Abstract
A genome-wide association study among Europeans related polymorphisms of the TLR locus at 4p14 and the FCGR2A locus at 1q23.3 to Helicobacter pylori serologic status. We replicated associations of 4p14 but not 1q23.3 with anti-H. pylori antibodies in 1,402 Finnish males. Importantly, our analysis clarified that the phenotype affected by 4p14 is quantitative level of these antibodies rather than association with seropositivity per se. Additionally, we annotated variants at 4p14 as expression quantitative trait loci associated with TLR6/10 and FAM114A1. Our findings suggest that 4p14 polymorphisms are linked to host immune response to H. pylori infection but not to its acquisition.
Introduction
Chronic Helicobacter pylori infection is causally associated with gastritis, gastroduodenal ulcer disease, gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma.1 A genome-wide association study (GWAS) of anti-H. pylori serologic status among Europeans identified inverse associations with single nucleotide polymorphism (SNPs) in the toll-like receptor (TLR) locus at 4p14 and the Fcγ receptor 2a (FCGR2A) locus at 1q23.3.2 Comparing anti-H. pylori immunoglobulin G (IgG) antibody levels in the highest quartile vs. lower levels, the 4p14 associations were strongly significant (top-ranked SNP, rs10004195; P=1.4e-18) and the 1q23.3 associations were borderline (top-ranked SNP, rs368433; P=2.1e-8). In contrast, there were no genome-wide significant associations with anti-H. pylori antibody levels in a GWAS among Mexican-Americans.3 To extend the previous findings among Caucasians, we evaluated associations of anti-H. pylori IgG with 4p14 and 1q23.3 loci in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC).
Results and discussion
Among ATBC participants, rs10004195-A at 4p14 was inversely associated with anti-H. pylori antibody levels in the highest 25%. The per-allele odds ratio (OR) was 0.61 (95% confidence interval (CI)=0.47–0.79; P=2.2e-4), consistent with the previous report. In contrast, seropositivity (73% of participants) was not associated with rs10004195 (OR=1.00; 95% CI=0.79–1.27; P=9.9e-1). Indeed, the minor allele frequency (MAF) of rs10004195 among seronegative individuals (MAF=0.15) was intermediate between the subjects with the highest 25% of antibodies (MAF=0.11) and all other seropositives (MAF=0.17) (Supplementary Table 1).
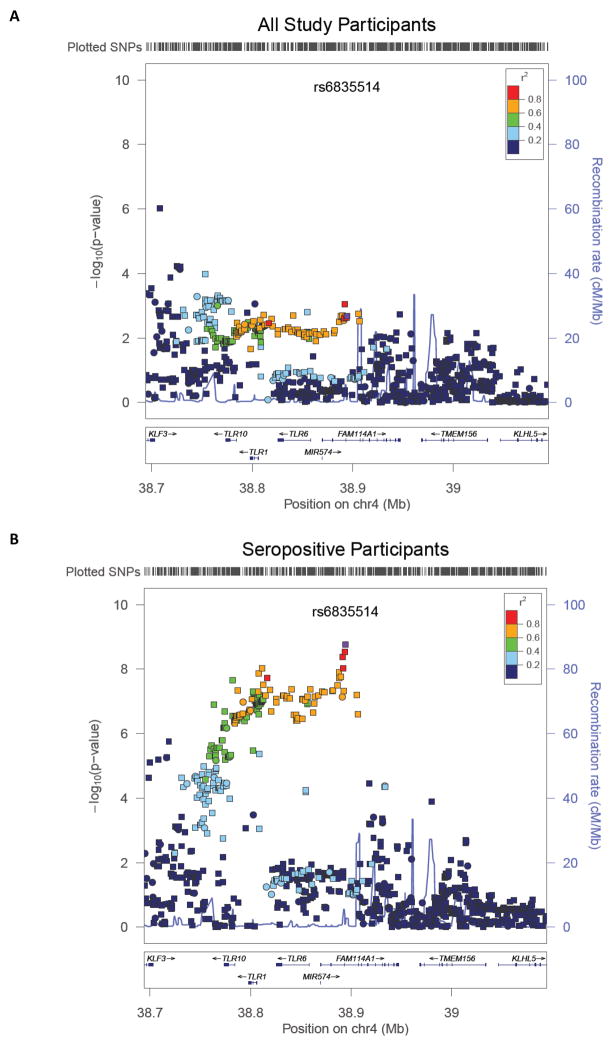
We also found associations of 4p14 variants with continuous anti-H. pylori antibody levels. Notably, the statistical significance as well as magnitude of effects was accentuated when we restricted our analysis to seropositive individuals (Figure 1). For example, the per-allele beta coefficients of rs10004195-A were −0.15 (standard error (SE)=0.05; P=3.4e-3; PFDR = 4.0e-2) and −0.20 (SE=0.04; P=3.8e-7; PFDR = 4.8e-6) among all and seropositive participants, respectively. The strongest signal was observed at rs6835514 (MAF=0.17; beta=−0.23; SE=0.04; P=1.6e-9; PFDR = 1.9e-6), which was in moderate linkage disequilibrium (LD) with rs10004195 (r2=0.62). Fifty-one nearby SNPs within moderate to high LD (r2 >0.6) of rs6835514 had p-values ranging from 4.7e-7 (PFDR = 5.6e-6) to 3.0e-9 (PFDR = 1.9e-6) (Supplementary Table 2). In analyses controlling for rs6835514, the effect of rs10004195 did not remain significant (beta=−0.03; SE=0.06; P=5.5e-1), indicating that the two SNP associations with IgG levels are not independent.
Figure 1.
4p14 locus (−log10 P) associations with anti-H. pylori antibodies estimated among (A) all and (B) seropositive ATBC participants.
Genomic region was defined as ±200 kb surrounding the index SNP (rs6835514, purple). Circles and squares indicate genotyped and imputed SNPs, respectively. Figure was generated with LocusZoom version 1.1 (http://csg.sph.umich.edu/locuszoom/) using Hg18/HapMap Phase II CEU as genome build/LD population.
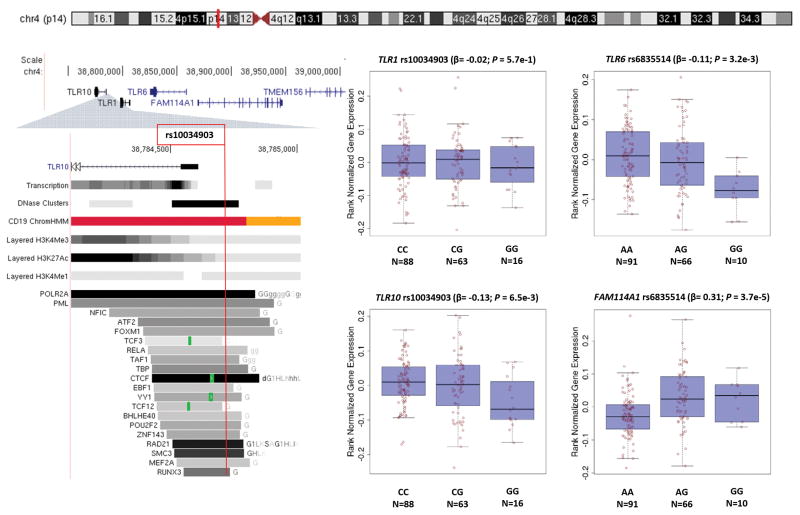
Furthermore, we explored putative functional effects of these 52 4p14 SNPs based on publically available data. Except for rs4833095 (Asn248Ser) and synonymous rs5743614 in TLR1, all other variants were located in intronic or non-coding regions. However, many of these remaining SNPs fall within proximal or distal regulatory elements (Supplementary Table 3). Using the Roadmap ChromHMM track, we found 7 promoter SNPs and 8 enhancer SNPs in CD19-positive primary blood cells. In ENCODE cell lines, 17 SNPs were mapped to DNaseI hypersensitive regions and 42 SNPs altered binding motifs of at least one transcription factor. GTEx expression quantitative trait loci (eQTL) data on whole-blood samples identified multiple SNPs significantly correlated (P<0.01) with mRNA transcript levels of TLR6/10 or FAM114A1 but not with TLR1 (Supplementary Table 3). The low IgG allele (G) of rs6835514 was inversely associated with mRNA levels of TLR6 (beta=−0.11; P=3.2e-3) and positively associated with FAM114A1 expression (beta=0.31; P=3.7e-5) (Figure 2). Of particular interest, rs10034903, which was mapped to an active promoter of TLR10 as well as a transcription factor binding site, appeared to be a significant eQTL for both TLR10 and FAM114A1 (Figure 2). Similar to rs6835514, the low IgG allele (G) of rs10034903 was associated with decreased mRNA expression of TLR10 (beta=−0.13; P=6.5e-3) and increased mRNA expression of FAM114A1 (beta=0.27; P=8.0e-4).
Figure 2.
Selected functional annotations of 4p14 locus SNPs.
NIH Epigenomics Roadmap and ENCODE data were screened using the UCSC Genome Browser to track transcription levels in GM12878 (ENCODE) and regulatory elements, including DNaseI hypersensitivity cluster (open chromatin structure; gray box indicating the extent of the hypersensitive region with shading proportional to the maximum signal strength observed in any cell line) from 125 cell types (ENCODE), Roadmap Chromatin State Segmentation using a Hidden Markov Model (ChromHMM) from CD19 Primary Cells (Promoter [Red] and Enhancer [Orange]), layered core histone marks H3K4Me3, H3K27Ac, and H3K4Me1 in GM12878 (ENCODE), and transcription factor (TF) binding site (gray box with shading proportional to the maximum signal strength; green highlight indicating the highest scoring site of a canonical motif for the corresponding TF) identified by ChIP-seq (ENCODE) experiments. GTEx data on 168 whole-blood samples were analyzed with box plots and regression statistics for expression quantitative trait loci (eQTL). The genomic location of rs10034903 is shown by the red vertical line.
Although roles in pathogen recognition and innate immunity have been well established,4 little is known about TLR1/6/10 with respect to H. pylori response.5, 6 In a recent report, heterodimeric TLR2/TLR10 was suggested to mediate H. pylori lipopolysaccharide recognition in activating the NF-kB signaling pathway.7 Additionally, a growing number of studies suggest genetic polymorphisms in TLR genes are associated with infectious disease susceptibility8. For example, rs5743618 (Ser602Ile of TLR1; r2=0.80 among CEU) in high LD with rs6835514 (r2=0.80 among CEU) has been associated with susceptibility to tuberculosis9, Chlamydia trachomtis infection10 and leprosy11; while rs4833095 (Asn248Ser of TLR1; r2=0.85 with rs6835514 among CEU) has been associated with Atopobium vaginae infection12 and placental malaria13.
Intriguingly, 4p14 has also been implicated as a susceptibility locus for IgE-mediated allergic sensitization and for hay fever-related asthma; minor alleles were associated with decreased levels of IgE14 and decreased risk of asthma15 in Caucasians, in parallel with our finding of an inverse association with anti-H. pylori antibody levels. However, three TLR SNPs included in the current report were inconsistently associated with active H. pylori infection determined by 13C-urea breath test in a Chinese population.16 Further studies are warranted of these genetic polymorphisms in relation to target gene regulation and disease consequences. Fine mapping studies are also needed to pinpoint the functionally relevant causal variants.
As for the borderline association reported for 1q23.3, we did not find qualitative or quantitative associations with anti-H. pylori antibodies. In particular, rs368433 was not associated with the highest quartile of antibody levels (OR=1.05; 95% CI=0.72–1.52; P=8.1e-1) nor with seropositivity overall (OR=0.84; 95% CI=0.58–1.23; P=3.8e-1) (Supplementary Table 1). Moreover, there were no significant associations with continuous IgG levels among either all participants or seropositives only (Supplementary Figure 1). Based on our observed 0.06 MAF of rs368433, estimated power to detect the previously reported 0.73 OR at a 5% significance level was 71% among all individuals and 80% among seropositives.
In conclusion, we confirmed the association of the 4p14 locus with anti-H. pylori antibodies among Caucasians, and clarified the phenotype affected by these polymorphisms. Our findings suggest that the 4p14 locus may modulate intensity of host immune response rather than acquisition of H. pylori infection per se. The clinical significance of higher levels of antibody to H. pylori remains to be determined; conflicting associations with either increased17, 18 or decreased19, 20 gastric cancer risk have been reported. Antigen specificity of the 4p14 locus associations should also be examined in future studies. These findings await extension to other ethnic/racial groups with differences in exposure patterns, bacterial strain pathogenicity, host genetic characteristics and population burdens of H. pylori-associated diseases.
Materials and methods
Our study included participants from the ATBC, a randomized, double-blind, placebo-controlled trial conducted 1985–1993 in 29,133 Finnish male smokers aged 50 to 69 years.21 Participants completed questionnaires at enrollment and serum samples were collected and stored at −70°C for future analyses. The current study included 1,402 participants who had both genotyping and H. pylori serology data available.
Antibodies to H. pylori were measured by enzyme-linked immunosorbent22, 23, 24, 25 and multiplex bead-based assays26, 27, as described previously. In order to combine measurements based on different technologies, we standardized levels using laboratory-specific means and standard deviations. Detailed information about genotyping and quality control was published previously28, 29. Briefly, a genome-wide scan was performed with Illumina HumanHap550/610 arrays. Imputation was performed using the hidden-Markov model algorithm implemented in MACH, based on HapMap CEU reference panel build 36, R22.
We defined candidate regions as ±200 kb from the previously reported top SNPs,2 rs10004195 at 4p14 (chr4:38584724–38984724, Hg19) and rs368433 at 1q23.3 (chr1:161284210–161684210, Hg19). We additionally included SNPs located in flanking regions of rs6835514 (chr4:38694380–39094380, Hg19), the most significant 4p14 SNP in our linear regression analysis. After quality control, 1,380 SNPs at 4p14 and 1,127 SNPs at 1q23.3 were available from genotyping or imputed data. Average call rate for the genotyped SNPs was 1 and average quality score for imputed SNPs (Rsq) was 0.92.
To refine the phenotype affected by gene polymorphisms, we tested several definitions of H. pylori serologic status. First, we used the same definition as the previous report in Caucasians, which compared individuals with IgG levels in the highest 25% to individuals in the other 75%. Second, to determine whether the loci are associated with H. pylori acquisition, we compared seropositives to seronegatives. Lastly, we analyzed IgG levels as a continuous variable. We assumed an additive genetic model with number of minor alleles as a predictor, using 10-year age groups and genotying principal components as covariables. We used logistic regression for dichotomous outcome variables and linear regression for the continuous outcome variable. Adjustment for multiple comparisons at 4p14 was performed by the false discovery rate (FDR) based on 1,380 SNPs, ignoring the high correlation among the tested SNPs. Analyses were conducted using SAS v9.3 (SAS Institute Inc., Cary, NC) and R v3.1.3. Statistical power was estimated with CaTS (http://csg.sph.umich.edu/abecasis/CaTS/index.html).
We focused functional annotation on the 52 SNPs located in moderate to high (r2>0.6) LD with rs6835514. The UCSC Genome browser (http://genome.ucsc.edu) was used to confirm genomic regions and to screen NIH Epigenomics Roadmap (http://www.roadmapepigenomics.org/) and ENCODE (http://genome.ucsc.edu/ENCODE/) tracks. HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) and RegulomeDB (http://regulomedb.org/) were also used to confirm SNP functions and to compile summary results.
To map promoter, enhancer, polycomb-repressed or heterochromatin regions, we used the chromatin state segmentation by Hidden Markov Model (ChromHMM) track from Roadmap reported for CD19 primary cells (presumably, circulating B-lymphocytes). DNase cluster assigned by DNase I hypersensitive assay results from 125 cell types, transcription factor binding sites defined by chromatin immunoprecipitation sequencing for 161 factors, and transcription levels determined by RNA-seq in GM12878 were tracked using ENCODE.
To identify putative target genes regulated by SNPs, we compiled eQTL results assessed in whole blood samples (n=168) from Genotype-Tissue Expression (http://www.gtexportal.org/home/). Linear regression was conducted for the 52 SNPs on log and quantile normalized RNA-seq levels of four genes to which any of these SNPs were mapped, including TLR1 (ENSG00000174125.3), TRL6 (ENSG00000174130.8), TLR10 (ENSG00000174123.6), and FAM114A1 (ENSG00000197712.7). Covariables included three genotyping principle components, 15 peer factors and sex. Based on the number of genes tested, our Bonferroni-corrected significance threshold was P=0.012 (0.05/4 genes).
Informed consents were obtaind from all participants. The study was approved by IRBs of the National Public Health Institute of Finland and the US National Cancer Institute.
Supplementary Material
Supplementary Figure 1: 1q23.3 locus (−log10 P) associations with anti-H. pylori antibodies estimated among (A) all and (B) seropositive ATBC participants.
Genomic region was defined as ±200 kb surrounding the index SNP (rs368433, purple). Circles and squares indicate genotyped and imputed SNPs, respectively. Figure was generated with LocusZoom version 1.1 (http://csg.sph.umich.edu/locuszoom/) using Hg18/HapMap Phase II CEU as genome build/LD population.
Supplementary Table 1: Associations of previously reported top SNPs with H. pylori serologic status in ATBC participants (n=1,402)
Associations of anti-H. pylori IgG antibody levels with 4p14 SNPs among all or seropositive ATBC participants
Functional annotation of SNPs within moderate to high LD (r2>0.6) of rs6835514
Acknowledgments
The authors thank the principal investigators of the ATBC trial and its multiple sub-studies for access to the genotyping and serology data on study participants.
Fund: This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics. Additionally, this research was supported by U.S. Public Health Service contracts N01-CN-45165, N01-RC-45035, N01-RC-37004 and HHSN261201000006C from the National Cancer Institute, Department of Health and Human Services.
Footnotes
Conflict of interests: All authors declare no competing interests.
References
- 1.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature reviews Cancer. 2002 Jan;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 2.Mayerle J, den Hoed CM, Schurmann C, Stolk L, Homuth G, Peters MJ, et al. Identification of genetic loci associated with Helicobacter pylori serologic status. Jama. 2013 May 8;309(18):1912–20. doi: 10.1001/jama.2013.4350. [DOI] [PubMed] [Google Scholar]
- 3.Rubicz R, Yolken R, Drigalenko E, Carless MA, Dyer TD, Kent J, Jr, et al. Genome-wide genetic investigation of serological measures of common infections. European journal of human genetics: EJHG. 2015 Mar 11; doi: 10.1038/ejhg.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signalling complexes. Nature reviews Immunology. 2014 Aug;14(8):546–58. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 5.Fukata M, Abreu MT. Role of Toll-like receptors in gastrointestinal malignancies. Oncogene. 2008 Jan 7;27(2):234–43. doi: 10.1038/sj.onc.1210908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wurfel MM, Hawn TR. Genetic variants associated with susceptibility to Helicobacter pylori. Jama. 2013 Sep 4;310(9):976. doi: 10.1001/jama.2013.194762. [DOI] [PubMed] [Google Scholar]
- 7.Nagashima H, Iwatani S, Cruz M, Jimenez Abreu JA, Uchida T, Mahachai V, et al. Toll-like Receptor 10 in Helicobacter pylori Infection. The Journal of infectious diseases. 2015 May 14; doi: 10.1093/infdis/jiv270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skevaki C, Pararas M, Kostelidou K, Tsakris A, Routsias JG. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious diseases. Clinical and experimental immunology. 2015 May;180(2):165–77. doi: 10.1111/cei.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YX, Jiang TT, Yang XY, Xue Y, Wang C, Liu JY, et al. Toll-Like Receptor-1,-2, and-6 Polymorphisms and Pulmonary Tuberculosis Susceptibility: A Systematic Review and Meta-Analysis. PloS one. 2013 May 14;8(5) doi: 10.1371/journal.pone.0063357. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor BD, Darville T, Ferrell RE, Kammerer CM, Ness RB, Haggerty CL. Variants in Toll-like Receptor 1 and 4 Genes Are Associated With Chlamydia trachomatis Among Women With Pelvic Inflammatory Disease. Journal of Infectious Diseases. 2012 Feb 15;205(4):603–9. doi: 10.1093/infdis/jir822. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong SH, Gochhait S, Malhotra D, Pettersson FH, Teo YY, Khor CC, et al. Leprosy and the Adaptation of Human Toll-Like Receptor 1. Plos Pathog. 2010 Jul;6(7) doi: 10.1371/journal.ppat.1000979. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verstraelen H, Verhelst R, Nuytinck L, Roelens K, De Meester E, De Vos D, et al. Gene polymorphisms of Toll-like and related recognition receptors in relation to the vaginal carriage of Gardnerella vaginalis and Atopobium vaginae. Journal of reproductive immunology. 2009 Jan;79(2):163–73. doi: 10.1016/j.jri.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Hamann L, Bedu-Addo G, Eggelte TA, Schumann RR, Mockenhaupt FP. The toll-like receptor 1 variant S248N influences placental malaria. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2010 Aug;10(6):785–9. doi: 10.1016/j.meegid.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Bonnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC, et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nature genetics. 2013 Aug;45(8):902–6. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira MA, Matheson MC, Tang CS, Granell R, Ang W, Hui J, et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. The Journal of allergy and clinical immunology. 2014 Jun;133(6):1564–71. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang FB, Li ZX, Wang YM, Zhang L, Ma JL, Zhou T, et al. Toll-like receptor 1 and 10 polymorphisms, Helicobacter pylori susceptibility and risk of gastric lesions in a high-risk Chinese population. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2015 Apr;31:263–9. doi: 10.1016/j.meegid.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida T, Kato J, Inoue I, Yoshimura N, Deguchi H, Mukoubayashi C, et al. Cancer development based on chronic active gastritis and resulting gastric atrophy as assessed by serum levels of pepsinogen and Helicobacter pylori antibody titer. International journal of cancer Journal international du cancer. 2014 Mar 15;134(6):1445–57. doi: 10.1002/ijc.28470. [DOI] [PubMed] [Google Scholar]
- 18.Yanaoka K, Oka M, Yoshimura N, Mukoubayashi C, Enomoto S, Iguchi M, et al. Risk of gastric cancer in asymptomatic, middle-aged Japanese subjects based on serum pepsinogen and Helicobacter pylori antibody levels. International journal of cancer Journal international du cancer. 2008 Aug 15;123(4):917–26. doi: 10.1002/ijc.23571. [DOI] [PubMed] [Google Scholar]
- 19.Tatemichi M, Sasazuki S, Inoue M, Tsugane S, Group JS. Clinical significance of IgG antibody titer against Helicobacter pylori. Helicobacter. 2009 Jun;14(3):231–6. doi: 10.1111/j.1523-5378.2009.00681.x. [DOI] [PubMed] [Google Scholar]
- 20.Yamaji Y, Mitsushima T, Ikuma H, Okamoto M, Yoshida H, Kawabe T, et al. Weak response of Helicobacter pylori antibody is high risk for gastric cancer: a cross-sectional study of 10,234 endoscoped Japanese. Scand J Gastroentero. 2002 Feb;37(2):148–53. doi: 10.1080/003655202753416795. English. [DOI] [PubMed] [Google Scholar]
- 21.Wang YY, Simpson JA, Wluka AE, Urquhart DM, English DR, Giles GG, et al. Reduced rates of primary joint replacement for osteoarthritis in Italian and Greek migrants to Australia: the Melbourne Collaborative Cohort Study. Arthritis Res Ther. 2009;11(3) doi: 10.1186/ar2721. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koshiol J, Flores R, Lam TK, Taylor PR, Weinstein SJ, Virtamo J, et al. Helicobacter pylori seropositivity and risk of lung cancer. PloS one. 2012;7(2):e32106. doi: 10.1371/journal.pone.0032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolzenberg-Solomon RZ, Blaser MJ, Limburg PJ, Perez-Perez G, Taylor PR, Virtamo J, et al. Helicobacter pylori seropositivity as a risk factor for pancreatic cancer. Journal of the National Cancer Institute. 2001 Jun 20;93(12):937–41. doi: 10.1093/jnci/93.12.937. English. [DOI] [PubMed] [Google Scholar]
- 24.Abnet CC, Kamangar F, Dawsey SM, Stolzenberg-Solomon RZ, Albanes D, Pietinen P, et al. Tooth loss is associated with increased risk of gastric non-cardia adenocarcinoma in a cohort of Finnish smokers. Scand J Gastroentero. 2005 Jun;40(6):681–7. doi: 10.1080/00365520510015430. English. [DOI] [PubMed] [Google Scholar]
- 25.Limburg PJ, Stolzenberg-Solomon RZ, Colbert LH, Perez-Perez GI, Blaser MJ, Taylor PR, et al. Helicobacter pylori seropositivity and colorectal cancer risk: A prospective study of male smokers. Cancer Epidem Biomar. 2002 Oct;11(10):1095–9. English. [PubMed] [Google Scholar]
- 26.Yu G, Murphy G, Michel A, Weinstein SJ, Mannisto S, Albanes D, et al. Seropositivity to Helicobacter pylori and risk of pancreatic cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013 Dec;22(12):2416–9. doi: 10.1158/1055-9965.EPI-13-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy G, Michel A, Taylor PR, Albanes D, Weinstein SJ, Virtamo J, et al. Association of seropositivity to Helicobacter species and biliary tract cancer in the ATBC study. Hepatology. 2014 Dec;60(6):1963–71. doi: 10.1002/hep.27193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nature genetics. 2009 Sep;41(9):986–90. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. American journal of human genetics. 2009 Nov;85(5):679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: 1q23.3 locus (−log10 P) associations with anti-H. pylori antibodies estimated among (A) all and (B) seropositive ATBC participants.
Genomic region was defined as ±200 kb surrounding the index SNP (rs368433, purple). Circles and squares indicate genotyped and imputed SNPs, respectively. Figure was generated with LocusZoom version 1.1 (http://csg.sph.umich.edu/locuszoom/) using Hg18/HapMap Phase II CEU as genome build/LD population.
Supplementary Table 1: Associations of previously reported top SNPs with H. pylori serologic status in ATBC participants (n=1,402)
Associations of anti-H. pylori IgG antibody levels with 4p14 SNPs among all or seropositive ATBC participants
Functional annotation of SNPs within moderate to high LD (r2>0.6) of rs6835514