Summary
The tetrameric M2 protein from influenza A conducts protons into the virus upon acid activation of its His37 tetrad and is a proven drug target. Here, in studies of full-length M2 protein solubilized in native-like liquid-crystalline lipid bilayers, a pH titration monitored by solid-state nuclear magnetic resonance revealed a clustering of the first three His37 pKas (6.3, 6.3, and 5.5). When the +2 state of the tetrad accepts a third proton from the externally exposed portion of the channel pore and releases a proton to the internally exposed pore, successful proton conductance is achieved, but more frequently the tetrad accepts and returns the proton to the externally exposed pore, resulting in a futile cycle. Both dynamics and conformational heterogeneity of the His37 tetrad featuring short hydrogen bonds between imidazolium-imidazole pairs are characterized, and the heterogeneity appears to reflect oligomeric helix packing and the extent of transmembrane helical bending around Gly34.
Graphical abstract
Introduction
While structural biology often aims for high resolution, meaning unique, precisely determined atomic positions, functional characterizations have increasingly implicated conformational heterogeneity and exchange dynamics. For membrane proteins, this is further complicated by the structural influence of membrane mimetic environments used to model the protein's native environment in experimental samples. These difficulties often force structural biologists to modify the native protein sequence via truncations, insertions, or site-specific modifications. In studies of the M2 proton channel, all of these tools have been used to gain structural and functional insights into this important drug target. Over the past 15 years truncated versions of M2 have been studied by solid-state nuclear magnetic resonance (ssNMR) (Andreas et al., 2010, 2015; Cady et al., 2010; Hu et al., 2007; Nishimura et al., 2002; Sharma et al., 2010) and solution NMR (Pielak and Chou, 2010; Schnell and Chou, 2008), electron spin resonance (ESR) (Nguyen et al., 2008; Thomaston et al., 2013), and crystallography (Acharya et al., 2010; Stouffer et al., 2008) utilizing a wide variety of detergent and lipid environments. With only a couple of exceptions (Tian et al., 2002, 2003) the full-length protein has not been studied until recent years (Liao et al., 2013; Miao et al., 2012, 2013) as the molecular and structural methodologies and technologies have advanced. Here, studies of the full-length tetrameric M2 (M2FL) protein by ssNMR is presented, demonstrating single-site resolution in this tetrameric complex (46 kDa) in liquid-crystalline lipid bilayers.
All the M2 structures in the PDB are from the transmembrane (TM) domain (M2TM, residues ∼22–46) or the conductance domain (M2CD, residues ∼22–62), and all show a left-handed tetrameric helical bundle for the TM domain, with one TM helix from each subunit. Details of the helical tilt, crossing point, and dimensions of the aqueous pore vary, resulting in differences in drug-binding sites and side-chain conformations for the His37 tetrad that is responsible for shuttling protons across this barrier (Zhou and Cross, 2013b). One helical turn below the histidine residues is a tetrad of Trp41 residues that acts as the primary gate for proton conductance (Tang et al., 2002). To the C terminus of the TM helix is an amphipathic helix that has been shown to interact with the interfacial region of the lipid bilayer (Sharma et al., 2010; Tian et al., 2003) and that appears to have a role in viral budding (Rossman et al., 2010; Rossman and Lamb, 2011). The G34A M2TM crystal structure identified water molecules in the N-terminal pore above the His tetrad as well as two waters between the His and Trp tetrads (Acharya et al., 2010) that are important for the interpretation of spectral results.
Through these multiple structures, the influence of the protein's environment for structure, dynamics, and function has become clearer not only for M2 but also for this important and very common class of membrane proteins (Zhou and Cross, 2013a, 2013b). While in solution NMR and X-ray diffraction studies the proteins are placed in detergent micelles and crystal lattices, in ESR and ssNMR studies the proteins are solubilized in lipid bilayers possessing dramatic gradients in dielectric constant and hydrophobicity, a defined hydrophobic thickness, and a lateral pressure profile that is unmatched by the detergent environments. Hong and coworkers have pioneered the use of model raft-like lipid environments (dipalmitoylphosphatidylcholine, dipalmitoylphosphatidylethanolamine, egg sphingomyelin, and cholesterol in a molar ratio of 28:21:21:30) for studies of M2 (Hu et al., 2011). These lipids model the bulk lipid environment of the Influenza viral coat. Griffin and coworkers have primarily used diphytanoylphosphatidylcholine (DPhPC) bilayers, a lipid from Archaea membranes that has two 3,7,11,15-tetramethylhexadecanoic acyl chains (Andreas et al., 2010). This lipid has the advantage that its phase transition temperature is exceptionally low (less than −120°C) (Lindsey et al., 1979) so that ssNMR studies at low temperature can still be performed in the liquid-crystalline phase. Our studies over the past 5 years have mostly been in lipid bilayers of DOPC (dioleoylphosphatidylcholine) and DOPE (dioleoylphosphatidylethanolamine), i.e. lipids that form liquid-crystalline bilayers, for it is well recognized now that M2 is not in the raft-like bulk lipid environment of the viral coat, but because it induces membrane curvature during viral budding it is more closely associated with the periphery of the raft-like domains, in contrast to hemagglutinin and neuraminidase that cluster in the raft-like domain (Rossman et al., 2010; Rossman and Lamb, 2011).
The first pH titration of the M2TM domain His37 tetrad (Hu et al., 2006) concluded that there was cooperative proton binding for the first two charges (pKas of 8.2), a third charge (pKa of 6.3) to activate the channel, followed by a fourth charge (pKa <5). Hong and coworkers have characterized the pKas for the M2TM domain in the raft-like lipids at 7.6, 6.8, 4.9, and 4.2 (Hu et al., 2011), and recently using a construct of 21–97 in raft-like lipids Hong and coworkers obtained pKas of 7.1, 7.1, and 5.4. Griffin and coworkers conducted a pH titration of M2CD in DPhPC lipids in which the first two His37 pKas were 7.5 and the second two pKas 4.5 (Colvin et al., 2014). Despite the differences in construct and lipid environment, the first two pKas appear to be above the pH of the endosome that activates the M2 channel, suggesting that the active state has either +2 or +3 charges for the His tetrad. Consequently, there appears to be consensus that there is cooperative proton binding for the first two protons, as originally suggested by Hu et al. (2006). It is clear that if a charge is distributed over two imidazole rings and hydrogen-bonded waters, that charge-charge repulsion in the histidine tetrad can be minimized, especially if the rings form imidazole-imidazolium hydrogen-bonded pairs (Dong et al., 2013).
Here, we report magic-angle spinning (MAS) ssNMR spectroscopy of selectively His37-labeled M2FL protein in lipid bilayers. Contrary to M2TM and M2CD models involving discrete conformations (Andreas et al., 2015; Pielak and Chou, 2010; Schnell and Chou, 2008; Sharma et al., 2010), our M2FL NMR spectra reveal significant conformational heterogeneity, especially under acid pH conditions whereby the channel becomes active. The spectra at each pH can be interpreted as arising from distinct groups of conformations that are in exchange on the millisecond-to-second timescales. One group of conformations features a pair of short hydrogen bonds between His37 imidazole and imidazolium. Another group of conformations feature the His37 tetrad in the +3 charge state, with dipolar couplings between multiple imidazolium and imidazole sites and between imidazolium and water. The heterogeneous conformational states have considerable lifetimes both at −10°C and at +23°C, suggesting that the conformational heterogeneity originates from side-chain rotameric states in the oligomeric helix packing. Proton conductance occurs through many futile proton transfers between the His37 tetrad and the surrounding water molecules and occasional productive shuttling.
Results and Discussion
Significant Conformational Heterogeneity Accompanies pH Titration
The pH titration of M2FL illustrated by a set of His37 side-chain 15N spectra at pH values between 4.5 and 8.8 (Figure 1) is complex, reflecting the sophisticated chemistry associated with proton conductance. At pH 8.8, five protonated nitrogen resonances are observed between 155 and 175 ppm in the 1D 15N spectrum, representing four τ and one π tautomer of the His37 side chains and indicating that the tetrameric protein has variable populations of τ and π tautomers at high pH. Two intense nonprotonated nitrogen resonances are also observed between 248 and 253 ppm. At pH 6.6, the protonated 15N resonances below 160 ppm become weak and the remaining protonated resonances start to broaden, with some spectral intensity observed in a broad envelope between 175 and 200 ppm. Likewise, some nonprotonated intensity is observed in a broad envelope between 248 and 230 ppm. At lower pH these broad spectral components grow in intensity.
Figure 1. pH Titration of the His37-Labeled M2FL at-10°C Using 1D 15N MAS NMR Spectra at 600 MHz for 1H and 12.0 kHz.
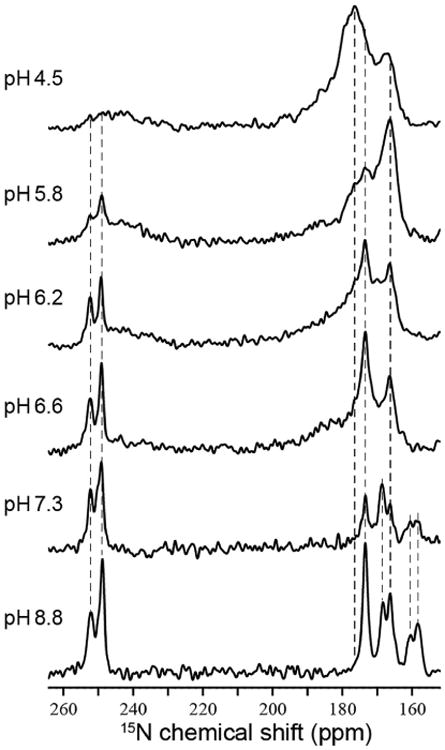
To optimize the signal-to-noise ratio for both NH and Nnon-H signals, low-pH samples used longer cross-polarization contact time than high pH samples during data collection. Dashed lines are drawn to help show spectral changes or lack thereof in the frequencies as a function of pH. Contact times: pH 4.5, 5 ms; pH 5.8, 5 ms; pH 6.2,5 ms; pH 6.6, 4 ms; pH 7.3, 2.5 ms; pH 8.8, 1.6 ms. See Figure S1 for a plot of the pH titration.
The aromatic/aliphatic and aromatic/aromatic regions of the 13C-13C correlation (CC) spectrum at pH 7.3 (Figure 2A) superficially suggest a dimer-of-dimer conformation as described by Griffin and coworkers (Andreas et al., 2010, 2012) and as suggested by the conductance domain structures in lipid bilayers (Andreas et al., 2015; Sharma et al., 2010). However, the asymmetry of some of these resonances suggests the possibility of additional heterogeneity. At pH 6.6 new resonances in the CC spectrum start to appear, reflecting charged His37 residues (Figure 2B), and by pH 6.2 numerous cross-peaks between the histidyl resonances are present in the aromatic/aromatic region (Figure 2C). At pH 5.8, cross-peaks that reflect distance restraints and conformational exchange are even stronger (Figure 2D).
Figure 2. Aromatic/Aliphatic and Aromatic/Aromatic Regions of 2D 13C-13C Correlation Spectra with 50-ms Mixing Time Using PARIS Irradiation Recorded at −10°C on His37-Labeled M2FL as a Function of pH.
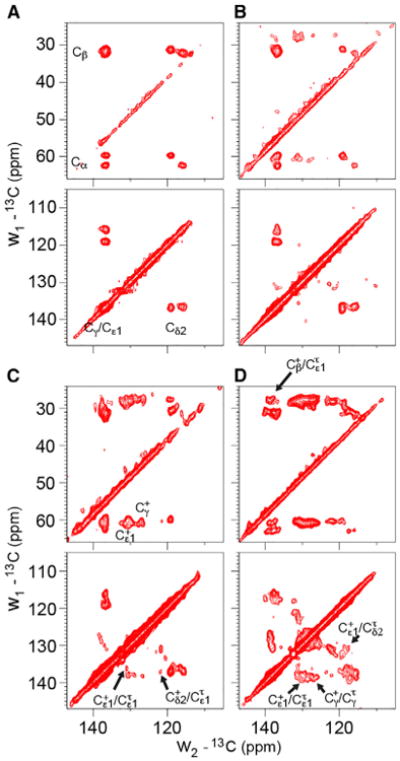
(A) pH 7.3. Apparent doubling of the backbone Cα, Cβ and side-chain Cδ2, Cγ/Cε1 resonances; only neutral τ and π tautomers are present.
(B) pH 6.6. Charged H37 resonances are first observed at this pH. Cγ and Cε1 resonances of the charged tautomers are resolved.
(C) pH 6.2. Heterogeneity is apparent in the contour shapes for both charged and neutral species. Some of the Cδ2 resonances are missing in the aliphatic/aromatic spectrum. A subset of cross-peaks between neutral and charged H37 are indicated with arrows in the aromatic-aromatic region.
(D) pH 5.8. Cross-peaks between neutral and charged H37 are becoming prominent at this pH, and heterogeneity is clear in the contoured line shapes.
In the 15N-13C correlation (NC) spectrum at pH 7.3 (Figure 3A), there is no evidence of charged histidine residues, in agreement with the CC results, confirming that the multiple protonated nitrogen resonances in Figure 1 represent only neutral τ and π tautomers at pH 7.3. A single π species is prominent in this NC spectrum (cross-peaks at 168/127 and 250/119). At least three τ species are observed, with Cδ2 (115–119 ppm) and Cε1 (135–138 ppm) resonances correlating with the protonated Nε2 resonances in the 159–175 ppm region. At pH 6.6 and 6.2 (Figures 3B and 3C), the NC spectra show the emergence of multiple broad resonances, between 175 and 200 ppm and between 248 and 232 in the 15N dimension, that extend considerably into the gap between the protonated and nonprotonated nitrogen resonances observed in the high-pH spectra. These resonances are consistent with broad resonance intensities seen in the 1D 15N spectra at the same pH (Figure 1).
Figure 3. 2D 15N-13C zf-TEDOR Spectra of His37-Labeled M2FL at Different pH Values Collected at −10°C.
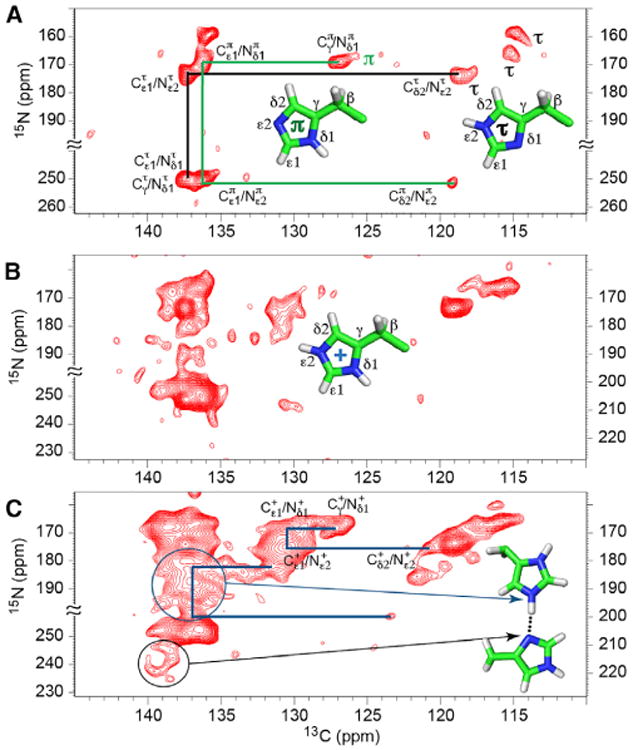
The total TEDOR mixing time of 1–1.3 ms was used to obtain one-bond 15N-13C correlations. Structures of the π, τ, and charged states of the His residues are displayed.
(A) pH 7.3. Three τ and one π are labeled. Only the assignments for one τ (black) and the observed π (green) tautomers are traced in the figure. The assignments of the other τ tautomers can be similarly traced.
(B and C) The pH 6.6 (B) and pH 6.2 (C) spectra were obtained with 15N frequency folded about 228.1 ppm (pH 6.2) and 227.3 ppm (pH 6.6). “≈” indicates bounds of the folded frequency range. The scales on the lower left and on the right are for nonprotonated and protonated nitrogens, respectively. The presence of charged histidine resonances is evident, and considerable heterogeneity is apparent among the neutral tautomer resonances. In (C), examples of traced resonance patterns for charged tautomers (blue lines) are illustrated as in (A) for the neutral tautomers. Resonances generated from multiple imidazolium-imidazole hydrogen-bonded nitrogen sites are circled. The black circle includes multiple nonprotonated nitrogen resonances associated with these hydrogen bonds, while the blue circle includes multiple resonances associated with the protonated nitrogen resonances associated with these hydrogen bonds.
Our NMR data do not allow for detailed structural characterization of the His37 tetrad at high pH (where the channel is nonconducting), due to the complex mixture of tautomeric species suggesting heterogeneity among the M2 tetramers. However, it is anticipated that a relatively stable conformation for the high pH His37 tetrad involves two pairs of τ-τ hydrogen bonds, each between a protonated Nε2 and a nonprotonated Nδ1. This is reminiscent of the dimer-of-dimer conformation that was initially proposed for the +2 charge state of the His37 tetrad (Hu et al., 2006). These hydrogen bonds between His side chains may undergo partner exchange in the tetramer, as proposed in 2010 (Sharma et al., 2010).
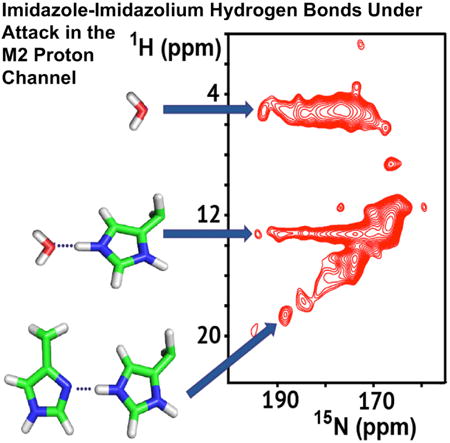
At lower pH much more can be described. In Figure 4 the 1H-15N correlation (HN) spectrum at pH 6.2 shows a few well-resolved resonances with exceptionally high 1H frequencies (up to 19 ppm) and high protonated 15N frequencies (up to 193 ppm). Previously Hong and coworkers published similar spectra, except that 15N frequencies were not observed above 182 ppm and 1H frequencies were not observed above 16 ppm at pH 4.5 (Hong et al., 2012). More specifically, spectra obtained at pH 6.0 by these authors showed the 15N resonances restricted to 172 ppm and 1H resonances restricted to less than 13 ppm, with no cross-peaks to water resonances at 5 ppm, in contrast to what we observe here. Whether this is a result of the M2TM construct or the viral membrane environment is not clear. As described above, the 1D 15N spectrum (Figure 1) and NC spectrum (Figure 3) at pH 6.2 also display the same high-frequency components for the protonated 15N resonances. These resonances are likely from Nε2 sites of charged His37 residues that feature lengthened NH covalent bonds and participate in short hydrogen bonds. The nonprotonated nitrogen resonances extending to 232 ppm as shown in the lower pH spectra of Figures 1 and 3 suggest slight protonation of the nominally nonprotonated sites. Together, these results present direct evidence for the formation of short imidazolium-imidazole hydrogen bonds as originally proposed in 2006 (Hu et al., 2006). To distinguish between homogeneous and heterogeneous broadening for the two broad-resonance envelopes in the 1D 15N spectra, 15N spin-echo spectra were obtained at pH 6.2 with echo times up to 5 ms (Figure S2). Homogeneous broadening would have led to a substantial loss in spectral intensity at the longer echo times, but this was not observed and, therefore, the broadening is heterogeneous, thereby reinforcing the dramatic conformational complexity that is already implicated by the multiple resonances in the NC and HN spectra.
Figure 4. 2D 1H-15N HETCOR Spectrum of the pH 6.2 Sample of His37-Labeled M2FL.
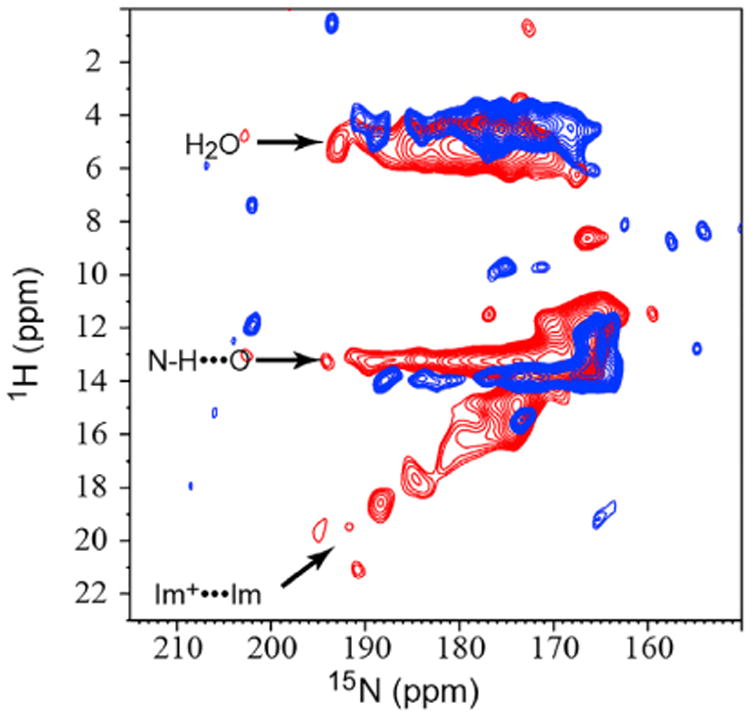
Blue: −10°C; red: +23°C. The 15N chemical shifts that range up to 193 ppm are coupled to multiple 1H frequencies that range up to 18.5 ppm. The ∼5 ppm 1H resonances reflect water protons and the resonances at ∼13 ppm resonances reflect the NH protons hydrogen bonded to water. The linear correlation between the high 1H and 15N frequencies reflect multiple short hydrogen-bonded states between imidazolium and imidazole. At +23°C, resonance intensities are greatly reduced due to a loss of sensitivity, but high 15N frequencies are present, indicating that the imidazolium-imidazole hydrogen-bonded states are still present. Support for the heterogeneous interpretation of these resonances is supported by Figure S2.
The complexity of the spectra, and the difficulty to obtain relaxation data for the array of nitrogen resonances between 155 and 252 ppm with varying relaxation rates, present challenges for accurately determining the His37 pKas from spectral intensities. We therefore took a heuristic approach based on the NC, CC, and 1D 15N spectra: the charges on the His37 tetrad were estimated to be 0 at pH 8.8 and 7.3, 0.5 ± 0.2 at pH 6.6, 1.4 ± 0.2 at pH 6.2, 2.2 ± 0.2 at pH 5.8, and 2.9 ± 0.2 at pH 4.5. Fitting these pH-dependent charges to a titration model yields apparent pKas of 6.3 ± 0.1, 6.3 ± 0.1, and 5.5 ± 0.3 (Figure S1). The first two pKas are significantly lower than in the first characterization from the M2TM construct (Hu et al., 2006). In this early study the calculation for the first pKas was largely based on the disappearance of the nonprotonated nitrogen resonances, which may have had a lot more to do with exchange dynamics than protonation of the histidine residues. For the M2FL protein, it is clear that the nonprotonated resonances are still at full strength at pH 7.3. While the pH-dependent data here were obtained at −10°C, in Figure 4 an HN spectrum at +23°C shows that the chemistry at −10°C and +23°C is similar. The rapid rise in the spectral intensities for the charged species below pH 7.3 shows that there is little opportunity for a singly charged His37 tetrad. The absence of a +1 charge state means that the binding of the first two protons to the His37 tetrad is cooperative with a Hill coefficient of 2, consistent with our early result (Hu et al., 2006) and other recent results (Colvin et al., 2014; Liao et al., 2015). The resulting +2 charge state, as suggested by the protonated and nonprotonated 15N chemical shifts as well as the 1H chemical shifts, contains short imidazolium-imidazole hydrogen bonds with lowered barriers for proton crossing between Nε2 and Nδ1. The sharing of a proton between Nε2 and Nδ1 sites permits significant delocalization of the charge and a reduction in the charge-charge repulsion between the two imidazolium-imidazole dimers of the His37 tetrad in the low-dielectric environment near the center of the lipid bilayer (Dong et al., 2013). The proximity of the third pKa value to the first two pKa values means that the +2 and +3 states coexist over a significant pH range. At pH 6.2, where the channel should become activated, our titration model (see Figure S2) estimates that the M2 tetramers have a 34% probability to be in the neutral state, a 55% probability to be in the +2 state, and an 11% probability to be in the +3 state.
Extensive Dynamics and Chemical Exchange Accompany Channel Activation
The HN spectrum at pH 6.2 (Figure 4) shows correlated resonances that indicate extensive dynamics and chemical exchange. The highly protonated 15Nε2 frequencies associated with short imidazolium-imidazole hydrogen bonds are coupled not only to the covalently bonded 1H that also participates in a short hydrogen bond (with chemical shifts up to 19 ppm), but also to 1H frequencies at 5 and 13 ppm. The 13 ppm chemical shift may be assigned to an imidazolium NH that is hydrogen bonded to water or to a carbonyl group, i.e., a species in which the short imidazolium-imidazole hydrogen bond is broken, presumably by protonation of the imidazole through attack by water or hydronium ion (leading to the +3 state). The fact that these resolved 1H resonances are not averaged suggests that the exchange rate between these states is slow on the timescale of the 1H frequency separation (∼3 ms), but fast compared to the 15N T1 relaxation rate (estimated to be 5 s). Moreover, the 5 ppm chemical shift can be assigned to a water proton bound to the same 15Nε2, implying that, once the imidazole-imidazolium hydrogen bond is broken, the Nε2H proton site can exchange with water at a rate that is slow compared with that of the 1H frequency separation (∼2 ms) and fast compared with that of the 15N T1 relaxation rate. This frequent proton exchange with water is the reason for our word choice of “short” instead of “strong” to describe these hydrogen bonds. At +23°C, the HN spectrum no longer shows the high-frequency 1H resonances (most likely the result of reduced sensitivity due to the short 1H T1ρ), but the His37 conformational heterogeneity and dynamics associated with these frequencies are still present, as indicated by the same high frequencies for the protonated 15N sites. Consequently, the short hydrogen-bonded states characterized at low temperature reflect the chemistry that occurs at (near) physiological temperatures. Furthermore, the proton exchange rate with water following the breaking of a short hydrogen bond is restrained to the same range as at lower temperature.
It should be noted that not all of the protonated nitrogen resonance frequencies exchange with water. The lowest frequencies in this spectrum at 166 ppm do not exchange. These protonated resonances are associated with Nε2 of the τ tautomer of the imidazolium-imidazole hydrogen-bonded pair. This assignment for the 166-ppm resonance is confirmed by the still strong 166-ppm resonance in the pH 4.5 spectrum (Figure 1), where the uncharged tautomers are in the minority and almost all are hydrogen bonded to charged tautomers, as confirmed by loss of the sharp nonprotonated resonances. The τ tautomer has its Nδ1 site involved in the imidazolium-imidazole hydrogen bond, while its Nε2 site is sequestered near the backbone in the inter-His/Trp tetrad space away from the few waters in this region. Indeed, it was hypothesized that this Nε2H would form an interaction with one of the indole rings (Hu et al., 2006; Sharma et al., 2010).
The CC spectrum at pH 6.2, and particularly that at pH 5.8, utilizing a mixing time of 50 ms, provides insights into the structure and dynamics of the charged His37 tetrad (Figure 2). Multiple cross-peaks between charged and τ tautomers are observed. These include Cε1(+)-Cδ2(τ) (130.7/117.7 ppm) and Cδ2(+)-Cε1/Cγ(τ) (122.3/138 ppm), consistent with the corresponding intradimer distances of 6.1 Å and 4.5/4.7 Å, respectively, in the PDB: 2L0J structure of the histidine tetrad with a +2 charge (Hu et al., 2006; Sharma et al., 2010). Despite the relatively short mixing time and the dynamics of these imidazole rings, the very limited 13C labeling permits the observation of relatively long distances. Another cross-peak was observed between imidazolium Cβ and imidazole Cε1 (27.5/136.2 ppm), marginally consistent with an interdimer distance of 6.6 Å. In addition, cross-peaks between charged and neutral tautomers involving just Cε1 (131/137 ppm) and just Cγ sites (127/138 ppm) can be explained by either intradimer distances of 4.3 and 6.0 Å, respectively from PDB: 2L0J, or chemical exchange between the charged and τ tautomers resulting from deprotonation/protonation. If this is due to chemical exchange, the exchange rate must be rapid on the mixing timescale of 50 ms and slow compared with the frequency separation that is on the order of 1 kHz (or 1-ms timescale). These timescales are consistent with the observed exchange processes from the HN spectra.
Conformational Heterogeneity in the His37 Tetrad
Remarkably the various imidazolium-imidazole hydrogen-bonded conformations identified by different 15N and 1H frequencies do not interchange on a timescale that would average the protonated or nonprotonated 15N frequencies or the various 1H frequencies at −10°C. In other words, the breaking and reforming of these short hydrogen bonds result in the same set of frequencies, otherwise there would be averaging of the frequencies in the HN spectra. The stability of the structural geometries that result in the specific set of 1H/15N/13C frequencies is significant. At a minimum the 15N frequencies are not averaged and their separation is as small as 200 Hz, implying lifetimes >5 ms. The ranges for the 15N and 1H frequencies suggest that these geometries differ by only a few tenths of an angstrom. These differences are unlikely to arise from variations in His37 side-chain or backbone torsion angles, since variations in these torsion angles would be unlikely to produce multiple conformational states with geometries differing in tenths of an angstrom and having lifetimes longer than 5 ms.
A second possible source for the varying hydrogen-bonding geometries is the variations in backbone torsion angles. While backbone hydrogen bonds of TM helices are strengthened in the low dielectric environment of lipid bilayers (Kim and Cross, 2002; Page et al., 2008), helix bending around Gly34 has been implicated by ssNMR spectroscopy (Hu et al., 2007; Li et al., 2007) and observed in molecular dynamics simulations (Yi et al., 2009). It is conceivable that several backbone helical conformations, differing in the extent of bending around Gly34 and possibly associated with the packing of the bulky Trp41 side chains in the pore, would produce a variety of ways to harbor the His37 tetrad. At the pH where the +2 state for the His tetrad dominates, it is now clear that imidazolium-imidazole hydrogen bonds dominate and, hence, dimer-of-dimer structure occurs. In the diphytanoyl lipids used by Griffin and coworkers (Andreas et al., 2010, 2012) the dimer-of-dimer structure appears to be evident throughout most of the TM helices. However, it is not clear that variation in backbone structure alone can account for the many distinct imidazolium-imidazole geometries observed here.
A third possible source is different packing geometries for the helices, i.e., the variation arises at the quaternary structure level. Indeed, it has been observed previously that over the course of a couple of days all of the backbone amides in the TM helix exchange with D2O, whereas many of the amide sites in the amphipathic helix do not (Tian et al., 2003). While this suggests rotational excursions on a much longer timescale than what is observed here as heterogeneous interactions in the His tetrad, it does suggest dynamics at the helix-helix interface and the potential for structural heterogeneity on much shorter timescales. Most of the interhelical interactions in the TM domain are dominated by van der Waals interactions. Multiple large hydrophobic residues are involved in these interfaces such that different rotameric states could lead to slight differences in the helix-helix separation. Such packing geometries, perhaps in concert with varying helix bending, may generate the observed structural heterogeneity for the histidine tetrad. Indeed, in PDB: 2L0J Leu38 residues at the helix-helix interface are found in two different rotamers, and correspondingly there is a 0.1-Å difference in distance between the His37 Cα carbons. These results have implications for other oligomeric membrane proteins. Many such proteins have increased stability by having more than one TM helix per monomer, but still the assumption to date for many of the oligomeric membrane protein structures in the PDB has been to apply rotational symmetry in structural calculations. As illustrated here, oligomeric asymmetry may lead to unique and heterogeneous native chemistry.
Conductance Mechanism
Despite the chemical complexity of the histidine states, the concept for the proton conductance mechanism is clear: there is no opportunity for a water wire through the His37 tetrad—instead, the His residues are protonated and deprotonated. However, the chemistry of the His37 residues appears to be highly dependent on the protein's environment and/or on the protein construct. Here, with the full-length protein we have shown that the His37 residues have three pKas clustered within a single pH unit so that the protein can initiate proton transport at a pH close to the pH at which the tetrad first takes on a charge. In other words, once an observable concentration of the +2 charge state is achieved with each hydrogen-bonded imidazolium-imidazole pair sharing a charge, there is an opportunity for one of the pairs to obtain a second charge from a hydronium ion in the aqueous pore exposed to the external surface of the virus. For a productive transfer of the proton to the viral interior (Figure 5), this pair of residues must donate a proton to water in the space between the His37 tetrad and Trp41 tetrad, and the proton must further travel down a water wire formed when the tryptophan gate is opened.
Figure 5. Functional Role of the His37 Tetrad from the Studies of the Full-Length M2 Protein.
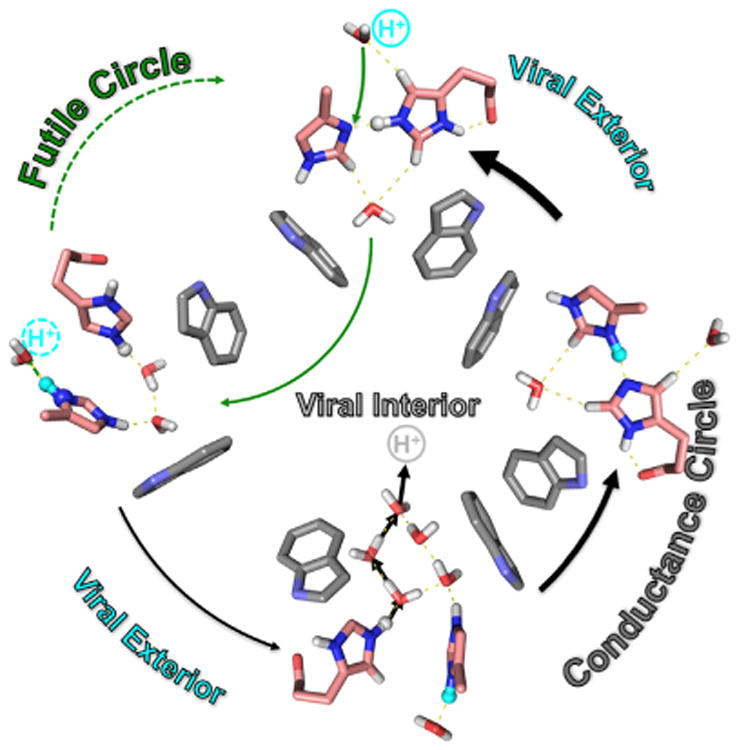
Two cycles can be described involving the protonation of the imidazolium-imidazole dimers and the deprotonation of a pair of imidazoliums. The futile cycle results when the protonation and deprotonation is from and to the same aqueous compartment. A productive conductance cycle occurs when protonationis from one compartment (externally exposed) and deprotonation is to the other compartment (internally exposed). Once the Trp41 gate opens, a water wire can carry the proton into the viral interior and the imidazolium and imidazole return to their starting hydrogen-bonded configuration.
Here, the imidazolium-imidazole dimers have been observed and short hydrogen bonds documented for these dimers. Furthermore, dynamics and proton exchange document the attack of a hydronium ion resulting in a +2 charge for the dimer (+3 state for the histidine tetrad). The charge-charge repulsion for a charged pair results in the exposure of Nε2Hs from both rings to the aqueous space between the His and Trp tetrads, while the Nδ1Hs of one ring is sequestered near the polypeptide backbone and the other is exposed to the waters in the external aqueous pore. If this later Nδ1H is captured by water, the imidazolium-imidazole dimer is reformed and a futile cycle results with no proton conductance. However, if a water in the space between the His and Trp tetrads captures a proton from one of the protonated Nε2 sites the imidazolium-imidazole dimer can again reform, with the result that a proton has been transported across the His tetrad. When the Trp gate opens, transport is completed into the viral interior. Release of an Nε2H to a water below the histidine tetrad directly leads to an imidazolium-imidazole dimer with the shared proton covalently bonded to the Nδ1 site instead of the Nε2 site. The shared proton is then expected to rapidly move across a low-energy barrier to covalently bond with the Nε2 site, thereby recovering the initial, more stable dimer configuration.
For a viral protein that has been exquisitely evolved to perform multiple biological functions, is the spectral, chemical, dynamic, and structural complexity of the His 37 tetrad functionally important? Several positive arguments can be readily made. For example, the conformational heterogeneity can be anticipated to facilitate water access. Indeed, it may be that the slow rate of proton exchange is the result of limited water access to nitrogen sites sequestered by the formation of the imidazolium-imidazole dimers. The variations in helix-helix packing and helix bending that likely generate the conformational heterogeneity of the His37 tetrad may also produce a range of water access, with the subpopulation having a high degree of water access primed for hydronium ion attack. In addition, the conformational heterogeneities in the +2 and +3 charge states suggest that there may be significant conformational overlap between these two states at the tertiary and quaternary levels. This conformational overlap allows for the third proton to jump easily between the +2 and +3 states. There is no evidence here that protons are selecting specific conformational states during the conductance cycle; instead, conformational heterogeneity appears to facilitate the cycling between charged states.
The structure, dynamics, and function of M2 are dependent on the construct (M2TM [∼22–46], M2CD [∼22–64], or the M2FL 97 residue protein) and the environment in which the protein is studied (detergent micelles, detergent crystal lattice, and various lipid bilayers). In a mixture of lipids that models the bulk of the viral coat, the antiviral drug amantadine does not block the pore of the M2CD construct (Cady et al., 2011) and neither do the antiviral drugs bind in the pore of M2CD solubilized in dihexanoylphosphatidylcholine micelles (Schnell and Chou, 2008). In DPhPC lipid bilayers of M2CD there is only a pair of resonances representing protonated nitrogens and a pair of resonances representing nonprotonated nitrogens over the entire pH range from 8.5 to 4.5 (Colvin et al., 2014), and yet two pKas were identified at pH 7.6 without any change in chemical shift. Clearly the chemistry of the His tetrad is sensitive to both the construct and its environment. While it has become well known that detergent-based environments can lead to substantial structural perturbations of membrane proteins, it is emphasized here that the lipid composition can lead to significant structural and functional perturbations, i.e., the physical properties of the membrane may influence the conformation or set of conformational states for the “active site,” i.e., the His37 tetrad, in the M2 protein. Our titration results here with the M2FL in liquid-crystalline lipid bilayers at −10°C with some additional data at room temperature present very different results that are consistent with the activation of the channel near pH 6 (Wang et al., 1995).
The implications of the conformational heterogeneity in the M2 protein tetramer are manifold. There is no simple or even elegant picture for proton conductance as we, and others, have been striving to achieve. Proton conductance is known to occur via a mechanism whereby the protons from the viral exterior bind to His37 followed by a side-chain conformational rearrangement and release via a water wire into the viral interior. Here, it is demonstrated that multiple conformational states can accept a proton for transport, and consequently, conformational selection does not seem to apply. Instead, we hypothesize that the conformational heterogeneity facilitates both water access to relatively sequestered nitrogen sites in the imidazole-imidazolium dimers and the cycling between different charged states. It is also known that the proton conductance of this channel is remarkably inefficient, perhaps a few hundred protons per second (Lin and Schroeder, 2001; Mould et al., 2000). Here, it is shown at −10°C that the short hydrogen bonds are broken at a rate that is on the millisecond to slightly submillisecond timescale, and at room temperature this rate does not appear to change substantially and does not appear to limit proton conductance. Instead, it is the Val27 and Trp41 gates that are recognized as the ultimate rate limiter (Zhou, 2011).
Many helical membrane proteins form oligomers that are stabilized by weak quaternary interactions, such as M2. Therefore, it can be expected that there will be significant conformational heterogeneity including rotational asymmetry, even if the average structure possesses rotational symmetry. Indeed, for M2FL rare helical rotational excursions have been observed through H-D exchange experiments (Tian et al., 2003). While these excursions were observed on the 104- or 105-s timescale, it suggests that far less dramatic helical motions may occur on much shorter timescales, such as those observed here. Based on the weak, primarily van der Waals interactions that hold the tetramer together, we should expect some heterogeneity in the quaternary structure; such heterogeneity would modulate the imidazole-imidazolium hydrogen-bond geometry, as we observe in both the HN and NC correlation spectra. As with M2, it can also be anticipated that details of the lipid environment and truncated constructs may have significant effects on the conformational states of other oligomeric proteins. These effects are likely to be more significant with smaller helical bundles as opposed to larger ones. In any event, it can be anticipated that the rotational symmetry that is typical of homomeric structures such as those of K+ channels, while representative of an average, may not do justice to the range of conformational states that no doubt exist, are likely influenced by the lipid environment, and have functional implications.
Conclusions
The unique histidine tetrad takes on multiple charges even though it is immersed in the lipid bilayer with only a single helical shell separating it from the low dielectric of the lipid bilayer interstices. In the +2 state, each charge is distributed over an entire imidazole-imidazolium dimer through a short hydrogen bond. In the +3 state, charge-charge repulsion is mitigated by interactions with waters, a carbonyl of the helical backbone, and potentially an indole ring from the Trp41 tetrad, resulting in proton shuttling across the histidine tetrad and possibly into the viral interior. The conformational heterogeneity uncovered here may facilitate access of waters and hydronium ions to the imidazole-imidazolium dimers and cycling between the charged states. The characterization of the charged states, the dynamics induced by water, and the conformational heterogeneity induced by malleable helix-helix packing and helix bending around the highly conserved Gly34 residue has led to rich detailing of the proton conductance mechanism for the M2 channel of influenza A, and suggests similar heterogeneity in other oligomeric membrane proteins.
Experimental Procedures
Protein Expression, Purification, and Reconstitution
M2FL (H57,90Y) having a single histidine residue (His37) was expressed in Escherichia coli. Bacterial cultures (25 ml) were transferred into two 700-ml M9 media flasks (with 2 g 13C-glucose, 1 g 15NH4Cl) and grown at 37°C until OD600 ∼0.7. 300 ml of M9 media with unlabeled amino acids (0.5 g Leu, 0.3 g Phe, 0.3 g Tyr, 0.3 g Trp) was added to each 700-ml culture for reverse-labeling His and suppressing the rest of the aromatic amino acids. The bacteria were grown at 22°C until OD600 ∼0.75 before 0.4 mM isopropyl β-D-1-thiogalacto-pyranoside was added to each culture to induce protein expression, which was carried out for 17 hr at 22°C. Benzonase nuclease (Novagen) was added to the cultures prior to French Press lysis of the cells followed by centrifugation and resuspension in buffer with 3% Empigen. The supernatant was filtered with a 0.2-μm filter and 20 mM imidazole, and was buffered to pH 8.0 before loading onto the Ni-affinity column using an AKTA-FPLC system (GE Healthcare). The column was washed in three steps followed by elution with 20 mM Tris, 0.5% octyl-β-glucoside (OG), and 500 mM imidazole (pH 8.0). The typical protein yield was ∼40 mg/l. The His-tag was removed by mixing M2 with tobacco etch virus protease in a ratio of 10:1 (w/w) followed by dialysis using a 3.5-kDa cutoff dialysis bag. Any precipitate was removed by centrifugation before mixing with Ni-resin. The flow-through contained the M2 free of its His-tag. The collected protein was concentrated by Speedvac and dialyzed in 20 mM Tris (pH 8.0). Finally, the protein in OG and Tris buffer was stored at 4°C.
M2FL was reconstituted in DOPC/DOPE (Avanti Polar Lipids, 4:1, mol/mol) in a protein to lipid molar ratio of 1:30. The protein-lipid-OG mixture was dialyzed in a 3.5-kDa cutoff dialysis bag to remove detergent. Final dialysates for the samples were 10 mM Tris (pH 8.8); 10 mM Tris (pH 7.3); 10 mM phosphate/citrate (pH 6.6 and 6.2); 10 mM citrate (pH 5.8 and 4.5). For the high-pH samples, Amberlite was added for the last two buffer changes to remove final traces of detergent. After dialysis the liposomes were pelleted by ultracentrifugation at 420,000 × g for 8 hr at 5°C. The proteoliposome pellet containing approximately 10 mg of M2FL protein was packed into a 3.2-mm thin-wall MAS rotor (36 μl sample volume).
Solid-State NMR Spectroscopy
MAS ssNMR spectra were acquired on a Bruker Avance 600.1-MHz NMR spectrometer using an NHMFL 3.2-mm Low-E triple-resonance biosolids MAS probe (Gor'kov et al., 2007; McNeill et al., 2009). The 13C chemical shifts were referenced to the carbonyl carbon resonance of glycine at 178.4 ppm relative to DSS (sodium 2,2-dimethyl-2-silapentane-5-sulfonate), and the 15N chemical shift reference was calculated by the known relative frequency ratio between DSS (13C) and liquid ammonia (15N). In all 13C-13C correlation experiments a 50-ms mixing time was used during which a PARIS irradiation (Weingarth et al., 2010) was applied. 1D 15N spectra (collected at −10°C, from calibration curve) were acquired with a Lee-Goldberg (LG) cross-polarization (CP) sequence (Fu et al., 2004). During CP, the 1Hs were spin-locked along the magic angle by the LG sequence. For the 2D15N-13Czf-TEDOR experiments, the total TEDOR mixing time of 1 ms was used to obtain one-bond 15N-13C correlations (Jaroniec et al., 2002). To minimize the experimental time for the pH 6.6 and 6.2 spectra, the indirect dimension was reduced through folding the spectral window by setting the 15N offset to 191.7 ppm (pH 6.2) and 190.9 ppm (pH 6.6), resulting in the spectra being folded about 228.1 ppm (pH 6.2) and 227.3 ppm (pH 6.6). The 2D 1H-15N HETCOR spectra were obtained under 12.2-kHz MAS. In the indirect dimension, 1H homonuclear decoupling was achieved using the phase-ramped frequency-switched LG sequence (Fu et al., 2004) with 71.5 kHz radiofrequency irradiation, corresponding to 123.7 kHz decoupling amplitude at the magic angle. Ashort contact time of 500 μs was used to transfer magnetization from 1H to 15N, during which the 1H magnetization was spin-locked along the magic angle to minimize long-range 1H-15N transfers. The 1H chemical shift was referenced to adamantane (1.63 ppm relative to tetramethylsilane). Resonance assignments for the 13C and 15N experiments were achieved using the correlation experiments described above and a set of 15N-filtered experiments that were recently described Fu and coworkers (Miao et al., 2014).
Supplementary Material
Highlights.
Unique His37 tetrad chemistry in full-length M2 proton channel defines function
His37 imidazolium-imidazole short hydrogen bonds exchange with water
Multiple His37 tetrad conformations reflect heterogeneous tetrameric helix packing
Hydrogen-bond exchange leads to both futile and proton conducting cycles
Acknowledgments
This work was supported by NIH Grant AI023007. The NMR experiments were performed at the National High Magnetic Field Laboratory, which is supported by a National Science Foundation Cooperative Agreement (DMR-1157490) with the State of Florida.
Footnotes
Supplemental Information: Supplemental Information includes two figures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2015.09.011.
Author Contributions: Y.M. carried out protein expression, purification, and reconstitution, and ssNMR sample preparation. Y.M. and R.F. performed all NMR experiments, NMR data processing, and spectral assignments. The interpretation of the data and the writing of the manuscript involved all of the authors.
References
- Acharya R, Carnevale V, Fiorin G, Levine BG, Polishchuk AL, Balannik V, Samish I, Lamb RA, Pinto LH, DeGrado WF, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc Natl Acad Sci USA. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas LB, Eddy MT, Pielak RM, Chou J, Griffin RG. Magic angle spinning NMR investigation of influenza A M2(18-60): support for an allosteric mechanism of inhibition. J Am Chem Soc. 2010;132:10958–10960. doi: 10.1021/ja101537p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas LB, Eddy MT, Chou JJ, Griffin RG. Magic-angle-spinning NMR of the drug resistant S31N M2 proton transporter from influenza A. J Am Chem Soc. 2012;134:7215–7218. doi: 10.1021/ja3003606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas LB, Reese M, Eddy MT, Gelev V, Ni QZ, Miller EA, Emsley L, Pintacuda G, Chou JJ, Griffin RG. Structure and mechanism of the influenza a M2 dimer of dimers. J Am Chem Soc. 2015 doi: 10.1021/jacs.5b04802. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S, Wang T, Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein. J Am Chem Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin MT, Andreas LB, Chou JJ, Griffin RG. Proton association constants of His 37 in the Influenza-A M218-60 dimer-of-dimers. Biochem. 2014;53:5987–5994. doi: 10.1021/bi5005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Yi M, Cross TA, Zhou HX. Calculations and validation of the pH-dependent structures of the His37-Trp41 quartet, the heart of acid activation and proton conductance in the M2 protein of Influenza A virus. Chem Sci. 2013;4:2776–2787. doi: 10.1039/C3SC50293G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R, Hu J, Cross TA. Towards quantitative measurements in solid-state CPMAS NMR: a Lee-Goldburg frequency modulated cross-polarization scheme. J Magn Reson. 2004;168:8–17. doi: 10.1016/j.jmr.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Gor'kov PL, Chekmenev EY, Li C, Cotten M, Buffy JJ, Traaseth NJ, Veglia G, Brey WW. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J Magn Reson. 2007;185:77–93. doi: 10.1016/j.jmr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Hong M, Fritzsching KJ, Williams JK. Hydrogen-bonding partner of the proton-conducting histidine in the influenza M2 proton channel revealed from 1H chemical shifts. J Am Chem Soc. 2012;134:14753–14755. doi: 10.1021/ja307453v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath DD, Vijayvergiya V, Cross TA. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci USA. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Schmidt-Rohr K, Hong M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J Am Chem Soc. 2011;134:3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. J Am Chem Soc. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- Kim S, Cross TA. Uniformity, ideality, and hydrogen bonds in transmembrane alpha-helices. Biophys J. 2002;83:2084–2095. doi: 10.1016/S0006-3495(02)73969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SY, Fritzsching KJ, Hong M. Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR. Protein Sci. 2013;22:1623–1638. doi: 10.1002/pro.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SY, Yang Y, Tietze D, Hong M. The influenza M2 cyto-plasmic tail changes the proton-exchange equilibria and the backbone conformation of the transmembrane histidine residue to facilitate proton conduction. J Am Chem Soc. 2015;137:6067–6077. doi: 10.1021/jacs.5b02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey H, Petersen NO, Chan SI. Physiochemical characterization of 1,2-diphytanoyl-sn-glycerol-3-phosphocholine in model membrane systems. Biochim Biophys Acta. 1979;555:147–167. doi: 10.1016/0005-2736(79)90079-8. [DOI] [PubMed] [Google Scholar]
- McNeill SA, Gor'kov PL, Shetty K, Brey WW, Long JR. A low-E magic angle spinning probe for biological solid state NMR at 750 MHz. J Magn Reson. 2009;197:135–144. doi: 10.1016/j.jmr.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Qin H, Fu R, Sharma M, Can TV, Hung I, Luca S, Gor'kov PL, Brey WW, Cross TA. M2 proton channel structural validation from full-length protein samples in synthetic bilayers and E. coli membranes. Angew Chem Int Ed Engl. 2012;51:8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Cross TA, Fu R. Identifying inter-residue resonances in crowded 2D C-C chemical shift correlation spectra of membrane proteins by solid-state MAS NMR difference spectroscopy. J Biomol NMR. 2013;56:265–273. doi: 10.1007/s10858-013-9745-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Cross TA, Fu R. Differentiation of histidine tautomeric states using (15)N selectively filtered (13)C solid-state NMR spectroscopy. J Magn Reson. 2014;245:105–109. doi: 10.1016/j.jmr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould JA, Li HC, Dudlak CS, Lear JD, Pekosz A, Lamb RA, Pinto LH. Mechanism for proton conduction of the M2 ion channel of influenza A virus. J Biol Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- Nguyen PA, Soto CS, Polishchuk A, Caputo GA, Tatko CD, Ma C, Ohigashi Y, Pinto LH, DeGrado WF, Howard KP. pH-induced conformational change of the influenza M2 protein C-terminal domain. Biochem. 2008;47:9934–9936. doi: 10.1021/bi801315m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Kim S, Zhang L, Cross TA. The closed state of a H+ channel helical bundle: combining precise orientational and distance restraints from solid state NMR. Biochem. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- Page RC, Kim S, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielak RM, Chou JJ. Solution NMR structure of the V27A drug resistant mutant of influenza A M2 channel. Biochem Biophys Res Comm. 2010;401:58–63. doi: 10.1016/j.bbrc.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- Thomaston JL, Nguyen PA, Brown EC, Upshur MA, Wang J, DeGrado WF, Howard KP. Detection of drug-induced conformational change of a transmembrane protein in lipid bilayers using site-directed spin labeling. Protein Sci. 2013;22:65–73. doi: 10.1002/pro.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Tobler K, Lamb RA, Pinto LH, Cross TA. Expression and initial structural insights from solid-state NMR of the M2 proton channel from influenza A virus. Biochem. 2002;41:11294–11300. doi: 10.1021/bi025695q. [DOI] [PubMed] [Google Scholar]
- Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarth M, Bodenhausen G, Tekely P. Broadband magnetization transfer using moderate radio-frequency fields for NMR with high static fields and spinning speeds. Chem Phys Lett. 2010;488:10–16. [Google Scholar]
- Yi M, Cross TA, Zhou HX. Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc Natl Acad Sci USA. 2009;106:13311–13316. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX. A theory for the proton transport of the influenza virus M2 protein: extensive test against conductance data. Biophys J. 2011;100:912–921. doi: 10.1016/j.bpj.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Ann Rev Biophys. 2013a;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Cross TA. Modeling the membrane environment has implications for membrane protein structure and function: influenza A M2 protein. Protein Sci. 2013b;22:381–394. doi: 10.1002/pro.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


