Summary
Human cells express natural antiviral proteins, such as APOBEC3G (A3G) that potently restrict HIV replication. As a counter defense, HIV encodes the accessory protein Vif, which binds A3G and mediates its proteasomal degradation. Our structural knowledge on how Vif and A3G interact is very limited since a co-structure is not available. We identified specific points of contact between Vif and A3G by using functional assays with full-length A3G, patient-derived Vif variants and HIV forced evolution. These anchor points were used to model and validate the Vif-A3G interface. The resultant co-structure model shows that the negatively charged β4-α4 A3G loop, which contains primate-specific variation, is the core Vif binding site and forms extensive interactions with a positively charged pocket in HIV Vif. Our data present a functional map of this viral-host interface and opens new avenues for targeted approaches to block HIV replication by obstructing the Vif-A3G interaction.
Graphical abstract
Introduction
APOBEC3G (A3G) is a member of the human APOBEC3 family of seven cytidine deaminases (A3A to A3H) that act as restriction factors of HIV (Harris et al., 2003; Mangeat et al., 2003; Sheehy et al., 2002; Zhang et al., 2003). In turn, HIV counteracts human A3G by expressing the accessory Vif protein, which mediates the proteasomal degradation of A3G by recruiting an E3 ubiquitin ligase complex (Marin et al., 2003; Sheehy et al., 2003; Yu et al., 2003). A3G consists of two deaminase domains: the catalytically inactive N-terminal domain contains the Vif binding site whereas the C-terminal domain has deaminase activity (Hache et al., 2005; Navarro et al., 2005). Despite the recently solved structures of Vif and the N-terminal domain of A3G, no Vif-A3G co-structure exists to date (Guo et al., 2014; Kouno et al., 2015).
Strong reciprocal selection shaped the Vif-A3G interface during primate evolution and lentiviral restriction by A3G is species specific. Previous studies showed that the A3G β4-α4 loop is important for its Vif-mediated degradation. This loop contains three residues 128-DPD-130 that are variable among primates and confers a species-specific barrier for transmission (Figure 1A)(Bogerd et al., 2004; Bulliard et al., 2009; Compton and Emerman, 2013; Compton et al., 2012; Huthoff and Malim, 2007; Letko et al., 2013; Mangeat et al., 2003; Schröfelbauer et al., 2004; Xu et al., 2004). For example, human A3G-128D and African green monkey (agm) A3G-128K are both efficiently counteracted by the Vif of their cognate lentiviruses, HIV-1 and SIVagm, respectively. This phenotype can be fully reversed by changing A3G-128D of the human A3G to a lysine, indicating that Vif specifically binds A3G at this position (Bogerd et al., 2004; Mangeat et al., 2003; Schröfelbauer et al., 2004; Xu et al., 2004). In addition, gorillas encode A3G-129Q, which confers resistance to SIVcpz, HIV-1 and HIV-2 Vif (Letko et al., 2013). The block to infection associated with the gorilla A3G-129Q is lost by “humanizing” the gorilla A3G to 129P (D'Arc et al., 2015; Letko et al., 2013).
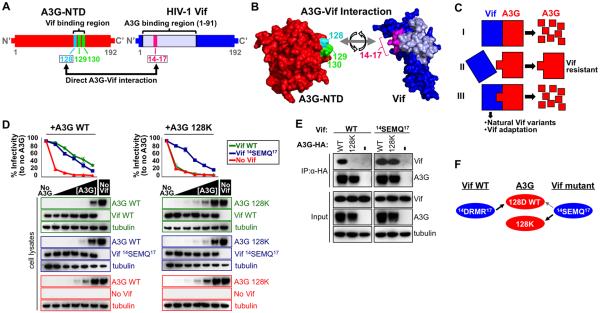
Figure 1. HIV-1 Vif and APOBEC3G amino acid pair mapping to determine the Vif-A3G interface.
(A) A3G amino acids 128, 129 and 130 in the N-terminal domain confer sensitivity to Vif-mediated degradation. A large part of the N-terminal part of Vif (1–91) is implicated in counteracting A3G. A3G-128 directly interacts with Vif-14-17 (See also Table S1).
(B) The interaction between A3G-128 and Vif-14-17 is insufficient to accurately model the Vif-A3G interaction.
(C) Overview of the Vif-A3G mapping approach. Vif binds A3G and leads to its proteasomal degradation (scenario I). Specific A3G mutations in the Vif-A3G interface abrogate Vif binding and, hence, the Vif-resistant A3G mutant is not degraded by Vif (Scenario II). By identifying Vif variants that can counteract the Vif-resistant A3G, one can pinpoint which Vif residues specifically interacts with the Vif-resistant A3G (Scenario III).
D) Increasing levels of WT A3G (0, 5, 10, 25, 50, 100 ng, left panel) and A3G-128K (right panel) were co-transfected with HIV-ΔVif and in the presence of WT NL4-3 Vif or Vif-14-SEMQ-17. Viral infectivity was assessed by TZM-bl reporter cells. 293T cell lysates were analyzed by western blot.
(E) WT and 14-SEMQ-17 NL4-3 Vifs were co-transfected with WT A3G or A3G-128K and immuno-precipitated with αHA-beads. Proteins were analyzed by western blot.
(F) Our data indicates that A3G-128 is in close proximity to Vif-14-17.
Numerous Vif residues throughout the N-terminal part of Vif have been implicated in counteracting A3G (See Figure 1A and summary in Table S1). Most notably mutating Vif amino acids 22, 26, 40-44 and 70 specifically abrogates A3G degradation, indicating that these Vif residues are required for A3G recognition (Summarized in Table S1). Interestingly, these residues are not implicated in degrading A3C, A3F and A3H, suggesting that Vif uses distinct binding sites for different APOBEC3 proteins (as reviewed in (Desimmie et al., 2014; Salter et al., 2014)).
In contrast, our knowledge on specific Vif-A3G interactions is more limited. Only one study demonstrated a direct point of interaction between Vif and A3G (Schröfelbauer et al., 2006). A human A3G-D128K mutant cannot be counteracted by HIV-1 Vif, but is efficiently degraded by SIVagm Vif. Mutating HIV-1 Vif 14-DRMR-17 to 14-SEMQ-17 enabled the mutant HIV-1 Vif to degrade A3G-128K, suggesting that Vif-14-17 and A3G-128 interact (Figure 1B) (Schröfelbauer et al., 2006). However, a single point of contact between Vif and A3G is not sufficient to correctly orient the two proteins (Figure 1B).
Structural approaches such as NMR or crystallography are traditionally used to solve protein-protein interfaces, but face technical limitations with protein complexes that are difficult to purify such as HIV Vif and A3G. We hypothesized that the Vif-A3G interface could be mapped using viral restriction as a read-out. This approach has the advantage of relying on full-length, functional viral and host proteins. It is based on the disruption of the Vif-A3G interface by specific A3G mutations and the subsequent identification Vif mutations that specifically restore viral rescue (Figure 1C). We successfully employed this strategy to identify three specific Vif-A3G points of contact. The resultant Vif-A3G co-structure model indicates that the β4-α4 A3G loop fits into a well-defined pocket on the surface of Vif and explains why this loop contains extensive primate-specific variation.
Results
Intracellular protein structure probing to determine the Vif-A3G interface
We propose that the interface between host defense proteins such as A3G and the corresponding lentiviral antagonist Vif can be accurately modeled by using A3G antiviral activity as a read-out (Described in Figure 1C). HIV-1 Vif binds A3G, which leads to its proteasomal degradation (Scenario I, no antiviral activity). Specific A3G mutations in the Vif-A3G interface abrogate Vif binding and A3G is not degraded (Scenario II, strong antiviral activity). Screening natural Vif variants or by adapting HIV to counteract the Vif-resistant A3G allows identification of Vif mutants that accommodate the A3G mutant (Scenario III, no antiviral activity). We anticipate that the accommodating Vif residue has opposite properties (e.g., charge or size), compared to the A3G mutation. These functional Vif-A3G interaction pairs can be used as anchor points to model the Vif-A3G interface with three contact points being required to correctly orient both proteins. Of note, only A3G and Vif mutants that are functionally active and and thus are properly folded were used in this study. All A3G mutants restricted HIV in the absence of Vif and all Vif mutants counteract either a WT A3G, a mutant A3G or A3F.
First point of contact: A3G-128 interacts with Vif-14-17
For the first point of contact we built on the observation that the HIV Vif-resistant mutant A3G-128K is counteracted by a Vif 14-SEMQ-17 mutant (Schröfelbauer et al., 2006). We confirmed this observation by testing the restriction of WT NL4-3, NL4-3 Vif-14-SEMQ-17 and NL4-3 ΔVif with increasing amounts of WT A3G and the mutant A3G-128K. In the absence of Vif, A3G potently restricted HIV and infectivity was rescued by degradation of A3G in the presence of WT Vif and Vif-14-SEMQ-17 (Figure 1D, left panel). However, A3G-128K was only counteracted by Vif 14-SEMQ-17 (Figure 1D, right panel) and not by WT Vif, which is in full agreement with the previous study (Schröfelbauer et al., 2006). We speculate that the large positively charged arginines in WT Vif 14-DRMR-17 specifically clash with the positively charged lysine of A3G-128K, which is compensated for by the smaller negatively charged glutamate in Vif 14-SEMQ-17. Of note, Vif 14-SEMQ-17 was also able to counteract WT A3G-128D, possibly due to the smaller size of the glutamate and aspartate, respectively. Together, our data indicate that the 14-17 stretch in Vif is close to A3G-128. Co-immunoprecipitation (co-IP) assays with WT A3G and A3G-128K with WT Vif and Vif-14-SEMQ-17 showed that WT Vif was only efficiently pulled down with WT A3G while Vif 14-SEMQ-17 efficiently interacted with both WT A3G and A3G-128K (Figure 1E). Taken together, our data indicate that residues Vif 14-17 are in close proximity to A3G-128 (Figure 1F).
Second point of contact: A3G-125R confers resistance to HIV-1 Vif
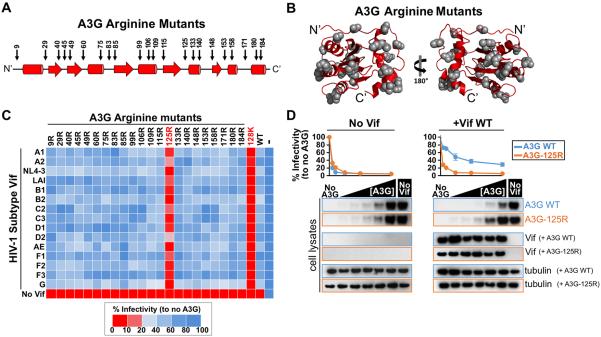
To identify A3G residues that confer resistance to Vif-mediated degradation, we performed an arginine scan of surface exposed α-helixes, β-strands and loops of the N-terminal domain of A3G (Figure 2A and 2B). We chose arginine- over the commonly used alanine-mutagenesis because a bulky and charged arginine would more efficiently perturb the interaction with Vif. We tested the antiviral activity and Vif sensitivity of 22 A3G arginine mutants using a HIV subtype Vif panel (Figure 2A) (Binka et al., 2012). In the absence of Vif, all A3G mutants restricted HIV to levels similar to that of WT A3G and A3G-128K, indicating they are fully functional (Figure 2C). While most of the A3G mutants were efficiently counteracted by the Vif variants, the mutant A3G-125R was resistant to Vif, similar to the A3G-128K variant (Figure 2C). A3G residue 125, although close to residue 128, has not previously been implicated in resistance to Vif. Of note, only one Vif of HIV subtype D (D2) could partially counteract A3G-125R (Figure 2C). Titrating WT A3G and A3G-125R restrict to similar levels in the absence of Vif (Figure 2D, left panel). However, only WT A3G was counteracted by Vif, whereas A3G-125R was fully resistant to Vif mediated degradation (Figure 2D, right panel).
Figure 2. A3G-125R confers resistance to HIV-1 Vif.
(A) 24 Arginines were individually introduced in the N-terminal domain of A3G at the indicated positions.
(B) The arginine mutants were modeled onto the predicted N-terminal domain of A3G.
(C) Single cycle infectivity assay with the A3G arginine mutants, A3G-128K, WT A3G and a panel of subtype Vifs. The average infectivity values of a triplicate transfection are shown as a heatmap where no restriction is indicated in blue and potent restriction is shown in red.
(D) Single cycle infectivity assays with increasing amount of WT A3G and A3G-125R (0, 5, 10, 25, 50, 100 ng) in the absence (left panel) or in the presence of NL4-3 Vif (right panel). 293T cell lysates were analyzed by western blot.
Second point of contact: Vif amino acids 19 and 22 determine activity against A3G-125R
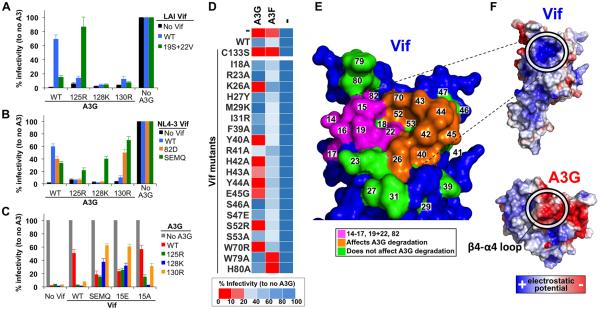
We next designed a series of experiments to find a Vif variant that counteracts A3G-125R. We therefore cloned a panel of 66 HIV subtype B Vif alleles from 35 chronically infected, untreated participants of the Amsterdam Cohort Studies into full-length NL4-3 (Kootstra et al., 2007). The variants differed by 7–16% at the amino acid level and were tested in single cycle infectivity assays in the presence of WT A3G or A3G-125R. In the absence of Vif (ΔVif), WT A3G potently restricted HIV, but was efficiently counteracted by all Vif variants (Figure 3A, top). In contrast, the majority of Vif variants failed to counteract A3G-125R, but some Vifs were able to counteract A3G-125R (Figure 3A, bottom). A comparison between the infectivity values in the presence of WT A3G and A3G-125R showed that 14 out of 66 Vifs had activity against A3G-125R (Figure 3B). An analysis of the amino acid differences between the 52 inactive and 14 active Vif variants by VESPA (Rose and Korber, 2000) pointed to Vif position 19 as being the most different (Figure 3C). An amino acid weblogo representation showed that Vifs unable to counteract A3G-125R encode either Vif-19R or Vif-19K (Figure 3D) (R: 45x and K: 7x). In contrast, the Vifs that are active against A3G-125R more often encode Vif-19S (S: 9/14, R: 4/14 and K: 1/14). However, the Vif activity cannot be fully explained by only Vif residue 19 as five Vifs with activity against A3G-125R encode an arginine or lysine at this position (e.g., R: 4/5 and K: 1/5). We therefore shifted our attention to the nearby Vif position 22, which was also variable (Figure 3D). The Vifs that failed to counteract A3G-125R typically encoded Vif-19R and Vif-22K (19R-22K: 40/52; 19K-22K: 7/52, 19R-22N: 2/52, 19R-22H: 3/52), while Vifs with activity against A3G-125R never encoded bulky and charged residues such as arginine and lysine at both positions (19S−22K: 9/14, 19R-22N: 2/14, 19R-22H: 1/14, 19R-22V: 1/14 and 19K-22N: 1/14; Figure 3D and Figure S1). These functional data suggest that the presence of two bulky positively charged arginines and lysines at Vif positions 19 and 22 would be incompatible with counteracting A3G-125R. Conversely, Vif counteracts A3G-125R if one of these two Vif residues (19 or 22) was not an arginine or a lysine and preferably encodes Vif-19S or Vif-22N, 22H or 22V (Figure S1). Vif residues 19 and 22 are in close proximity to the 14-17 stretch and potentially represent a second point of contact between Vif and A3G (Figure 3E).
Figure 3. Vif screening identifies Vif-positions 19 and 22 to confer activity to A3G-125R.
(A) 66 patient-derived Vifs and LAI, NL4-3 and an inactive Vif were cloned into full-length NL4-3 and were tested in single cycle infectivity assays in the presence of WT A3G (top panel) or A3G-125R (bottom panel).
(B) The infectivity values in the presence of A3G-125R were divided by the infectivity values measured in the presence of WT A3G. The Vifs that can (in green) or cannot (in black) counteract A3G-125R are indicated.
(C) The frequency of the most abundant amino acid at each position of the Vifs able to counteract A3G-125R was divided by the amino acid frequency of the inactive Vifs using VESPA (Rose and Korber, 2000).
(D) Weblogo plot of amino acids 14–27 of the Vifs that fail to counteract A3G-125R (top) and that of the Vifs that efficiently counteract A3G-125R (bottom). See also Figure S1.
(E) Vif positions 19 and 22 are indicated in green on the Vif crystal structure. Both residues are in close proximity to Vif-14-17 (pink).
We next performed “loss of function” and “gain of function” experiments using Vifs that can or cannot counteract A3G-125R. We selected four Vifs with activity against A3G-125R, encoding either a lysine at Vif positions 19 or an arginine at position 22 and we introduced a second arginine or lysine at position 19 or 22 (Figure 4A). The WT patients' Vif variants degraded and counteracted A3G-125R, but the mutant Vifs lost activity against A3G-125R (Figure 4B, left). Importantly, the loss of activity of the Vif mutants was specific for A3G-125R since activity against WT A3G remained unchanged (Figure 4B, right).
Figure 4. A3G position 125 interact with Vif amino acids 19 and 22.
(A) Four patient-derived Vifs that counteract A3G-125R and do not encode an arginine or lysine at both position 19 and 22 were mutated to 19R and 22K.
(B) The four patient Vifs and their respective mutants were tested in single cycle infectivity assays in the presence of A3G-125R (left panel) and WT A3G (right panel) and 293T cell lysates were analyzed by western blot.
(C) LAI Vif, which encodes 19R and 22K and does not counteract A3G-125R, was mutated at positions 19 and 22 with the indicated residues. Antiviral activity of these Vif mutants was tested in the presence of A3G-125R (D, left panel) or WT A3G (D, right panel). 293T cell lysates were analyzed by western blot.
(E) Immuno-precipitations were performed with WT LAI Vif and its 19 and 22 mutants and WT A3G or A3G-125R. Proteins were analyzed by western blot.
(F) Combined, our data show that Vif-19+22 interact with A3G-125. See also Figure S2.
For the “gain of function” experiments we used HIV-1 LAI Vif, which encodes an arginine and a lysine at positions 19 and 22, respectively, and did not counteract A3G-125R (Figures 2C and 3B). We mutated positions 19 to a serine and 22 to a valine individually or in combination (Figure 4C). LAI WT Vif showed little activity against A3G-125R, but LAI Vif-19S and Vif-22V displayed increased A3G-125R degradation (Figure 4D, left). Moreover, the double mutant, Vif-19S+22V, efficiently degraded and counteracted A3G-125R (Figure 4D). The effect of the single mutations at positions 19 and 22 of Vif was specific for A3G-125R and the double mutant completely lost its activity against WT A3G (Figure 4D, right), indicating specific interactions between Vif positions 19+22 and A3G position 125. Co-IP experiments showed that individual or combined Vif mutations at positions 19 and 22 improved the association of Vif with A3G-125R (Figure 4E). In contrast the Vif Vif-19S−22V double mutant showed reduced interaction with WT A3G; an observation that is in good agreement with its inability to degrade WT A3G (Figure 4D).
Our observation that Vif-19S+22V degraded A3G-125R, but not WT A3G indicates that the interaction between these residues is specific. Human and chimpanzee A3G always encode 125Y and the combined absence of arginines or lysines at both Vif positions 19 and 22 (e.g., Vif 19S+22V) is never observed in natural HIV-1 and SIVcpz Vif sequences (Figure S2A). The subtype D Vif variant (Vif D2, Figure 2C), which had partial activity against A3G-125R was the only Vif in the subtype panel to encode Vif-19S (Figure S2B).
Combined, our data indicate a specific interaction between Vif residues 19+22 and the A3G amino acid at position 125.
Third point of contact: A3G position 130 interacts with Vif position 82
To obtain the third specific Vif-A3G pair we focused on A3G position 130 that was shown to be important for Vif-mediated degradation (Bulliard et al., 2009; Compton and Emerman, 2013; Compton et al., 2012; Huthoff and Malim, 2007). In the absence of Vif, both WT and the A3G-130R mutant potently restricted HIV (Figure 5A) and indeed A3G-130R resisted Vif-mediated degradation and efficiently inhibited HIV (Figure 5A). To find Vif variants that naturally counteract A3G-130R, we first tested our subtype and patient-derived Vif panels against A3G-130R. However, all tested Vifs failed to counteract A3G-130R (data not shown).
Figure 5. A3G position 130 interacts with Vif position 82.
(A) HIVΔVif was co-transfected with either WT A3G or A3G-130R in the presence or absence of NL4-3 Vif. Infectivity was measured by infecting TZM-bl reporter cells. 293T cell lysates were analyzed by western blot.
(B) Schematic overview of the lentiviral A3G expression cassette. The transduced cells were lysed and probed for A3G protein expression. The asterisk indicates the inactive A3G, which contains two inactivating mutations in both catalytic sites (E67A+E259A). The non-permissive T-cell lines H9 and MT2 both endogenously express A3G.
(C) The SupT1 cell lines expressing WT A3G, the inactive A3G deaminase mutant, A3G-130R and A3G-128K were infected with NL4-3 and supernatants were harvested each day and checked for infectivity on TZM-bl reporter cells. Error bars represent standard deviations of three separate infections.
(D) SupT1 cells expressing A3G-130R were infected with NL4-3 and maintained for 8 weeks and Vif was PCR amplified and sequenced from proviral DNA isolated 6 weeks and 8 weeks post infection. The chromatograms show a mixed population of Vif-82G/D after 6 weeks and Vif-82D represent the majority of viruses 8 weeks post infection.
(E) Vif position 82 is indicated in green on the Vif crystal structure and is in close proximity to Vif-14-17 and Vif19+22 (pink).
(F) WT and 82D Vifs were tested together with A3G-130R and WT A3G in single cycle infectivity assays. 293T cell lysates were analyzed by western blot.
(G) Vif-82D and Vif-82K were cloned into full-length NL4-3 and viruses were produced in 293T and used to infect SupT1 cells expressing empty vector, WT A3G or A3G-130R and supernatants were harvested each day and checked for infectivity on TZM-bl reporter cells. Error bars represent standard deviations of three separate infections.
(H) Immuno-precipitations were performed with WT NL4-3 Vif and Vif-82D and WT A3G or A3G-125R and proteins were analyzed by western blot.
(I) Combined, our data show that WT Vif-82G and Vif-82K interact with A3G-130D and Vif-82D specifically interacts with A3G-130R indicating that Vif-82 interact with A3G-130. See also Figure S3.
As an alternative approach to acquire Vif mutants that can counteract A3G-130R we conducted forced evolution experiments. We generated stable cell lines of permissive SupT1 T-cells expressing A3G, GFP and puromycin N-acetyl-transferase separated by P2A and T2A “self-cleaving” peptides (Figure 5B). The combined expression of A3G with GFP and puromycin N-acetyl-transferase ensures that all puromycin-resistant cells express GFP and A3G. The four cell lines expressed WT A3G, a catalytically inactive A3G (E67A+E259A), A3G-130R and A3G-128K to levels comparable to endogenously expressed A3G in the non-permissive T-cell lines H9 and MT2 (Figure 5B). Of note, the A3G proteins are slightly larger due to the addition of the P2A peptide. The cell lines were infected with HIV-1 NL4-3 and replication was monitored over two weeks. HIV replication was robust on the cells expressing WT A3G and the A3G catalytic mutant, whereas viral replication was restricted on cells expressing A3G-130R and A3G-128K (Figure 5C). Long-term culturing of the infected A3G-130R expressing cells showed syncytia after 8 weeks, indicative of productive viral replication and revealed the emergence of a viral strain encoding a Vif-G82D substitution (Figure 5D). Of note, no other Vif changes were observed.
Vif position 82 is close to positions 14-17 and 19+22, indicating that Vif position 82 could represent a direct contact point for A3G position 130 (Figure 5E). Single cycle infectivity assays showed that Vif-82D displayed improved activity against A3G-130R, while its activity against WT A3G was slightly reduced compared to the WT Vif (Figure 5F).
We next tested the fitness of Vif-82D in T-cells expressing WT A3G or A3G-130R. We included Vif-82K to test whether the positively charged lysine would further obstruct binding with A3G-130R. All three viruses replicated similarly in the absence of A3G (Figure 5G, top). WT and Vif-82K viruses replicated efficiently in the presence of WT A3G, while Vif-82D showed delayed replication (Figure 5G, middle). In contrast, NL4-3 Vif-82D was the only virus able to replicate efficiently on SupT1 expressing A3G-130R (Figure 5G, bottom). Interestingly, the Vif-82K virus was even more restricted on A3G-130R expressing cells than WT HIV (Figure 5, bottom). This suggests that identically charged amino acids at A3G position 130 and Vif position 82 perturbate the Vif-A3G interaction and can be restored by introducing residues with opposite charges.
Co-IPs showed a diminished interaction between A3G-130R and WT Vif, which was restored for Vif-82D (Figure 5H). In contrast, both Vif variants interacted efficiently with WT A3G. The importance of WT Vif-82G is further underscored by its extreme conservation among all HIV-1 and SIVcpz Vifs (Figure S2). Combined, our data indicate that A3G position 130 interacts with Vif position 82.
Vif-A3G contact points specificity and mapping the A3G-binding surface of Vif
To confirm the specificity of the three points of interaction and exclude indirect structural changes we assessed the infectivity of HIV produced in the presence of different A3G and Vif variants. Vif-19S+22V only counteracted A3G-125R, but not WT A3G, A3G-128K and A3G-130R indicating that the interaction between A3G-125 and Vif-19+22 is specific (Figure 6A). Also Vif-82D specifically counteracted A3G-130R efficiently, but failed to rescue infectivity with A3G-125R or A3G-128K, suggesting a specific interaction (Figure 6B). However, Vif-14-SEMQ-17 was able to counteract all three A3G mutants, which indicates an aspecific effect (Figure 6C). Because we hypothesized that A3G-128D and A3G-128K would interact with Vif-15R and Vif-15E, respectively, we tested two additional Vif mutants, Vif-15E and Vif-15A, for their activity against the A3G mutant panel. Indeed, Vif-15E counteracted the different A3G mutants similarly to Vif-14-SEMQ-17, indicating that only Vif position 15 plays a role in binding to A3G positions 125, 128 and 130 (Figure 6C). Vif-15A could not counteract A3G-128K, while maintaining activity against A3G-125R and A3G-130R, indicating that the negative charge of the glutamate is required for the interaction with A3G-128K (Figure 6C). Combined, our data suggest that in the Vif-A3G interface A3G-125 must be close to Vif residues 19 and 22, A3G-130 is close to Vif-82 and Vif-15 is in close proximity to A3G-128, but also close to A3G-125 and A3G-130 (Summarized in Table S2).
Figure 6. Vif-A3G contact points specificity and mapping the A3G-binding surface of Vif.
(A, B, C) HIVΔVif was co-transfected with either WT A3G, A3G-125R, A3G-128K or A3G-130R in the presence or absence of Vif WT, Vif-19S+22V, Vif-82D, Vif-14-SEMQ-17, Vif-15E and Vif-15A. Infectivity was measured by infecting TZM-bl reporter cells.
(D) Vif residues around Vif-14-17, 19+22 and 82 were mutated to the indicated amino acids and were tested against A3G and A3F in single cycle infectivity assays. The average infectivity values of a triplicate transfection are shown as a heatmap where no restriction is indicated in blue and potent restriction is shown in red.
(E) The results from (A) are mapped onto the Vif crystal structure, Vif-14-17, 19+22 and 82 are indicated in pink, the Vif residues important for counteracting A3G are indicated in orange and Vif residues that not affect counteracting A3G are shown in green. Vif residues 79 and 80 are specifically required for counteracting A3F.
(F) The electrostatic potential of Vif and A3G were analyzed by ABPS tool in PYMOL using standard settings.
We next performed extensive mutagenesis of Vif residues located in close structural proximity to the Vif residues 14–17, 19+22 and 82 to further define the A3G binding site of A3G. Importantly, Vif mutants were tested against A3G as well as A3F to minimize erroneous conclusions caused by either Vif misfolding or failure to interact with E3 ligase components. Only Vif mutations that resulted in a selective loss of activity against A3G or A3F, but not both, were interpreted as being specific binding sites. Mutating Vif residues 26, 40, 42, 43, 44, 45, 52 and 70 resulted in a loss of A3G recognition, but not against A3F, which indicates that these residues are specifically involved in A3G binding, which is in agreement with previous studies (Figure 6D and E, indicated in orange) (Chen et al., 2009; Dang et al., 2009; He et al., 2008; Mehle et al., 2007; Russell and Pathak, 2007; Simon et al., 2005). Vif residues 40-YRHHY-44 have been previously implicated to be specifically required for counteracting A3G (Russell and Pathak, 2007). Mutating Vif at positions 18, 23, 27, 29, 31, 39, 41, 46, 47, 53 did not affect A3G or A3F counteraction, suggesting that these residues are not interacting with A3G or A3F (Figure 6E, indicated in green). Mutating Vif positions 79 or 80 resulted in a specific loss of activity against A3F, but not A3G suggesting that these residues are required for interaction with A3F (Figure 6E) (He et al., 2008). Combined our data show that the Vif region required for counteracting A3G forms a defined pocket that consists of Vif residues 14-17, 19+22 and 82 on one side and Vif residues 26, 40, 42, 43, 44, 45 and 70 on the opposite side (Figure 6E).
Electrostatic forces play a crucial role in interactions between proteins (Baker et al., 2001). We therefore mapped the electrostatic potentials of both Vif and A3G. Interestingly the Vif pocket has a positive electrostatic potential, whereas the A3G β4-α4 loop has a negative electrostatic potential, which would favor a direct interaction between Vif and A3G (Figure 6F).
The β4-α4 A3G loop fits in the positively charged pocket of Vif
To predict how Vif and A3G interact, we applied protein docking with the crystal structure of Vif and a N-terminal domain model of A3G based on the A3C crystal structure. The A3G model we used is structurally similar to the recently published A3G N-terminal domain NMR structure (Kouno et al., 2015) (Figure S4A). Vif-A3G structures were predicted using ZDock software (Pierce et al., 2014) with constraints to align the three Vif-A3G contact points and predictions were individually inspected. Out of 23 predictions, 5 very similar structures fit our three Vif-A3G contact points (Figure 7A and Figure S4B). In these models A3G-128K is in close proximity to Vif-14DRMR-17 with Vif-15 interacting with A3G-128 (Figure 7B), which is in agreement with the specificity experiments (Figure 6A, B and C). Furthermore, A3G-125Y is positioned between Vif-19N and Vif-22K and A3G-130D is next to Vif-82D.
Figure 7. The β4-α4 A3G loop fits in the positively charged pocket of Vif.
(A) The model of the N-terminal domain of A3G was based on the A3C crystal structure and was submitted together with the Vif crystal structure to ZDOCK protein docking software. Structures were visually inspected and a structure that fits our experimental data is shown. See also Figure S4.
(B) Individual close ups of the Vif-A3G structure model show a close interaction with Vif-14-17 with A3G-128 (left), A3G-128 is located between Vif-19+22 (middle) and A3G-130 and Vif-82 are in close proximity (right).
(C) Individual close ups of the Vif-A3G structure model show a close interaction with Vif-15R with A3G-125, A3G-128 and A3G-130.
(D) Schematic overview of the relative distances between A3G residues 125, 128 and 130 and Vif residues 15, 19, 22 and 82.
(E) A close up of the Vif-A3G structure model indicates that the A3G β4-α4 loop between fits in the Vif pocket flanked by Vif-14-17 and 19+22 on one and Vif-40-44 on the other side.
(F) The docked Vif-A3G model is shown in context with the E3-ligage components elongin B, elongin C and the transcription co-factor CBF-β from the published Vif-crystal structure (4N9F). The structure for full-length cullin5 was predicted from the available structural data for cullin1 and cullin5. Spheres representing approximate sizes and locations for the A3G-CTD, Rbx2 and E2 ubiquitin conjugating enzyme are shown in red, purple and green, respectively. Ubiquitin is shown in white.
(G) Overview of primate-specific A3G polymorphisms in the A3G β4-α4 loop and their sensitivity to Vif proteins from different primates. The three amino acids at A3G positions 128, 129 and 130 are indicated. African green monkeys encode different A3G loop variants depending on their species subtypes. The red squares indicate that the Vif protein can not counteract the specific primate A3G and the white squares represent efficient counteraction by Vif.
A closer inspection of our Vif-A3G structure model shows that Vif-15 is in close proximity to A3G positions 125, 128 and 130 (Figure 7C), which explains the why Vif-15E efficiently counteracted A3G-125R, A3G-128K and A3G-130R (Figure 6C). We speculate that A3G-128 can only interact with Vif position 15 and that their interaction is efficient as long as their charges are opposite. In contrast, A3G-125 and A3G-130 have more structural flexibility and can both interact with Vif position 15, but can only individually interact with Vif-19+22 and Vif-82, respectively (Figure 7C and 7D). Our model reveals that the β4-α4 A3G loop, which contains A3G positions 125, 128 and 130, fits in the Vif pocket but also that other A3G β4-α4 loop residues, such as 124, 127, 129 and 131 interact with Vif residues lining the pocket, underscoring the importance of the A3G β4-α4 loop as the core binding site of Vif (Figure 7E). By using our Vif-A3G structure model we can detect other potential points of contact between Vif and A3G, such as the A3G loops between α-helix-1 and β-strand-1, α-helix-2 and β-strand-2 and β-strand-3 and α-helix-3 (Figure S4C and D) which is in good agreement with a recent A3G NMR study (Kouno et al., 2015) (Figure S4E).
We next integrated our Vif-A3G interaction model in the context of the entire E3-ligase complex by using the crystal structure of Vif, CBF-β, Elongin B and C and the N-terminal part of cullin5 and modeled the missing C-terminal part of Cullin 5 based on an existing Cullin 1 crystal structure (Goldenberg et al., 2004; Guo et al., 2014). Because the orientation of the C-terminal A3G domain to the N-terminal domain remains unknown we represent the C-terminal A3G domain as a sphere placed at the end of the N-terminal A3G domain. This shows that A3G binding to Vif does not overlap with other proteins such as CBF-β and Elongin C binding (Figure 7F). Furthermore, our model predicts that the C-terminal domain of A3G is oriented towards the currently unknown E2 ligase, which is indeed the main target for ubiquitination (Iwatani et al., 2009).
In summary, the identification of three specific points of contact between Vif and A3G in combination with extensive Vif mutagenesis allowed us to generate an “in vivo” Vif-A3G protein interaction model.
Discussion
Despite extensive research on HIV-1 Vif and A3G, a co-structure of this viral host interface remains elusive. We mapped the Vif-A3G interface by disrupting Vif-A3G interactions and identifying Vif mutations that specifically restore viral rescue. The strength of our approach are that full-length intracellular proteins in functional assays. Our approach identified three points of contact and generated a Vif-A3G interaction model, which explains previous reports of Vif mutagenesis.
Our Vif-A3G structure model indicates that the A3G β4-α4 loop fits into a well-defined pocket on the surface of Vif. A3G loop β4-α4 residues 128, 129 and 130 have been under positive selection during primate evolution and primate-specific variants can block the transmission of SIV or HIV from other primates (Bogerd et al., 2004; Compton and Emerman, 2013; Compton et al., 2012; Letko et al., 2013; Mangeat et al., 2004; Schröfelbauer et al., 2004; Xu et al., 2004). The primate specific β4-α4 loop changes and their effects on SIV/HIV restriction are summarized in Figure 7G. The Vif proteins of SIV or HIV counteract the A3G of their natural host but the viruses are restricted by other primate A3G variants with β4-α4 loop changes. Most notably gorilla A3G, which encodes 129Q is resistant to all SIV and HIV variants except SIVgor (Letko et al., 2013). Our interaction model underscores that the A3G β4-α4 loop represents the core Vif binding site and may explain why species-specific variation in this relatively small loop determines primate-species tropism.
We chose to use an N-terminal A3G domain homology model based on an A3C crystal structure because all APOBEC3 domains are structurally similar. Very recently, the NMR-structure of the Vif-binding N-terminal domain of A3G was solved (Kouno et al., 2015). However, the N-terminal A3G domain used in this study lacks antiviral activity due to the 25 substitutions or deletions needed to increase its solubility (Kouno et al., 2015). Our modeled A3G N-terminal domain is nonetheless in good agreement with this NMR structure (Kouno et al., 2015) (Figure S4A, top panel). Of note, the β4-α4 loop, containing A3G-125, 128 and 130 varies considerably between NMR structures, suggesting some structural flexibility (Kouno et al., 2015) (Figure S4A, bottom panel).
Previous studies identified numerous amino acids in Vif to be important for binding to A3G (Figure 1A and Table S1). However, our Vif mutagenesis and Vif-A3G interface model suggest that the A3G binding site of Vif is limited to a well- defined pocket located on one side of Vif (Figure 6A). We speculate that some of the Vif residues located outside of this defined Vif pocket area are not involved in A3G binding, but lead to Vif misfolding or affect e.g. binding to CBF-β (Table S1). Therefore, Vif mutants should also be tested against other APOBEC3 proteins, such as A3F that uses another Vif binding site in order to ensure functionality.
Our model also informs on interactions of SIV Vifs with primate A3G. SIV Vifs are very different from HIV-1 Vif and it is unknown whether they adopt a similar protein structure. However, two potential Vif-A3G contact points between African green monkey and sooty mangabey A3G and Vif proteins of their respective SIVs were previously described. African green monkey subspecies show variation at A3G position 130 (A3G-130D or A3G-130H). SIV adaptation in an African green monkey encoding A3G-130H selected for a specific C84Y Vif change, (Compton et al., 2012; Compton et al., 2013), which is very close to the HIV-1 Vif-82D adaptation to A3G-130R (Figure 5). Of note, the HIV-1 Vif 14-SEMQ-17 mutant did not counteract A3GAGM, suggesting that other polymorphisms between African green monkey and human A3G could affect Vif binding (Schröfelbauer et al., 2006). Macaque A3G contains an Y59LR variation in the β2-α2 loop rendering macaque A3G resistant to SIV from sooty mangabeys (Krupp et al., 2013). SIV from sooty mangabeys adapted to macaque A3G-Y59LR by acquiring a Vif-G17E change, which is in close proximity to the β2-α2 loop in our Vif-A3G model. Combined, these data fit in our Vif-A3G structure model and indicate that the Vif-A3G interface is likely conserved among primates and their cognate viruses.
Our Vif-A3G structure model will assist in the future generation of a Vif-A3G co-structure determined by crystallography or NMR, which is required to determine detailed molecular interactions between specific Vif and A3G amino acids and their potential structural rearrangements that occur after Vif binding to A3G. Our approach to decipher the structural interface can be applied to other proteins that are difficult to analyze by classical structural approaches. Furthermore, our model could guide future studies towards the discovery of small molecule interventions suitable to inhibit HIV replication by perturbing the Vif-A3G interface.
EXPERIMENTAL PROCEDURES
Amsterdam Cohort Vif variants
The Amsterdam Cohort Studies on HIV infection and AIDS have been conducted in accordance with the ethical principles set out in the declaration of Helsinki and all participants provided written informed consent. The study was approved by the Academic Medical Center institutional Medical Ethics Committee of the University of Amsterdam. HIV-1 Vif was cloned from 35 ACS participants at late time points during the course of infection (Kootstra et al., 2007). Vif was amplified as described previously (Ooms et al., 2013). The GenBank accession numbers for Vif sequences reported in this study are KT860442-KT860507.
Plasmids
The C-terminally FLAG-tagged Vif expression plasmid encoding NL4-3, LAI, and HIV-1 subtype Vifs were previously described (Binka et al., 2012). A3G mutants were C-terminally triple HA-tagged and cloned into ptr600 expression vector as previously described (Ooms et al., 2012). Point mutagenesis of Vif and A3G was performed by standard overlapping PCR. The Vif Amsterdam Cohort variants were cloned into a modified NL4-3 molecular clone with the Integrase and Vif ORFs separated (Sakurai et al., 2004; Stern et al., 2010). In addition, a stop codon in frame with Vpr was inserted after Vif to prevent any production of Vpr.
HIV Single Cycle Infectivity Assays
For titration experiments, increasing amounts of A3G-3xHA expression plasmids (0, 10, 25, 50, 100ng) and Vif plasmids (50ng) were co-transfected with NL4-3 ΔVif (500ng) in 293T cells as previously described (Letko et al., 2013). In all other assays, 20ng A3G or 50ng A3F was used. Culture medium was replaced 24-hours post-transfection and supernatants were collected 48 hours post-transfection and infectivity of analyzed by infecting TZM-bl reporter cells as previously described (Letko et al., 2013). Average relative infectivity values and their standard deviations were calculated from representative triplicate transfections. Proteins were analyzed by western blot as previously described (Letko et al., 2013).
Co-immuno-precipitation
HEK 293T cells were co-transfected with FLAG-Vif expression plasmids (50ng), HA-A3G expression plasmid (200ng) and GST stuffer DNA (250ng) in a 24-well format. Cells were lysed two days post-transfection in a mild lysis buffer (1% triton X-100 in 1x PBS and EDTA-free protease inhibitor cocktail, Roche). The cleared lysates were incubated with EZ-View anti-HA beads (Sigma) at 4°C for two hours. Beads were washed 4 times in mild lysis buffer. Proteins were eluted and analyzed by western blot as previously described (Letko et al., 2013).
Lentiviral vectors and Cell transduction
A3G-P2A-maxGFP cassettes were constructed by overlap PCR and cloned into the EF1-T2A-puro lentivector backbone (SystemBio). Lentiviral vectors were produced in 293T cells according to the manufacturer's recommendations. SupT1 cells were transduced and placed under puromycin (0.5 μg/mL) 2 days later. After transduction and selection, cell lysates were analyzed for A3G expression using an A3G-specific antibody (C17, NIH AIDS reagents).
HIV-1 replication assay
Viral stocks were produced as described (Ooms et al., 2013). 0.5×106 SupT1 cells were infected in triplicate (MOI 0.01) in the presence of 5 μg/ml polybrene in a 24-well format (1.5 ml). 200 ul of the supernatants were collected every day and cultures were replenished with fresh media. After two weeks, supernatants were used to infect TZM-bl reporter cells. Error bars represent standard deviations of three separate infections.
HIV-1 forced evolution
A 25 ml flask with 5.0×106 SupT1 cells expressing A3G-130R was infected with NL4-3 (MOI 1.0). Cells were cultured and cellular DNA was isolated using the DNeasy DNA isolation kit. Proviral Vif was amplified by PCR and sequenced as previously described (Ooms et al., 2013).
Molecular Docking and Visualization
The structure for A3G amino acids 4–189 was predicted from the A3C crystal structure (3VOW,(Kitamura et al., 2012)) and A3G sNTD structure (2MZZ,(Kouno et al., 2015)) using the SwissModel server. Vif (49NF,(Guo et al., 2014)) and A3G-NTD structures were submitted to the Z-Dock server (Pierce et al., 2014). Proteins structures were visualized using The PyMOL Molecular Graphics System (v.1.3 Schrödinger, LLC.). Poisson-Blotzmann electrostatic potential maps for Vif and A3G-NTD structures were produced using the PDB2PQR server (Dolinsky et al., 2004) and the ABPS toolkit in PyMol (Baker et al., 2001). A full-length Cullin5 structure was predicted from available data for Cullin1 and Cullin5 using the SwissModel server (Goldenberg et al., 2004). Full-length Cullin5 was then structurally aligned to the Cullin5 fragment in the Vif-E3 crystal structure (49NF) using SwissPDBViewer.
Supplementary Material
ACKNOWLEDGMENTS
The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (HIV NL4-3: #114, NL4-3 ΔVif: #2481, SupT1: #100 and TZM-bl: #8129. This work was funded, in part, by NIH/NIAID grants AI064001, AI089246 and AI090935. This study was performed as part of the Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Centre of the University of Amsterdam and Sanquin Blood Supply Foundation (http://www.amsterdamcohortstudies.org/). The ACS is part of the Netherlands HIV Monitoring Foundation and financially supported by the Netherlands National Institute for Public Health and the Environment. We are greatly indebted to all cohort participants for their continuous participation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS M.O., V.S and M.L. designed experiments. M.L. and M.O. performed experiments. T.B and N.K provided the Amsterdam Cohort Vif samples. M.L. and M.O. and V.S. wrote the paper. All authors participated in the study concept and gave comments on the manuscript.
REFERENCES
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binka M, Ooms M, Steward M, Simon V. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. Journal of virology. 2012;86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard Y, Turelli P, Rohrig UF, Zoete V, Mangeat B, Michielin O, Trono D. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. Journal of virology. 2009;83:12611–12621. doi: 10.1128/JVI.01491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, He Z, Wang T, Xu R, Yu XF. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. Journal of virology. 2009;83:8674–8682. doi: 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Emerman M. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS pathogens. 2013;9:e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Hirsch VM, Emerman M. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell host & microbe. 2012;11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Malik HS, Emerman M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368:20120496. doi: 10.1098/rstb.2012.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arc M, Ayouba A, Esteban A, Learn GH, Boue V, Liegeois F, Etienne L, Tagg N, Leendertz FH, Boesch C, et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1343–1352. doi: 10.1073/pnas.1502022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Zhou T, York I.a., Zheng Y-H. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. Journal of virology. 2009;83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. Journal of molecular biology. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic acids research. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. The Journal of biological chemistry. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Harris R, Bishop K, Sheehy A. DNA Deamination Mediates Innate Immunity to Retroviral Infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. Journal of molecular biology. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. Journal of virology. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan D, Liu L. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proceedings of the National Academy of Sciences. 2009 doi: 10.1073/pnas.0906652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nature structural & molecular biology. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- Kootstra NA, Navis M, Beugeling C, van Dort KA, Schuitemaker H. The presence of the Trim5alpha escape mutation H87Q in the capsid of late stage HIV-1 variants is preceded by a prolonged asymptomatic infection phase. AIDS. 2007;21:2015–2023. doi: 10.1097/QAD.0b013e3282effa87. [DOI] [PubMed] [Google Scholar]
- Kouno T, Luengas EM, Shigematsu M, Shandilya SM, Zhang J, Chen L, Hara M, Schiffer CA, Harris RS, Matsuo H. Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nature structural & molecular biology. 2015 doi: 10.1038/nsmb.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp A, McCarthy KR, Ooms M, Letko M, Morgan JS, Simon V, Johnson WE. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS pathogens. 2013;9:e1003641. doi: 10.1371/journal.ppat.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M, Silvestri G, Hahn BH, Bibollet-Ruche F, Gokcumen O, Simon V, Ooms M. Vif proteins from diverse primate lentiviral lineages use the same binding site in APOBEC3G. Journal of virology. 2013;87:11861–11871. doi: 10.1128/JVI.01944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. The Journal of biological chemistry. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature medicine. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mehle A, Wilson H, Zhang C, Brazier AJ, McPike M, Pery E, Gabuzda D. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. Journal of virology. 2007;81:13235–13241. doi: 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, König R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Ooms M, Brayton B, Letko M, Majdak S, Pilcher CD, Hecht F, Barbour J, Simon V. HIV-1 adaptation to human APOBEC3H haplotypes. Cell host & microbe. 2013 doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Ooms M, Krikoni A, Kress AK, Simon V, Munk C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. Journal of virology. 2012;86:6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Russell R.a., Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. Journal of virology. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A, Jere A, Yoshida A, Yamada T, Iwamoto A, Adachi A, Fujita M. Functional analysis of HIV-1 vif genes derived from Japanese long-term nonprogressors and progressors for AIDS. Microbes and infection / Institut Pasteur. 2004;6:799–805. doi: 10.1016/j.micinf.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Salter JD, Morales GA, Smith HC. Structural insights for HIV-1 therapeutic strategies targeting Vif. Trends in biochemical sciences. 2014;39:373–380. doi: 10.1016/j.tibs.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröfelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröfelbauer B, Senger T, Manning G, Landau NR. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. Journal of virology. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Letters to Nature. 2002;418:4–8. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature medicine. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS pathogens. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MA, Hu C, Saenz DT, Fadel HJ, Sims O, Peretz M, Poeschla EM. Productive replication of Vif-chimeric HIV-1 in feline cells. Journal of virology. 2010;84:7378–7395. doi: 10.1128/JVI.00584-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan M.a., Strebel K, Pathak VK. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.