Abstract
Background and Objective:
Chlamydophila psittaci is a lethal bacterium that causes endemic avian chlamydiosis, and respiratory psittacosis. Laboratory diagnosis of Chlamydophila psittaci is difficult by culture. This study was design to investigate the presence of Chlamydophila psittaci in collected pharyngeal swabs from asyptomatic pigeons by PCR.
Materials and Methods:
Pharyngeal samples from pigeons with no symptoms of disease (n=280) were collected during hot and cold seasons in different parts of Ahvaz. DNA was extracted from specimens and subjected to PCR targeting pmp genes and 16s–23s rRNA intergenic spacer of Cp. psittaci and chlamydiales specific primers.
Results:
Of 280 samples 2 (0.7%) harbor were positive for chlamydiales (16s–23s intergenic spacer) and Cp. psittaci specific genes (pmp gene).
Conclusions:
In this research the pigeons were asymptomatic carriers for Cp. psittaci in their respiratory discharges. These results suggest that Cp. psittaci infection of human can occur in very close and continuous contact with pigeons.
Keywords: Chlamydophila psittaci, Pigeon, PCR
INTRODUCTION
The Chlamydiaceae family is composed of a group of obligate intracellular bacteria that are found worldwide. This family is classified in two genera: the genus Chlamydia (including the species C. muridarum, C. suis and C. trachomatis) and the genus Chlamydophila (including the species Cp. abortus, Cp. caviae, Cp. felis, Cp. pecorum, Cp. pneumoniae and Cp. psittaci) (1). Chlamydophila psittaci (Cp. psittaci), can infect 465 avian species in 30 avian orders, with at least 153 species in the order Psittaciformes (2). This bacterium can be transmitted as metabolically inactive particles called elementary bodies (EBs) (3) from pet birds to humans (4). Transmission of this atypical respiratory pathogen can occur through direct contact with infected birds, birds’ feces, the cloacae, nasal discharges, conjunctiva secretions causing respiratory disease in both mammals and birds (5). Transmission of Cp. psittaci from birds to humans, particularly to people at high risk like veterinarians, bird breeders and animal shopkeepers, has been reported before (6–8).
Chlamydophila psittaci has seven known genotypes (A-F and E/B) (9) and two non avian genotypes (M56 and WC). All genotypes can be transmitted to humans and cause psittacosis or parrot fever. Genotypes are distinguished by sequencing of the outer membrane protein A (ompA) gene (10). Cp. psittaci has the ability to remain infectious in the environment for months, presenting a variety of public health issues, including economically devastating outbreaks in poultry farms and occasionally severe pneumonia in humans (11). Chlamydiosis’ symptoms in humans are variable, ranging from no clinical signs at all to severe systemic disease (12).
According to OIE (World Organization for Animal Health) recommendations, avian chlamydiosis diagnosis requires either isolation and identification of the organism, or demonstration of a four-fold increase in specific humoral antibody, as well as typical clinical signs (13). Amongst the tools available to demonstrate chlamydial presence in the host, PCR is an applicable technique due to its high specificity and sensitivity. This method presents the advantages of being simple, fast, easy to standardize and more suitable and safer than culture for processing large numbers of specimens (16).
The available current conventional PCR protocols for detection of avian species use single copy genes such as the ompA gene (2–11,14) or ribosomal RNA genes (16S–23S) (15) as amplification targets. More recently, a new set of primers (CpsiA/CpsiB) targeting conserved pmp (polymorphic membrane protein) genes family of C. abortus has been suggested as an effective and improved tool for PCR detection of avian chlamydiosis (16).
This family of genes, unique to Chlamydiaceae, was highlighted in all the genomes sequenced to date. Even though their function is still unknown, Pmp proteins are predicted to be localized in the outer membrane. In this study, the use of the CpsiA/CpsiB primers was extended to represent chlamydial strains of the six major serovars (serovars A–F) in the samples collected in field.
The aim of this study was to determine the presence of Cp. psittaci in pigeon population in Ahvaz city, Khuzestan province by amplification of chlamydiales and Cp. psittaci species specific genes in PCR.
MATERIALS AND METHODS
Sampling and DNA extraction.
In the present study, 280 samples of pharyngeal swab from chuana fossa of pigeons in transport medium (18) in different areas of Ahvaz were collected during two seasons (hot and cold) and analyzed by PCR for detection of pmp genes of Cp. psittaci. Genomic DNA was extracted from swab samples. Pharyngeal swab material in transport medium were homogenized in 0.5 ml TE buffer (10mM Tris-hydrochloric acid, 1mM EDTA, pH 8) supplemented with dithiothreithol (2 percent) and boiled for 10 minutes. After centrifugation, aliquots of 50 to 100 μl of the supernatant were used in the PCR (19). The extracted DNA of each sample was kept frozen at −20°C until used. Cp. psittaci strain 6BC was used as positive control and a negative DNA control was performed by adding 1 μl of sterile ultrapure deionized water.
Gene amplification.
cIGS-1f/cGIS-2r (20) and CpsiA/CpsiB (2–16) protocols, which target 16S–23S rRNA intergenic spacer (chlamydiales) and pmp genes (Cp. psittaci), respectively, were used in this study. Genomic DNA purified from biological samples was firstly submitted to a CpsiA/CpsiB PCR amplification including Cp. psittaci 6BC strain DNA in the mix (equivalent to 100 CFU per reaction) in order to assess the presence of PCR inhibitors in these samples. For checking the DNA extraction protocol 16S–23S rRNA intergenic spacer and 18s rRNA primers were used for prokaryotic and eukaryotic ribosomal RNA, respectively (Table 1).
Table 1:
Genes studied and specific primers in this research
| Reference | Size(bp) | Sequence | Gene |
|---|---|---|---|
| Borel et al. 2006a | 352 | Forward: CAA GGT GAG GCT GAT GAC5′- Reverse: TCG CCT KTC AAT GCC AAG5′- |
Chlamydiales (16-23S spacer) |
| Laroucau et al. 2001 | 300 | Forward: ATG AAA CAT CCA GTC TAC TGG5′- Reverse: TTG TGT AGT AAT ATT ATC AAA5′- |
Chlamydophila psittaci (gene pmp) |
| Binnicker et al. 2004 | 488 | Forward: TCA AGA ACG AAA GTC GGA GG5′- Reverse: GGA CAT CTA AGG GCA TCA CA5′ |
eukaryotic rRNA18 s |
RESULTS
Using prokaryotic and eukaryotic ribosomal RNA specific primers, several DNA extracted from pharyngeal swabs and white blood cells were examined separately in PCR.
Out of 280 pharyngeal samples, 2 (0.71%) were positive for Cp. psittaci infection using PCR. Analysis of PCR products for presence of pmp gene of Cp. psittaci on agarose gel revealed a 300 bp fragment (Fig. 1). The positive control showed the expected amplification product specific for Cp. psittaci (300 bp). The chlamydials specific primers (cIGS-1f/cGIS-2r) amplified a 352bp fragment in positive samples and the positive control (Fig. 2). Since positive results have been obtained by PCR from asymptomatic birds, it has been concluded that pharyngeal swab is suitable for detection of chlamydial infections. The relation between season, age, and gender with PCR results have been shown in Table 2.
Fig. 1.
Results of the PCR assay for identification of Cp.psittaci gene pmp:
− : negative control
+ : positive control strains of Cp. psittaci
Numbers 1,2: positive samples with pmp gene (300bp)
Numbers 3,4,5,6,7,8: negative samples without pmp gene
LA: 100bp ladder.
Fig. 2.
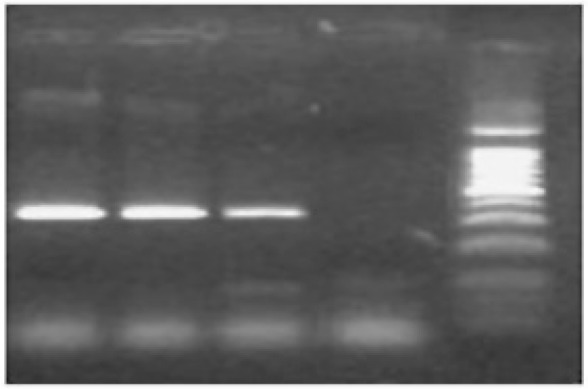
Results of the PCR assay for identification of chlamydials 16-23S spacer gene.
- - Ladder 100bp
- - Negative control
- - Positive control strains of Cp. psittaci (352bp)
- - Two positive samples with 16-23S spacer gene.
Table 2.
Analysis of results based on gender, age and season
|
PCR |
Season |
PCR |
Age |
PCR |
Gender | |||
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | |||
| 138 | 2 | Hot | 167 | 2 | 1> | 156 | 1 | Male |
| 140 | - | Cold | 111 | - | 1< | 122 | 1 | Female |
| 278 | 2 | Total | 278 | 2 | Total | 278 | 2 | Total |
In this study the pigeons were asymptomatic carriers for Cp. psittaci in their respiratory excretions.
DISCUSSION
The Chlamydiaceae are etiological agents of many important human and animal diseases (21). Chlamydophila psittaci is a causative agent of psittacosis, systemic diseases in psittacine birds which can be acute, protracted, chronic, or subclinical manifestation that represents the most important animal chlamydiosis of zoonotic character (22). Furthermore, from 1941 to 2003, 78 cases in humans (23) and from 2005 to 2009, 66 human cases of psittacosis were reported to the Centers for Disease Control and Prevention (CDC) due to contact with feral pigeons (2–5). Polymerase chain reaction has been documented as a highly sensitive method for the detection of Cp. psittaci (25,26). CpsiA/CpsiB, which were primarily designed from the Cp. abortus pmp genes (27) and described as being able to detect four different avian strains (2–16), are suitable for amplification of the 7 pmp related Cp. psittaci 6BC genes (data not shown). The total number of pmp genes for Cp. psittaci is still unknown. With a set of primers such as CpsiA/CpsiB, which targets several genomic fragments, the bacterial dilution effect is limited. Besides, the short size of the PCR products is an additional advantage for achieving better sensitivity results. All of the samples were amplified with CpsiA/CpsiB primers. On clinical samples, designed primers are the best among those tested for detection of Cp. psittaci by simple conventional PCR. RFLP experiments performed using PCR fragments amplified with the CpsiA/CpsiB primers gave promising results, demonstrating that these primers may provide an interesting tool for molecular typing when the bacterium cannot be grown from pathological samples (2–17). In several studies, polymerase chain reaction and PCR-based methods were used to detect chlamydial DNA in pharyngeal, faecal, genital secretion samples and semen (28). A nested PCR-enzyme immunoassay (PCR-EIA) was developed to detect the Cp. psittaci outer membrane protein A (ompA) gene in pharyngeal swabs. The ompA nested PCR-EIA was more sensitive than the 16S-rRNA based nested PCR and isolation, revealing 105 out of 200 (52.5%) positives against 13 and 74 for the latter two tests, respectively. Teankum et al. examined the prevalence of chlamydial infection in semen and genital tracts of boars. They evaluated suitable PCR assays with bacteriology, LPS-ELISA, and immunohistochemistry (IHC). Three Chlamydias pecific PCR assays targeting the 16S rRNA gene and the flanking IGS were compared. DNA dilution experiments revealed that 16Sig and IGS-S Chlamydia-specific PCRs were at least 10-fold more sensitive than the IGS-L PCR and serological and immunohistochemistry techniques (29).
Olsen et al. (1998) examined 312 fecal samples from 18 bird species to investigate to what extent wild passerine birds are carriers of Cp. psittaci. By using the PCR technique and subsequent DNA sequencing, they demonstrated Cp. psittaci DNA in fecal samples from nine (2.9%) birds of six different species. In this study, Cp. psittaci DNA was detected in 2 (1.4%) of 280 pharyngeal samples. The technique offers advantages over other testing methods in that it is fairly rapid and does not require organisms to be present in large numbers or to be viable (30).
Heddema et al. determined the prevalence and genotype of Cp. psittaci in fresh fecal samples from feral pigeons in Amsterdam, The Netherlands. The prevalence was 7.9% overall (26/331; 95% confidence interval, 5 to 11). Ten genotyped PCR-positive samples were all genotype B (31). Also, Sareyyupoglu et al. investigated the shedding of Cp. psittaci in fecal samples from 47 cage birds using PCR testing in Turkey. Following PCR with Cp. psittaci specific primers, 43 (91.5%) samples were determined to harbour-specific DNA (32). Doosti and Arshi (2011) determined the prevalence of Chlamydophila psittaci in pigeon feces in Iran using PCR assay. DNA was extracted from 445 fecal samples of pigeons. The prevalence of this pathogen was 14.3% in the region of this study (33).
In a study conducted by the Pantchev et al. (2009), real-time PCR assays revealed 71 cases (92.2%) of Cp. psittaci infection in specimens from parrots and parakeets. Therefore, a suitable method was designed to research on chlamydials infections (34), The difference observed in the results can be related to host species (wild or domestic), physiological status (with or without symptoms), maintenance (free or cage), sampling, technique and geographical area.
In conclusion, we detected a lower prevalence of Cp. psittaci DNA from pharyngeal sample of pigeons with PCR assay in Ahvaz, which indicates the risk for veterinarians, employees and operators of aviaries and pet shops, and pet owners regarding exposure to the disease. This is the first study in Iran regarding investigation of shedding of Cp. psittaci in pharyngeal samples using PCR. We hope it will provide an alternative and easier method for the routine diagnostic laboratories working in the field of avian medicine.
ACKNOWLEDGEMENT
We thank Prof. Baris Sareyyupoglu (Department of Microbiology, Faculty of Veterinary Medicine, Ankara, Turkey) for his kind support in providing us the control Cp. psittaci DNA.
REFERENCES
- 1. Everett KD, Andersen AA. Identification of nine species of the Chlamydiaceae using PCR-RFLP. Int J Syst Bacteriol 1999; 49 (Pt 2); 803–813. [DOI] [PubMed] [Google Scholar]
- 2. Vanrompay D, Harkinezhad T, van de Walle M, Beeckman D, van Droogenbroeck C, Verminnen K, et al. Chlamydophila psittaci transmission from pet birds to humans. Emerging Infectious Diseases 2007; 13: 1108– 1110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Binet R, Maurelli AT. Frequency of development and associated physiological cost of azithromycin resistance in Chlamydia psittaci 6BC and C. trachomatis L2. Antimicrob Agents Chemother 2007; 51: 4267– 4275 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnston WB, Eidson M, Smith KA, Stobierski MG. Compendium of chlamydiosis (psittacosis) control, 1999. Psittacosis Compendium Committee, National Association of State Public Health Veterinarians. J Am Vet Med Assoc 1999; 214: 640– 646 . [PubMed] [Google Scholar]
- 5. Smith K.A., Bradley K.K., Stobierski M.G., Tengelsen L.A. Compendium of measures to control Chlamydophila psittaci (formerly Chlamydia psittaci) infection among humans (psittacosis) and pet birds. J Am Vet Med Assoc 2005; 226: 532– 539 . [DOI] [PubMed] [Google Scholar]
- 6. Huminer D, Samra Z, Weisman Y, Pitlik S. Family outbreaks of psittacosis in Israel. Lancet 1988; 2 (8611): 615– 618. [DOI] [PubMed] [Google Scholar]
- 7. Hinton DG, Shipley A, Galvin JW, Harkin JT, Brunton RA. Chlamydiosis in workers at a duck farm and processing plant. Aust Vet J 1993; 70; 174–176. [DOI] [PubMed] [Google Scholar]
- 8. Saito T, Ohnishi J, Mori Y, Iinuma Y, Ichiyama S, Kohi F. Infection by Chlamydophilia avium in an elderly couple working in a pet shop. J Clin Microbiol 2005; 43: 3011– 3013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geens T, Desplanques A, Van Loock M, Bonner BM, Kaleta EF, Magnino S. Sequencing of the Chlamydophila psittaci ompA gene reveals a new genotype, E/B, and the need for a rapid discriminatory genotyping method. J Clin Microbiol 2005; 43; 2456–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vanrompay D, Butaye P, Sayada C, Ducatelle R, Haesebrouck F. Characterization of avian Chlamydia psittaci strains using omp1 restriction mapping and serovar-specific monoclonal antibodies. Res Microbiol 1997; 148: 327– 333 . [DOI] [PubMed] [Google Scholar]
- 11. Kaltenboeck B, Kousoulas KG, Storz J. Detection and strain differentiation of Chlamydia psittaci mediated by a twostep polymerase chain reaction. J Clin Microbiol 1991; 29 : 1969– 1975 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersen AA, Vanrompay D. Avian chlamydiosis. Rev Sci. Tech 2000; 19: 396– 404 . [PubMed] [Google Scholar]
- 13. OIE Manual of diagnostic test and vaccines for terrestrial animals (mammals, birds and bees). 2004, Fifth edition. [PubMed] [Google Scholar]
- 14. Sayada C, Andersen AA, Storey CC, Milon A, Hashimoto N, Hirai K, et al. Usefulness of omp1 restriction mapping for avian Chlamydia psittaci isolate differentiation. Res Microbiol 1995; 146: 155–165. [DOI] [PubMed] [Google Scholar]
- 15. Everett KD, Andersen AA. The ribosomal intergenic spacer and domain I of the 23S rRNA gene are phylogenetic markers for Chlamydia spp. Int J Syst Bacteriol 1997; 47: 461– 473 . [DOI] [PubMed] [Google Scholar]
- 16. Laroucau K, Souriau A, Rodolakis A. Improved sensitivity of PCR for Chlamydophila using pmp genes. Vet Microbiol 2001; 82: 155– 164 . [DOI] [PubMed] [Google Scholar]
- 17. Laroucau K, Trichereau A, Vorimore F, Mahe AM. A pmp genes-based PCR as a valuable tool for the diagnosis of avian chlamydiosis. Vet Microbiol 2006; 121: 150– 157 . [DOI] [PubMed] [Google Scholar]
- 18. Quinn PJ, Carter ME, Markey B, Carter GR. Clin Vet Microbiol 2004, Appendix 2. pp: 624. [Google Scholar]
- 19. Hewinson RG, Rankin SES, Bevan BJ, Field M, Woodward MJ. Vet Rec 1991; 128: 129–130. [DOI] [PubMed] [Google Scholar]
- 20. Borel N, Mukhopadhyay S, Kaiser C, Sullivan ED, Miller RD, Timms P, et al. Tissue Micro-Array (TMA) analysis of normal and persistent Chlamydophila pneumonia infection. BMC Infect Dis 2006a; 6: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bush RM, Everett KD. Molecular evolution of the Chlamydiaceae. Int J Syst Evol Microbiol 2001: 51: 203–220. [DOI] [PubMed] [Google Scholar]
- 22. Goellner S, Schubert E, Liebler-Tenorio E, Hotzel H, Saluz HP, Sachse K. Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect Immun 2006; 74: 4801– 4808 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haag-Wackernagel D, Moch H. Health hazards posed by feral pigeons. J Infect 2004; 48: 307– 313 . [DOI] [PubMed] [Google Scholar]
- 24. Huminer D, Pitlik S, Kitayin D, Weissman Y, Samra Z. Prevalence of Chlamydia psittaci infection among persons who work with birds. Isr J Med Sci 1992; 28: 739– 741 . [PubMed] [Google Scholar]
- 25. Takashima I, Imai Y, Itoh N, Kariwa H, Hashimoto N. Polymerase chain reaction for the detection of Chlamydia psittaci in the feces of budgerigars. Microbiol Immunol 1996; 40: 21– 26 . [DOI] [PubMed] [Google Scholar]
- 26. Trevejo RT, Chomel BB, Kass PH. Evaluation of the polymerase chain reaction in comparison with other diagnostic methods for the detection of Chlamydia psittaci. J Vet Diagn Invest 1999; 11: 491– 496 . [DOI] [PubMed] [Google Scholar]
- 27. Longbottom D, Russell M, Jones G, Lainson F, Herring A. Identification of a multigene family coding for the 90 kDa proteins of the ovine abortion subtype of Chlamydia psittaci. FEMS Microbiol Lett 1996;142: 277– 281 . [DOI] [PubMed] [Google Scholar]
- 28. Van Loock M, Verminnen K, Messmer TO, Volckaert G, Goddeeris BM, Vanrompay D. Use of a nested PCR-enzyme immunoassay with an internal control to detect Chlamydophila psittaci in turkeys. BMC Infectious Diseases 2005; 5: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Teankum K, Pospischil A, Janett F, Bürgi E, Brugnera E, Hoelzle K, Polkinghorne A, Weilenmann R, Zimmermann DR, Borel N. Detection of chlamydiae in boar semen and genital tracts. Vet Microbiol 2006; 116: 149– 157 . [DOI] [PubMed] [Google Scholar]
- 30. Olsen B, Persson K, Broholm KA. PCR detection of Chlamydia psittaci in faecal samples from passerine birds in Sweden monoclonal antibodies. Res Microbiol 1998; 148: 327– 333 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heddema ER, Beld MGHM, de Wever B, Langerak AAJ, Pannekoek Y, Duim B. Development of an internally controlled real-time PCR assay for detection of Chlamydophila psittaci in the LightCycler 2.0 system. Clin Microbiol Infect 2006; 12: 571– 575 . [DOI] [PubMed] [Google Scholar]
- 32. Sareyyupoglu B, Cantekin Z, Bas B. Chlamydophila psittaci DNA detection in the faeces of cage birds. Zoonoses Public Health 2007; 54: 237– 242 . [DOI] [PubMed] [Google Scholar]
- 33. Doosti A, Arshi A. Determination of the Prevalence of Chlamydia psittaci by PCR in Iranian Pigeons. Int J Biol 2011; 3, 4 : 79–82. [Google Scholar]
- 34. Pantchev P, Sting R, Bauerfeind R, Tyczka J, Sachse K. New real-time PCR tests for species-specific detection of chlamydophila psittaci and chlamydophila abortus from tissue samples. Vet J 2009; 181: 145– 150 . [DOI] [PubMed] [Google Scholar]



