggesting independent regulation and metabolic pathways, while still being susceptible to hepatic-directed reduction during inflammation [56]. Some negative correlates are components of lipoprotein particles, involved in anti-inflammation (APOA1/2), antioxidant functions (PON1, PON3, and PCYOX1) and reverse cholesterol transport (PLTP, CLU and LCAT) (all P <1.0x10-4). These observations coupled with those of increased abundance in pro-inflammatory serum amyloid A (SAAs) apoliporoteins with AGP support considerable alterations in plasma lipoprotein composition during homeostatic regulation of inflammation [57]. RBP4, TTR, AHSG, and APOA1/2 are well-known negative acute phase proteins [10] that are consistent with known redistributions of vitamin A, calcium, phosphate and lipids during inflammation [57–60].
In our analysis, a small decrement in AGP per two-fold increment in relative protein abundance identifies proteins that are likely to decrease markedly during acute inflammation. The smallest decrease in AGP (20–24%) was observed with a two-fold rise in sex hormone binding globulin (SHBG), insulin-like growth factor (IGF) acid labile subunit and IGF binding protein (BP) 3 (all P <1.0x10-5), consistent with expected reductions in insulin-like growth factor 1 and androgens during inflammation [10, 61]. IGFALS and IGFBP3 form a ternary complex with most plasma IGFs which regulate somatic growth and development [62, 63]. SHBG binds and regulates circulating androgens and estrogens [64]. Population studies have revealed inverse associations between SHBG and inflammatory markers and adiposity-related early onset of puberty among girls [65, 66]. Observed inverse associations between AGP and hepatic proteins may reflect metabolic adaptation to altered endocrine signaling in response to inflammation.
Proteins that serve as components or regulators of the extracellular matrix (ECM) were negatively associated with inflammation [67]. Our PCA results revealed that variance in the abundance of these proteins differed from those of hepatic origin, possibly due to differences in intravascular concentration between classic plasma and extravascular proteins. Lumican (LUM), cartilage oligomeric matrix protein (COMP), tetranectin (CLEC3B), osteomodulin (OMD), and collagen α-1 and -3 type VI (COL6A1/3) are enriched in cartilage, bone matrix, skeletal muscle and adipose tissues [68–72]. Beyond structural and functional components of ECM, many cell surface molecules or enzymatic proteins are involved in penetrating the vascular endothelial cells, regulation of pericellular proteolysis of ECM, and cell migration into inflamed tissues, critical to tissue repair and turnover [73–81]. Our results suggest that proteins that maintain integrity of the ECM are down-regulated, possibly reflecting metabolic rebalancing between host defense and healing mechanisms [82]. These proteins are particularly important in chronic inflammatory conditions that commonly accompany degradation of connective tissue [83–86].
Our findings corroborate those in -omics studies in animals that have examined changes in gene or protein expression levels of acute phase proteins during inflammation. Yoo et al. showed in mice that 898 out of 8,551 protein-encoding genes (~7%) in the hepatic transcriptome were altered, equally up and down, by endotoxin-induced inflammation [87]. Similarly, we observed that half of the population plasma inflammasome was primarily hepatic in origin, equally divided across positive and negative correlates of AGP, and possibly reflecting a need to maintain protein equilibria in the vascular compartment [44]. Kelly-Spratt et al. reported that a third of ~500 plasma proteins detected in mice increased or decreased by more than 1.25 fold in response to induced-inflammation [22]. We observed that a large fraction (~20%) of our measured plasma proteome covaried with inflammation, at a false discovery rate below 1% (S1 and S2 Tables). The study also showed that induced-inflammation reduced the abundance of proteins involved in ECM and collagen network remodeling [22], which were also similarly observed in the present study.
The diversity of plasma proteins observed to be associated with AGP in this ambient population offer an unbiased view of inflammation. With this large-scale and untargeted approach, we identified potential networks of interaction and a large number of candidate biomarkers. Defining the plasma inflammasome with respect to AGP, an established index of subclinical inflammation, enabled us to identify proteins that are likely relevant to the homeostatic response to inflammation. Results of additional analyses in relation to plasma CRP concentration and children’s recent morbidity offer further support of the validity of the population plasma inflammasome indexed by AGP. While our unit of estimated protein amount, i.e., relative abundance, restricts ability to directly apply these findings, biomarkers of strongest association can be considered candidates for absolute quantification to expand the repertoire of biomarkers that reflect diverse mechanisms and possibly sources inflammation. However, with a cross-sectional design, we could not infer metabolic proximity or causality of association among proteins. We also did not have information about specific parasitic or bacterial infection or environmental toxins that could reveal greater specificity of protein associations with causal agents. We depleted, but did not completely remove, plasma samples of 6 highly abundant proteins, and the employed iTRAQ technology could not quantify low abundant cytokines and chemokines which mediate inflammation. These limitations reveal challenges of profiling a whole plasma inflammasome of a likely vast dynamic range in protein abundance. However, it may be promising to investigate and integrate less abundant proteins from different spectrums of abundance to build a more complete profile of a population plasma inflammasome. Lastly, although many proteins are involved in non-specific response to inflammation, the findings of this study will be most likely generalizable to populations living in areas where undernutrition, infections and environmental hazards are common.
Conclusions
This study provides evidence of strong association between an index biomarker of chronic inflammation and proteins of host defense, nutrient and hormonal metabolism and tissue remodeling. It is tempting to speculate that the low-grade inflammation seen in this study of young children could reflect mild pathological processes early in life, and thus risk, of adult chronic diseases of rising prominence in impoverished societies of South Asia.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The Johns Hopkins Nutriproteomics Research Team, in addition to co-authors, includes Margia Arguello, Raghothama Chaerkady, Hongie Cui, Lauren R. DeVine, Jaime Johnson, Robert O’Meally, Subarna K. Khatry, Ashika Nanayakkara-Bind, Hee-Sool Rho, Sudeep Shrestha and Fredrick Van Dyke. We thank C. Conover Talbot, Jr. for assistance with the HUGO gene annotation.
Data Availability
The dataset of inflammatory protein biomarkers reported in this manuscript can be made available to researchers who qualify as co-investigators by making a request to the Johns Hopkins Nutritproteomics Investigative Team (c/o KP West Jr,kwest1@jhu.edu). This restriction exists because biospecimens for this analysis were collected from a cohort of children actively being followed in Nepal whose parents consented with an understanding that all personal information about their son/daughter would remain secure with study investigators and not be shared with anyone not working for the research project.
Funding Statement
This work was supported by: 1. The Bill and Melinda Gates Foundation, Seattle, WA, USA. Grants OPP5241 (Yiwu He, Senior Program Officer) and OPP614 (Ellen Piwoz, Senior Program Officer), http://www.gatesfoundation.org/. 2. The Office of Health, Infectious Diseases and Nutrition, US Agency for International Development, Washington, DC. Grant number (HRN-A-00-97-00015-00), http://www.usaid.gov/who-we-are/organization/bureaus/bureau-global-health. 3. The Sight and Life Global Nutrition Research Institute, Baltimore, MD, USA, http://www.sightandlife.org/ and http://www.jhsph.edu/departments/international-health/news/dsm-sight-and-life.html. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kushner I. The phenomenon of the acute phase response. Ann N Y Acad Sci. 1982;389:39–48. [DOI] [PubMed] [Google Scholar]
- 2. Ceciliani F, Giordano A, Spagnolo V. The systemic reaction during inflammation: the acute-phase proteins. Protein Pept Lett. 2002;9(3):211–223. [DOI] [PubMed] [Google Scholar]
- 3. Kumar V, Abbas AK, Aster JC. Inflammation and Repair In: Kumar V, Abbas AK, Aster JC, editors. Robbins Basic Pathology. 9th ed Philadelphia, U.S.: ELSEVIER Saunders; 2012. [Google Scholar]
- 4. Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140(6):871–882. 10.1016/j.cell.2010.02.029 [DOI] [PubMed] [Google Scholar]
- 5. Mal-Ed Network Investigators. The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis. 2014;59 Suppl 4:S193–206. 10.1093/cid/ciu653 [DOI] [PubMed] [Google Scholar]
- 6. Prendergast AJ, Rukobo S, Chasekwa B, Mutasa K, Ntozini R, Mbuya MN, et al. Stunting is characterized by chronic inflammation in Zimbabwean infants. PLoS One. 2014;9(2):e86928 10.1371/journal.pone.0086928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez L, Cervantes E, Ortiz R. Malnutrition and gastrointestinal and respiratory infections in children: a public health problem. Int J Environ Res Public Health. 2011;8(4):1174–1205. 10.3390/ijerph8041174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmed S, Moore SE, Kippler M, Gardner R, Hawlader MD, Wagatsuma Y, et al. Arsenic exposure and cell-mediated immunity in pre-school children in rural bangladesh. Toxicol Sci. 2014;141(1):166–175. 10.1093/toxsci/kfu113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turner PC. The molecular epidemiology of chronic aflatoxin driven impaired child growth. Scientifica (Cairo). 2013;2013:152879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340(6):448–454. [DOI] [PubMed] [Google Scholar]
- 11.AmiGO2. acute-phase response (GO:0006953): the Gene Ontology; 2014. 2.1.4. Available: http://amigo.geneontology.org/amigo/term/GO:0006953/.
- 12. Whicher T, Bienvenu J, Price CP. Molecular biology, measurement and clinical utility of the acute phase proteins. Pure and Applied Chemistry. 1991;63(8):1111–1116. [Google Scholar]
- 13. Schulze KJ, Christian P, Wu LS, Arguello M, Cui H, Nanayakkara-Bind A, et al. Micronutrient deficiencies are common in 6- to 8-year-old children of rural Nepal, with prevalence estimates modestly affected by inflammation. J Nutr. 2014;144(6):979–987. 10.3945/jn.114.192336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thurnham DI, McCabe GP. Influence of infection and inflammation on biomarkers of nutritional status with an emphasis on vitamin A and iron In: World Health Organization, editor. Priorities in the Assessment of Vitamin A and Iron Status in Populations. Geneva, Switzerland: World Health Organization; 2012. p. 63–80. [Google Scholar]
- 15. Raiten DJ, Sakr Ashour FA, Ross AC, Meydani SN, Dawson HD, Stephensen CB, et al. Inflammation and Nutritional Science for Programs/Policies and Interpretation of Research Evidence (INSPIRE). J Nutr. 2015;145(5):1039S–1108S. 10.3945/jn.114.194571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ceciliani F, Pocacqua V. The acute phase protein alpha1-acid glycoprotein: a model for altered glycosylation during diseases. Curr Protein Pept Sci. 2007;8(1):91–108. [DOI] [PubMed] [Google Scholar]
- 17. Cole RN, Ruczinski I, Schulze K, Christian P, Herbrich S, Wu L, et al. The plasma proteome identifies expected and novel proteins correlated with micronutrient status in undernourished Nepalese children. J Nutr. 2013;143(10):1540–1548. 10.3945/jn.113.175018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuasa I, Umetsu K, Suenaga K, Robinet-Levy M. Orosomucoid (ORM) typing by isoelectric focusing: evidence of two structural loci ORM1 and ORM2. Hum Genet. 1986;74(2):160–161. [DOI] [PubMed] [Google Scholar]
- 19. Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. [DOI] [PubMed] [Google Scholar]
- 20. Kalmovarin N, Friedrichs WE, O'Brien HV, Linehan LA, Bowman BH, Yang F. Extrahepatic expression of plasma protein genes during inflammation. Inflammation. 1991;15(5):369–379. [DOI] [PubMed] [Google Scholar]
- 21. Petrilli V, Papin S, Tschopp J. The inflammasome. Curr Biol. 2005;15(15):R581 [DOI] [PubMed] [Google Scholar]
- 22. Kelly-Spratt KS, Pitteri SJ, Gurley KE, Liggitt D, Chin A, Kennedy J, et al. Plasma proteome profiles associated with inflammation, angiogenesis, and cancer. PLoS One. 2011;6(5):e19721 10.1371/journal.pone.0019721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saha S, Harrison SH, Chen JY. Dissecting the human plasma proteome and inflammatory response biomarkers. Proteomics. 2009;9(2):470–484. 10.1002/pmic.200800507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qian WJ, Jacobs JM, Camp DG 2nd, Monroe ME, Moore RJ, Gritsenko MA, et al. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5(2):572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calder PC, Ahluwalia N, Albers R, Bosco N, Bourdet-Sicard R, Haller D, et al. A consideration of biomarkers to be used for evaluation of inflammation in human nutritional studies. Br J Nutr. 2013;109 Suppl 1:S1–34. 10.1017/S0007114512005119 [DOI] [PubMed] [Google Scholar]
- 26. Christian P, Khatry SK, Katz J, Pradhan EK, LeClerq SC, Shrestha SR, et al. Effects of alternative maternal micronutrient supplements on low birth weight in rural Nepal: double blind randomised community trial. BMJ. 2003;326(7389):571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stewart CP, Christian P, Schulze KJ, Leclerq SC, West KP Jr., Khatry SK. Antenatal micronutrient supplementation reduces metabolic syndrome in 6- to 8-year-old children in rural Nepal. J Nutr. 2009;139(8):1575–1581. 10.3945/jn.109.106666 [DOI] [PubMed] [Google Scholar]
- 28. Stewart CP, Christian P, LeClerq SC, West KP Jr., Khatry SK. Antenatal supplementation with folic acid + iron + zinc improves linear growth and reduces peripheral adiposity in school-age children in rural Nepal. Am J Clin Nutr. 2009;90(1):132–140. 10.3945/ajcn.2008.27368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herbrich SM, Cole RN, West KP Jr., Schulze K, Yager JD, Groopman JD, et al. Statistical inference from multiple iTRAQ experiments without using common reference standards. J Proteome Res. 2013;12(2):594–604. 10.1021/pr300624g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harville DA. Maximum Likelihood Approaches to Variance Component Estimation and to Related Problems. Journal of the American Statistical Association. 1977;72(358):320–338. [Google Scholar]
- 31. Robinson GK. That BLUP is a good thing: the estimation of random effects. Stat Sci. 1991;6:15–32. [Google Scholar]
- 32. Luo R, Zhao H. Protein quantitation using iTRAQ: Review on the sources of variations and analysis of nonrandom missingness. Stat Interface. 2012;5(1):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gower JC, Lubbe SG, Roux NJL. Understanding Biplots: Wiley; 2011. [Google Scholar]
- 34. Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43(Database issue):D1079–1085. 10.1093/nar/gku1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Binder JX, Pletscher-Frankild S, Tsafou K, Stolte C, O'Donoghue SI, Schneider R, et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford). 2014;2014:bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38(Database issue):D492–496. 10.1093/nar/gkp858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, et al. The GOA database: Gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015;43(Database issue):D1057–1063. 10.1093/nar/gku1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. UniProt C. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014;42(Database issue):D191–198. 10.1093/nar/gkt1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130 10.1186/gb-2009-10-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85(9):660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thurnham DI, Mburu AS, Mwaniki DL, De Wagt A. Micronutrients in childhood and the influence of subclinical inflammation. Proc Nutr Soc. 2005;64(4):502–509. [DOI] [PubMed] [Google Scholar]
- 42. Nakamura H, Yuasa I, Umetsu K, Nakagawa M, Nanba E, Kimura K. The rearrangement of the human alpha(1)-acid glycoprotein/orosomucoid gene: evidence for tandemly triplicated genes consisting of two AGP1 and one AGP2. Biochem Biophys Res Commun. 2000;276(2):779–784. [DOI] [PubMed] [Google Scholar]
- 43. Kushner I, Mackiewicz A. Acute phase proteins as disease markers. Dis Markers. 1987;5(1):1–11. [PubMed] [Google Scholar]
- 44. Mackiewicz A, Kushner I, Baumann H. Acute Phase Proteins: Molecular Biology, Biochemistry, and Clinical Applications. Florida: CRC Press; 1993. [Google Scholar]
- 45.Samols D, Agrawal A, Kushner I. Acute Phase Proteins. Cytokine references online [Internet]. 2002. Available: https://www.researchgate.net/profile/Irving_Kushner/publication/228050478_Acutephase_Proteins/links/00b7d51597e03b5192000000.pdf.
- 46. Navitsky RC, Dreyfuss ML, Shrestha J, Khatry SK, Stoltzfus RJ, Albonico M. Ancylostoma duodenale is responsible for hookworm infections among pregnant women in the rural plains of Nepal. J Parasitol. 1998;84(3):647–651. [PubMed] [Google Scholar]
- 47. Chowdhury R, Huda MM, Kumar V, Das P, Joshi AB, Banjara MR, et al. The Indian and Nepalese programmes of indoor residual spraying for the elimination of visceral leishmaniasis: performance and effectiveness. Ann Trop Med Parasitol. 2011;105(1):31–35. 10.1179/136485911X12899838683124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coles CL, Sherchand JB, Khatry SK, Katz J, Leclerq SC, Mullany LC, et al. Nasopharyngeal carriage of S. pneumoniae among young children in rural Nepal. Trop Med Int Health. 2009;14(9):1025–1033. 10.1111/j.1365-3156.2009.02331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Groopman JD, Egner PA, Schulze KJ, Wu LS, Merrill R, Mehra S, et al. Aflatoxin exposure during the first 1000 days of life in rural South Asia assessed by aflatoxin B(1)-lysine albumin biomarkers. Food Chem Toxicol. 2014;74:184–189. 10.1016/j.fct.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thakur JK, Thakur RK, Ramanathan A, Kumar M, Singh SK. Arsenic Contamination of Groundwater in Nepal—An Overview. Water. 2010;3(1):1–20. [Google Scholar]
- 51. Hochepied T, Berger FG, Baumann H, Libert C. Alpha(1)-acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 2003;14(1):25–34. [DOI] [PubMed] [Google Scholar]
- 52. Wang X, Abraham S, McKenzie JA, Jeffs N, Swire M, Tripathi VB, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature. 2013;499(7458):306–311. 10.1038/nature12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. O'Donnell LC, Druhan LJ, Avalos BR. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol. 2002;72(3):478–485. [PubMed] [Google Scholar]
- 54. Ramirez VP, Gurevich I, Aneskievich BJ. Emerging roles for TNIP1 in regulating post-receptor signaling. Cytokine Growth Factor Rev. 2012;23(3):109–118. 10.1016/j.cytogfr.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fey GH, Gauldie J. The acute phase response of the liver in inflammation. Prog Liver Dis. 1990;9:89–116. [PubMed] [Google Scholar]
- 56. Aldred AR, Schreiber G. The Negative Acute Phase Proteins In: Mackiewicz A, Kushner I, Baumann H, editors. Acute Phase Proteins Molecular Biology, Biochemistry, and Clinical Applications: Taylor & Francis; 1993. p. 21–38. [Google Scholar]
- 57. Khovidhunkit W, Kim MS, Memon RA, Shigenaga JK, Moser AH, Feingold KR, et al. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J Lipid Res. 2004;45(7):1169–1196. [DOI] [PubMed] [Google Scholar]
- 58. Moody BJ. Changes in the serum concentrations of thyroxine-binding prealbumin and retinol-binding protein following burn injury. Clin Chim Acta. 1982;118(1):87–92. [DOI] [PubMed] [Google Scholar]
- 59. Rosales FJ, Ross AC. Acute inflammation induces hyporetinemia and modifies the plasma and tissue response to vitamin A supplementation in marginally vitamin A-deficient rats. J Nutr. 1998;128(6):960–966. [DOI] [PubMed] [Google Scholar]
- 60. Jahnen-Dechent W, Heiss A, Schafer C, Ketteler M. Fetuin-A regulation of calcified matrix metabolism. Circ Res. 2011;108(12):1494–1509. 10.1161/CIRCRESAHA.110.234260 [DOI] [PubMed] [Google Scholar]
- 61. Tsilidis KK, Rohrmann S, McGlynn KA, Nyante SJ, Lopez DS, Bradwin G, et al. Association between endogenous sex steroid hormones and inflammatory biomarkers in US men. Andrology. 2013;1(6):919–928. 10.1111/j.2047-2927.2013.00129.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Le Roith D, Bondy C, Yakar S, Liu JL, Butler A. The somatomedin hypothesis: 2001. Endocr Rev. 2001;22(1):53–74. [DOI] [PubMed] [Google Scholar]
- 63. Baxter RC. Insulin-like growth factor binding proteins in the human circulation: a review. Horm Res. 1994;42(4–5):140–144. [DOI] [PubMed] [Google Scholar]
- 64. Anderson DC. Sex-hormone-binding globulin. Clin Endocrinol (Oxf). 1974;3(1):69–96. [DOI] [PubMed] [Google Scholar]
- 65. Liao CH, Li HY, Yu HJ, Chiang HS, Lin MS, Hua CH, et al. Low serum sex hormone-binding globulin: marker of inflammation? Clin Chim Acta. 2012;413(7–8):803–807. 10.1016/j.cca.2012.01.021 [DOI] [PubMed] [Google Scholar]
- 66. Pinkney J, Streeter A, Hosking J, Mohammod M, Jeffery A, Wilkin T. Adiposity, chronic inflammation, and the prepubertal decline of sex hormone binding globulin in children: evidence for associations with the timing of puberty (earlybird 58). J Clin Endocrinol Metab. 2014;99(9):3224–3232. 10.1210/jc.2013-3902 [DOI] [PubMed] [Google Scholar]
- 67. Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326(5957):1216–1219. 10.1126/science.1176009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Byron A, Humphries JD, Humphries MJ. Defining the extracellular matrix using proteomics. Int J Exp Pathol. 2013;94(2):75–92. 10.1111/iep.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wilson R. The extracellular matrix: an underexplored but important proteome. Expert Rev Proteomics. 2010;7(6):803–806. 10.1586/epr.10.93 [DOI] [PubMed] [Google Scholar]
- 70. Wewer UM, Ibaraki K, Schjorring P, Durkin ME, Young MF, Albrechtsen R. A potential role for tetranectin in mineralization during osteogenesis. J Cell Biol. 1994;127(6 Pt 1):1767–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Khan T, Muise ES, Iyengar P, Wang ZV, Chandalia M, Abate N, et al. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol Cell Biol. 2009;29(6):1575–1591. 10.1128/MCB.01300-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Urciuolo A, Quarta M, Morbidoni V, Gattazzo F, Molon S, Grumati P, et al. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat Commun. 2013;4:1964 10.1038/ncomms2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Arora PD, Glogauer M, Kapus A, Kwiatkowski DJ, McCulloch CA. Gelsolin mediates collagen phagocytosis through a rac-dependent step. Mol Biol Cell. 2004;15(2):588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, et al. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci. 2001;114(Pt 15):2755–2773. [DOI] [PubMed] [Google Scholar]
- 75. Fukasawa K, Fujii H, Saitoh Y, Koizumi K, Aozuka Y, Sekine K, et al. Aminopeptidase N (APN/CD13) is selectively expressed in vascular endothelial cells and plays multiple roles in angiogenesis. Cancer Lett. 2006;243(1):135–143. [DOI] [PubMed] [Google Scholar]
- 76. Hotchkiss KA, Basile CM, Spring SC, Bonuccelli G, Lisanti MP, Terman BI. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp Cell Res. 2005;305(1):133–144. [DOI] [PubMed] [Google Scholar]
- 77. Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J Endocrinol. 2011;209(2):139–151. 10.1530/JOE-10-0377 [DOI] [PubMed] [Google Scholar]
- 78. Korpos E, Wu C, Sorokin L. Multiple roles of the extracellular matrix in inflammation. Curr Pharm Des. 2009;15(12):1349–1357. [DOI] [PubMed] [Google Scholar]
- 79. Man XY, Finnson KW, Baron M, Philip A. CD109, a TGF-beta co-receptor, attenuates extracellular matrix production in scleroderma skin fibroblasts. Arthritis Res Ther. 2012;14(3):R144 10.1186/ar3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. O'Brien P O'Connor BF. Seprase: an overview of an important matrix serine protease. Biochim Biophys Acta. 2008;1784(9):1130–1145. 10.1016/j.bbapap.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 81. Yucel G, Oro AE. Cell migration: GSK3beta steers the cytoskeleton's tip. Cell. 2011;144(3):319–321. 10.1016/j.cell.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schreiber G. Synthesis and secretion of APP In: Glaumann H, Theodore P, Redman C, editors. Plasma protein secretion by the liver. London: Academic Press; 1983. [Google Scholar]
- 83. Redlich K, Smolen JS. Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 2012;11(3):234–250. 10.1038/nrd3669 [DOI] [PubMed] [Google Scholar]
- 84. Ademowo OS, Staunton L, FitzGerald O, Pennington SR. Biomarkers of inflammatory arthritis and proteomics In: Stanilova SA, editor. Genes and Autoimmunity—Intracellular Signaling and Microbiome Contribution: InTech; 2013. p. 237–267. [Google Scholar]
- 85. Delano MJ, Moldawer LL. The origins of cachexia in acute and chronic inflammatory diseases. Nutr Clin Pract. 2006;21(1):68–81. [DOI] [PubMed] [Google Scholar]
- 86. Tseng S, Reddi AH, Di Cesare PE. Cartilage Oligomeric Matrix Protein (COMP): A Biomarker of Arthritis. Biomark Insights. 2009;4:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yoo JY, Desiderio S. Innate and acquired immunity intersect in a global view of the acute-phase response. Proc Natl Acad Sci U S A. 2003;100(3):1157–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The dataset of inflammatory protein biomarkers reported in this manuscript can be made available to researchers who qualify as co-investigators by making a request to the Johns Hopkins Nutritproteomics Investigative Team (c/o KP West Jr,kwest1@jhu.edu). This restriction exists because biospecimens for this analysis were collected from a cohort of children actively being followed in Nepal whose parents consented with an understanding that all personal information about their son/daughter would remain secure with study investigators and not be shared with anyone not working for the research project.