Abstract
Wound healing is an essential physiological process for tissue homeostasis, involving multiple types of cells, extracellular matrices, and growth factor/chemokine interactions. Many in vitro studies have investigated the interactions between cues mentioned above; however, most of them only focused on a single factor. In the present study, we design a wound healing device to recapitulate in vivo complex microenvironments and heterogeneous cell situations to investigate how three types of physiologically related cells interact with their microenvironments around and with each other during a wound healing process. Briefly, a microfluidic device with a micropillar substrate, where diameter and interspacing can be tuned to mimic the topographical features of the 3D extracellular matrix, was designed to perform positional cell loading on the micropillar substrate, co-culture of three types of physiologically related cells, keratinocytes, dermal fibroblasts, and human umbilical vein endothelial cells, as well as an investigation of their interactions during wound healing. The result showed that cell attachment, morphology, cytoskeleton distribution, and nucleus shape were strongly affected by the micropillars, and these cells showed collaborative response to heal the wound. Taken together, these findings highlight the dynamic relationship between cells and their microenvironments. Also, this reproducible device may facilitate the in vitro investigation of numerous physiological and pathological processes such as cancer metastasis, angiogenesis, and tissue engineering.
I. INTRODUCTION
Wound healing is an essential physiological process that is important for tissue homeostasis.1 The wound healing process, particularly in skin, is of great significance to be clearly elucidated, because skin is the largest organ in the body and performs many critical roles such as barrier protection from physical or chemical insults, sensory functions, and regulation of homeostasis.2 Damaged skin cannot sustain these functions and numerous complications may occur such as infection or fluid loss.3 Cutaneous wound healing requires precise coordination of epithelialization, dermal repair, angiogenesis, and numerous types of cells, including epithelial cells, fibroblasts, immune cells, and endothelial cells, to invade the wound bed to fill in and repair the tissue.4 However, the complicated mechanism of interactions among these cells and each role in cutaneous wound healing have not yet been investigated thoroughly.
Recently, on the basis of conventional in vivo (e.g., minipig5) and in vitro (e.g., scratch assay6 and transwell assay7) models, a great deal of studies has greatly enhanced the understanding of cell interactions during wound healing process. For example, dermal fibroblasts would be activated and differentiated into myofibroblasts when co-cultured with dermal microvascular endothelial cells and then migrated to heal the wound.6 Contacting with fibroblasts stimulated the migration and proliferation of keratinocytes during wound healing.7 In return, the expression and synthesis of type I collagen (the predominant form of collagen in fibroblasts in human skin) was regulated by keratinocyte-releasable factors.8 However, the conventional in vivo model is laborious and costly. Also, it always causes ethical or legal concerns. In addition, most of the conventional in vitro studies are always based on 2D environments in which cells grow and migrate on a flat surface, showing little consideration of in vivo physiological cellular microenvironment. In fact, cells grow in a 3D environment that embed in extracellular matrices and varies in composition, density, and stiffness and migrate through complex topographical features.9–11
There has been evidence that cells on substrates composed of pillars or pits exhibited spindle shape and pseudopodial protrusions, more akin to the in vivo situation.12 Micropillar technology has shown a great promise for medical implants or sensors in recent years.13 Lots of works have demonstrated that micropillar substrate could affect cell behavior in terms of cell adhesion,13–16 proliferation,13,15,17 morphology13,15,18–21 migration,15,18,22,23 differentiation19,24 and even gene expression,16,20 which may account for at least part of the striking differences between cells grown in vivo and in vitro. In addition, there have been numerous strategies for studying wound healing, among which the most widely used method is the scratch assay.6,25–28 In the scratch assay, a “wound” is generated by a pipette through removing a stripe of cells on a confluent cell monolayer. However, this method is manual and very tedious, which limits the ability to reproducibly perform the wound healing assay with multiple samples, and the resulting data may be subjective and inaccurate. Additionally, various factors in this assay, such as small molecules released from injured cells, the remnant debris of detached cells, and the sudden availability of free space, all possibly contribute to heal the wound, which always causes to fail to analyze the cues independently.11,29 Another common way used to create an in vitro wound is laminar flow, in which a wound is made by trypsin digestion. However, trypsin digestion is always utilized to make wound in mono-culture. When being applied to co-culture of heterogeneous cell types, it is difficult to operate and realize a reproducible wound with accurate scale of lesion. In addition, trypsin digestion can damage adjacent cells and induce secretion of inflammation factors, which might render the wound healing process more complex.30–32
In the current study, we present a wound healing device (Figure 1) that could investigate cell response to the substrate with topographical cues, as well as interactions between heterogeneous cells during wound healing, which is physiological relevance to the realistic situation in vivo. Using this device, we first realized the co-culture of three types of wound healing-related cells: HaCaT (a type of human skin epithelial cells), CCC-ESF-1 (a type of human dermal fibroblasts), and human umbilical vein endothelial cell (HUVEC) (a type of human umbilical vein endothelial cells that is widely used in many skin vascularization studies).6,25,33–36 Distinct responses of these three types of cells to the micropillar substrate with different pillar sizes were observed, and the cell morphology, cytoskeleton organization, and nuclear shape were analyzed quantitatively. Afterwards, an identical injury-free skin wound was created by peeling off an integrated stencil. The in vitro wound healing process was spatiotemporally tracked, and the interactions among these cells were dynamically analyzed.
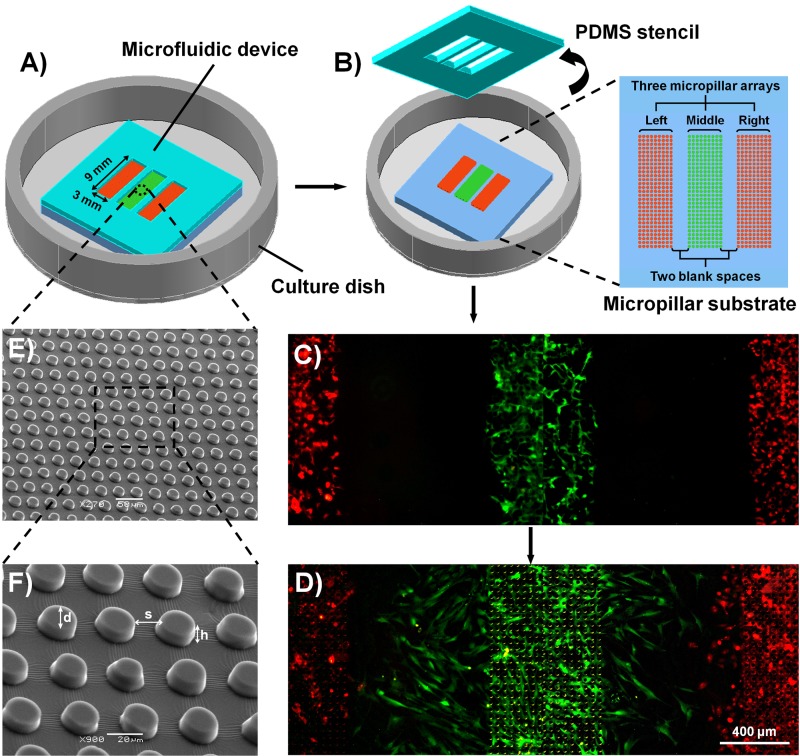
FIG. 1.
Schematic illustration of the experiment procedures and characterization of the microfluidic device. (a) The microfluidic device used in the current study, which was composed of four layers, a PDMS stencil, a PDMS micropillar substrate, a thin PDMS membrane (not shown), and a polystyrene culture dish. The PDMS stencil was sealed to the micropillar substrate, and each type of cells was seeded into the appropriate region (from left to right, HaCaT, ESF-1, and HUVEC cells, respectively). (b) The PDMS stencil was peeled off after the cells were attached and cultured on the micropillar substrate for 24 h. (c) A typical fluorescence image of well-distributed cells after the stencil was just peeled off. To visualize clearly the coexistence of cells, HaCaT (left), ESF-1 (middle), and HUVEC (right) cells were specifically stained using CellTracker Orange, CellTracker Green, and CellTracker Orange, respectively. (d) A typical fluorescence image of cells after wounding, corresponding to 36 h. (e) SEM image of the micropillar substrate. (f) Enlarged image of the square in the dotted lines in (e).
II. EXPERIMENTAL
A. Materials and reagents
RTV 615 polydimethylsiloxane (PDMS) prepolymer and curing agent were purchased from Momentive Performance Materials (Waterford, NY); surface-oxidized silicon wafers from Shanghai Xiangjing Electronic Technology Ltd. (Shanghai, China); AZ 50XT photoresist and developer from AZ Electronic Materials (Somerville, NJ, USA); Poly-l-lysine (PLL), acridine orange (AO), propidium iodide (PI), and Hoechst 33258 from Sigma-Aldrich (MO, USA); and cell culture medium, fetal bovine serum (FBS), TRITC-phalloidin, CellTracker Green CMFDA, and CellTracker Orange CMRA from Gibco Invitrogen Corporation (CA, USA). All solvents and other chemicals were purchased from local commercial suppliers and were of analytical reagent grade, unless otherwise stated. All solutions were prepared using ultra-purified water supplied by a Milli-Q system (Millipore®).
B. Device design and fabrication
In the current study, PDMS was used to fabricate the device because of its easy fabrication, low cost, plasticity, practical scalability, nontoxicity, and thermal stability. Generally, the device utilized for this study comprises four layers: a PDMS stencil, a PDMS micropillar substrate, a thin PDMS membrane (not shown), and a polystyrene culture dish (Figures 1(a) and 1(b)). The PDMS micropillar substrate was fabricated using soft lithography.37–39 First, the patterns were designed using AutoCAD software. Generally, the micropillar substrate had three regions (3 mm wide, 9 mm long, and separated by 800 μm space) and each region consisted of an array of cylindrical pillars. The pillars had a height of 10 μm, a diameter of 15, 18 or 21 μm and were arranged in squares with pillar interspaces of 15, 18, and 21 μm, respectively. The patterns were printed onto transparent films (MicroCAD Photomask Ltd., Suzhou, China) and used as a photomask. Then, a mold was fabricated in a single step under UV light using AZ 50XT photoresist. Before fabricating the PDMS micropillar substrate, the mold was exposed to trimethylchlorosilane vapor for 3 min. Then, a well-mixed PDMS pre-polymer (RTV 615 A and B in 10:1 ratio) was poured onto the mold and placed in a Petri dish to yield a 1 mm-thick layer. After degassing, the mold was baked for 30 min at 80 °C. Afterwards, the PDMS layer was peeled off from the mold, trimmed, cleaned, and placed in a polystyrene culture dish coated (3000 rpm, 45 s, ramp 15 s) with PDMS pre-polymer (RTV 615 A and B in 15:1 ratio) that had been cured for 15 min in the oven (80 °C).
To fabricate the PDMS stencil, a well-mixed PDMS prepolymer (RTV 615 A and B in 5:1 ratio) was spin coated onto a blank silicon wafer (300 rpm, 60 s). After baked for 30 min at 80 °C, the thin PDMS layer (nearly 500 μm thick) was peeled off from the silicon wafer. Then, rectangular openings (3 mm wide, 9 mm long, and separated by 800-μm space) were made by artificial hand cut. The PDMS stencil was made a little larger than that of the bottom PDMS micropillar substrate. From the extra part of the PDMS stencil, it can be easily peeled off from the micropillar substrate by using a sterile forcep when making wound. After that, the PDMS stencil was aligned to the micropillar substrate under a stereomicroscope to ensure that the edge of the PDMS strip in 800 μm width was right near the border of the micropillars. The whole device was then baked at 80 °C for 24 h. This baking time can make sure that the micropillar substrate and PDMS stencil were sealed together to prevent liquid leakage and cell cross-contamination, as well as to easily peel off the PDMS stencil from the micropillar substrate.
C. Cell culture
Human embryonic skin fibroblasts CCC-ESF-1 (described as ESF-1 in the next study) and the immortalized human keratinocyte cell line HaCaT were both obtained from Chinese Academy of Medical Sciences (Beijing, China). HUVECs were obtained from the Chinese Academy of Sciences (Shanghai, China). ESF-1 and HUVEC cells were both cultured using Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Grand Island, NY) supplemented with 10% FBS (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. HaCaT cells were cultured using Minimum Essential Medium with Earle's Balanced Salts (MEM-EBSS, Invitrogen, Grand Island, NY) supplemented with 10% FBS (Invitrogen), 100 units/ml penicillin, and 100 mg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. The cells were normally passaged at a ratio of 1:2 every 2 days to maintain them in the exponential growth phase. Before use, they were harvested through trypsinization with 0.25% trypsin (Invitrogen) in Ca2+- and Mg2+- free Hanks' balanced salt solution (CMF-HBSS) at 37 °C. Trypsinization was stopped by the addition of fresh supplemented medium, and cell suspension was then centrifuged at a rotational speed of 1000 rpm for 5 min. After that, the cells were resuspended in supplemented medium for use.
D. Cell seeding, co-culturing, and wounding
The device was first sterilized with UV light for 2 h and then coated with poly(l-lysine) (0.1 mg/ml in a borate-buffered saline solution) over night.40 After rinsing thrice with DMEM, different types of cells were seeded into their corresponding regions for co-culturing, namely, HaCaT cells in the left region, ESF-1 cells in the middle region, and HUVECs in the right region (Figures 1(c) and 1(d)). To find the optimum seeding density for co-culturing during the follow-up wound healing assay, a seeding density test of each type of cells was performed by using different cell densities (1 × 105 cells/ml, 5 × 105 cells/ml, and 1 × 106 cells/ml).11,29 It should be noted that during this step, the liquid surface must be maintained flat to ensure the cells were uniformly distributed when the cell suspension was added to each region. The device was then placed in a humidified atmosphere with 5% CO2 at 37 °C for 2 h to allow cell attachment. Cell culture medium was added to submerge the device to supply culture nutrients to the cells. The stencil was gently peeled off with forceps after the cells were cultured for typically 24 h, and then the cells were allowed to migrate freely to heal the wound.
E. Cell staining
Assessment of cell adhesion was performed using the AO/PI double-staining protocol.39 After removing the growth medium and washing with PBS, the AO/PI staining solution (10 μg/ml each in PBS) was introduced into the cell culture region, and the staining process was performed for 10 min at room temperature. Then, PBS was introduced for 10 min as a final rinse. In these procedures, the dead cells were stained red by fluorescent dye PI, whereas the living cells were stained green by the fluorescent dye AO. For clear visualization of the cell morphology, cytoskeleton organization, and nuclei shape, the actin filament and nuclear staining was also performed. Briefly, the cells were fixed using 4% paraformaldehyde for 10 min at room temperature after washing thrice with PBS. The cultures were permeabilized with PBS containing 0.2% Triton X-100 for 30 min. Then, the cultures were incubated at 37 °C for 20 min with TRITC-phalloidin (100 nM in PBS) for actin filament staining and another 10 min in PBS containing Hoechst dye (H33258 fluorochrome, 0.5 μg/ml) for nuclear staining. To clearly visualize the coexistence of HaCaT, ESF-1, and HUVEC cells in the wound healing assay, the cells were specifically stained using CellTracker Green CMFDA (10 μmol/l in DMEM) or CellTracker Orange CMRA (10 μmol/l in DMEM) before the seeding process, according to the manufacturer's instructions (Invitrogen).
F. Scanning electron micrograph (SEM) observation
SEM was used to characterize the pillar morphology and observe the cells adhered on the substrate.13,41 Cells were cultured on the micropillar substrate for 24 h, fixed with 4% paraformaldehyde for 1 h, and then further rinsed thrice with PBS. Dehydration was performed by rinsing the samples through graded ethanol/water mixtures (50%, 70%, 80%, 90%, and 100%, each step for 10 min). Ethanol was slowly exchanged successively by amyl acetate and isoamyl acetate. Finally, samples were dried by using the critical point method and then sputter-coated by a thin layer of gold.39
G. Quantitative analysis of cell morphology
To study how the micropillars affect cells' morphology, cytoskeleton organization, and nuclear shape, a low cell density was seeded for each pillar array. After 24 h culturing, a quantitative analysis of the cell morphology was performed by measuring the projected spreading areas, aspect ratio, and circularity of cells on each substrate. Generally, cell areas, perimeters, and aspect ratio were analyzed using software Image-Pro Plus 6.0 and were used to calculate circularity, as a measure of the degree of branching in cell shape, following the studies reported previously.12,18 The circularity was defined as 4πA/P2, where A is the projected spreading area of the cell, and P is the perimeter.
H. Microscopy and image analysis
An inverted microscope (Olympus, CKX41) with a CCD camera (QIMAGING, Micropublisher 5.0 RTV) and a mercury lamp (Olympus, U-RFLT50) was used to acquire phase contrast and fluorescence images. Cell numbers adhered to the micropillar substrate were counted using Image-Pro Plus 6.0 (IPP 6.0) software (Media Cyternetics, Silver Spring, MD). Time lapse images of cell migration were obtained at 0, 4, 12, 24, and 36 h, and cell migration distances were measured using IPP 6.0 software. SPSS 12.0 (SPSS, Inc.) was employed to perform data statistical analysis. Each experiment was repeated at least three times, and the results, including the error bars in the graphs, were given as the mean ± standard deviation.
III. RESULTS AND DISCUSSION
A. Device design, fabrication, and characterization
In the current study, we described a microfluidic device that could be used for the investigation of cell response to various patterned micropillar substrates, and for modeling an identical injury-free wound by simply peeling off an integrated PDMS stencil (Figure 1). Generally, the device was composed of four layers, a PDMS stencil, a PDMS micropillar substrate, a thin PDMS membrane (not shown), and a polystyrene culture dish (Figures 1(a) and 1(b)). The micropillar substrate (3 mm wide and 9 mm long) consisted of three sets of micropillar arrays, separated by two blank spaces (800 μm in width). The heights of the micropillars in different arrays were all 10 μm. From left to right, the micropillar diameter and edge to edge spacing in each array were 15, 18, and 21 μm and 15, 18, and 21 μm, respectively. The specific geometric dimensions of micropillars in the array were chosen to expect to deform the cell morphology because they are close to cells sizes.12,13,23 For clarity, the diameter of the micropillars and spacing between neighbor micropillars in the different arrays were denoted as d15, d18, and d18 μm and s15, s18, and s21 μm, respectively, in the next description. SEM image showed that the micropillars were well arranged on the substrate (Figures 1(e) and 1(f)). The PDMS stencil on the micropillar substrate corresponding to the pattern of the micropillar substrate, contained three blanks (3 mm wide and 9 mm long), and the distance between adjacent blanks was 800 μm, indicating that the maximum cell migration distance was 800 μm. By using the PDMS stencil, the edge of the PDMS strip in 800-μm width was right aligned at the border of the micropillars, so that when the PDMS stencil was peeled off, the border of the micropillars was just the starting line from which cells migrated. In addition, each type of cells could be just located at their right position (Figure 1(c)) and no liquid leakage or cell cross contamination occurred. The device was bonded in a polystyrene culture dish by a pre-coated PDMS membrane for supplying culture medium.
B. Optimization of cell seeding density for the co-culture study
To find an optimized cell seeding density for the study of cell migration during wound healing assay, each type of cells at different densities (1 × 105 cells/ml, 5 × 105 cells/ml, and 1 × 106 cells/ml) was, respectively, seeded into their culture regions. To clearly observe cell coverage rate, after 24-h culturing, cells were stained using AO/PI. The results showed that after 24-h incubation, none of these cells seeded at 1 × 105 cells/ml and 5 × 105 cells/ml could completely fill the migrating region (Figures S1–S3 in the supplementary material).51 ESF-1 cells seeded at 1 × 106 cells/ml reached 80% confluence (Figures S2 and S4 in the supplementary material)51 on the d18-μm micropillar substrate. At the same cell density, HUVEC cells almost completely covered the seeding area (more than 90% confluence), regardless of the sizes of micropillars (Figures S3 and S4 in the supplementary material).51 However, HaCaT cells seeded at 1 × 106 cells/ml were far from covering the whole migration region and the cell coverage rate was nearly 30% (Figures S1 and S4 in the supplementary material),51 so a high seeding density of 3 × 106 cells/ml was used for HaCaT cells in the next experiments, intending to completely cover the corresponding region. Based on a comprehensive analysis of these results, HaCaT cells at 3 × 106 cells/ml, ESF-1 and HUVEC cells at 1 × 106 cells/ml were, respectively, used for the next wound healing assay. In addition, we found that cells seeded at a low concentration of 1 × 105 cells/ml were sparsely distributed and suitable for the study of cell morphology, cytoskeleton organization, and nuclear shapes on the micropillar substrates.
C. Effect of the micropillars on cell attachment and proliferation
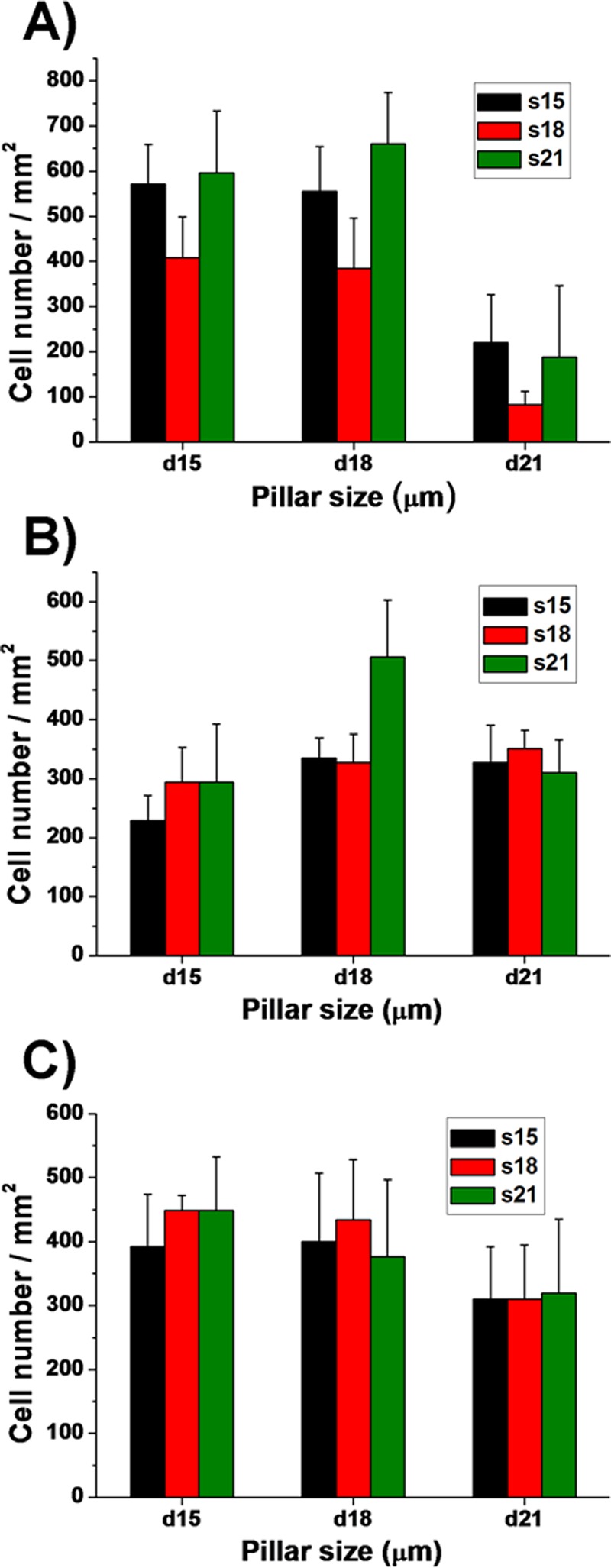
During the seeding density optimization test, we found that different types of cells showed different adhesion performance on various micropillar substrates. Both HaCaT and HUVEC cells showed preferential adhesion to d15 and d18-μm pillars rather than to the d21-μm pillars, regardless of the interspacing between micropillars, especially for HaCaT cells (Figure 2, and Figures S5 and S6 in the supplementary material51). From the statistical data, we could clearly see that there was almost 400–600 HaCaT cells/mm2 on the d15 and d18-μm pillar substrates, only 100–250 HaCaT cells/mm2 on the d21-μm pillar substrate at the same seeding density of 1 × 106 cells/ml (Figure 2(a)). Similarly, HUVEC cells on the d21-μm pillar substrate were about 150 cells/mm2 (Figure 2(c)), less than those on the d15 and 18-μm pillar substrates. In addition, few HaCaT cells were attached on the s18-μm micropillar substrate, in comparison with the other spacing (Figure 2(a)). Compared with HaCaT and HUVEC-C cells, fibroblast ESF-1 showed a rather different response to these micropillars. ESF-1 cells preferred to adhere to d18 and d21-μm micropillar substrates, rather than to the d15-μm pillar substrate (Figure 2(b) and Figure S7 in the supplementary material51). These results may be due to the different cell sizes and origin.42 According to what we have measured, the mean diameter of ESF-1 cells is 22 μm and almost in a round shape after just trypsinization, which is larger than HaCaT and HUVEC cells. So the d15-μm micropillar substrate may be not suitable for ESF-1 cells to attach. These results may also indicate that cells could send local protrusions to probe the physical properties of the environment and later anchor themselves on the substrates that are suitable for their growth and proliferation.42,43 In general, we found that the d18-μm micropillar substrate could prompt more cell adhesion and proliferation than the other substrates. In addition, cells on the d18-μm micropillar substrate showed higher cell viability and better morphology than those on the other substrates. As a result, we chose the d18-μm micropillar substrate to do the follow-up wound healing studies.
FIG. 2.
Quantitative assessment of the cell attachment on the different micropillar substrates. Histograms present cell numbers of (a) HaCaT, (b) ESF-1, and (c) HUVEC cells. Cell numbers were obtained by manual counting of AO stained cells. For clarity, the different diameters (15, 18, and 21 μm) and spacing (15, 18, and 21 μm) of the micropillars were denoted as d15, d18, and 21 μm and s5, s18 and s21 μm, respectively.
D. Effect of the micropillars on cell morphology, cytoskeleton organization, and nuclear shape
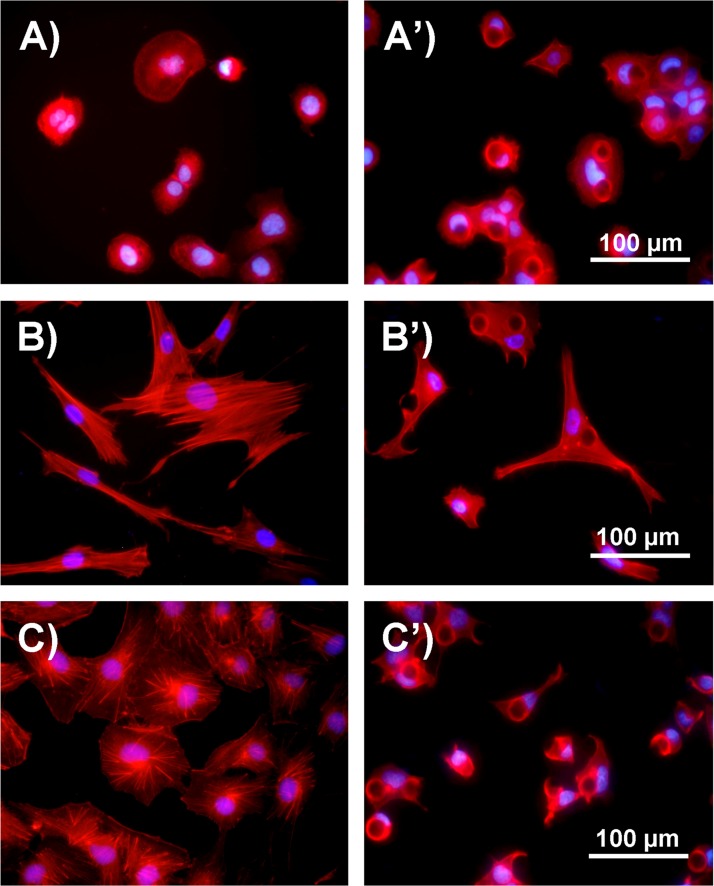
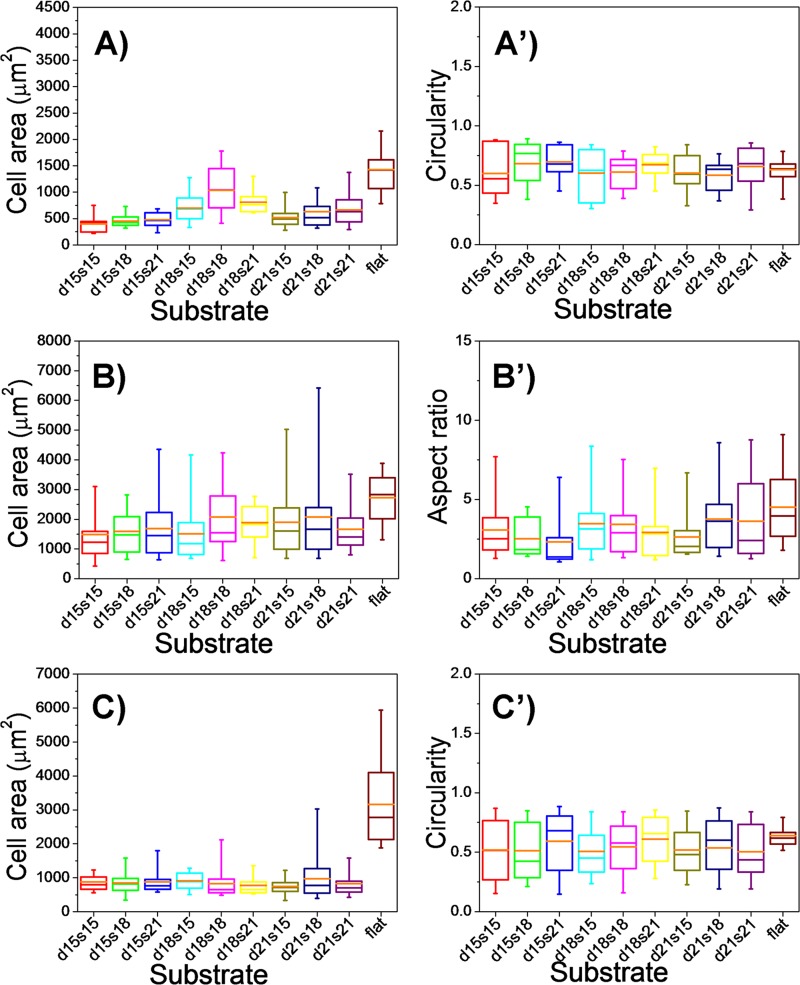
From the study above, we found that all the three types of cells showed characteristic morphologies after 24-h culture on different micropillar substrates. Generally, cells were less spread out on the micropillar substrates compared with those on the flat surface (Figure 3), which was further confirmed by the quantitative data (Figures 4(a), 4(b), and 4(c)). The cell areas on the micropillar substrates were calculated by those projecting to the two-dimensional flat surface, which were much smaller than those on the flat surface, indicating cell morphology response to the topographical surface feature.12,18 Cells on the micropillar substrates mainly exhibited two kinds of morphologies. One irregularly spread in the gaps of micropillars and the other adhered on the top of the micropillars and most exhibited a round or cap-like shape (Figures S8–S10 in the supplementary material).51 This phenomenon was also verified by the statistic data (Figures 4(a')–4(c')). The circularity value of cells on the micropillar substrates showed a much broader range than that on the flat substrate, and it was obviously that there were mainly two concentrated circularity values among all the cells on the micropillar substrates, one at near 0.3 and the other at near 0.8 (Figure S11 in the supplementary material).51 Concerning the cells cultured on different micropillar substrates, cells on the d18s18-μm micropillar substrate always had a larger spreading area than those on the other micropillar substrates. In addition, the circularity value of HaCaT and HUVEC cells on the s21-μm micropillar substrate was always larger than those on the s15- and s18-μm micropillar substrates, and large proportion of cells on the s21-μm micropillar substrate showed a round shape (Figures S8 and S10 in the supplementary material).51 For ESF-1 cells, the mean value of aspect ratio on the flat surface was larger than that on the pillar patterned substrates (Figure 4(b')). The reason is that cells on the micropillar substrates extended lots of branches according to the micropillar pattern, while cells on the flat substrate were just in elongated shape (Figures 3(b) and 3(b')).
FIG. 3.
Comparison of cell morphology and cytoskeleton after 24-h culture on the flat and micropillar substrates. (a) Fluorescence image of HaCaT cells cultured on the flat substrate. (a') Fluorescence image of HaCaT cells cultured on the d18s18-μm micropillar substrate. (b) Fluorescence image of ESF-1 cells cultured on the flat substrate. (b') Fluorescence image of ESF-1 cells cultured on the d18s18-μm micropillar substrate. (c) Fluorescence image of HUVEC cells cultured on the flat substrate. (c') Fluorescence image of HUVEC cells cultured on the d18s18-μm micropillar substrate. Cells were stained for actin filament (red) and nuclei (blue).
FIG. 4.
Quantitative analysis of cell morphology on different substrates, including the flat substrate and micropillar substrates with different diameters (15, 18, and 21 μm) and spacing (15, 18, and 21 μm). (a) Spreading area and (a') circularity of HaCaT cells on different substrates. (b) Spreading area and (b') aspect ratio of ESF-1 cells on different substrates. (c) Spreading area and (c') circularity of HUVEC cells on different substrates. Orange bars within box plots indicate the mean values.
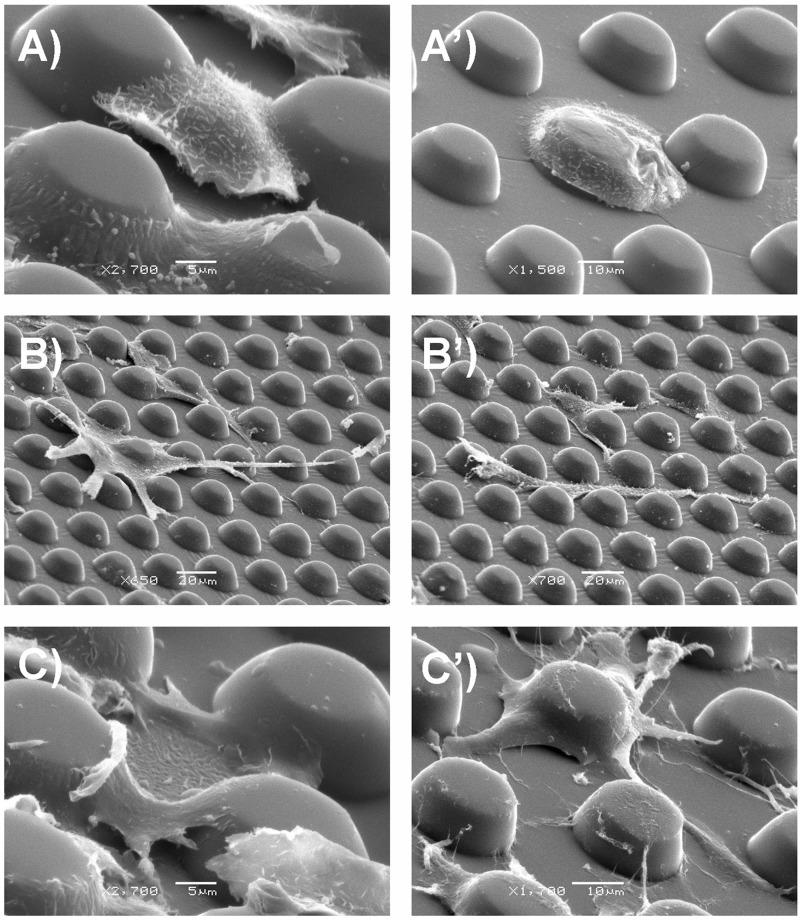
SEM observation showed that micropillar interspaces also have influence on cell positioning and spreading. On the small spacing (15 μm) pillar surface, cells tended to suspend from pillar to pillar (HaCaT and HUVEC cells) or locate on the top of pillars and cover two or three pillars, extending multidirectional protrusions (ESF-1 cells) and anchoring to the surrounding pillars (Figures 5(a)–5(c)). In contrast, on the large spacing (21 μm) pillar surface, cells inclined to adhere to one pillar first and then extend down to the flat part around the pillars (HaCaT cells), extend cell protrusions to the surrounding pillars (HUVEC cells), or just spread bypassing the pillars (ESF-1 cells) (Figures 5(a')–5(c')). The result indicated that it was difficult for the cells on the micropillars with large interspacing to establish connection with the others on the surrounding micropillars. In addition, we also observed thick lamellipodia (Figure S12A in the supplementary material)51 and filopodia (Figures S12B and S12C in the supplementary material)51 for cells (especially for ESF-1 and HUVEC cells) on the micropillar substrates, in contrast to the large and thin lamellipodia for that on the flat substrate (Figure S12D in the supplementary material),51 suggesting that these cell protrusions had a close relationship with cell positioning and migration.44
FIG. 5.
SEM images of the three types of cells on d18s15-μm (a)–(c) and d18s21-μm (a')–(c') micropillar substrates. (a) and (a') HaCaT cells. (b) and (b') ESF-1 cells. (c) and (c') HUVEC cells.
In general, pronounced modifications in the cytoskeleton organization, nuclear orientation, and shape were observed for the cells cultured on the micropillar substrates, compared with those on the flat surface. On the flat substrate, cells presented many prominent, highly organized, and uniformly distributed stress fibers which were either radially arranged from the cell bodies (HaCaT and HUVEC cells) or aligned parallel to the long axis of the cells (ESF-1 cells). However, on the micropillar substrates, cells showed disorganized, multidirectional, and obscure fibers which were arranged according to the pattern of the micropillars, and many intracellular actin rich rings that outlined the top surface of the upright micropillars with which cells were in contact. One reason for the presence of these actin thick rings may be that during spreading, the cell body intended to wrap around neighboring pillars from the side walls that they came across (Figure S13 in the supplementary material);51 another possible reason may be that cell plasma covering on the micropillars is thinner than that of the other parts. Concerning the cell nuclei, on the flat substrate, nuclei were almost distributed in the center of cell body, while the orientation of those on the micropillar substrate were strongly influenced by the location of the surrounding micropillars, which were mainly located in the gaps of micropillars (Figure 3). Furthermore, in contrast to the round or oval nuclei shapes on the flat substrate, nuclei of cells on the micropillar substrates took on many distinct shapes, including crescent, cross-like, and dumbbell, which was deformed by the micropillars to match features of the underlying surface topography, as previously investigated.45–47 This phenomenon was obviously observed for HaCaT and HUVEC cells cultured on the micropillar substrates (Figures S14 and S15 in the supplementary material).51 However, only a few of ESF-1 cell nuclei were deformed slightly (Figure S16 in the supplementary material).51
In addition, we also observed distinct cell responses to micropillars with different heights. HUVEC cells cultured on the pillar substrates with a low height (just tiny bumps on the substrates) showed regular cell shapes with some prominent and ordered stress fibers, but no nucleus deformation (Figure S17 in the supplementary material),51 which was more alike to those on the flat surfaces, but quite different from those on the micropillar substrate with a high height (e.g., 10 μm), implying that the height of the micropillars had an effect on cell morphology, cytoskeleton organization, and nuclear shapes. In summary, cells are less spread out on the micropillar substrate compared with those on the plane, indicating that cell spreading was confined by the micropillars. Different types of cells showed different responses to the micropillars with different sizes because of their different cell sizes and morphologies. Actin cytoskeleton would be rearranged according to the underlying surface topography which accounted for the dramatic cell morphology changes.42,48 Nuclei deformation is most likely the result of a balance between the rigidity of the nucleus and the force that the cytoskeleton is able to exert on it.23 All together, these results confirmed that cells could interact with the micropillar substrates and respond dynamically and spatiotemporally to the surrounding extracellular matrix.
E. Cell-cell interactions during wound healing
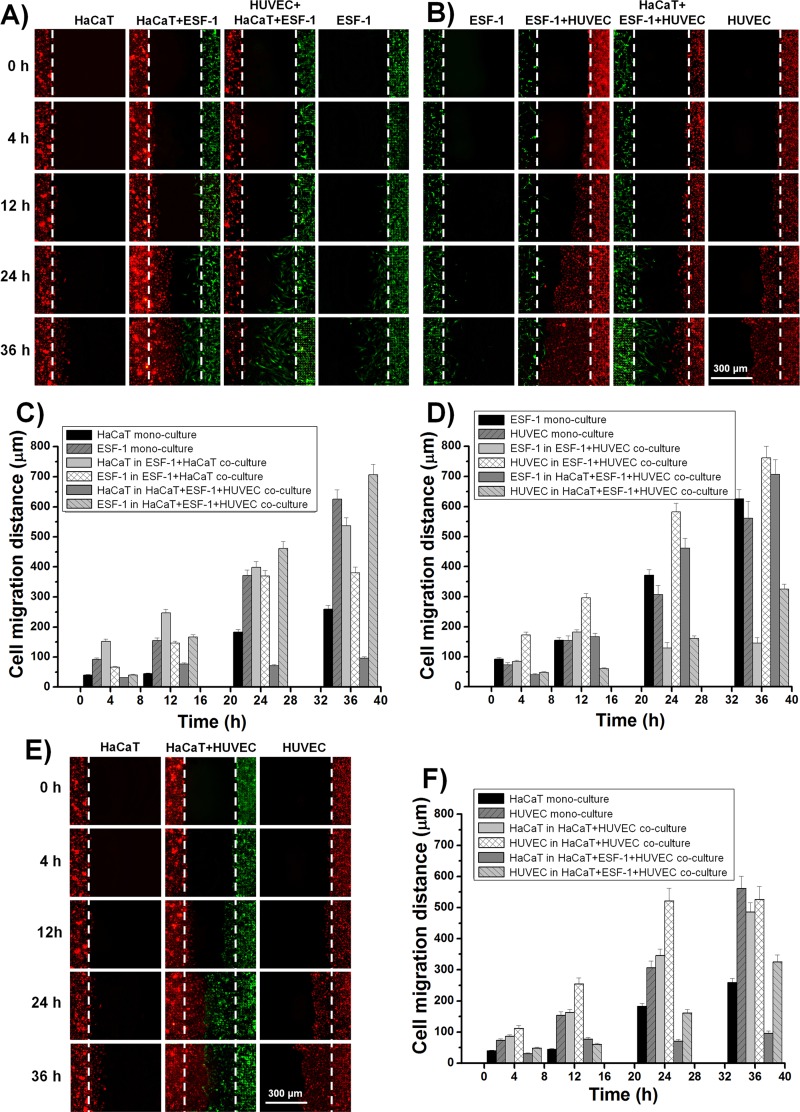
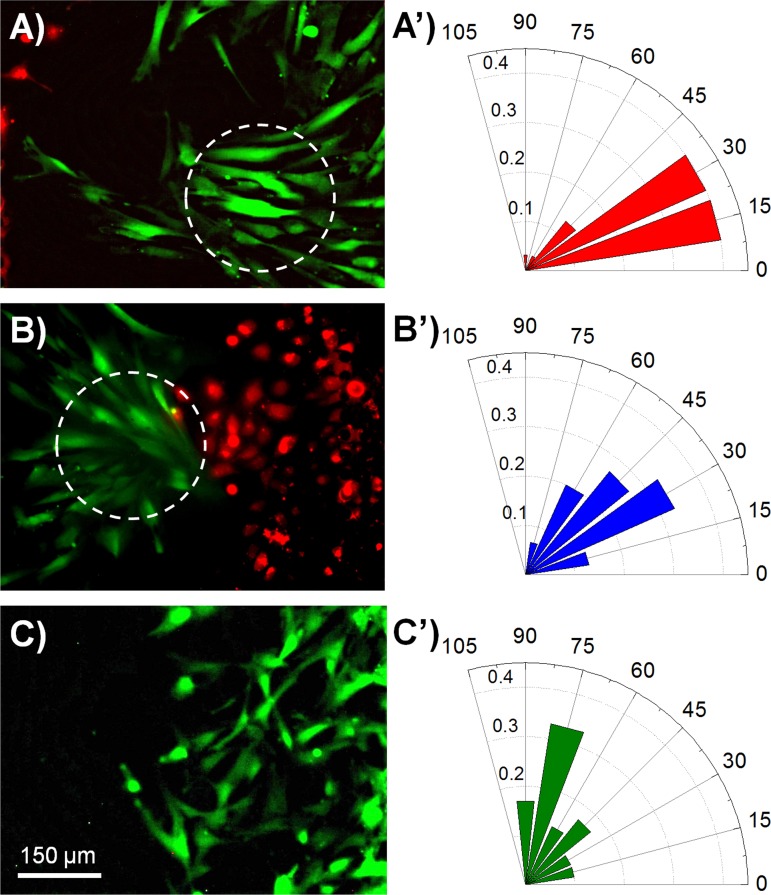
To clearly distinguish different types of cells and dynamically follow their tracks when they were co-cultured, in this part of study, cells were stained with different CellTracker dyes (HaCaT cells were stained with CellTracker Orange, ESF-1 cells with CellTracker Green, and HUVEC cells were stained with CellTracker Orange or CellTracker Green) before cell loading (Figures S18A–S18C in the supplementary material).51 Afterwards, cells were trypsinized, adjusted to the appropriate density (HaCaT, 3 × 106 cells/ml, ESF-1 and HUVEC cells, 1 × 106 cells/ml) and then seeded into the appropriate regions (HaCaT in the left region, ESF-1 in the middle region, and HUVEC in the right region of the device). Simultaneously, six control groups were also carried out, namely, mono-culture of each type of cells, co-culture of HaCaT and ESF-1 cells (HaCaT in the left region and ESF-1 in the middle region of the device), co-culture of ESF-1 and HUVEC cells (ESF-1 in the middle region and HUVEC in the right region of the device), and co-culture of HaCaT and HUVEC cells (HaCaT and HUVEC in any two adjacent regions of the device). Thus, we could independently and comparatively study the cell-cell interaction among these cells during the in vitro wound healing process (Figures 6(a), 6(b), and 6(e)). After 24-h culturing, cells mainly covered their corresponding regions (Figures S18D–S18F in the supplementary material).51 Then, the PDMS stencils were gently peeled off and cells were allowed to migrate freely towards the wound (Figure S19 in the supplementary material).51 Different time points (0, 4, 12, 24, and 36 h)27 were selected to quantitatively analyze cell migration. After 36 h, the 800-μm injury-free wound was almost healed in the co-culture groups. Cell migration distance (the distance between the leading edge and initial margin)49 was calculated under various conditions (Figures 6(c), 6(d), and 6(f)). For clear and comparative data analysis, we divided these tests into three groups: (i) HaCaT and ESF-1 mono-culture and their co-culture (Figures 6(a) and 6(c)), (ii) ESF-1 and HUVEC mono-culture and their co-culture (Figures 6(b) and 6(d)), and (iii) HaCaT and HUVEC mono-culture and their co-culture (Figures 6(e) and 6(f)). For the first group, the migration distance of HaCaT cells during HaCaT and ESF-1 co-culture was much longer than that of HaCaT in mono-culture at each time point, confirming that the proliferation and migration of keratinocytes were enhanced when co-cultured with fibroblasts,7 while the migration distance of ESF-1 cells during HaCaT and ESF-1 co-culture was close to that in the ESF-1 mono-culture except at 36 h (Figure 6(c)). With regard to the second group, ESF-1 cells during ESF-1 and HUVEC co-culture migrated slower than or equal to the mono-cultured ESF-1 cells (Figure 6(d)), while HUVEC cells in the ESF-1 and HUVEC-C co-culture migrated much faster than the mono-cultured HUVEC cells. The result indicates that fibroblasts stimulate the proliferation and migration of endothelial cells.6 Concerning the third group, both HaCaT and HUVEC cells in the HaCaT and HUVEC co-culture migrated much faster than each of them in their mono-culture (Figures 6(e) and 6(f)). However, in the co-culture of HaCaT, ESF-1, and HUVEC cells, the migration distances of both HaCaT and HUVEC cells were much shorter than each of them in mono-culture, whereas the migration distance of ESF-1 cells was much longer than that in the ESF-1 mono-culture, which indicated that fibroblasts accounted for a large proportion in the cells to heal the wound and made a great contribution to wound healing.6 Although this is inconsistent with the above results in two types of cell co-culture, it is possibly close to the real soft tissue wound healing process in vivo.4 These different results are mainly due to the cell-cell interactions. In detail, we observed distinct migration characteristics during wound healing, that is, cells exhibited directional migration when they were co-cultured (Figures S20 and S21 in the supplementary material).51 This phenomenon was especially obvious for ESF-1 cells. ESF-1 cells co-cultured with HaCaT and HUVEC cells gathered into a cluster and then migrated towards the wound following the leader cells, almost in the horizontal direction (Figures 7(a) and 7(b)); however, when ESF-1 cells were mono-cultured, they just migrated diffusively and random towards the wound (Figure 7(c)). This was specifically confirmed by computing the angle between the long axes of the cell and the horizontal line (Figures 7(a')–7(c')).50 Summarily, as the results showed, the inducible motility of one kind of cells occurred in the presence of another heterogeneous cells located at the opposite side of the contact region.
FIG. 6.
Dynamic cell migration under different conditions after the wound. (a) Dynamic migration of HaCaT and ESF-1 cells in their mono-culture and co-culture. (b) Real-time migration of ESF-1 and HUVEC cells in their mono-culture and co-culture. (e) Dynamic migration of HaCaT and HUVEC cells in their mono-culture and co-culture. (c), (d), and (f) Quantitative analysis of cell migration distances during (a), (b), and (e). The initial leading edges (shown as white dashed lines) represent baselines for the migration assay.
FIG. 7.
Comparison of the migration direction of ESF-1 cells in the co-culture and mono-culture during the wound healing. (a) Fluorescence image of ESF-1 cells during HaCaT, ESF-1, and HUVEC cell co-culture, showing ESF-1 cells migrated towards HaCaT cells. (b) Fluorescence image of ESF-1 cells during HaCaT, ESF-1, and HUVEC cell co-culture, showing that ESF-1 cells migrated towards HUVEC cells. (c) Fluorescence image of ESF-1 cells during their mono-culture, showing that ESF-1 cells migrated towards the wound. (a')–(c') The orientation of ESF-1 cells in the co-culture and mono-culture during the wound healing, corresponding to (a), (b), and (c), respectively. The direction of each bar in the rose plots indicates the angular ESF-1 cell orientation, whereas the magnitude of each bar shows the fraction of cells with the indicated ESF-1 cell orientation.
In addition, during the wound healing process in vitro, we also noticed different cell behaviors between the cells on the micropillar substrates and on the flat surfaces (Figure S22 in the supplementary material).51 Clearly, when ESF-1 cells transmigrated from the micropillars onto the flat surface, their morphology took a dramatic change and further, the long axis of the cells which had just moved out from the micropillars were either perpendicular or parallel to the wound. This phenomenon is consistent with in vivo wound healing process during which cells transmigrated through the 3D extracellular matrices to heal the wound.12 In addition, HUVEC cells that appeared to be crawling onto the micropillars from the flat surface were captured to take on a more 3D state, which was in accordance with the previous study.12
IV. CONCLUSIONS
In this study, we presented a microfluidic device for investigating cells response to the substrate with topographic cues and the interactions between heterogeneous cells. Using this device, we realized positional cell loading, co-culture of heterotypic cells, and controlled study of cell response to the substrate and cell-cell interactions. Concerning the cell response to substrates, our findings confirmed that cell attachment, morphology, cytoskeleton organization, and nucleus shape are all strongly affected by the micropillar substrates. Afterwards, a wound-healing assay was conducted by co-culturing three types of physiologically related cells. The results suggested that epithelial cells, fibroblasts, and endothelial cells showed collaborative relationship to heal the wound. When the three types of cells were co-cultured together, fibroblasts made a greater contribution than the other two types of cells. In addition, cell motion was more directional in the co-culture condition. We believe that this work is helpful for the understanding of the wound healing process. Using this method, patterning of different types of cells on various types of substrates can be realized and the cells could sense and respond not only to the signals of other kinds of cells but also to the physical aspects of the substrate, such as stiffness and topographical cues, which is more akin to the in vivo situation. Additionally, this device could be downsized to compatible with different kinds of cell culture plates and applied for various biological researches such as embryogenesis, immune response, and tissue engineering. The further study based on the device is under way in our laboratory.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Grant Nos. 21175107 and 21375106) and the Fundamental Research Funds for the Central Universities (Grant No. Z109021423).
References
- 1. Shaw T. J. and Martin P., J. Cell Sci. 122, 3209–3213 (2009). 10.1242/jcs.031187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xie Y., Rizzi S. C., Dawson R., Lynam E., Richards S., Leavesley D. I., and Upton Z., Tissue Eng., Part C 16, 1111–1123 (2010). 10.1089/ten.tec.2009.0725 [DOI] [PubMed] [Google Scholar]
- 3. Moulin V., Auger F. A., Garrel D., and Germain L., Burns 26, 3–12 (2000). 10.1016/S0305-4179(99)00091-1 [DOI] [PubMed] [Google Scholar]
- 4. Singer A. J. and Clark R., N. Engl. J. Med. 341, 738–746 (1999). 10.1056/NEJM199909023411006 [DOI] [PubMed] [Google Scholar]
- 5. Fu X. B., Fang L. J., Li H. H., Li X. K., Cheng B. O., and Sheng Z. Y., Wound Repair Regener. 15, 540–548 (2007). 10.1111/j.1524-475X.2007.00262.x [DOI] [PubMed] [Google Scholar]
- 6. Oberringer M., Meins C., Bubel M., and Pohlemann T., Biol. Cell 99, 197–207 (2007). 10.1042/BC20060116 [DOI] [PubMed] [Google Scholar]
- 7. Wang Z., Wang Y., Farhangfar F., Zimmer M., and Zhang Y., PLoS One 7, e40951 (2012). 10.1371/journal.pone.0040951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghaffari A., Kilani R. T., and Ghahary A., J. Invest. Dermatol. 129, 340–347 (2008). 10.1038/jid.2008.253 [DOI] [PubMed] [Google Scholar]
- 9. Cukierman E., Pankov R., and Yamada K. M., Curr. Opin. Cell Biol. 14, 633–640 (2002). 10.1016/S0955-0674(02)00364-2 [DOI] [PubMed] [Google Scholar]
- 10. Guilak F., Cohen D. M., Estes B. T., Gimble J. M., Liedtke W., and Chen C. S., Cell Stem Cell 5, 17–26 (2009). 10.1016/j.stem.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poujade M., Grasland-Mongrain E., Hertzog A., Jouanneau J., Chavrier P., Ladoux B., Buguin A., and Silberzan P., Proc. Natl. Acad. Sci. U. S. A. 104, 15988–15993 (2007). 10.1073/pnas.0705062104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghibaudo M., Trichet L., Le Digabel J., Richert A., Hersen P., and Ladoux B., Biophys. J. 97, 357–368 (2009). 10.1016/j.bpj.2009.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baker D. W., Liu X., Weng H., Luo C., and Tang L., Biomacromolecules 12, 997–1005 (2011). 10.1021/bm1013487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehnert D., Wehrle-Haller B., David C., Weiland U., Ballestrem C., Imhof B. A., and Bastmeyer M., J. Cell Sci. 117, 41–52 (2004). 10.1242/jcs.00836 [DOI] [PubMed] [Google Scholar]
- 15. Nematollahi M., Hamilton D. W., Jaeger N. J., and Brunette D. M., J. Biomed. Mater. Res. A 91A, 149–157 (2009). 10.1002/jbm.a.32202 [DOI] [PubMed] [Google Scholar]
- 16. Su W.-T., Liao Y.-F., Lin C.-Y., and Li L.-T., J. Biomed. Mater. Res. A 93A, 1463–1469 (2010). 10.1002/jbm.a.32643 [DOI] [PubMed] [Google Scholar]
- 17. Ng C. K. M. and Yu K. N., Biointerphases 7, 1–8 (2012). 10.1007/s13758-012-0021-2 [DOI] [PubMed] [Google Scholar]
- 18. Frey M. T., Tsai I. Y., Russell T. P., Hanks S. K., and Wang Y., Biophys. J. 90, 3774–3782 (2006). 10.1529/biophysj.105.074526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu J., Wang Y., Yang M. T., Desai R. A., Yu X., Liu Z., and Chen C. S., Nat. Methods 7, 733–736 (2010). 10.1038/nmeth.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mussig E., Schulz S., Spatz J. P., Ziegler N., Tomakidi P., and Steinberg T., Eur. J. Cell Biol. 89, 315–325 (2010). 10.1016/j.ejcb.2009.08.004 [DOI] [PubMed] [Google Scholar]
- 21. Ghibaudo M., Di Meglio J., Hersen P., and Ladoux B., Lab Chip 11, 805–812 (2011). 10.1039/C0LC00221F [DOI] [PubMed] [Google Scholar]
- 22. Miyoshi H., Adachi T., Ju J., Lee S. M., Cho D. J., Ko J. S., Uchida G., and Yamagata Y., Biomaterials 33, 395–401 (2012). 10.1016/j.biomaterials.2011.09.094 [DOI] [PubMed] [Google Scholar]
- 23. Booth-Gauthier E. A., Du V., Ghibaudo M., Rape A. D., Dahl K. N., and Ladoux B., Integr. Biol. 5, 569–577 (2013). 10.1039/c3ib20231c [DOI] [PubMed] [Google Scholar]
- 24. Steinberg T., Schulz S., Spatz J. P., Grabe N., Mussig E., Kohl A., Komposch G., and Tomakidi P., Nano Lett. 7, 287–294 (2007). 10.1021/nl062271z [DOI] [PubMed] [Google Scholar]
- 25. Oberringer M., Meins C., Bubel M., and Pohlemann T., J. Mol. Histol. 39, 37–47 (2008). 10.1007/s10735-007-9124-3 [DOI] [PubMed] [Google Scholar]
- 26. Demirovic D. and Rattan S. I. S., Biogerontology 12, 437–444 (2011). 10.1007/s10522-011-9326-7 [DOI] [PubMed] [Google Scholar]
- 27. Seltana A., Basora N., and Beaulieu J., Wound Repair Regener. 18, 114–122 (2010). 10.1111/j.1524-475X.2009.00554.x [DOI] [PubMed] [Google Scholar]
- 28. Tremel A., Cai A., Tirtaatmadja N., Hughes B. D., Stevens G. W., Landman K. A., and O'Connor A. J., Chem. Eng. Sci. 64, 247–253 (2009). 10.1016/j.ces.2008.10.008 [DOI] [Google Scholar]
- 29. Xie Y., Zhang W., Wang L., Sun K., Sun Y., and Jiang X., Lab Chip 11, 2819–2822 (2011). 10.1039/c0lc00562b [DOI] [PubMed] [Google Scholar]
- 30. van der Meer A. D., Vermeul K., Poot A. A., Feijen J., and Vermes I., Am. J. Physiol. Heart Circ. Physiol. 298, H719–H725 (2010). 10.1152/ajpheart.00933.2009 [DOI] [PubMed] [Google Scholar]
- 31. Felder M., Sallin P., Barbe L., Haenni B., Gazdhar A., Geiser T., and Guenat O., Lab Chip 12, 640–646 (2012). 10.1039/C1LC20879A [DOI] [PubMed] [Google Scholar]
- 32. Murrell M., Kamm R., and Matsudaira P., PLoS One 6, e24283 (2011). 10.1371/journal.pone.0024283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schoop V. M., Mirancea N., and Fusenig N. E., J. Invest. Dermatol. 112, 343–353 (1999). 10.1046/j.1523-1747.1999.00524.x [DOI] [PubMed] [Google Scholar]
- 34. Shephard P., Martin G., Smola-Hess S., Brunner G., Krieg T., and Smola H., Am. J. Pathol. 164, 2055–2066 (2004). 10.1016/S0002-9440(10)63764-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Witte R. P. and Kao W. J., Biomaterials 26, 3673–3682 (2005). 10.1016/j.biomaterials.2004.09.054 [DOI] [PubMed] [Google Scholar]
- 36. Sorrell J. M., Baber M. A., and Caplan A. I., Cells Tissues Organs 186, 157–168 (2007). 10.1159/000106670 [DOI] [PubMed] [Google Scholar]
- 37. Lotters J. C., Olthuis W., Veltink P. H., and Bergveld P., J. Micromech. Microeng. 7, 145–147 (1997). 10.1088/0960-1317/7/3/017 [DOI] [Google Scholar]
- 38. Sniadecki N. J. and Chen C. S., Method Cell Biol. 83, 313–328 (2007). 10.1016/S0091-679X(07)83013-5 [DOI] [PubMed] [Google Scholar]
- 39. Liu W., Li L., Wang J., Tu Q., Ren L., Wang Y., and Wang J., Lab Chip 12, 1702–1709 (2012). 10.1039/c2lc00034b [DOI] [PubMed] [Google Scholar]
- 40. Mazia D., Schatten G., and Sale W., J. Cell Biol. 66, 198–200 (1975). 10.1083/jcb.66.1.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. du Roure O., Saez A., Buguin A., Austin R. H., Chavrier P., Siberzan P., and Ladoux B., Proc. Natl. Acad. Sci. U. S. A. 102, 2390–2395 (2005). 10.1073/pnas.0408482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anselme K. and Bigerelle M., Int. Mater. Rev. 56, 243–266 (2011). 10.1179/1743280411Y.0000000001 [DOI] [Google Scholar]
- 43. Dickinson L. E., Rand D. R., Tsao J., Eberle W., and Gerecht S., J. Biomed. Mater. Res. A 100A, 1457–1466 (2012). 10.1002/jbm.a.34059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang C., Czech L., Gerboth S., Kojima S., Scita G., and Svitkina T., PLoS Biol. 5, e317 (2007). 10.1371/journal.pbio.0050317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davidson P. M., Özçelik H., Hasirci V., Reiter G., and Anselme K., Adv. Mater. 21, 3586–3590 (2009). 10.1002/adma.200900582 [DOI] [Google Scholar]
- 46. Davidson P., Fromigué O., Marie P., Hasirci V., Reiter G., and Anselme K., J. Mater. Sci.: Mater. Med. 21, 939–946 (2010). 10.1007/s10856-009-3950-7 [DOI] [PubMed] [Google Scholar]
- 47. Pan Z., Yan C., Peng R., Zhao Y., He Y., and Ding J., Biomaterials 33, 1730–1735 (2012). 10.1016/j.biomaterials.2011.11.023 [DOI] [PubMed] [Google Scholar]
- 48. Doroudian G., Curtis M. W., Gang A., and Russell B., Biochem. Biophys. Res. Commun. 430, 1040–1046 (2013). 10.1016/j.bbrc.2012.11.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang X. W., Li L., Tu Q., Wang J. C., Liu W. M., Wang X. Q., Ren L., and Wang J. Y., Microfluid. Nanofluid. 10, 1333–1341 (2011). 10.1007/s10404-011-0766-9 [DOI] [Google Scholar]
- 50. Weiger M. C., Vedham V., Stuelten C. H., Shou K., Herrera M., Sato M., Losert W., and Parent C. A., PLoS One 8, e58859 (2013). 10.1371/journal.pone.0058859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.See supplementary material at http://dx.doi.org/10.1063/1.4936927E-BIOMGB-9-017506 for the detailed description of the additional experimental setup and results.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4936927E-BIOMGB-9-017506 for the detailed description of the additional experimental setup and results.