Abstract
Background and Objective:
The genus Xanthomonas is composed of phytopathogenic bacterial species. In addition to causing crops diseases, most of the Xanthomonas species especially Xanthomonas campestris produce xanthan gum via an aerobic fermentation process. Xanthan gum is, an important exopolysaccharide from Xanthomonas campestris, mainly used in the food, petroleum and other industries. the purpose of this study was assessment of relationship between genetic diversity and xanthan production in Xanthomonas spp.
Materials and Methods:
In this study 15 strains of Xanthomonas spp. which had previously been isolated from soils of vegetable farms, were discriminated from each other using Enterobacterial Repetitive Intergenic Consensus (ERIC) PCR and 16S rDNA sequencing methods. Xanthan production of strains was measured in 250 ml flask. The results of ERIC PCR and xanthan production was compared.
Results:
ERIC-PCR patterns not only could differentiate all Xanthomonas campestis from the control i.e. Xanthomonas translucent but also discriminate strains of Xanthomonas to three clusters with 40% similarity based on Jaccard’s coefficient. This clustering of the strains was in agreement with other characteristics including xanthan production and biochemical features.
Discussion:
The results showed that genomic fingerprinting conferred adequate genetic data for discriminating between strains of the species Xanthomonas campestris. The data indicated a partial relationship between ERIC-PCR patterns and xanthan production by the strains.
Conclusion:
Further development of experiments may result in making good prediction about xanthan production capability of the Xanthomonas strains on the basis of ERIC-PCR method.
Keywords: Genetic diversity, Xanthomonas, Xanthan, ERIC-PCR
INTRODUCTION
The genus Xanthomonas is composed of phytopathogenic bacterial species that cause diseases in diverse crops and results in declined yields. Strains of the genus also can produce xanthan gum through an aerobic fermentation (1). Xanthan gum is a high molecular weight anionic polysaccharide. This polymer displays appropriate properties: high viscosity at low concentrations, pseudoplasticity, and insensitivity to a wide range of temperature, pH and electrolyte variations. Because of its special rheological properties, xanthan is used in food, cosmetics, pharmaceuticals, paper, paint, textiles, adhesives and oil and gas industry (2). The flow characteristics of xanthan, coupled with its stability to salts and extremes of pH, gives it a technical advantage over most polymers used in drilling. The greatest potential for xanthan gum appears to lay in the enhanced oil recovery operations (3).
“Many reports showed high level of polymorphism among Xanthomonas species affecting their biological and fermentation properties”. Genomic fingerprinting by PCR amplification, with primers specific to the highly conserved repetitive elements such as the 35–40 bp repetitive extragenic palindromic (REP) sequence, and the 124–127 bp enterobacterial repetitive intergenic consensus (ERIC) have been used successfully to characterize a large number of bacteria and distinguish closely related strains (4–6).
The screening of Xanthomonas strains with potential in xanthan production industry is a continuous need. Although the use of genomic fingerprinting techniques to study variability in microorganisms is fairly common, there are few reports on screening associated with gum production, especially in relation to xanthan production. The objectives of this study were to: (i) assess genetic diversity and evaluate the use of rep-PCR to differentiate among some Iranian Xanthomonas strains, and (ii) investigate the correlation between the genetic diversity and xanthan production capabilities of the strains.
MATERIALS AND METHODS
Bacterial strains.
Fifteen isolates of the genus Xanthomonas were used in this study. The isolates were isolated in our previous study from soil of four farms gone under cultivation of cauliflower and cabbage (Brassica oleraceae) crops in Tehran and Alborz Provinces, Iran (7). The disease is not widespread in Iran, and the strains were isolated during an intensive soil screening program. The isolates and their geographical origins are listed in Table 1. Pure cultures of the bacteria were maintained on Yeast Malt (YM) agar slants at 4 °C and transferred to fresh medium every 14 days to prevent strains from losing their production capability (8).
Table 1.
The Xanthomonas isolates used in this study and their geographical origins and xanthan production capabilities
| Numbers assigned to isolates | Isolates | Province | Field | Crops | Apparent viscosity of broth (cP) | Raw xanthan (g l−1) |
|---|---|---|---|---|---|---|
| 1 | X. translucens SAM 0402 | Tehran | 1 | Cauliflower (Brassica oleracea) | ∼1a | ∼0 a |
| 2 | X. campestris SAM 0302 | 1 | 960b | 9.69b | ||
| 3 | X. campestris SAM 0401 | 1 | 870b | 9.34b | ||
| 4 | X. campestris SAM 0301 | 1 | 904b | 9.68b | ||
| 5 | X. campestris SAM 3301 | Alborz | 2 | Red Cabbage (Brassica oleracea) | 1426c | 11.92c |
| 6 | X. campestris SAM 3302 | 2 | 1524c | 11.42c | ||
| 7 | X. campestris SAM 3303 | 2 | 1526c | 11.25c | ||
| 8 | X. campestris SAM 4101 | 3 | Green Cabbage (Brassica oleracea) | 1532c | 11.61c | |
| 9 | X. campestris SAM 4204 | 4 | 1242c | 10.73c | ||
| 10 | X. campestris SAM 4205 | 4 | 412b | 7.12b | ||
| 11 | X. campestris SAM 4210 | 4 | 1514c | 12.10c | ||
| 12 | X. campestris SAM 4213 | 4 | 1554c | 11.56c | ||
| 13 | X. campestris SAM 4215 | 4 | 1547c | 11.61c | ||
| 14 | X. campestris SAM 4217 | 4 | 1525c | 11.96c | ||
| 15 | X. campestris SAM 4220 | 4 | 1505c | 11.05c |
These results were significantly different (p<0.05).
Xanthan production.
Actively growing cells from 24-h slant cultures of each isolate were inoculated to test tubes containing YM broth. The cultures were incubated at 28 °C overnight and then transferred to 100-ml flasks containing 20 ml of YM broth. After incubation at 28 °C on an orbital shaker at 140 rpm, the inocula were added to 250-ml flasks containing 50 ml of production medium. The composition of the medium was the same as synthetic medium introduced by Roseiro (9). Following incubation at 28°C and 140 rpm for 72 h, the apparent viscosity of fermentation broth was measured at room temperature using a Brookfield system viscometer (Anton Paar, DV1) with spindle number 3 at 60 rpm. Raw xanthan was precipitated with 1.5 volumes of isopropyl alcohol and 0.5 g l−1 NaCl and dried in an oven. The experiments were carried out in four replicates.
Genomic fingerprinting.
Bacteria were cultured in a liquid medium containing (g l−1) Trypton 10, NaCl 5 and yeast extract 5; pH 7.2. Total genomic DNA was extracted from overnight cultures by the method of Boucher (10). ERIC-PCR. Primer sequences corresponding to ERIC (ERICIR [5′-ATGTAAGCTCCTGGGGATTCAC-3′] and ERIC2 [5′-AAGTAAGTGACTGGGG TGAGCG-3′] were used (11). The PCR was performed with 25-μl volumes containing the Taq DNA polymerase buffer 1X, 200 mM dNTP, 0.4 mM primer, and 1.0 U of Taq DNA polymerase. Fifty nanograms of bacterial DNA or approximately 10,000 bacterial cells were used as the template. PCR amplification was performed with a PEQLAB Thermocycler using the following cycles: 1 initial cycle at 95°C for 5 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min with ERIC primers, and extension at 72°C for 1 min with a single final extension cycle at 72°C for 10 min and store at 4°C.
A 6 to 8 μl aliquot of amplified PCR product was separated by gel electrophoresis on 1.5% agarose gels in 0.5 X TBE buffer (12) for 5 h at 70 V, stained with ethidium bromide, and photographed on a UV transilluminator Ingenious LBR model.
The PCR reaction was carried out in two separate trials. Samples from each isolate loaded on gel electrophoresis plate and results were compared.
Bacterial identification.
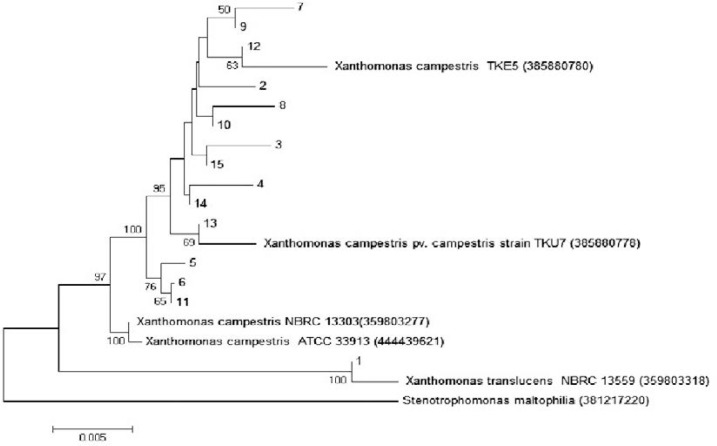
In this study Xanthomonas species were identified by sequencing of 16S rDNA region using universal primers including 27F and 1492R. Total genomic DNA was extracted by phenol – chloroform method. PCR was performed using universal primers and small amount of products were loaded on 1% agarose gel to detect a sharp single band, the remaining PCR product was sequenced. Sequencing results were assessed by bioEdit version 7.0.9.0 program and phylogenetic tree was constructed by MEGA5 program based on neighbor joining method (Fig. 1).
Fig. 1.
Phylogenetic tree constructed with neighbor joining method. Our isolates were numbered 1 to 15 and Stenotrophomonas maltophilia numbered 16 and used as out group.
Data analysis.
The results obtained from the xanthan production experiments were analyzed statistically by one-way analysis of variance and Tukey test with 95% confidence level using Minitab (version 15.2) software. ERIC-PCR amplification products were listed as discrete character states per strain (presence or absence of band). Jaccard’s similarity coefficients between each strain were calculated using the SIMQUAL program in NTSYS-pc, version 2.02e, and these data were subjected to UPGMA cluster analysis by use of the SAHN program of NTSYS (13).
RESULTS
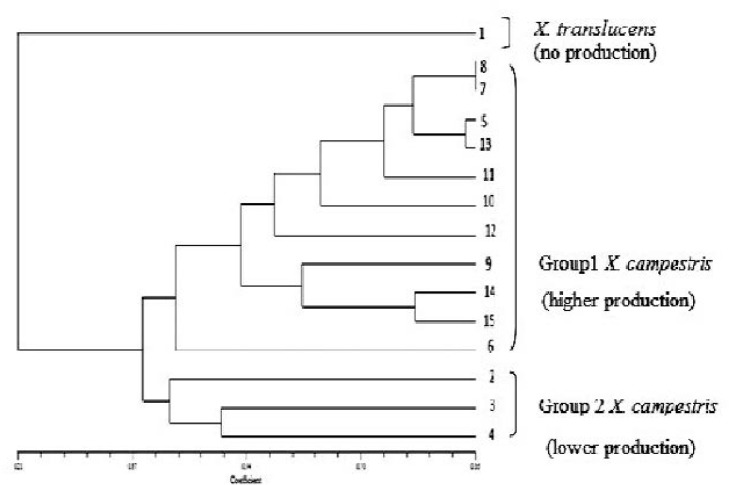
Average of apparent viscosity of culture broth and amount of raw xanthan obtained from the Xanthomonas isolates is shown in Table 1. From the xanthan production results, it is realized that the isolates can be placed in three statistically significant groups: isolate 1 not producing xanthan, isolates 2, 3, 4 and 10 which produced an average broth viscosity of 786 centipoises (cp) and average raw xanthan concentration of 8.96 g l−1, and a third group with higher xanthan production capability. The last group includes isolates (5–9 and 11–15) which produced average broth viscosity and raw xanthan concentration of 1490 cP and 11.49 g l−1 respectively.
Result of 16S rRNA gene sequencing showed 14 isolates including isolates numbered 2 to 15 were Xanthomonas campestris and isolate 1 was Xanthomonas translucens. Similarity percent data from comparing to selected strain on NCBI data base are given in Table 2 and phylogenetic tree is shown in Fig. 3. The result of reproducibility tests confirmed the ability of this method as fingerprinting technique.
Table 2.
Results of genotypic and phenotypic characterization for identification of the isolates
| Isolate number in this study | The most similar registered strain in NCBI database according to 16S rDNA identity | Max. Identity* | Nitrate reduction | Max. Temperature (°C) | Max. NaCl (%) | Citrate, Propionate | Acid from sucrose | Growth at %10 glucose | Acid from selected sugars** | Lactose, Raffinose | Manitol | Lecithinase |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | X. translucens NBRC13559 | 99 | + | 42 | 5 | − | − | − | − | − | − | + |
| 2 | X. campestris NBRC 13303 | 100 | − | 39 | 5 | + | + | + | + | + | − | + |
| 3 | X. campestris TKU7 | 99 | − | 39 | 5 | + | + | + | + | + | + | + |
| 4 | X. campestris TKU7 | 100 | − | 38 | 5 | + | + | + | + | + | + | + |
| 5 | X. campestris NBRC 13303 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
| 6 | X. campestris NBRC 13303 | 99 | − | 38 | 4 | + | + | + | + | − | − | − |
| 7 | X. campestris TKU7 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
| 8 | X. campestris TKU7 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
| 9 | X. campestris TKU7 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
| 10 | X. campestris TKE5 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
| 11 | X. campestris NBRC 13303 | 99 | − | 38 | 4 | + | + | + | + | − | − | − |
| 12 | X. campestris TKU7 | 100 | − | 37 | 4 | + | + | + | + | − | − | − |
| 13 | X. campestris TKU7 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
| 14 | X. campestris TKE5 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
| 15 | X. campestris TKE5 | 100 | − | 38 | 4 | + | + | + | + | − | − | − |
Max identity for 16S rRNA gene sequence similarity in NCBI data base
D- Xylose, D- Mannose, D- Fructose, Cellobiose, Maltose
Fig. 3.
Dendrogram based on the cluster analysis (UPGMA) of the estimate of genetic similarity by ERIC-PCR drawn with NTSYS-pc, version 2.02e. The numbers 1–15 indicates the isolates 1–15. Group 1 X. campestris: High and acceptable production. Group 2 X.campestris : less than 10 g/l production and less than 1×103cP viscosity. The strain 1 was, identified as X. translucens, isolated from the same place and used as negative control.
ERIC-PCR.
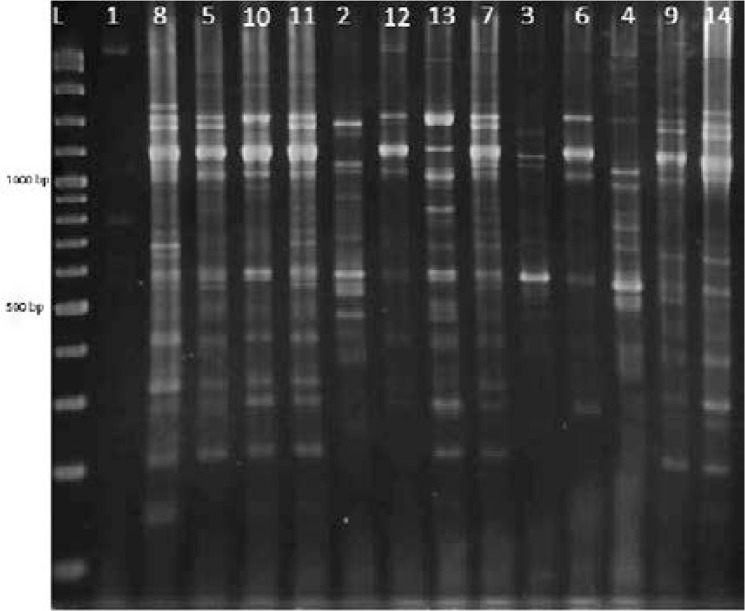
ERIC-PCR yielded 5 to more than 21 PCR products, ranging in size from 100 bp to over 5 kb. There were differences in the concentration of some amplified fragments as well as in the occurrence of numerous polymorphic bands. Differences among strains were assessed visually on the basis of the migration patterns of PCR products (Fig. 2). UPGMA clustering separated all the isolates into different groups at a cut off of 86% similarity (Fig. 3). Three clusters were clearly separated with 40% similarity. This clustering was in agreement with xanthan production capability groups except for the isolate 10.
Fig. 2.
Patterns of ERIC-PCR on agarose gel electrophoresis. Lanes 1–15 include the isolates of 1–15 and Lane L indicates DNA ladder 100bp plus.
DISCUSSION
In this study we have demonstrated Entrobacterial Repetitive Intergenic Consensus as ERIC sequences are present in the genome of variant Xanthomonas strains, confirming and extending the conclusion of Versalovic et al. (5), de Bruijn (4), and Louws et al. (6). The Xanthomonas isolates used in this study were isolated previously from two distinct geographical locations with different cultivated crops. Based on 16S rDNA sequences, all the isolates are closely related to X. campestris, except for the isolate 1 that belongs to X. translucens (Table 1). According to Louws et al. (6) and Barak and Gilbertson (14) rep-PCR can prove intraspecific variability among strains of heterogeneous pathovars of X. campestris. Thus, great genetic diversity among the isolates used in this study was expected and is consistent with previous studies and further confirms that X. campestris is heterogeneous.
After performing ERIC-PCR fingerprinting, a total of 21 distinctive bands ranging in size between 150 and 2500 bp was found among the analyzed isolates. Arshiya et al. (15) . Arshiya et al. (15) found bands ranging in size between 100 and 3,000 bp based on BOX, ERIC and REP profiles of 56 different Xanthomonas axonopodis pv. citri,, Although the number of distinctive bands found in our work is lower than that reported in other studies, we have to take into account that of the above authors worked with strains representing several Xanthomonas pathovars. We have also demonstrated that the ERIC-PCR protocols are rapid molecular characterization of Xanthomonas bacteria. The ERIC-PCR protocol clearly has the potential to differentiate Xanthomonas strains, including those pathovars not easily differentiated by other phenotypic and phylogenetic techniques (6). Our results showed partial relationship between ERIC-PCR patterns and xanthan production. Isolate 1 placed in a separate group has no xanthan production. Isolates 2, 3 and 4 have similar xanthan production about 9.5 g l−1 and other isolates except isolate 10 have about 11.5 g l−1 xanthan productions according to different geographical regions (Table 1). The results of this study were different with the results of Mayer et al. (1) that showed there were no relations between AFLP and xanthan production. This result can represent superiority of REP-PCR over AFLP method that was agreement with Rademaker et al. resuts(16). The data presented here suggest that ERIC-PCR should also be a useful tool for assessment correlation between geographical region and xanthan production with ERIC-PCR patterns. Samples from the same or near sites showed similar patterns (Fig 1) of ERIC-PCR and clustered in the same group ( Fig 2 ). ERIC-PCR analysis could cluster strains based on their geographical origin. Similar results were found by Massomo et al. (17) and Zhai et al. (18). However, contrary results have been obtained by Jensen et al. (19), Arshiya et al. (15) and Zaccardelli et al. (20).
CONCLUSION
ERIC-PCR was a good method for assessment of genetic diversity among Xanthomonas bacteria and the relation of them with xanthan production. If this method is developed by further strains it may predict ability of a strain to produce more and better quality of xanthan based on ERIC-PCR method.
REFERENCES
- 1. Mayer L, Silva WP da, Moura AB, Vendruscolo CT. AFLP analysis of Xanthomonas axonopodis and Xanthomonas arboricola strains used in xanthan production studies reveal high levels of polymorphism. Braz J Microbiol 2010; 55: 741– 748 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jana AK, Ghosh P. Stimulation of xanthan production by Xanthomonas campestris using citric acid. World J Microbiol Biotechnol 1997; 13: 261– 264 . [Google Scholar]
- 3. Rottava I, Batesini G, Silva MF, Lerin L, De Oliveira D, Padilha FF, Toniazzo G, et al. Xanthan gum production and rheological behavior using different strains of Xanthomonas sp. Carbohydr Polym 2009; 77: 65– 71 . [Google Scholar]
- 4. Louws FJ, Fulbright DW, Stephens CT, Bruijn FJ. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol 1994; 60: 2286– 2295 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Versalovic J, Schneider M, de Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence based PCR (REP-PCR). Meth Cell Mol Biol 1994; 5: 25–40 [Google Scholar]
- 6. De Brujin FJ, Rademaker JLW, Schneider M, Rossbach U, Louws FJ. REP-PCR Genomic fingerprinting of plant-associated bacteria and computer-assisted phylogenetic analyses. APS Press; 1996; 497–502. [Google Scholar]
- 7. Soudi MR, Alimadadi N, Ghadam P. Minimal phenotypic test for simple differentiation of Xanthomonas campestris from other yellow-pigmented bacteria isolated from soil. Iran J Microbiol 2011; 3: 84– 91 . [PMC free article] [PubMed] [Google Scholar]
- 8. García-Ochoa F, Santos VE, Casas JA, Gómez E. Xanthan gum: production, recovery, and properties. Biotechnol Adv 2000; 18: 549– 579 . [DOI] [PubMed] [Google Scholar]
- 9. Roseiro JC, Esgalhado ME, Amaral-Collaço, Emery A. Medium development for xanthan production. Process Biochem 1992; 27: 167– 175 . [Google Scholar]
- 10. Boucher CA, Barberis P, Demery DA. Transposon mutagenesis of Pseudomonas solanacearum: Isolation of Tn5-induced avirulent mutants. J Gen Microbiol 1985; 131: 2449–2457. [Google Scholar]
- 11. Versalvoice J, Kouetch T, Lupski JR. Distribiution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genome. Nucleic Acids Res 1991; 19: 6823– 6831 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor New York: laboratory Press, Cold spring Harbor; 1989. [Google Scholar]
- 13. Pooler M R, Ritchie D F, Hartung JS. Genetic relationships among strains of Xanthomonas fragariae based on random amplified polymorphic DNA PCR, repetitive extragenic palindromic PCR, and enterobacterial repetitive intergenic consensus PCR data and generation of multiplexed PCR primers useful for the identification of this phytopathogen. Appl Environ Microbiol 1996; 62: 3121– 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barak JD, Gilbertson RL. Genetic diversity of Xanthomonas campestris pv. vitians, the causal agent of bacterial leaf spot of lettuce. Phytopathology 2003; 93: 596– 603 . [DOI] [PubMed] [Google Scholar]
- 15. Arshiya M, Suryawanshi A, More D, Mushtaq M, Baig V. Repetitive PCR based detection of Genetic Diversity in Xanthomonas axonopodis pv citri Strains. Journal of Applied Biology and Biotechnology 2014; 2: 17– 22 . [Google Scholar]
- 16. Rademaker J.L, Hoste B, Louws FJ, Kersters K, Swings J, Vauterin L, Vauterin P, de Bruijn FJ. Comparison of AFLP and REP-PCR genomic fingerprinting with DNA–DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol 2000; 50: 665–677 [DOI] [PubMed] [Google Scholar]
- 17. Massomo SMS, Nielsen H, Mabagala RB, Giese KM, Hockenhull J, Mortensen CN. Identification and characterization of Xanthomonas campestris pv. campestris strains from Tanzania by pathogenicity tests, biolog, rep – PCR and fatty acid methyl ester analysis. Eur J Plant Pathol 2003; 109: 775– 789 . [Google Scholar]
- 18. Zhai J, Luo Y, Zheng D, Huang X. Evaluation of Genetic Diversity of highly virulent strains of Xanthomonas campestris pv. malvacearum by REPPCR fingerprinting. J Phytopathol 2010; 158: 764– 768 . [Google Scholar]
- 19. Jensen LB, Garcia-Migura L, Sanchez-Valenzuela AJ, Løhr M, Hasman H, Aarestrup FM. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Meth 2010; 80: 25– 43 . [DOI] [PubMed] [Google Scholar]
- 20. Zaccardelli M, Campanile F, Moretti C, Buonaurio R. Short communication characterization of Italian populations of Xanthomonas campestris pv. Campestris using primers based on DNA repetitive sequences. J Plant Pathol 2008; 90: 375– 381 . [Google Scholar]