Abstract
In recent years the zebrafish has become a popular model system to study organ development and disease. To facilitate these studies, genetic tools are required which allow to modify and manipulate gene expression in organs of interest. Here we describe a zebrafish 2.3kb glutamyl aminopeptidase (enpep) promoter fragment, and show that it can drive gene expression specifically in the kidney during early and late development. We established a stable transgenic line using this promoter fragment that has specific GFP expression in pronephric ducts and tubules starting at 20 hours post-fertilization.
Introduction
Glutamyl aminopeptidase is a small membrane bound metalloprotease that cleaves N-terminal aspartyl and glutamyl residues from peptides (Dijkman et al., 2006). Its best-known target is Angiotensin II (Ang II) a component of the angiotensin-renin system (RAS) (Cogolludo et al., 2005; Reaux et al., 2001). The RAS system plays a role in the regulation of blood pressure and is also important for the development of the mammalian kidney (Dijkman et al., 2006).
The zebrafish pronephric kidney forms in a stepwise fashion. The complete pronephric duct is present around 24 hours post fertilization (hpf), the tubules, which connect the duct to the glomerulus form slightly later at around 30hpf. The midline glomerulus derives from bilateral primordia that later fuse in the middle of the larvae. Glomerular filtrate can be detected beginning around 48 hpf (Drummond et al., 1998; Dressler, 1999; Drummond, 2003; Wingert et al., 2007; Drummond, 2000).
Although the zebrafish only develops a pronephros and mesonephros, and not a metanephros, as do mammals, many of the pathways involving induction and differentiation of kidney development are conserved with amniotes (Drummond, 2005; Dressler, 1999; Wingert et al., 2007). Furthermore mutations of the zebrafish orthologs of human genes responsible for polycystic kidney disease cause a similar phenotype in zebrafish larvae (Sun et al., 2004; Drummond, 2005; Low et al., 2006; Weber S et al., 2008; Obara et al., 2006; Bahadori et al., 2003; Hostetter CL et al., 2003).
Here we describe a 2.3kb glutamyl aminopeptidase promotor sequence which can drive strong expression of GFP in the zebrafish pronephric duct and tubules but not the glomerulus. We established a line stably expressing GFP under control of this promoter (Tg(enpep:GFP)) and demonstrated persistent transgene activation in adult fish. These results will aid to study kidney development in zebrafish as well as genetic manipulation to influence kidney development and function.
Results
To find promoters that have the potential to drive expression in absorptive epithelia, such as is present in the intestine and kidney, we searched the zebrafish gene expression database (http://zfin.org/cgi-bin/webdriver?MIval=aa-xpatselect.apg (Thisse, B. and Thisse, C., 2004)) for genes showing strong expression restricted to these organs. One gene (zfin id: ch211-146m5.2, im:7152184, http://zfin.org/cgi-bin/webdriver?MIval=aa-fxallfigures.apg&OID=ZDB-PUB-040907-1&fxallfig_probe_zdb_id=ZDB-EST-050309-369 ) showed strong specific expression. BlastN analysis revealed that this gene is identical to a predicted gene with high similarity to glutamyl aminopeptidase (enpep) (XM_679675, also called aminopeptidase A or differentiation antigen gp160). A BlastX analysis using this DNA sequence showed that the translated protein was very similar to mouse and human ENPEP proteins (identities 60%, positives 76%, score=1108 bits; identities 56%, positives 73 %, score=1067 bits, respectively). A TBLASTN with the human protein against the zebrafish nr database gave only good hits for the above sequences and a zebrafish gene predicted to be an ortholog of Aminopeptidase N, albeit with much lower degree of amino acid identity (34%). This suggests that XM_679675 is indeed the orthologue of human ENPEP.
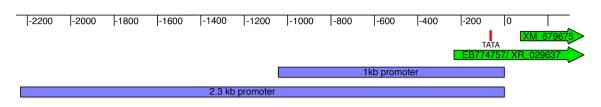
To find the zebrafish enpep promoter we analyzed 2.3 kbp sequence upstream of XM_679675 derived from the zebrafish genomic BAC clones zC200O8 and zC146M5 sequenced by the Sanger center. We used the softberry FPROM program (http://softberry.com/berry.phtml?topic=fprom&group=programs&subgroup=promoter) to identify possible transcription start sites. This identified a possible promotor and TATA box located 108 bp and 136 bp upstream, respectively of the first bp of XM_679675. The XM_679675 sequence starts with an ATG, which is predicted to be the translational start. Accordingly, we chose the sequence 108 bp upstream of XM_679675 as the 3′ end of the promoter (Fig. 1). However, a subsequent entry in the GenBank data base (accession number EB774757) led to the identification of a new predicted gene, XR_029637, that extended 200 bp in the 5′ direction from XM_679675. Because we cannot be certain that additional transcribed sequence related to this gene will be subsequently identified, the transcription initiation site of the zebrafish enpep gene is not clearly defined at this time.
Figure 1.
Promoter organization of the predicted zebrafish enpep gene: Blue: 2.3 kbp and 1 kbp promoter constructs. Green: predicted enpep coding sequence. XM_679675 was the original synthetic construct, XR_029637 is the most recent prediction derived from the zebrafish genome sequencing project. Red: TATA box predicted by F-Prot software.
To test if the 2.3kb fragment containing the presumptive enpep promoter could drive expression in the developing kidney and intestine we amplified it from genomic DNA and cloned it into the pT2KXIG in vector, replacing the original EF1p promoter (Kawakami and Shima, 1999; Kawakami et al., 1998). We thus created a GFP-reporter construct flanked by two Tol2 transposase sites. These sites greatly increase the integration rate into the zebrafish genome when co-injected with transposase mRNA (Kawakami, 2004; Kawakami et al., 2004). We injected this construct and the transposase mRNA into the first cell of newly fertilized zebrafish eggs. In most of the injected eggs we could see strong GFP expression in the pronephric duct and tubules starting around 28 hpf. This expression persisted throughout embryonic and larval development.
To see if the construct was stably integrated in the genome we grew the fish to adulthood and mated them with wild type fish. 5 out of 7 of the egglays derived from these matings showed uniform expression throughout the pronephric tubules and duct in 10- 20% of the embryos, consistent with mosaic germline integration of the transgene (Fig. 2).The expression started around 20hpf and got stronger till 28hpf. We could not detect expression in the intestine in any of the lines examined. One cross showed weak GFP expression in trunk muscle cells. Larvae of one of the informative crosses were raised to establish stable transgenic lines with GFP expression that was restricted to the kidney (Tg(enpep:GFP)).
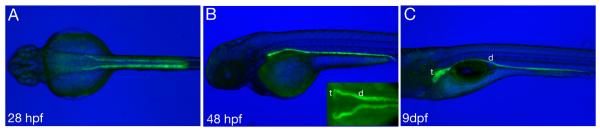
Figure 2.
GFP expression driven by the 2.3kb enpep promoter: Fluorescent GFP image superimposed on bright field image. GFP expression is detectable at 28 hpf in the pronephric duct (A, dorsal view). At 48 hpf (B, lateral view, inset dorsal view) expression is present in both duct (d) and tubules (t). Expression persists at 9 dpf (C) when the tubules are more prominent and form several loops.
We next asked if a shorter 1 kbp promoter fragment derived from the larger 2.3 kbp fragment could drive reporter gene expression in the developing pronephros (Fig 1). In contrast to the previous promoter, the F1 offspring of injected larvae had noticeable GFP expression in trunk and head in addition to the pronephros expression. We assume that the 1 kb promoter fragment lacks an inhibitory regulatory element which results in weak activation of the reporter in cells that do not normally express enpep.
To determine whether the 2.3 kbp enep promoter fragment was active in all cells of the pronephric duct and tubule, as well as the glomerulus, which is less easily identified in live embryos, we analyzed histological sections of transgenic larvae immunostained with antibodies that recognize GFP and laminin. As shown in Fig. 3 all cells of the pronophric tubules and ducts expressed GFP, we could also detect GFP expression in the cells resembling podocytes of the glomerulus that connect to the ducts. GFP expression was also detectable in the adult kidney in a comparable pattern as in the larvae. No GFP expression was detectable in the intestine at 3 or 6dpf (Fig. 3).
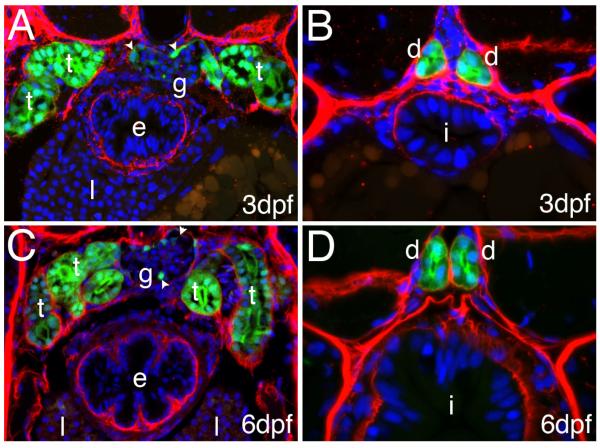
Figure 3.
GFP expression in kidney tubules and ducts: Cross section of 3dpf (A, B) and 6 dpf (C, D) Tg(enpep:GFP) larva stained with anti-GPF (green) and anti-Laminin (red) antibodies: GFP expression can be detected in both pronephric tubules (“t”, A, B) and ducts (“d”, C, D) as well as in podocyte like cells (arrowheads) of the glomerulus (“g” A, B). At 6 dpf several loops of the tubules are present (C). No expression is present in the intestine (i) esophagus (e), or liver (l).
Discussion
Here we show that a 2.3 kbp promoter fragment from the enpep coding sequence can drive specific GFP expression in the pronephric tubules and ducts of zebrafish larvae. The expression in the pronephros recapitulates the enep expression previously recorded by RNA in situ hybridization, with the exception of the intestinal epithelium. We assume that additional promoter elements needed to drive intestinal expression are not included in the 2.3kb fragment we identified.
The transgenic line we established will help studying the development of the zebrafish pronephros in wild type and mutant larvae and will be useful for establishing zebrafish models of human kidney diseases. This line could also help to screen for pharmacological reagents and genes capable of modulation enpep expression in the kidney, thus influencing activation of Angiotensin II.
Material and Method
Identifying the enpep upstream sequence
We used the Sanger BLAST server (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/d_rerio) to identify 5 kb upstream of XM_679675 from the clone zC200O8 (acc.# CR559943, zC146M5: acc# BX005193). Tests for translated sequence were done using NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) and TIGR (http://tigrblast.tigr.org/tgi/) BLAST. For promotor prediction we used softberry fprom (http://www.softberry.com/berry.phtml?topic=fprom&group=programs&subgroup= promoter). The identified promoter sequence has the gene bank accession number XXXX.
Amplification and cloning of the promoter
The 2.3kb fragment was amplified from Tü genomic DNA with primer containing BamH1 and Xho1 adapters with Clontech Advantage 2 polymerase. The primer sequences are 5′: AT-CTCGAG-AAATCACTTTGCCTTTTTCCAGTTGTCA; 3′: AT-GGATCC-GAACCCACACACTCTCCTCACGCTTC. The 5′ primer for the 1kb fragment is TGCAGTCTAGACACCGGAAGCTGG. The fragment was initially cloned in Promega pGem T Easy vector. After verification of the sequence it was cut out with BamH1 and Xho1 and cloned into pT2KXIG in. pT2KXIG in was prepared by cutting out the EF1p promotor and the intron with the same enzymes.
Injections of DNA construct
Injections were done into the first cell of zebrafish embryos. Transposase mRNA was prepared from pCS-TP after Not1 digestion using the Ambion mMessage Machine sp6 kit. mRNA was purified with chloroform/phenol according to the protocol. DNA was prepared with Qiagen midi prep kit. 20ng/ul DNA was co-injected with 20ng/ul transposase mRNA in water.
Fish maintenance and image acquisition
Fish were raised as described (Haffter et al., 1996) at 28.5C. Live Images were taken with an Olympus MVX10 fluorescent dissecting scope. Images of sections were taken with a BX61 fluorescent Microscope.
Immunohistochemistry
For fluorescent staining whole larvae were fixed with 4% PFA (PBS) for 4h, frozen in MeOH and permeabilized by Collagenase digestion in PBS. Larvae were washed with PBST and blocked with 10% goat serum/ PBST. Embryos were labeled with anti-laminin (1:50 Sigma, L9393) and Alexa 488 anti-GFP (1:200 Molecular Probes, A21311) antibody. After washing larvae were incubated with secondary antibody (alexa 568 anti mouse, Molecular Probes).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature
- Bahadori R, Huber M, Rinner O, Seeliger MW, Geiger-Rudolph S, Geisler R, Neuhauss SC. Retinal function and morphology in two zebrafish models of oculo-renal syndromes. Eur J Neurosci. 2003;18:1377–1386. doi: 10.1046/j.1460-9568.2003.02863.x. [DOI] [PubMed] [Google Scholar]
- Cogolludo A, Pérez-Vizcaíno F, Tamargo J. New insights in the pharmacological therapy of arterial hypertension. Curr Opin Nephrol Hypertens. 2005;14:423–427. doi: 10.1097/01.mnh.0000168334.09454.1c. [DOI] [PubMed] [Google Scholar]
- Dijkman HB, Assmann KJ, Steenbergen EJ, Wetzels JF. Expression and effect of inhibition of aminopeptidase-A during nephrogenesis. J Histochem Cytochem. 2006;54:253–262. doi: 10.1369/jhc.5A6815.2005. [DOI] [PubMed] [Google Scholar]
- Dressler GR. Kidney development branches out. Dev Genet. 1999;24:189–193. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<189::AID-DVG1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Drummond I. Making a zebrafish kidney: a tale of two tubes. Trends Cell Biol. 2003;13:357–365. doi: 10.1016/s0962-8924(03)00124-7. [DOI] [PubMed] [Google Scholar]
- Drummond IA, Majumdar A, Hentschel H, Elger M, Solnica-Krezel L, Schier AF, Neuhauss SC, Stemple DL, Zwartkruis F, Rangini Z, Driever W, Fishman MC. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- Drummond IA. The zebrafish pronephros: a genetic system for studies of kidney development. Pediatr Nephrol. 2000;14:428–435. doi: 10.1007/s004670050788. [DOI] [PubMed] [Google Scholar]
- Drummond IA. Kidney development and disease in the zebrafish. J Am Soc Nephrol. 2005;16:299–304. doi: 10.1681/ASN.2004090754. [DOI] [PubMed] [Google Scholar]
- Hostetter CL, Sullivan-Brown JL, Burdine RD. Zebrafish pronephros: a model for understanding cystic kidney disease. Dev Dyn. 2003;228:514–522. doi: 10.1002/dvdy.10371. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene. 1999;240:239–244. doi: 10.1016/s0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT, Kane ME, Obara T, Weimbs T. Polycystin-1, STAT6, and P100 Function in a Pathway that Transduces Ciliary Mechanosensation and Is Activated in Polycystic Kidney Disease. Dev Cell. 2006;10:57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Obara T, Mangos S, Liu Y, Zhao J, Wiessner S, Kramer-Zucker AG, Olale F, Schier AF, Drummond IA. Polycystin-2 immunolocalization and function in zebrafish. J Am Soc Nephrol. 2006;17:2706–2718. doi: 10.1681/ASN.2006040412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaux A, Fournie-Zaluski MC, Llorens-Cortes C. Angiotensin III: a central regulator of vasopressin release and blood pressure. Trends Endocrinol Metab. 2001;12:157–162. doi: 10.1016/s1043-2760(01)00381-2. [DOI] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004 [Google Scholar]
- Weber S, Taylor JC, Winyard P, Baker KF, Sullivan-Brown J, Schild R, Knüppel T, Zurowska AM, Caldas-Alfonso A, Litwin M, Emre S, Ghiggeri GM, Bakkaloglu A, Mehls O, Antignac C, ESCAPE Network. Schaefer F, Burdine RD. SIX2 and BMP4 mutations associate with anomalous kidney development. J Amer Soc Nephr. 2008;19:891–903. doi: 10.1681/ASN.2006111282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Selleck R, Yu J, Song HD, Chen Z, Song A, Zhou Y, Thisse B, Thisse C, McMahon AP, Davidson AJ. The cdx Genes and Retinoic Acid Control the Positioning and Segmentation of the Zebrafish Pronephros. PLoS Genet. 2007;3:e189. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]