Abstract
Antibodies are key molecules in the fight against infections. Although previously thought to mediate protection solely in the extracellular environment, recent research has revealed that antibody-mediated protection extends to the cytosolic compartment of cells. This postentry viral defense mechanism requires binding of the antibody to a cytosolic Fc receptor named tripartite motif containing 21 (TRIM21). In contrast to other Fc receptors, TRIM21 shows remarkably broad isotype specificity as it does not only bind IgG but also IgM and IgA. When viral pathogens coated with these antibody isotypes enter the cytosol, TRIM21 is rapidly recruited and efficient neutralization occurs before the virus has had the time to replicate. In addition, inflammatory signaling is induced. As such, TRIM21 acts as a cytosolic sensor that engages antibodies that have failed to protect against infection in the extracellular environment. Here, we summarize our current understanding of how TRIM21 orchestrates humoral immunity in the cytosolic environment.
Keywords: TRIM21, Fc-receptor, antibodies, isotype, intracellular immunity, virus
Introduction
Antibodies are a crucial part of the immune response toward invading pathogens such as viruses, and the induction of so-called neutralizing antibodies is a primary goal of vaccination (1). In addition, there is great interest in the development of broadly neutralizing antibodies specific for major human pathogens such as human immunodeficiency virus (HIV) and influenza virus (2,3). Neutralization of viruses by antibodies is predicted to depend on high-affinity binding to specific epitopes of surface-exposed viral proteins that are required for binding to target cell receptors (4). Such antibodies are thought to function according to the occupancy model that requires binding of a critical number of antibodies to a viral particle in such a way that most or all neutralizing epitopes are occupied (5). This may occur independently, or in concert with other antibody-mediated effector functions such as antibody-dependent cellular phagocytosis or antibody-dependent cellular cytotoxicity (6,7). These effector functions are induced upon binding of antibody–virus immune complexes to classical Fc γ receptors (FcγRs) expressed on the surface of hematopoietic cells such as natural killer (NK) cells, macrophages, and dendritic cells, which results in clearance and induction of T-cell responses (8). Neutralizing antibodies also prevent infection in concert with non-classical Fc receptors such as the neonatal Fc receptor (FcRn) and the polymeric Ig receptor (pIgR) (9,10). While FcRn mediates bidirectional transport of IgG across mucosal epithelial surfaces (11–15), pIgR mediates unidirectional transcytosis of IgA and IgM from the tissue into the luminal space (9). Transcellular transport of neutralizing but also non-neutralizing antibody has been shown to facilitate protection against viral infection (9,16–21).
Although neutralizing antibodies are produced during antiviral responses, the majority of the antibodies in the polyclonal response have no neutralizing activity by the classical definition (22,23). This is due to the fact that they bind internal viral epitopes released from infected lysed cells, or viral surface proteins that are not involved in viral attachment and entry into host cells (24). In addition, viruses are known to display immune-dominant non-neutralizing epitopes that can bias the polyclonal response toward a non-neutralizing phenotype (25–28). As such, neutralization of viruses by antibodies has until recently been assumed to be a solely extracellular or intravesicular event.
However, in recent years, it has become clear that the antiviral function of antibodies also extends into the cytosolic compartment of cells (29–31). This additional level of protection is orchestrated by the interferon (IFN)-inducible cytosolic Fc receptor tripartite motif containing-21 (TRIM21). Engagement of TRIM21 results in rapid postentry elimination of antibody–virus via recruitment of the proteasomal machinery (32,33), in a mechanism termed antibody-dependent cellular neutralization (ADIN). Concurrently, inflammatory signaling is also induced (34). Therefore, antibodies that have failed to block entry of a virus particle into the cell and that is not intercepted by antibody-mediated effector functions operating in the extracellular environment may still be protective in the cytosolic compartment, even though they are, by the classical definition, non-neutralizing. Instead, the cell takes advantage of TRIM21 to set up one last line of antiviral defense.
TRIM21 can be distinguished from other Fc receptors in two ways. Firstly, TRIM21 shows remarkably broad antibody specificity as it can activate both of its functions upon binding to IgG, IgM as well as IgA (32,35), while other Fc receptors display more restricted antibody isotype and subtype specificities (36–38). Secondly, TRIM21 is broadly expressed by cells of most histogenic linages (39), while expression of classical FcγRs is mainly restricted to hematopoietic cells (8). This suggests that a susceptible pathogen may be targeted by TRIM21 independently of the site of infection and local distribution of antibody isotypes.
TRIM21 is a multi-domain Fc receptor with ubiquitin ligase activity
The TRIM21–IgG interaction was first described in a yeast two-hybrid screen in a study that investigated the role of TRIM21 as an autoantigen in immune disorders such as systemic lupus erythematosus and Sjögren's syndrome, in which it is referred to as Ro52 or SS-A (40). In subsequent studies, TRIM21 was immunoprecipitated independently of antibody specificity, and the binding site for TRIM21 was postulated to be localized to the CH2–CH3 interface of Fc as it was found to compete with binding of Staphylococcus protein A and Streptococcus protein G (41,42). Furthermore, the corresponding binding site on TRIM21 was found localized to the C-terminal PRYSPRY domain as truncation of this domain resulted in loss of binding. Although the interaction between TRIM21 and IgG was initially thought to be irrelevant due to the topologically distinct localization of the two proteins, a specific role in antiviral defense was more recently described (32).
TRIM21 is an Fc receptor that is structurally unrelated to all other classes of Fc receptors (43,44). It is part of the TRIM family which consists of over 100 members in humans (45), with a diverse set of cellular roles including antiviral defense (46,47). One of the most studied members is TRIM5α, which mediates restriction of simian immunodeficiency virus via an antibody-independent mechanism (48). TRIM21 shares the same structural architecture as other TRIM proteins and consists of an N-terminal RING domain with E3 ubiquitin ligase activity, a B-box, and a central coiled-coil domain that is referred to as RBCC (43). It is, however, the C-terminal domain of TRIM proteins that determines ligand specificity and function, and in half of all known TRIM proteins this is a so-called PRYSPRY domain. The PRYSPRY domain of TRIM21 contains the antibody binding site, and is a globular fold comprising a β-sandwich of two antiparallel β-sheets connected by flexible loops (43), and is a fusion of PRY and SPRY elements which are of distinct evolutionary origin (49). Furthermore, TRIM proteins are known to form dimers or higher order structures via their coiled-coil domains and both heteromeric and homomeric TRIMs have been described (50). Crystallographic data of the TRIM25 coiled-coil have revealed that it has an antiparallel helical structure that places the N-terminal RING domains at opposite sides of the dimeric structure, while the C-terminal PRYSPRY domains are positioned at the center (51). Although a crystal structure of full-length TRIM21 has yet to be solved, the presence of the coiled-coil suggests that TRIM21 adopts a similar structural arrangement that would place its two PRYSPRY domains in close proximity to each other. Consistent with this, full-length TRIM21 has been shown to exist as a dimer in solution and form stable 1:1 complexes with human IgG1 (32). Thus, the two PRYSPRY domains of a dimeric TRIM21 molecule may bind simultaneously to one IgG Fc (32). This symmetrical mode of binding will allow TRIM21 to rapidly intercept incoming antibodies (32,34).
A molecular basis for the TRIM21–IgG interaction
A detailed understanding of the TRIM21–IgG interaction has been obtained from solving a co-crystal structure between the C-terminal PRYSPRY domain of human TRIM21 and an Fc fragment derived from human IgG1 (43). The complex reveals a 2:1 stoichiometry where a PRYSPRY domain binds to the interface between the CH2 and CH3 found on each side of the Fc. As such, the binding site for TRIM21 is distinct from that of the classical FcγRs and C1q in the lower hinge and CH2 domain (52–55), but overlaps with the binding site for FcRn (56,57) as well as bacterial and viral Fc receptors (58–60). In contrast to binding to FcγRs, neither TRIM21 nor FcRn binding to IgG is affected by removal of N297-linked glycans of the CH2 domains (42).
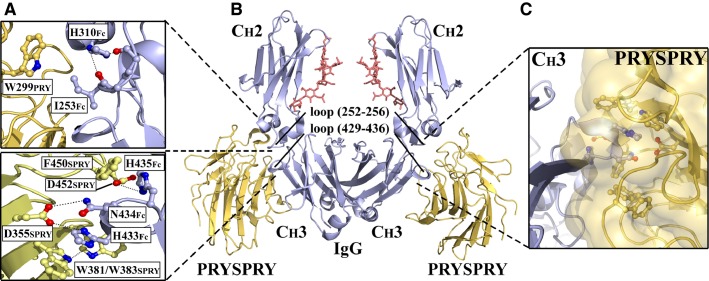
The core interaction site is formed between the SPRY element and the CH3 domain of IgG Fc (43,44). Here, the protruding and conserved Fc loop encompassing residues 429–436 is inserted into a deep hydrophobic pocket on the SPRY surface where the apex Fc residues H433, N434, and H435 (HNH motif) form a central hydrogen bond network surrounded by a hydrophobic shield of aromatic side chains that are engaged in aromatic stacking interactions. Specifically, H433Fc and N434Fc interact with D355SPRY located at the base of the SPRY binding pocket via hydrogen bonds, while H433Fc also interacts with W381SPRY via aromatic stacking, which may also involve W383SPRY. Furthermore, H435Fc forms a hydrogen bond with D452SPRY and stacks with F450SPRY. In addition, I253Fc of the CH2 loop encompassing residues 252–256 is involved in a hydrophobic interaction with W299PRY. An overview of the co-crystal complex and the interaction network is presented in Fig. 1.
Fig. 1.
Molecular basis for the TRIM 21–IgG interaction. Structural illustrations of the interaction between the human TRIM21 PRYSPRY (yellow) domain and the Fc part (blue) derived from human IgG1. The conserved CH2-loop (252–256) and the CH3-loop (429–436) of the Fc part are indicated while N297-linked glycans are shown in red. (A) Close-up view of the interactions formed between the CH2 domain of IgG1 and the PRY element (upper panel) and the CH3 domain and the SPRY element (lower panel). (B) The symmetric 2:1 complex in which two PRYSPRY domains bind one homodimeric Fc fragment at the CH2–CH3 interface. (C) An illustration showing insertion of conserved and protruding Fc-loop 429–436 into the hydrophobic binding pocket on the surface of TRIM21 PRYSPRY. The figure was prepared using PyMOL and based on the Protein Data Bank structure 2IWG (43).
In contrast to the strict pH dependence of the FcRn–IgG interaction, TRIM21 binds Fc pH independently, but is sensitive to high salt concentrations (43). The interaction has been shown to be highly conserved in mammals, and displays wide cross-species reactivity as both human and mouse TRIM21 bind IgG from a range of mammals (44). This is supported by inspection of the co-crystal structure between the mouse TRIM21 PRYSPRY domain and an Fc fragment derived from mouse IgG2a, which shows that the mouse and human mode of binding are highly similar (44).
The binding affinity between human IgG and the monomeric human PRYSPRY domain has been measured to 150 nM using isothermal titration calorimetry, while the murine interaction has an affinity of 500 nM, where the human interaction occurs with more negative enthalpy (43,44). However, the fact that full-length TRIM21 is a dimeric molecule has direct implications for its functional affinity to antibodies. The two PRYSPRY domains of a dimeric TRIM21 molecule may bind simultaneously to one IgG Fc as suggested by the crystal structures (32). Simultaneous engagement of both heavy chains likely explains the significantly increased interaction of dimeric TRIM21, yielding an affinity of 0.6 nM for the IgG–TRIM21 interaction as measured by fluorescence anisotropy (32). Notably, this represents an increase of more than 300-fold relative to monomeric affinity, which makes TRIM21 the highest affinity Fc receptor in humans.
TRIM21 has broad antibody isotype specificity
While Fc receptors are highly selective in regard to isotype and subtype binding, TRIM21 not only binds IgG but also IgM and IgA (32,35). Interestingly, the binding site for TRIM21 on IgA and IgM overlaps with that of Fc α receptor I (FcαRI), Fcα/μ receptor, pIgR, as well as several pathogen-produced Fc receptors (61–68). The binding affinities of IgM and IgA toward monomeric human PRYSPRY are considerably weaker, and have been measured to be 17 and 50 μM, respectively (32,35). However, despite relatively weak monomeric binding, the dimeric nature of full-length TRIM21 suggests that the functional affinities for IgM and IgA are much stronger (35). Assuming that the increase over monomeric affinity upon binding of full-length TRIM21 to IgG is similar for IgM and IgA, the binding strength may be in the sub-micrometer range during physiological conditions (35).
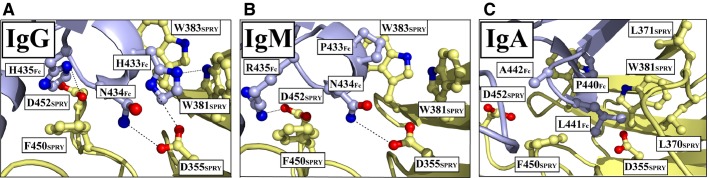
The weak monomeric binding of IgM and IgA is very likely due to the sequence variation in the stretch of amino acids corresponding to the CH3-loop 429–436 of IgG (35). Specifically, the apex HNH motif of IgG is PNR in IgM and PLA in IgA. However, binding to TRIM21 is still accommodated, as there is structural conservation of the Fc loop among the three isotypes. This has been addressed by molecular modeling showing that the structural loops in IgG and IgM superimpose closely at the secondary structure level allowing them to insert into the hydrophobic PRYSPRY binding pocket (35). Although the individual mutations H433A, N434A, N434D, and H435A in a murine IgG1 antibody (69) have been shown to abolish TRIM21 recognition, the specific replacements in IgM and IgA are predicted to form specific interactions with PRYSPRY residues (35).
In IgM, conservation of the central N434 is thought to be crucial, as its hydrogen bonding potential with D355SPRY is likely maintained, while replacement of H433Fc with proline will abolish its hydrogen bonding potential with D355SPRY and its ability to interact with two Fc residues simultaneously. However, the proline residue may still interact with W381SPRY and W383SPRY. Even though proline–tryptophan pairing is not a true aromatic stacking, it generates binding energy that is roughly double that of a hydrogen bond (70). Furthermore, the replacement of H435Fc with arginine in IgM abolishes its aromatic stacking potential with F450SPRY, but the arginine may still bind D452SPRY, given a favorable side chain conformation. In IgA, P440 corresponding to H433 in IgG is thought to have a similar role as in IgM. The presence of L441, the equivalent of N434 in IgG, cannot interact with D355SPRY, but is instead thought to form hydrophobic contacts with L370SPRY and L371SPRY, while A442Fc is predicted not to contribute in binding (35). Structural predictions of the interaction between IgM and IgA are shown in Fig. 2.
Fig. 2.
TRIM21 interacts with the Fc part of IgG, IgM, and IgA. (A) Close-up of the interaction network between the HNH motif of the IgG1 CH3 loop (429–436) and TRIM21 PRYSPRY. (B, C) Structural models predicting the interactions of TRIM21 with IgM and IgA. Amino acid residues found in IgM and IgA (PNR and PLA, respectively), were modeled into the IgG1–PRYSPRY structure. The figure was prepared using PyMOL and based on the Protein Data Bank structure 2IWG (35,43).
ADIN: antibody-dependent intracellular neutralization
During infection with a non-enveloped virus, antibodies that are attached to the viral capsid are also carried into the cytosolic compartment of target cells where they are recognized by TRIM21 (32,35) (Fig. 3). Subsequently, TRIM21 engagement targets the virus for proteasomal degradation before it has had time to replicate and spread. This was first demonstrated using human adenovirus type 5 (AdV5) carrying the green fluorescent protein reporter gene as a model pathogen, incubated with anti-AdV polyclonal serum from goat (32). Upon infection of HeLa cells, protection from infection was found to correlate with the level of TRIM21 expression, as depletion of TRIM21 gave high infection levels while upregulation of TRIM21 by IFN-α stimulation increased ADIN (32).
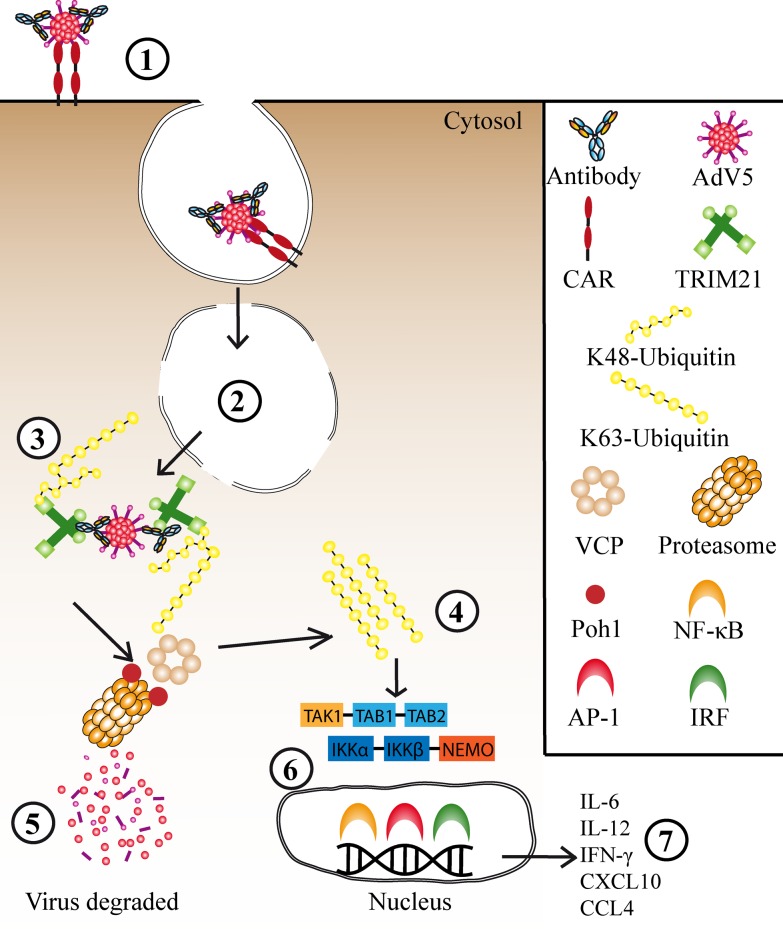
Fig. 3.
Cytosolic antiviral functions mediated by TRIM 21. (1) AdV5 in complex with antibodies interacts with CAR expressed on the surface of the target cell, which triggers endocytosis. (2) Exposure to acidic pH in the endosome induces a conformational change in the viral particle which results in membrane lysis and the AdV5 escapes into the cytosol bound to antibodies. Here, TRIM21 is rapidly recruited and binds to the Fc part of the antibodies leading to (3) sequential formation of K63 and K48-linked ubiquitin chains that result in targeting to VCP and the proteasomal machinery where Poh1 of the 19S regulatory subunit liberate (4) K63-linked ubiquitin chains and in turn is thought to activate the TAK1–TAB1–TAB2 and IKKα–IKKβ–NEMO enzyme complexes. (5) This results in degradation of the virus before it has time to replicate, and (6) activation of the transcription factor pathways NF-κB, AP-1, and IRF3/5/7 (7) accompanied by synthesis and secretion of pro-inflammatory cytokines and chemokines.
Following endocytosis via the coxsackie and adenovirus receptor (CAR), AdV5 in complex with non-neutralizing antibodies lyses the endosomal membrane and enters the cytosol where it is intercepted by TRIM21. Engagement of TRIM21 results in autoubiquitination in a manner that is dependent upon the E3 activity of its RING domain (32). Ubiquitination leads to recruitment of the proteasomal machinery and rapid degradation of the viral complex, in a process that is perturbed by the proteasomal inhibitor, epoxomicin. The degradation process has been demonstrated to be strictly dependent on the AAA ATPase v97/VCP (33). VCP is a multi-domain protein complex that interacts directly with multi-ubiquitin chains via its N-terminal domain and is capable of extracting proteins from larger complexes as well as unfolding them (71–78). Therefore, a general role for VCP in a preproteasomal degradation step has been suggested (79), and VCP was recently found to be recruited to stalled proteasomes (80). Considering that the intact AdV capsid measures 90 nm in diameter (81), and that the diameter of the proteasomal barrel is only 2 nm (82,83), such a predisassembly step is intuitively required (33). It is likely the challenging nature of the viral capsid rather than an intrinsic feature of ADIN that results in VCP dependence, as TRIM21 mediates the proteasomal turnover of ectopically expressed IgG Fc independently of VCP (33).
The fast rate of ADIN is crucial as it must occur before AdV reaches the nucleus for initiation of replication. This has been demonstrated using a fate-of-capsid assay, in which cellular levels of the major capsid protein, hexon, were degraded within 2 h of infection (32). The ADIN mechanism is strictly dependent on ubiquitination, in which TRIM21 itself is the substrate for attachment of ubiquitin (32,84). The ubiquitin machinery involves four enzymes: the ubiquitin activating (E1), the ubiquitin conjugating (E2), the substrate specifying ubiquitin ligase (E3), as well as the ubiquitin erasing deubiquitinase (DUB), of which E2 is responsible for the ubiquitin chain linkage topology (85–87). The E2 enzyme Ube2W was recently shown to mediate attachment of K63-linked monoubiquitin to TRIM21 (84). This makes TRIM21 accessible as a substrate for the E2 enzyme pair Ube2N/Ube2V2, resulting in extension of K63-linked ubiquitin chain in vitro. Furthermore, it was demonstrated that both Ube2W and Ube2N were required for attachment of both K63 and K48-linked ubiquitin chains to TRIM21 in cells, in which the attachment of K48-linked ubiquitin chains is thought to result in targeting to the proteasome (84). As TRIM21 undergo autoubiquitination, this could allow it to be effective against a diverse range of pathogens. This hypothesis is supported by the fact that antibody-coated beads transfected into cells become positive for ubiquitin, suggesting that any cytosolic particles bound by antibodies are potential targets for TRIM21-mediated ADIN.
Virus-associated IgM and IgA activate ADIN
During the initial stages of the antibody response, IgM is the main antibody isotype produced. IgM forms pentamers through association with the common joining chain (J-chain). IgA is the main isotype at mucosal barriers and the second most prevalent isotype in serum at 2–3 mg/ml compared to 12 mg/ml for IgG. Two subclasses of IgA exist, IgA1 and IgA2, and each is expressed as monomeric (mIgA), dimeric (dIgA), and secretory (S-IgA) forms (88). In serum, mIgA1 is the prevalent form while S-IgA2 dominates at mucosal sites (89,90). Immune complexes containing mIgA1 interact with FcαRI, which mediates phagocytosis, cellular cytotoxicity, antigen presentation, and cytokine release in professional cells, equivalent to the classical FcγRs for IgG (91). IgA and IgM bind the pIgR, which mediates transport from the tissue into the lumen of body cavities, and upon release they are associated with the pIgR-derived secretory component (SC) to form S-IgA and S-IgM (9). Interestingly, the SC-binding site on IgA overlaps with the binding site for TRIM21 (61), although SC is thought to associate with only one heavy chain in each IgA monomer (62). This may explain the weak affinity between S-IgA and the PRYSPRY domain (35).
During infection with AdV5, both IgM- and IgA-bound antibodies are recognized by TRIM21 and direct elimination of the virus via ADIN. This has been demonstrated with human IgM and IgA isolated from serum, and as observed for IgG, IFN-α stimulation increases neutralization in a TRIM21-dependent manner for both isotypes (32,35). Despite reduced affinity for S-IgA, virus-specific S-IgA from human serum was found to activate ADIN, as cellular depletion of TRIM21 partially restored viral infection. Although the presence of the SC appears to inhibit TRIM21 function, data suggest that the disulfide bonds between IgA and the SC may be broken upon exposure to the reducing environment of the cytosol, allowing attachment by TRIM21 (35). Interestingly, in a recent study, it was demonstrated that S-IgA forms trimers and tetramers in nasal secretions of healthy adults that contributed to enhanced neutralization potency (92). Such higher forms of S-IgA may likely reach the cytosol together with TRIM21-susceptible pathogens.
ADIN is a highly efficient neutralization mechanism
An intriguing feature of the ADIN mechanism is that only a few antibodies per virus are required for it to be effective as ADIN activity closely correlates with the expression level of TRIM21 rather than the extent of opsonization of the virus by a non-entry blocking antibody (69). Using a murine monoclonal IgG1 antibody (9C12) specific for the major capsid protein of AdV, hexon, a non-entry neutralizing epitope, together with manipulation of cellular TRIM21 levels using IFN-α or TRIM21 shRNA, allowed investigation of how antibody, TRIM21 and virus contribute to the efficiency of neutralization. In IFN-α-stimulated cells that expressed high levels of TRIM21, as few as 1.6 antibody molecules per virus were found to be sufficient for neutralization, while five times more antibody was required in unstimulated cells (69). Furthermore, ADIN was found to increase in a linearly incremental manner, demonstrating that each antibody associated with the virus contributes equally to its elimination upon binding to TRIM21 (69). This is in contrast to the occupancy model of entry neutralization, in which efficient prevention of infection can occur only after a critical number of antibodies occupy specific viral epitopes that are involved in cellular attachment and entry. Consequently, this results in a lag phase in neutralization curves when infection levels are plotted against antibody concentration until the critical number of antibodies required to block entry has been reached (93). This finding was supported by data demonstrating that polyclonal serum-containing antibodies specific for AdV5 required a higher antibody–virus stoichiometry for neutralization to occur when cellular TRIM21 was depleted, indicating a shift from cytosolic ADIN to entry blocking (69). Thus, when the polyclonal response contains few entry neutralizing antibodies, ADIN may be responsible for the majority of the neutralization activity. Low antibody–virus stoichiometry may also result in inefficient FcγR-mediated effector functions by immune cells as efficient phagocytosis requires the formation of immune complexes and cross-binding to cell-surface FcγRs. In addition, as TRIM21 also engages IgM and IgA, it is likely to contribute to early protection, and at the gate of entry of most viral pathogens, the mucosal barrier.
This however, does not mean that TRIM21-mediated protection becomes irrelevant in the later stages of infection when the amounts of entry neutralizing antibodies with high affinity are likely to increase. Infection by several viruses has been shown to occur even at saturating levels of entry neutralizing antibodies (93). The virus particles that still infect their target cells under these conditions are known as the persistent fraction. In the case of AdV5, the magnitude of the persistent fraction has been shown to correlate with the expression level of TRIM21, as a considerable increase in the persistent infection level was measured upon TRIM21 depletion (69). ADIN is also saturated at high viral loads, supporting the notion that TRIM21 levels are deterministic and suggesting that TRIM21 could be overcome by pathogens during high levels of viremia. As the efficacy of ADIN is also dependent on other cellular components such as VCP and the proteasome, the expression levels and rate of enzymatic turnover of these components are also likely to affect the ability of TRIM21 to induce ADIN (69).
Antibodies are DAMPs recognized by TRIM21
While extracellular detection of antibody–virus immune complexes by cell-surface-expressed FcγRs results in phosphorylation of their intracellular immunoreceptor tyrosine-based activation motif, triggering phagocytosis and immune activation (8), known mechanisms for detection of pathogens in the intracellular environment occur independently of antibodies and are instead dependent upon recognition of pathogen-associated molecular patterns. This is achieved via pathogen recognition receptors (PRRs) such as Toll-like receptors (94), and cytosolic receptors for viral nucleic acid such as retinoic acid-inducible gene-1 (RIG-1) and melanoma-differentiation associated protein 5 (Mda5) (95). The immune system can also be activated by tissue damage as a result of pathogen replication via the mislocalization of self-molecules. Such host derived factors are known as damage-associated molecular patterns (DAMPs) (96). Even though the concentration of IgG in human serum is as high as 12 mg/ml, it is excluded from the cytosolic compartment of cells during normal physiological circumstances. However, during infection, when antibodies of the IgG, IgM, and IgA isotypes enter the cytosol bound to a pathogen, they act like a DAMP upon TRIM21 recognition (34). Importantly, in addition to ADIN, engagement of TRIM21 induces an antiviral state by activating inflammatory responses. Unanchored K63-linked ubiquitin is an immune second messenger that activates the transcription factor pathways NF-κB, AP-1, IRF3, IRF5, and IRF7, via the enzyme complexes TAK1-TAB1-TAB2 (97,98) and IKKα–IKKβ–NEMO (99,100), and inhibition of these complexes prevents TRIM21-induced signaling (34). Whenever a ubiquitin-modified substrate is targeted to the proteasome, there are three DUBs in the 19S regulatory particle, Usp14/Ubp6, Uch37/UCHL5, and Rpn11/Poh1, that remove ubiquitin before substrate degradation (101). It was recently demonstrated that cells depleted of Poh1 were unable to activate NF-κB signaling in response to AdV5–IgG complexes, suggesting that Poh1 cleaves off TRIM21 K63-linked ubiquitin chains, which thereby activate an innate signaling response (84). This assure that TRIM21 only induces a signaling response if a virus antibody complex is targeted for proteasomal destruction, a mechanism that prevents ADIN from antagonizing signaling by rapid degradation of the virus, which thus synchronizes the activation of the two effector functions (84).
Activation of innate signaling pathways by TRIM21 induces rapid production of pro-inflammatory cytokines and chemokines, which has been shown to occur in both primary human lung fibroblasts and monocytes. Specifically, the cytokines IL-12, IL-6, and IFN-γ, as well as the chemokines CXCL10 and CCL4 were detected in response to IgG-coated AdV5. Furthermore, it was shown that inflammatory signaling was induced solely by TRIM21 upon detection of AdV5, and independently of known FcγRs and PRRs, as chemical inhibition of MyD88, TRIF, and the tyrosine kinase Syk or knockdown of cytosolic RIG-1 and Mda5 did not impact signaling (34).
Activation of TRIM21 sensing places the cell in an antiviral state. This involves increased expression of major histocompatibility complex class I (MHC I), as well as the activating MHC I-like NK2G2D ligands in non-hematopoietic cells (34). As TRIM21 engagement results in rapid degradation of viral proteins, as well as inflammatory signaling that upregulates the expression of MHC I, it is possible that peptides derived from the virus could be transported via the transporter associated with antigen processing (TAP) and loaded onto MHC I or NKG2D molecules. This would then make the cell a target for the cytotoxic effects of NK cells and CD8+ T cells; however, this has not yet been tested experimentally. In principle, the cell could ‘cure’ itself from infection by ADIN rather than being killed by effector lymphocytes. Induction of cell killing by lymphocytes requires clustering of MHC I molecules and T-cell receptors (102), and it may be speculated that the level of infection could be a factor that determines whether or not the cell would clear the virus solely via ADIN, or if T-cell help is required.
An important question is how the induction of signaling via TRIM21 is regulated. The immune system has evolved specific mechanisms to prevent excessive inflammation and such regulation must also be active in the context of TRIM21. For the classical FcγRs, this is achieved by regulation of the expression levels of activating receptors and the inhibitory FcγRIIb (8). As TRIM21 detection of cytosolic antibodies results in synchronized activation of both an effector and a signaling response, there must be strict regulation to avoid excessive inflammation at low pathogenic load. This may be a result of different requirements for activation of downstream ubiquitin-dependent processes, which must be related to how TRIM21 detects incoming antibody–virus complexes, and is potentially also self-limiting because the signaling stimulus (i.e. intracellular antibody) is also rapidly degraded by the effector mechanism (32).
TRIM21-susceptible pathogens
The full spectrum of pathogens that activates ADIN and TRIM21 signaling has yet to be determined. However, as both the effector and sensory functions of TRIM21 are strictly dependent on the presence of antibodies in the cytosol, the mechanism by which the pathogen enters the cell is crucial (103). Regarding viral infections, the main targets for TRIM21 are non-enveloped viruses, as enveloped viruses shed their outer membrane coat upon fusion with the plasma or endosomal membrane and deliver their nucleocapsids into the cytosol. Therefore, antibodies will be left behind at the cell membrane or inside endosomal vesicles. In line with this, the enveloped respiratory syncytial virus (RSV) is not detected by TRIM21 when added to cells in complex with anti-RSV antibodies (34).
Some non-enveloped viruses may also avoid TRIM21 detection due to their route of infection. For example, some rhinoviruses of the family Picornaviridae are thought to inject their genome through a pore in the endosomal membrane, thereby leaving their nucleocapsid and any attached antibody behind inside the endosome (104). In the case of AdV, its interaction with the cell-surface-expressed CAR triggers endocytosis (105). The virus then escapes endosomes and enters the cytosol by lysing the endosomal membrane, a process that relies on partial un-coating of the virus and exposure of an internal capsid protein that induces membrane damage (106). Once inside the cytosol, the sub-viral particle uses microtubule-associated motor proteins to reach the nuclear membrane (107). Subsequently, it docks to the nuclear pore complex through which the viral genome enters the nucleus (108). Therefore, any attached antibody is readily accessible for TRIM21 recognition. Viruses may also differ in their susceptibility to TRIM21 detection due to their behavior once they reach the cytosolic compartment (109,110). Although it is likely that some viruses have evolved to evade TRIM21 or even take advantage of TRIM21 as part of an un-coating mechanism, the rapid detection at a stage prior to synthesis of new viral proteins, as well as bridging by antibody, may make TRIM21 detection particularly difficult to antagonize (103). Indeed, neutralization of several non-enveloped viruses by few antibody molecules has been described for AdV, papillomaviruses, and poliovirus (111–119).
Importantly, TRIM21 sensing is not restricted to virus infection as it can activate antibody-dependent NF-κB signaling in response to the intracellular bacterium Salmonella enterica (34), which traffics between intracellular vesicles and the cytosol (120,121). Such TRIM21-dependent immune signaling has been shown to be even more potent upon infection with a Salmonella mutant (ΔSifA) that fails to maintain vesicle integrity, resulting in a greater proportion of bacteria in the cytosol (34). Although bacteria are presumably too large to be degraded by the proteasome, and ADIN elimination of AdV has been shown to happen independently of autophagy (32), a possible link between TRIM21 and the formation of autophagosomes in the context of Salmonella has been suggested (122).
The importance of TRIM21 for systemic protection
The fact that evolution has maintained strong binding between TRIM21 and antibodies across species points to its importance in host defense. Humoral immunity during the course of an infection occurs by a variety of different mechanisms, and the relative contribution of each will depend on the nature of the pathogen as well as the polyclonal antibody response (1,5,24). Therefore, to address whether or not TRIM21 makes a significant contribution to systemic protection in each case, this must be studied in the context of immune serum raised naturally against the pathogen. This has so far been investigated both in vitro and in vivo using the wildtype mouse adenovirus type-1 (MAV-1) (123,124) which causes hemorrhagic encephalitis in mice. To recover from acute MAV-1 infection, mice need a robust antibody response (125–127). The ability of anti-MAV-1 serum to neutralize MAV-1 was assessed in cells derived from wildtype or TRIM21 knockout mouse embryonic fibroblasts, and a significant proportion of neutralization was TRIM21-dependent (123). Furthermore, upregulation of TRIM21 expression by IFN stimulation increased the ADIN potency of both pooled serum and immune serum collected from individual mice. The importance of TRIM21 for MAV-1 protection in vivo was studied by comparing wildtype TRIM21+/+, TRIM21 heterozygote+/−, and TRIM21 knockout−/− C57BL/6 mice (124). Upon challenge with MAV-1, it was shown that naive mice lacking TRIM21 had a higher viral load and increased mortality within the first week of infection. As these experiments were performed in naive animals, protection was presumably a result of interaction between anti-MAV-1 IgM and TRIM21. In passive transfer experiments using antisera against MAV-1, it was demonstrated that survival closely correlated with TRIM21 expression, as transfer of serum was fully protective in wildtype, but not in TRIM21-deficient, mice. Heterozygous animals expressing intermediate levels of TRIM21 showed a haplo-insufficiency phenotype, with comparable mortality to mice homozygous null for TRIM21, but an average brain viral load that was intermediate between wildtype and knockout animals at the end point of the experiment. Moreover, in agreement with in vitro findings, IFN induction correlated with increased TRIM21 expression and reduced infection, highlighting the importance of TRIM21 for efficient ADIN activity. Early anti-serum and diluted late anti-serum was more dependent on TRIM21 for protection compared to non-diluted late anti-serum (124). This indicates that when other antibody-mediated mechanisms such as entry neutralization are incompletely protective, TRIM21 functions become increasingly important as they are able to operate under conditions of few antibodies per virus (69,124). In addition, the use of a fully replicative wildtype virus in these studies demonstrates that TRIM21 is effective during a spreading infection (123,124).
As pathogens enter the body through mucosal surfaces, it raises an intriguing question as to whether TRIM21 is activated in polarized epithelial cells. IgA and IgM bind pIgR, which mediates transport from the basolateral to the apical surface, and detection of viruses intracellularly may direct their transcytosis to the apical surface (9). In regard to IgG, FcRn mediates bidirectional transport of monomeric IgG or IgG-immune complexes across polarized cellular layers (11–15). FcRn has even been shown to facilitate neutralization in a specific case where a pH-dependent IgG1 antibody against influenza only neutralizes when the antibody-containing endosome undergoing FcRn transport fuses with a virus-containing acidic endosome (20). However, unpicking the antiviral contribution of immunoglobulin transport receptors like FcRn in vivo is complicated by their role in determining antibody serum half-life and biodistribution. An anti-HIV-1 IgG1 engineered for improved FcRn binding was shown to have increased serum half-life, enhanced mucosal localization and superior protection against intrarectal infection with simian-HIV (16), but the mechanistic correlation between these properties was not determined. While there are many examples of viruses hijacking antibody transport receptors to facilitate infection, whether or not FcRn and pIgR can directly prevent infection, or if there is crosstalk between these endosomal receptors and TRIM21 as they intercept viruses bound by antibody, remains to be addressed.
Conclusions and prospects
During an infection, TRIM21 detects antibodies that are carried by pathogens into the cytosol. Once antibodies are detected, TRIM21 mediates ADIN, a highly efficient viral elimination mechanism and concomitantly activates pro-inflammatory signaling resulting in a protective response. Although the complete spectrum of pathogens that are susceptible has yet to be determined, the fast rate of detection and the adaptor function of antibodies are believed to make antagonism on the part of the virus particularly difficult. It is therefore likely that diverse non-enveloped viruses are targets for TRIM21. TRIM21 has been shown to make a significant contribution to systemic protection, and thereby provide important immune defense alongside extracellular mechanisms. In addition, the ability of TRIM21 to interact with three antibody isotypes is a unique feature among known Fc receptors. This, together with the broad expression profile of TRIM21, allows it to function during all stages of an immune response at diverse sites.
Although the antibody binding properties and antiviral functions of TRIM21 have been well established, there are several questions regarding its biology that remain unanswered. Of particular interest is how the effector and signaling responses are regulated in order to avoid excessive inflammation. As both arms of TRIM21 function depend on a complex sequential ubiquitination process, which results in synchronized activation, regulation may depend on different activation thresholds for downstream ubiquitin-dependent processes. It will also be important to address how TRIM21 synergizes with other immune responses. For instance, ADIN may contribute to cross-presentation of viral peptides on MHC class I molecules, resulting in the recruitment of cytotoxic lymphocytes to the infected cell. While TRIM21 is promiscuous in its antibody binding, the functional implications of this are largely unexplored as are the isotype-specific responses that might be elicited. For instance, while TRIM21 binds all four subtypes of human IgG, it is not known whether their use during infection results in different responses. Moreover, as the four IgG subclasses differ significantly in their ability to interact with classical Fc receptors, TRIM21 activity could be altered indirectly. The main gateway for viral pathogens into the body is via mucosal surfaces. This raises intriguing questions as to whether or not TRIM21 makes a significant contribution to protection at these body sites, and whether it functionally intersects with the transepithelial transport of antibodies by FcRn and the pIgR. Recently, it was demonstrated that complement factor C3 attached to different non-enveloped viruses can be carried inside the cytosol during infection (128). Here, C3 detection resulted in proteasomal targeting of the virus and induction of inflammatory signaling in a process dependent upon mitochondrial antiviral-signaling protein (MAVS) (128). As such, it could be speculated that other intracellular receptors specific for other immune serum proteins with antiviral functions have yet to be discovered.
Acknowledgments
This work was supported in part by the Research Council of Norway through its Center of Excellence funding scheme (project number 179573). J. T. A. was supported by the Research Council of Norway (grant no. 230526/F20 and 179573/V40). S. F. was supported by the University of Oslo. L. C. J. and R. E. W. were supported by the Medical Research Council (UK; U105181010) and the European Research Council (281627−IAI).
References
- 1.Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 2.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 4.Dimmock NJ. Neutralization of animal viruses. Curr Top Microbiol Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 5.Parren PW, Burton DR. The antiviral activity of antibodies in vitro and in vivo. Adv Immunol. 2001;77:195–262. doi: 10.1016/S0065-2776(01)77018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 9.Brandtzaeg P. Secretory IgA: designed for anti-microbial defense. Front Immunol. 2013;4:222. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojas R, Apodaca G. Immunoglobulin transport across polarized epithelial cells. Nat Rev Mol Cell Biol. 2002;3:944–955. doi: 10.1038/nrm972. [DOI] [PubMed] [Google Scholar]
- 11.Dickinson BL, et al. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J Clin Invest. 1999;104:903–911. doi: 10.1172/JCI6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghetie V, Ward ES. Transcytosis and catabolism of antibody. Immunol Res. 2002;25:97–113. doi: 10.1385/IR:25:2:097. [DOI] [PubMed] [Google Scholar]
- 13.Spiekermann GM, et al. Receptor-mediated immunoglobulin G transport across mucosal barriers in adult life: functional expression of FcRn in the mammalian lung. J Exp Med. 2002;196:303–310. doi: 10.1084/jem.20020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He W, Ladinsky MS, Huey-Tubman KE, Jensen GJ, McIntosh JR, Bjorkman PJ. FcRn-mediated antibody transport across epithelial cells revealed by electron tomography. Nature. 2008;455:542–546. doi: 10.1038/nature07255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzaban S, et al. The recycling and transcytotic pathways for IgG transport by FcRn are distinct and display an inherent polarity. J Cell Biol. 2009;185:673–684. doi: 10.1083/jcb.200809122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko SY, et al. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514:642–645. doi: 10.1038/nature13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Palaniyandi S, Zeng R, Tuo W, Roopenian DC, Zhu X. Transfer of IgG in the female genital tract by MHC class I-related neonatal Fc receptor (FcRn) confers protective immunity to vaginal infection. Proc Natl Acad Sci USA. 2011;108:4388–4393. doi: 10.1073/pnas.1012861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sholukh AM, et al. Defense-in-depth by mucosally administered anti-HIV dimeric IgA2 and systemic IgG1 mAbs: complete protection of rhesus monkeys from mucosal SHIV challenge. Vaccine. 2015;33:2086–2095. doi: 10.1016/j.vaccine.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidji E, McDonagh S, Genbacev O, Tabata T, Pereira L. Maternal antibodies enhance or prevent cytomegalovirus infection in the placenta by neonatal Fc receptor-mediated transcytosis. Am J Pathol. 2006;168:1210–1226. doi: 10.2353/ajpath.2006.050482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai Y, et al. Intracellular neutralization of viral infection in polarized epithelial cells by neonatal Fc receptor (FcRn)-mediated IgG transport. Proc Natl Acad Sci USA. 2011;108:18406–18411. doi: 10.1073/pnas.1115348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burns JW, Siadat-Pajouh M, Krishnaney AA, Greenberg HB. Protective effect of rotavirus VP6-specific IgA monoclonal antibodies that lack neutralizing activity. Science. 1996;272:104–107. doi: 10.1126/science.272.5258.104. [DOI] [PubMed] [Google Scholar]
- 22.Roost HP, et al. Early high-affinity neutralizing anti-viral IgG responses without further overall improvements of affinity. Proc Natl Acad Sci USA. 1995;92:1257–1261. doi: 10.1073/pnas.92.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmann MF, et al. The role of antibody concentration and avidity in antiviral protection. Science. 1997;276:2024–2027. doi: 10.1126/science.276.5321.2024. [DOI] [PubMed] [Google Scholar]
- 24.Hangartner L, Zinkernagel RM, Hengartner H. Antiviral antibody responses: the two extremes of a wide spectrum. Nat Rev Immunol. 2006;6:231–243. doi: 10.1038/nri1783. [DOI] [PubMed] [Google Scholar]
- 25.Leung DT, et al. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumida SM, et al. Neutralizing antibodies to adenovirus serotype 5 vaccine vectors are directed primarily against the adenovirus hexon protein. J Immunol. 2005;174:7179–7185. doi: 10.4049/jimmunol.174.11.7179. [DOI] [PubMed] [Google Scholar]
- 27.Schrader JW, McLean GR. Location, location, timing: analysis of cytomegalovirus epitopes for neutralizing antibodies. Immunol Lett. 2007;112:58–60. doi: 10.1016/j.imlet.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 29.McEwan WA, James LC. TRIM21-dependent intracellular antibody neutralization of virus infection. Prog Mol Biol Transl Sci. 2015;129:167–187. doi: 10.1016/bs.pmbts.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Watkinson RE, McEwan WA, James LC. Intracellular antibody immunity. J Clin Immunol. 2014;34(Suppl):S30–S34. doi: 10.1007/s10875-014-0017-4. [DOI] [PubMed] [Google Scholar]
- 31.Randow F, MacMicking JD, James LC. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallery DL, McEwan WA, Bidgood SR, Towers GJ, Johnson CM, James LC. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci USA. 2010;107:19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauler F, Mallery DL, McEwan WA, Bidgood SR, James LC. AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. Proc Natl Acad Sci USA. 2012;109:19733–19738. doi: 10.1073/pnas.1210659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14:327–336. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bidgood SR, Tam JC, McEwan WA, Mallery DL, James LC. Translocalized IgA mediates neutralization and stimulates innate immunity inside infected cells. Proc Natl Acad Sci USA. 2014;111:13463–13468. doi: 10.1073/pnas.1410980111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogarth PM, Pietersz GA. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat Rev Drug Discov. 2012;11:311–331. doi: 10.1038/nrd2909. [DOI] [PubMed] [Google Scholar]
- 37.Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004;4:89–99. doi: 10.1038/nri1266. [DOI] [PubMed] [Google Scholar]
- 38.Bruhns P, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 39.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, Eversole T, Lee DJ, Sontheimer RD, Capra JD. Protein-protein interactions between native Ro52 and immunoglobulin G heavy chain. Scand J Immunol. 1999;49:620–628. doi: 10.1046/j.1365-3083.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- 41.Rhodes DA, et al. The 52 000 MW Ro/SS-A autoantigen in Sjogren's syndrome/systemic lupus erythematosus (Ro52) is an interferon-gamma inducible tripartite motif protein associated with membrane proximal structures. Immunology. 2002;106:246–256. doi: 10.1046/j.1365-2567.2002.01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol. 2007;44:2406–2414. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 43.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci USA. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol. 2012;770:27–37. doi: 10.1007/978-1-4614-5398-7_3. [DOI] [PubMed] [Google Scholar]
- 46.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 47.Rajsbaum R, Garcia-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakayama EE, Shioda T. Anti-retroviral activity of TRIM5 alpha. Rev Med Virol. 2010;20:77–92. doi: 10.1002/rmv.637. [DOI] [PubMed] [Google Scholar]
- 49.Rhodes DA, de Bono B, Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116:411–417. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napolitano LM, Meroni G. TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc Natl Acad Sci USA. 2014;111:2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fcgamma receptor in complex with Fc. J Biol Chem. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 53.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 54.Lu J, Chu J, Zou Z, Hamacher NB, Rixon MW, Sun PD. Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding. Proc Natl Acad Sci USA. 2015;112:833–838. doi: 10.1073/pnas.1418812112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Idusogie EE, et al. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J Immunol. 2000;164:4178–4184. doi: 10.4049/jimmunol.164.8.4178. [DOI] [PubMed] [Google Scholar]
- 56.Vaughn DE, Bjorkman PJ. Structural basis of pH-dependent antibody binding by the neonatal Fc receptor. Structure. 1998;6:63–73. doi: 10.1016/s0969-2126(98)00008-2. [DOI] [PubMed] [Google Scholar]
- 57.Oganesyan V, et al. Structural insights into neonatal Fc receptor-based recycling mechanisms. J Biol Chem. 2014;289:7812–7824. doi: 10.1074/jbc.M113.537563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 59.Kato K, et al. Model for the complex between protein G and an antibody Fc fragment in solution. Structure. 1995;3:79–85. doi: 10.1016/s0969-2126(01)00136-8. [DOI] [PubMed] [Google Scholar]
- 60.Sprague ER, Wang C, Baker D, Bjorkman PJ. Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol. 2006;4:e148. doi: 10.1371/journal.pbio.0040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis MJ, Pleass RJ, Batten MR, Atkin JD, Woof JM. Structural requirements for the interaction of human IgA with the human polymeric Ig receptor. J Immunol. 2005;175:6694–6701. doi: 10.4049/jimmunol.175.10.6694. [DOI] [PubMed] [Google Scholar]
- 62.Bonner A, Almogren A, Furtado PB, Kerr MA, Perkins SJ. The nonplanar secretory IgA2 and near planar secretory IgA1 solution structures rationalize their different mucosal immune responses. J Biol Chem. 2009;284:5077–5087. doi: 10.1074/jbc.M807529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pleass RJ, Dunlop JI, Anderson CM, Woof JM. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human fcalpha receptor (FcalphaR) CD89. J Biol Chem. 1999;274:23508–23514. doi: 10.1074/jbc.274.33.23508. [DOI] [PubMed] [Google Scholar]
- 64.Carayannopoulos L, Hexham JM, Capra JD. Localization of the binding site for the monocyte immunoglobulin (Ig) A-Fc receptor (CD89) to the domain boundary between Calpha2 and Calpha3 in human IgA1. J Exp Med. 1996;183:1579–1586. doi: 10.1084/jem.183.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woof JM. The human IgA-Fc alpha receptor interaction and its blockade by streptococcal IgA-binding proteins. Biochem Soc Trans. 2002;30:491–494. doi: 10.1042/bst0300491. [DOI] [PubMed] [Google Scholar]
- 66.Pleass RJ, Areschoug T, Lindahl G, Woof JM. Streptococcal IgA-binding proteins bind in the Calpha 2-Calpha 3 interdomain region and inhibit binding of IgA to human CD89. J Biol Chem. 2001;276:8197–8204. doi: 10.1074/jbc.M009396200. [DOI] [PubMed] [Google Scholar]
- 67.Ghumra A, et al. Structural requirements for the interaction of human IgM and IgA with the human Fcalpha/mu receptor. Eur J Immunol. 2009;39:1147–1156. doi: 10.1002/eji.200839184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Senior BW, Woof JM. Sites in the CH3 domain of human IgA1 that influence sensitivity to bacterial IgA1 proteases. J Immunol. 2006;177:3913–3919. doi: 10.4049/jimmunol.177.6.3913. [DOI] [PubMed] [Google Scholar]
- 69.McEwan WA, et al. Regulation of virus neutralization and the persistent fraction by TRIM21. J Virol. 2012;86:8482–8491. doi: 10.1128/JVI.00728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biedermannova L, Riley K, Berka K, Hobza P, Vondrasek J. Another role of proline: stabilization interactions in proteins and protein complexes concerning proline and tryptophane. Phys Chem Chem Phys. 2008;10:6350–6359. doi: 10.1039/b805087b. [DOI] [PubMed] [Google Scholar]
- 71.Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 72.Fu X, Ng C, Feng D, Liang C. Cdc48p is required for the cell cycle commitment point at Start via degradation of the G1-CDK inhibitor Far1p. J Cell Biol. 2003;163:21–26. doi: 10.1083/jcb.200307025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Janiesch PC, et al. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- 74.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 75.Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 76.Ramadan K, et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 78.Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- 79.Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, Young P. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J Mol Biol. 2009;394:732–746. doi: 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 80.Isakov E, Stanhill A. Stalled proteasomes are directly relieved by P97 recruitment. J Biol Chem. 2011;286:30274–30283. doi: 10.1074/jbc.M111.240309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 A resolution. Science. 2010;329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 83.Xie Y. Structure, assembly and homeostatic regulation of the 26S proteasome. J Mol Cell Biol. 2010;2:308–317. doi: 10.1093/jmcb/mjq030. [DOI] [PubMed] [Google Scholar]
- 84.Fletcher AJ, Mallery DL, Watkinson RE, Dickson CF, James LC. Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proc Natl Acad Sci USA. 2015;112:10014–10019. doi: 10.1073/pnas.1507534112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 87.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woof JM, Kerr MA. The function of immunoglobulin A in immunity. J Pathol. 2006;208:270–282. doi: 10.1002/path.1877. [DOI] [PubMed] [Google Scholar]
- 89.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 90.Kerr MA. The structure and function of human IgA. Biochem J. 1990;271:285–296. doi: 10.1042/bj2710285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Monteiro RC, Van De Winkel JG. IgA Fc receptors. Annu Rev Immunol. 2003;21:177–204. doi: 10.1146/annurev.immunol.21.120601.141011. [DOI] [PubMed] [Google Scholar]
- 92.Suzuki T, et al. Relationship of the quaternary structure of human secretory IgA to neutralization of influenza virus. Proc Natl Acad Sci USA. 2015;112:7809–7814. doi: 10.1073/pnas.1503885112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol. 2002;83(Pt 9):2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 94.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 95.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schaefer L. Complexity of danger: the diverse nature of damage-associated molecular patterns. J Biol Chem. 2014;289:35237–35245. doi: 10.1074/jbc.R114.619304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 100.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 101.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10:R110 003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 103.McEwan WA, Mallery DL, Rhodes DA, Trowsdale J, James LC. Intracellular antibody-mediated immunity and the role of TRIM21. BioEssays. 2011;33:803–809. doi: 10.1002/bies.201100093. [DOI] [PubMed] [Google Scholar]
- 104.Fuchs R, Blaas D. Uncoating of human rhinoviruses. Rev Med Virol. 2010;20:281–297. doi: 10.1002/rmv.654. [DOI] [PubMed] [Google Scholar]
- 105.Burckhardt CJ, Suomalainen M, Schoenenberger P, Boucke K, Hemmi S, Greber UF. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe. 2011;10:105–117. doi: 10.1016/j.chom.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 106.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leopold PL, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 108.Greber UF, Suomalainen M, Stidwill RP, Boucke K, Ebersold MW, Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brabec-Zaruba M, Pfanzagl B, Blaas D, Fuchs R. Site of human rhinovirus RNA uncoating revealed by fluorescent in situ hybridization. J Virol. 2009;83:3770–3777. doi: 10.1128/JVI.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brabec M, et al. Opening of size-selective pores in endosomes during human rhinovirus serotype 2 in vivo uncoating monitored by single-organelle flow analysis. J Virol. 2005;79:1008–1016. doi: 10.1128/JVI.79.2.1008-1016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Booy FP, Roden RB, Greenstone HL, Schiller JT, Trus BL. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 A resolution. J Mol Biol. 1998;281:95–106. doi: 10.1006/jmbi.1998.1920. [DOI] [PubMed] [Google Scholar]
- 112.Emini EA, Kao SY, Lewis AJ, Crainic R, Wimmer E. Functional basis of poliovirus neutralization determined with monospecific neutralizing antibodies. J Virol. 1983;46:466–474. doi: 10.1128/jvi.46.2.466-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Emini EA, Ostapchuk P, Wimmer E. Bivalent attachment of antibody onto poliovirus leads to conformational alteration and neutralization. J Virol. 1983;48:547–550. doi: 10.1128/jvi.48.2.547-550.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smith JG, Cassany A, Gerace L, Ralston R, Nemerow GR. Neutralizing antibody blocks adenovirus infection by arresting microtubule-dependent cytoplasmic transport. J Virol. 2008;82:6492–6500. doi: 10.1128/JVI.00557-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mandel B. The interaction of neutralized poliovirus with HeLa cells. I. Adsorption. Virology. 1967;31:238–247. doi: 10.1016/0042-6822(67)90167-5. [DOI] [PubMed] [Google Scholar]
- 116.Toogood CI, Crompton J, Hay RT. Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J Gen Virol. 1992;73(Pt 6):1429–1435. doi: 10.1099/0022-1317-73-6-1429. [DOI] [PubMed] [Google Scholar]
- 117.Varghese R, Mikyas Y, Stewart PL, Ralston R. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J Virol. 2004;78:12320–12332. doi: 10.1128/JVI.78.22.12320-12332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wetz K, Willingmann P, Zeichhardt H, Habermehl KO. Neutralization of poliovirus by polyclonal antibodies requires binding of a single IgG molecule per virion. Arch Virol. 1986;91:207–220. doi: 10.1007/BF01314281. [DOI] [PubMed] [Google Scholar]
- 119.Wohlfart CE, Svensson UK, Everitt E. Interaction between HeLa cells and adenovirus type 2 virions neutralized by different antisera. J Virol. 1985;56:896–903. doi: 10.1128/jvi.56.3.896-903.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Beuzon CR, Salcedo SP, Holden DW. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology. 2002;148(Pt 9):2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- 121.Birmingham CL, Brumell JH. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006;2:156–158. doi: 10.4161/auto.2825. [DOI] [PubMed] [Google Scholar]
- 122.Rakebrandt N, Lentes S, Neumann H, James LC, Neumann-Staubitz P. Antibody- and TRIM21-dependent intracellular restriction of Salmonella enterica. Pathog Dis. 2014;72:131–137. doi: 10.1111/2049-632X.12192. [DOI] [PubMed] [Google Scholar]
- 123.Watkinson RE, Tam JC, Vaysburd MJ, James LC. Simultaneous neutralization and innate immune detection of a replicating virus by TRIM21. J Virol. 2013;87:7309–7313. doi: 10.1128/JVI.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vaysburd M, et al. Intracellular antibody receptor TRIM21 prevents fatal viral infection. Proc Natl Acad Sci USA. 2013;110:12397–12401. doi: 10.1073/pnas.1301918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moore ML, McKissic EL, Brown CC, Wilkinson JE, Spindler KR. Fatal disseminated mouse adenovirus type 1 infection in mice lacking B cells or Bruton's tyrosine kinase. J Virol. 2004;78:5584–5590. doi: 10.1128/JVI.78.11.5584-5590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Welton AR, Gralinski LE, Spindler KR. Mouse adenovirus type 1 infection of natural killer cell-deficient mice. Virology. 2008;373:163–170. doi: 10.1016/j.virol.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Charles PC, Guida JD, Brosnan CF, Horwitz MS. Mouse adenovirus type-1 replication is restricted to vascular endothelium in the CNS of susceptible strains of mice. Virology. 1998;245:216–228. doi: 10.1006/viro.1998.9180. [DOI] [PubMed] [Google Scholar]
- 128.Tam JC, Bidgood SR, McEwan WA, James LC. Intracellular sensing of complement C3 activates cell autonomous immunity. Science. 2014;345:1256070. doi: 10.1126/science.1256070. [DOI] [PMC free article] [PubMed] [Google Scholar]