SUMMARY
Skeletal muscle fibers differentiate into specific fiber types with distinct metabolic properties determined by their reliance on oxidative phosphorylation (OXPHOS). Using in vivo approaches, we find that OXPHOS-dependent fibers, compared to glycolytic fibers, contain elongated mitochondrial networks with higher fusion rates that are dependent on the mitofusins Mfn1 and Mfn2. Switching of a glycolytic fiber to an oxidative IIA type is associated with elongation of mitochondria, suggesting that mitochondrial fusion is linked to metabolic state. Furthermore, we reveal that mitochondrial proteins are compartmentalized to discrete domains centered around their nuclei of origin. The domain dimensions are dependent on fiber type and are regulated by the mitochondrial dynamics proteins Mfn1, Mfn2, and Mff. Our results indicate that mitochondrial dynamics is tailored to fiber type physiology and provides a rationale for the segmental defects characteristic of aged and diseased muscle fibers.
Graphical Abstract
INTRODUCTION
Genetic evidence indicates that mitochondrial fusion is important for skeletal muscle function. Deletion of the mitofusins Mfn1 and Mfn2, pro-fusion proteins on the mitochondrial outer membrane, results in severe mitochondrial myopathy, including decreased muscle mass, exercise intolerance and lactic acidosis (Chen et al., 2010). In addition, some patients harboring mutations in Mfn2 or Opa1 (a pro-fusion protein on the mitochondrial inner membrane) manifest late-onset myopathy (Amati-Bonneau et al., 2008; Hudson et al., 2008; Rouzier et al., 2012). Finally, cultured fibers from dissociated flexor digitorum brevis muscle show mitochondrial fusion events that are sensitive to Mfn1 but not Mfn2 knockdown (Eisner et al., 2014).
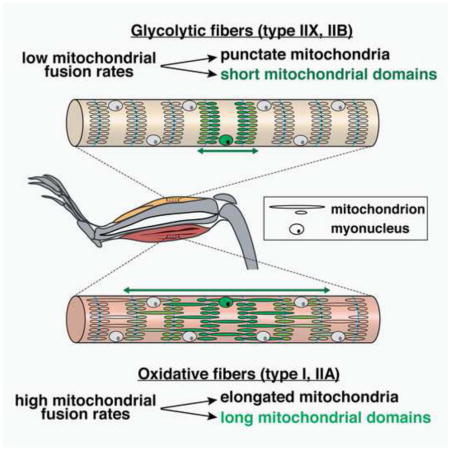
Skeletal muscle has specialized cell biological characteristics that raise important issues concerning the role of mitochondrial dynamics. First, every muscle contains a mixture of distinct fiber types that are specialized according to the work they perform (Pette and Staron, 2000). Oxidative or “slow-twitch” fibers have slow contraction rates, high mitochondrial content, increased reliance on oxidative phosphorylation (OXPHOS), high resistance to fatigue, and high representation in postural muscles. In contrast, glycolytic or “fast-twitch” fibers have rapid contractions, lower mitochondrial content, decreased reliance on OXPHOS, low resistance to fatigue, and high representation in muscle groups used for directional movement. In this study, we refer to four types of muscle fibers that form a continuum in terms of OXPHOS capacity, with I>IIA>IIX>IIB. We wondered whether the dynamics of the mitochondrial network varies according to muscle fiber type.
Second, each individual myofiber is a long, multinucleated cylindrical cell formed by the fusion of numerous myoblasts during development. Due to their extreme size and long, extended shape, intermixing of mitochondria may be constrained compared to cultured mammalian cells, where mitochondrial fusion results in rapid and extensive homogenization of the mitochondrial population (Legros et al., 2002; Mattenberger et al., 2003; Mishra and Chan, 2014). Histological data from aged and diseased fibers hint that regionalization of organelles can occur (Elson et al., 2002; Shoubridge et al., 1990; Wanagat et al., 2001), but there has been no direct demonstration and the factors controlling regionalization are unknown. To address these issues, we developed methods to measure mitochondrial fusion in intact muscle tissues and in specific muscle fiber types. We demonstrate that mitochondrial fusion rates correlate with oxidative capacity at the individual fiber level. We also developed a method to genetically label individual myonuclei in a stochastic manner, allowing us to uncover discrete mitochondrial domains within myofibers. The dimensions of these mitochondrial domains correlate with the oxidative capacity of the muscle fiber and are regulated by mitochondrial fusion and fission. Thus, mitochondrial dynamics is specialized according to fiber type within skeletal muscle and regulates homogenization of the organelle population along the fiber.
RESULTS
Mitochondrial fusion correlates with the metabolic state of muscle fiber types
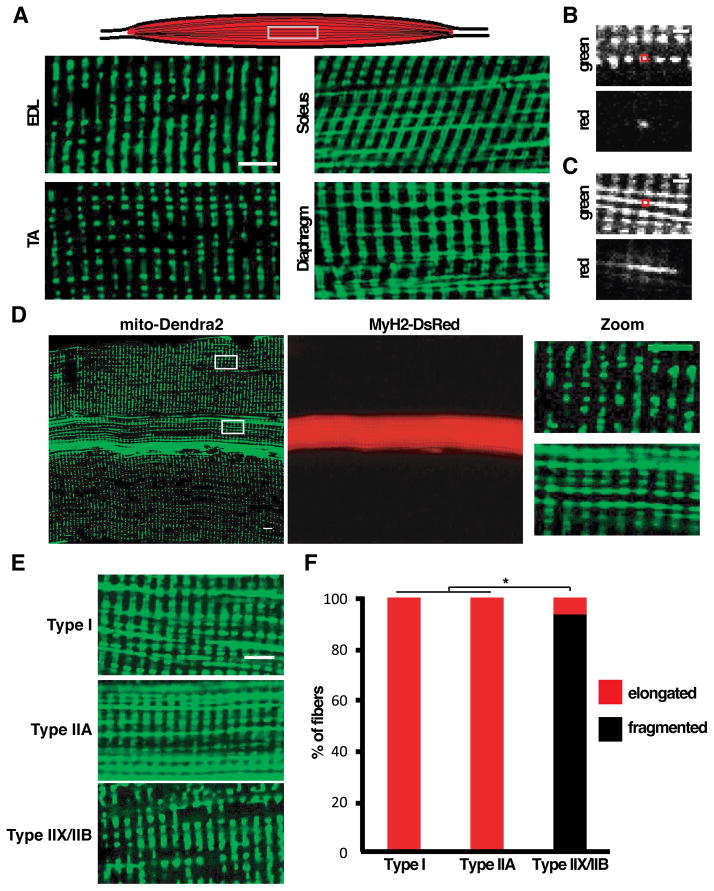
Using knock-in mice with the fluorescent protein Dendra2 targeted to the mitochondrial matrix (Pham et al., 2012), we first examined mitochondrial morphology in the extensor digitorum longus (EDL) and the tibialis anterior (TA), both fast-twitch muscles. In approximately 85% of fibers, we observed small block-like mitochondria arranged in rows and columns when viewed along the long axis of the fiber (Figure 1A). The architecture of these interfibrillar mitochondria is consistent with previous EM studies (Ogata and Yamasaki, 1997), and we have shown that each block is a mitochondrial dyad, with the z-lines of the sarcomeres running down the middle of the columnar blocks (Pham et al., 2012). In striking contrast, we observed dramatically different mitochondrial morphologies in fibers of the soleus and diaphragm, two slow-twitch muscles with high oxidative capacity. Here, columns of mitochondria are still present, but in addition, there are prominent longitudinal connections that span mitochondria from multiple sarcomeres (Figure 1A).
Figure 1. Mitochondrial morphology and connectivity correlates with fiber type in mouse skeletal muscle.
(A) Schematic of skeletal muscle showing orientation of images (top), and mitochondrial morphology in glycolytic (EDL, TA) and oxidative (soleus, diaphragm) muscles as visualized by expression of mito-Dendra2. Scale bar, 5 μm. (B) Photoconversion of mito-Dendra2 in an ex vivo EDL muscle. The red box indicates the photoconverted region. Scale bar, 2 μm. (C) Same as (B), but in soleus muscle. (D) Co-labeling of mitochondria (mito-Dendra2) and type IIA fibers (MyH2-DsRed transgene) in EDL muscle, with higher magnification images on the far right. Scale bar, 5 μm. (E) Representative images of mitochondrial morphology in type I, IIA and IIX/IIB fibers, visualized by expression of mito-Dendra2. Scale bar, 5 μm. (F) Distribution of mitochondrial profiles in Type I, IIA and IIX/IIB fibers. Greater than 30 fibers were scored per fiber type. *, p<0.01 (z-test).
Further analysis revealed that mitochondria in oxidative muscles are indeed interconnected along the longitudinal axis. Photoconversion of Dendra2 in an ex vivo preparation was used to probe mitochondrial connectivity. The punctate mitochondria in the predominant EDL fibers show no connections with neighbors along a row (Figure 1B). In the soleus, however, the longitudinal rows are partially continuous, as indicated by immediate diffusion of the photoconverted Dendra2 signal (Figure 1C). These results suggest a correlation between fiber type and mitochondrial morphology, with fibers of the more oxidative muscles having elongated and interconnected mitochondria. However, a minority of EDL fibers deviates from the predominant pattern of punctate mitochondria, instead having extended longitudinal mitochondria resembling those in the soleus (Figure 1D).
Because each muscle contains a mixture of distinct fiber types, we utilized transgenic mouse models that allow fluorescent labeling of type I, IIA and IIX/IIB fibers (see Experimental Procedures) (Chakkalakal et al., 2012b). As expected, the majority of EDL fibers were identified as glycolytic type IIX/IIB, and these have punctate mitochondria (Figure 1D, 1E, 1F, and S1A). However, ~15% of EDL myofibers were identified as type IIA (labeled by a MyH2-DsRed transgene), and these fibers always contained elongated mitochondria (Figures 1D, 1E, and 1F). This composition of fiber types is consistent with quantitative studies (Agbulut et al., 2003) showing that the mouse EDL consists of predominantly glycolytic type IIB and IIX fibers and a smaller population of oxidative IIA fibers. The mitochondria in the EDL type IIA fibers resemble those of soleus- or diaphragm-derived type I fibers (labeled by a MyH7-CFP transgene), which are also oxidative (Figures 1E, 1F, S1B, and S1C). These observations indicate that mitochondrial dynamics is specified in a fiber-autonomous manner, as opposed to being determined by the overall muscle type.
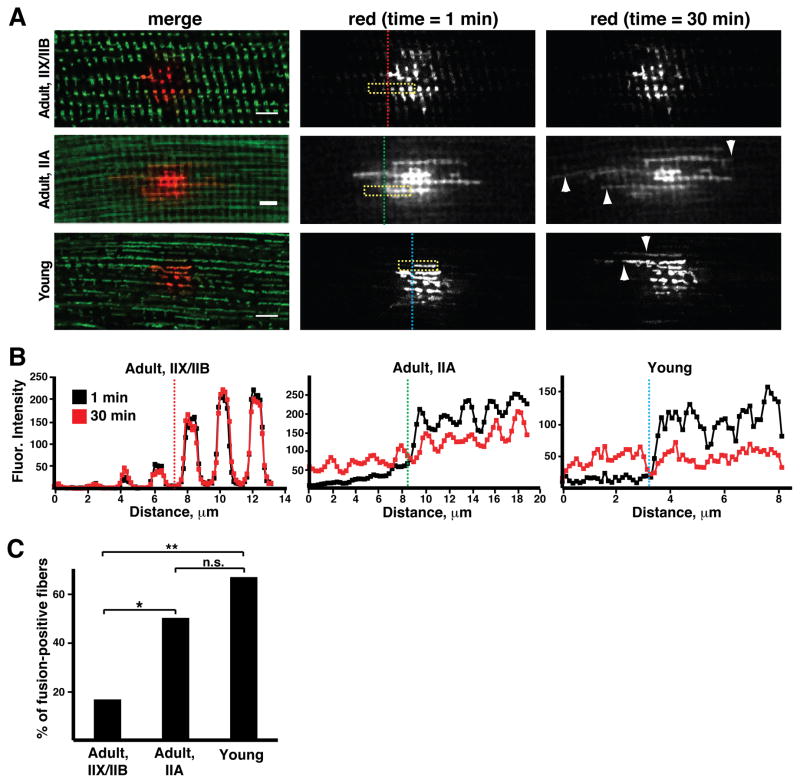
To directly monitor fusion rates, we coupled photoconversion of Dendra2-labeled mitochondria with time-lapse imaging of freshly dissected ex vivo preparations of muscle tissues. Mitochondrial fusion was sparse or undetectable in glycolytic IIX/IIB fibers (Figure 2A). Longitudinal profiles of fluorescence indicated that the organelles are well separated and did not fuse over a 30 minute period (Figure 2B). In contrast, the oxidative IIA fibers in the EDL showed much higher rates of mitochondrial fusion (Figures 2A and 2B). Fusion could be detected by a decrease in photoconverted signal in activated mitochondria and a simultaneous increase in signal in neighboring organelles (Figure 2B). In contrast to fusion events in cultured cells (Chen et al., 2003), these fusion events occurred without apparent mitochondrial movement. We conclude that the glycolytic fiber types (IIX and IIB) have punctate, isolated mitochondria with little fusion, whereas the oxidative fiber types (I and IIA) have elongated, interconnected mitochondria with higher rates of fusion (Figure 2C). These differences do not appear to be secondary to changes in the expression of fusion and fission proteins (Figure S1D).
Figure 2. Mitochondrial fusion is enhanced in oxidative and young muscle fibers.
(A) Time-lapse imaging of photoconverted mito-Dendra2 was used to follow mitochondrial fusion in ex vivo EDL muscle preparations from adult (3 month) and young (p10) animals. At time t=0 min, a subset of mitochondria were photoconverted. The merged images (left panels) indicate the differences in mitochondrial morphology. The photoconverted signal (red) was followed over 30 minutes. Longitudinal fusion events (arrowheads) were visualized by spread of signal to neighboring mitochondria. Yellow boxes indicate regions of interest analyzed in (B), and vertical dashed lines indicate the respective longitudinal positions. Scale bars, 5 μm. (B) Intensity of photoconverted signal at t=1 and 30 minutes in the regions of interest indicated by yellow boxes in (A). The longitudinal positions indicated by the vertical dashed lines have a counterpart in (A). Fusion events in IIA and young fibers are identified by the decrease in signal in the donating mitochondria on the right, coupled with increasing signal in neighboring mitochondria on the left. IIX/IIB fibers exhibit well-separated organelles with few or no (this case) fusion events. (C) Percentage of myofibers showing fusion events (over 30 minutes) for adult IIX/IIB, IIA and young myofibers. At least 10 trials were performed per fiber type. *, p<0.05; **, p<0.01; n.s., not significant (z-test).
The above results suggest that distinctions in mitochondrial dynamics are a terminal characteristic of differentiated fiber types. This prompted us to examine undifferentiated neonatal myofibers, where establishment of the adult myosin heavy chain expression pattern has not yet occurred (Agbulut et al., 2003). In 10–14 day old animals, all EDL fibers contained highly elongated mitochondria with high rates of mitochondrial fusion (Figure 2A–C), even though the majority of them (>85%) (Agbulut et al., 2003) will eventually develop into glycolytic IIX/IIB fibers with low fusion activity. Thus, fusion rates appear to be developmentally regulated and are particularly elevated in undifferentiated fibers prior to growth and specification.
Mfn1 and Mfn2 are required for mitochondrial elongation and fusion in type IIA fibers
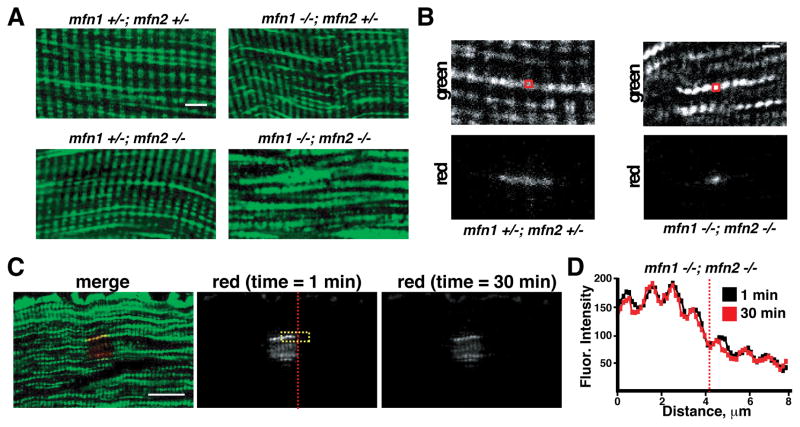
Previously, we showed that removal of Mitofusins 1 and 2 (Mfn1 and Mfn2) in skeletal muscle results in smaller muscles with lower mtDNA content (Chen et al., 2010). In that study, only fast-twitch muscles, such as the glycolytic tibialis anterior, were studied. The data in our current study suggests that the more oxidative fibers (where fusion rates are elevated) should also have a dependence on mitofusin function. To test this idea, we specifically examined type IIA fibers in MLC-Cre; mito-Dendra2cond; mfn1 −/loxP; mfn2 −/loxP animals. In the soleus of these animals, type IIA fibers are null for Mfn1 and Mfn2 and can be identified by expression of mito-Dendra2. Consistent with our previous study (Chen et al., 2010), we found that dual deletion of Mfn1 and Mfn2 caused compensatory proliferation of mitochondria (Figure 3A). This resulted in extensive interfibrillar mitochondria that were tightly packed between myofibrils. The mitochondria were aligned longitudinally and spanned the A-band between z-lines of the sarcomere. However, they did not appear interconnected, because the individual organelles were small and punctate (Figure 3B). In contrast, the single removal of Mfn1 or Mfn2 did not cause morphological changes in type IIA fibers (Figure 3A). Photoconversion experiments to probe mitochondrial connectivity confirmed that the mitochondria in Mfn1/2-deficient IIA fibers were discontinuous (Figure 3B) and exhibited no signs of content mixing (Figures 3C and 3D). Thus, Mfn1 and Mfn2 are together required in oxidative type IIA fibers to generate longitudinally connected mitochondria, and the presence of either protein alone is sufficient to maintain organelle morphology.
Figure 3. Deletion of Mfn1 and Mfn2 abrogates mitochondrial fusion in oxidative IIA muscle fibers.
(A) Representative images of mitochondrial morphology in type IIA fibers from animals of the indicated genotype. Scale bar, 5 μm. (B) Photoconversion of mito-Dendra2 in ex vivo IIA fibers from animals of the indicated genotype. The red boxes indicate the photoactivated regions. Scale bar, 2 μm. (C) Photoconversion experiment (as in Figure 2A) to follow fusion in an ex vivo IIA fiber from an mfn1 −/−,mfn2 −/− animal. Scale bar, 10 μm. (D) Same as Figure 2B, but in the IIA fiber shown in (C). The region quantitated is labeled by the yellow box in (C), and the longitudinal position is indicated by the dashed vertical line.
Mitochondrial morphology responds to changes in OXPHOS activity
Our results indicate that mitochondrial fusion is enhanced in type I and IIA fibers, which have high oxidative capacity. Neonatal myofibers, which also have high mitochondrial fusion activity (Figure 2), contain uniformly high oxidative activity, regardless of the terminal fiber type (Chen et al., 2010; Redenbach et al., 1988). We therefore wondered whether the distinctions in mitochondrial topology between muscle fibers of different types and developmental stages are a response to their distinct OXPHOS capacities.
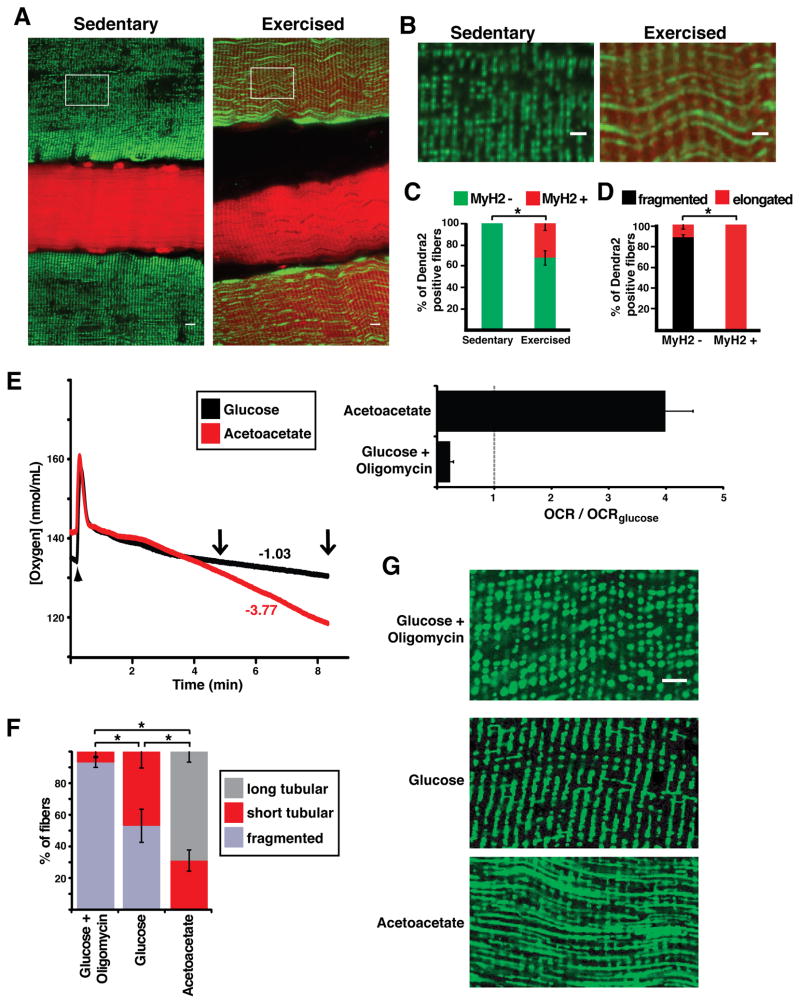
To test this hypothesis, we induced glycolytic fibers to respire more actively in an in vivo setting and examined whether this metabolic shift caused an accompanying change in mitochondrial morphology. In mice, endurance exercise is well known to induce increased respiratory capacity and fiber-type switching in certain hind-limb muscles, such as the plantaris muscle (Allen et al., 2001). We utilized cell lineage tracing to track the fate of IIX/IIB fibers by expression of Pvalb-Cre, which permanently activates a mito-Dendra2 expression cassette in these cells. Simultaneously, the type IIA fate was tracked by the expression of DsRed under control of the MyH2 promoter. Thus, using mice with both reporters allows IIA and IIX/IIB fiber types to be simultaneously monitored as a function of a 4-week running regimen. Prior to running, 0% of IIX/IIB fibers (marked by mito-Dendra2) were positive for the IIA DsRed marker (Figures 4A–C). This observation indicates that type IIX/IIB and type IIA fibers are non-overlapping cell types under our basal pre-exercise conditions. After 4 weeks of voluntary running, analysis of the plantaris muscle revealed a substantial increase in oxidative myofibers, due to fiber type switching from IIX/IIB to IIA (Figure 4A–C). Approximately 30% of the IIX/IIB-marked fibers were now also positive for the type IIA marker, a staining pattern never observed under sedentary conditions. In all of these “switched” fibers (expressing both the IIA and IIX/IIB markers), mitochondrial morphology was substantially elongated (Figures 4B and 4D). Thus, the exercise regimen promoted substantial fiber type switching that is associated with a dramatic change in mitochondrial structure.
Figure 4. Mitochondrial morphology in muscle fibers responds to changes in OXPHOS activity.
(A) Co-labeling of IIA fibers (MyH2-DsRed transgene; red) and mitochondria from IIX/IIB fibers (mito-Dendra2; green) in plantaris muscle from sedentary and exercised mice. The mito-Dendra2 marker is activated in IIA/IIB fibers by the Pvalb-Cre driver. Note that co-expression of these markers is found only in fibers from exercised animals. Scale bar, 10 μm. (B) Enlarged view of mitochondrial morphology from regions indicated in (A) (boxes). Scale bar, 5 μm. (C) Percentage of originally IIX/IIB fibers (Dendra2 positive) which are co-labeled with the IIA marker (MyH2-DsRed) in sedentary and exercised mice. At least 30 fibers were scored for 5 animals in each group. *, p<0.05 (z-test). (D) In exercised mice, percentage of originally IIX/IIB fibers (Dendra2 positive) which have elongated or fragmented mitochondria. Fibers were separated into two groups: those having switched to the IIA fiber type (MyH2-DsRed positive) and those that have not switched (MyH2-DsRed negative). *, p<0.01 (z-test). (E) Representative oxygen consumption rates (OCR) measured ex vivo in paired EDL muscles from the same animal (left). The chamber was closed at the time indicated by the arrowhead. Respective oxygen consumption rates (nmol/min/mL) are indicated for the time interval between the two arrows. (Right) Oxygen consumption rates, normalized to OCR in glucose-containing media. Experiments were performed in triplicate. (F) Percentage of IIX/IIB fibers having long tubular, short tubular or fragmented mitochondria, after overnight incubation in the indicated media. At least 30 fibers per muscle were counted, and experiments were performed in triplicate. *, p<0.01 (t-test). (G) Representative images of mitochondrial morphology from IIX/IIB fibers after overnight incubation in the indicated media. Mitochondria were visualized by expression of mito-Dendra2. Scale bar, 5 μm. Error bars indicate standard errors.
To further test the relationship between metabolism and mitochondrial dynamics, we induced EDL muscle to respire more actively ex vivo, where fiber type switching does not occur. We isolated individual EDL muscles from animals in which the IIX/IIB fibers were labeled with mito-Dendra2 and measured oxygen consumption in media containing glucose or acetoacetate. Similar to cultured fibroblasts (Mishra et al., 2014), EDL muscle placed in oxidative (acetoacetate-containing) media exhibited a several-fold higher rate of oxygen consumption and increased mitochondrial membrane potential as compared to muscle in glucose-containing media (Figure 4E, S2A–D). With glucose-containing media, the IIX/IIB fibers contain largely punctate mitochondria, with half of the fibers containing a few tubular mitochondria (Figures 4F and 4G). Addition of oligomycin, which inhibits complex V activity, resulted in fibers with completely fragmented mitochondria having no interconnections. With oxidative media, the majority of IIX/IIB EDL muscle fibers showed rows of interconnected, highly tubular mitochondria, a phenotype never found in IIX/IIB fibers under the glucose-containing condition or in vivo (Figures 4F and 4G). Photoconversion experiments indicate that the matrices of these rows of mitochondria are partially continuous (Figure S1E). Thus, the data suggest that mitochondrial morphology is not an obligate characteristic of fiber type but instead can dynamically respond to the OXPHOS activity of each individual fiber.
The length of mitochondrial domains in vivo correlates with myofiber type
Muscle fibers are long, multinucleated cells formed from the fusion of myoblasts. This unusual cell biology raises the issue of whether the mitochondria are well homogenized through extensive fusion, or whether the mitochondria are compartmentalized into discrete domains that are locally controlled by nearby nuclei. To distinguish between these two extreme models, we used mouse genetics to stochastically label a mitochondrial protein expressed by a single myonucleus (Figures 5A and 5B). During postnatal development, satellite cells (a resident stem cell population of muscular tissue) can differentiate and fuse with existing myofibers, thereby providing new nuclei and organelles to the growing fiber (Keefe et al., 2015). We reasoned that visualization of individual satellite cell fusion events and the extent of mitochondrial spreading should provide an indication of the underlying mitochondrial dynamics.
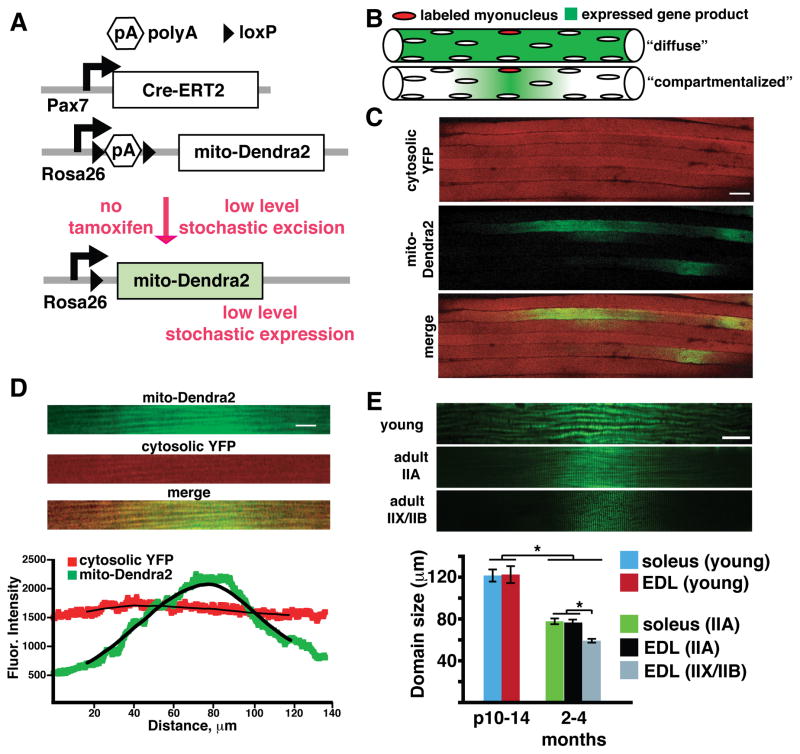
Figure 5. Mitochondria form domains in mouse skeletal muscle.
(A) Schematic for stochastic satellite cell labeling experiment. Tamoxifen-dependent Cre-ERT2 (driven by the Pax7 promoter) is expressed in satellite cells. In the absence of tamoxifen, leaky Cre activity occasionally excises a poly(A) stop cassette, allowing rare, stochastic expression of mito-Dendra2. (B) Two extreme models of mitochondrial homogenization in myofibers. Single myonuclei are randomly labeled by leaky activity of Cre-ERT2 recombinase in the absence of tamoxifen. The expressed fluorescent gene products may then be homogenized throughout the fiber (top), or compartmentalized to a “domain” surrounding the labeled myonucleus (bottom). (C) Low magnification view of cytosolic (cytosolic YFP, shown in red for clarity) and mitochondrial (mito-Dendra2, green) domains in EDL muscle. Scale bar, 50 μm. (D) Representative single mitochondrial domain (green) from a type IIB fiber. For comparison, note that the cytosolic YFP localization is homogeneous (red). Scale bar, 10 μm. (Bottom) Quantitation of fluorescence intensity of mito-Dendra2 and cytosolic YFP. (E) (Top) Representative mitochondrial domains from young, IIA and IIX/IIB fibers in wild-type mice. Scale bar, 20 μm. (Bottom) Quantitation of domain size in the indicated fiber types from wild-type mice. At least 30 domains per fiber type were quantified. *, p<0.001 (t-test). Error bars indicate standard errors.
We crossed the satellite-cell specific Pax7-CreERT2 driver (Lepper et al., 2009) to a floxed allele of mito-Dendra2 and cytosolic YFP (Figure 5A). In these animals, activation of Cre leads to mito-Dendra2 expression specifically in pre-fused satellite cells. The CreERT2 molecule is tamoxifen-dependent, and we expected that low levels of tamoxifen would be needed for stochastic activation. Surprisingly, even in the absence of tamoxifen, we occasionally detected mito-Dendra2 expression in single satellite cells on the fiber periphery, representing the pre-fused population (Figure S3A). In addition, we could detect post-fusion events, demonstrated by the presence of mito-Dendra2 or cytosolic YFP within an existing myofiber (Figures 5C and S3B). Myofibers were uniformly labeled with cytosolic YFP. In contrast, mito-Dendra2 expression was localized to small longitudinal regions, typically less than 100 μm in length (Figures 5C and D). These observations indicate that the CreERT2 activity is leaky enough to provide stochastic activation in a small subset of satellite cells. Moreover, mitochondrial proteins show regionalization to their nucleus of origin, in contrast to cytosolic proteins.
The mito-Dendra2 mitochondrial domains were randomly distributed throughout the tissue and typically shaped as single peaks with a Gaussian distribution of fluorescence intensity along the longitudinal axis (Figures 5D and S3C). Such peaks likely represent single fusion events between a satellite cell and a myofiber. Occasionally, we were able to find double and triple peaks, indicating fusion events that had occurred near one another (Figure S3C). Localization of mito-Dendra2 was uniform along the transverse axis, indicating that domains occur only in the longitudinal direction (Figures S3D, E). Using a conditional nuclear-targeted GFP allele, we could estimate the prevalence of labeled myonuclei within muscle fibers (Figure S3F). Less than 1% of total myonuclei were labeled, and >70% of the time, the nuclear GFP expression profile was consistent with the presence of a singly activated myonucleus.
Two lines of evidence indicate that these mitochondrial domains are stable over time, as opposed to being transient phenomena from recent satellite cell fusion events. First, in ex vivo culturing experiments, we did not observe significant changes in domain size or dissipation of the mito-Dendra2 signal over the course of 72 hours (Figures S4A, B). Thus, mitochondrial domains are stable under standard culturing conditions that maintain myofibers in a healthy and functional state (Casas et al., 2010; Eisner et al., 2014; Keire et al., 2013). Second, we observed a significant increase in the frequency of mitochondrial domains in aged animals, suggesting that domains are stably accumulating over time (Figures S4C, D). It is well known that Pax7-positive satellite cells decrease in abundance with age (Chakkalakal et al., 2012a; Collins et al., 2007; Fry et al., 2015; Shefer et al., 2006). If the observed mitochondrial domains were transient reflections of recent satellite cell fusion events, we would expect their frequency to decrease in aged animals.
Because interfibrillar mitochondria are localized to the Z-disc with little movement, we hypothesized that domain size would be at least partially dependent on the ability of neighboring organelles to undergo fusion and share protein products. To test this idea, we quantified the size of individual domains (calculated as the full width at half-maximum fluorescence intensity) in a large number of fibers, and classified them by fiber type. To simplify quantification, we restricted our analysis to domains with single peaks. In the adult EDL, type IIX/IIB fibers had the smallest domain dimension, approximately 60 μm (Figure 5E). The more oxidative IIA fibers, which we showed had more fusogenic mitochondria, had larger domains that approached 80 μm. Domain size in neonatal muscle was the largest, approximately twice the size of adult IIX/IIB fibers (Figure 5E). Because these results may be complicated by the increased abundance of Pax7-positive satellite cells in young animals, we specifically limited our analysis to mito-Dendra2 domains containing a single peak. Indeed, the same fiber types from different muscles showed similar domain size, and aged animals did not show any further decreases in domain size (Figures 5E and S4E). The correlation of mitochondrial domain size with the metabolic state of the fiber suggests that mitochondrial fusion is an important regulator of domain size, allowing OXPHOS-dependent fiber types to more efficiently homogenize their organelle population.
Mitofusins and Mff control the length of mitochondrial domains
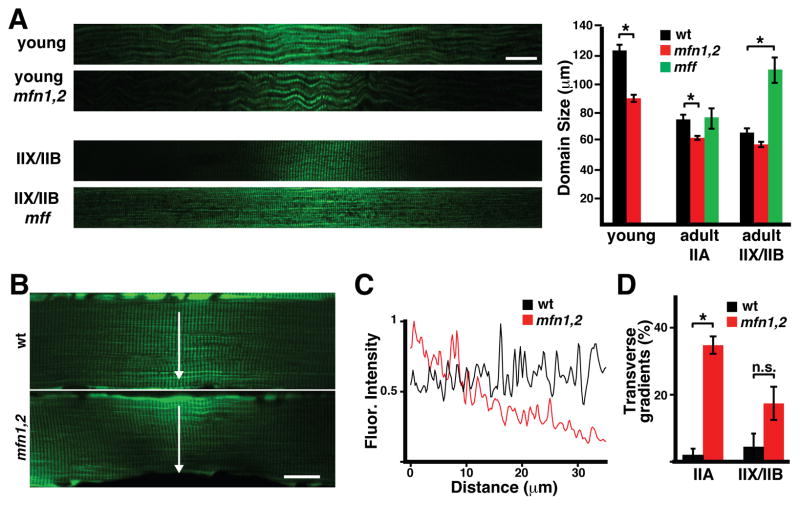
To definitively implicate mitochondrial dynamics in regulation of domain size, we utilized conditional alleles of Mitofusin 1 and Mitofusin 2. Combining these alleles with the Pax7-CreERT2 driver allows removal of the mitofusins in the few stochastically activated satellite cells, without affecting overall animal physiology. Domains from these animals showed smaller sizes, particularly in the young and oxidative fiber types (Figure 6A). Strikingly, some domains were extremely small in fusion-deficient animals, with limited spread of the Dendra2 signal even along the transverse axis of the fiber (Figures 6B and 6C). Such transverse gradients were present at much higher frequency in mutant animals, regardless of fiber type (Figure 6D). Thus, domain size and shape are dependent on the mitofusins. These substantial changes due to mitofusin deletion are remarkable, given that in this experimental system, the nuclei surrounding the mitochondrial domain have normal mitofusin expression. Mitochondrial fusion is known to require mitofusin function in both partners undergoing the fusion process (Koshiba et al., 2004). In contrast, deletion of mitochondrial fission factor (Mff) in the whole animal results in much larger domains, especially in the glycolytic IIX/IIB fibers (Figure 6A).
Figure 6. Mitochondrial domain size is regulated by Mfn1, Mfn2 and Mff.
(A) (Left) Representative mitochondrial domains from wild-type and mutant mice with the indicated fiber types and genotypes. Scale bar, 20 μm. (Right) Quantitation of domain lengths. At least 30 domains per group were quantified. *, p<0.001 (t-test). (B) Representative mitochondrial domains in wildtype and mfn1/2 IIA fibers. Note that the domain in the mfn1/2 fiber shows a gradient in the transverse direction (arrow, perpendicular to the long axis of the fiber). Scale bar, 20 μm. (C) Quantitation of mito-Dendra2 fluorescence intensity in the transverse direction (from B). (D) Percentage of mitochondrial domains showing transverse gradients, in the indicated fiber type and genotype. At least 30 domains per group were quantified. *, p<0.001; n.s., not significant (t-test). Error bars indicate standard errors.
DISCUSSION
Our results suggest that mitochondrial fusion rates are a defining characteristic between different muscle fiber types, similar to other mitochondrial properties, such as abundance in the sarcolemmal region and respiratory chain activity. Previous EM studies have suggested that “red” muscles may have an altered mitochondrial topology, with the mitochondria appearing more “string-like” and stretching across the A-band (Fujioka et al., 2013; Ogata and Yamasaki, 1997). Here, we have extended this analysis by correlating topology, interconnectivity and fusion with specific fiber types as defined by myosin heavy chain expression. The fiber types I and IIA have elongated mitochondria and higher rates of mitochondrial fusion compared to glycolytic muscle fibers. Therefore, the level of mitochondrial fusion correlates with the OXPHOS capacity of the fiber. We do not think that mitochondrial morphology plays a direct role in the determination of fiber type, as enhanced fusion ex vivo is not accompanied with type switching, and deletion of the mitofusins does not prevent formation of oxidative fibers (Chen et al., 2010). Instead, we suspect that mitochondrial morphology adapts to the specific functional needs of the fiber. Elongated mitochondrial networks have been associated with increased metabolic states (Gomes et al., 2011; Mitra et al., 2009; Tondera et al., 2009) and may facilitate maintenance of ATP levels in oxidative muscle fibers that require long-term energy production. One might expect that enhancing mitochondrial fusion (via Mfn1/2 overexpression, for instance) would prove beneficial, particularly for the function of oxidative fibers.
By activating mito-Dendra2 expression in specific myonuclei, we reveal that mitochondrial domains exist in vivo in muscle fibers. The dimensions of these domains are dependent on the fusion and fission processes, with increased fusion correlated with longer domains. In addition, we would expect a dependence on other biophysical parameters such as protein synthesis and turnover rates, as well as mRNA and protein diffusion constants. Domains have been visualized previously for Golgi and nuclear proteins (Hall and Ralston, 1989; Pavlath et al., 1989), as well as structural proteins such as dystrophin (Blaveri et al., 1999; Kinoshita et al., 1998), though the biophysical parameters underlying the sizes of these domains have not been explored.
Even though oxidative muscle fibers have higher rates of mitochondrial fusion, we find that all muscle fibers show mitochondrial compartmentalization, which implies that homogenization of mitochondria is relatively limited. We can suggest several reasons why it may be more challenging to homogenize mitochondria in skeletal muscle versus cultured cells. First, myofibers are extremely long cells, often several centimeters or longer in length. Second, mitochondria in muscle fibers are not known to move, thereby limiting interactions between organelles. And most importantly, the fiber is a syncytial cell in which numerous nuclei are spaced apart longitudinally, providing an intrinsically high degree of heterogeneity as compared with a singly nucleated cell. Our results suggest that mitochondrial proteomes in muscle fibers are determined largely by the nearest nuclei.
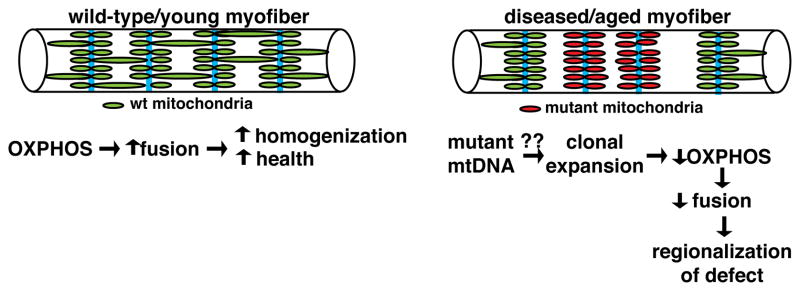
The phenomenon of mitochondrial domains, as well as their regulation by OXPHOS capacity, is likely to be relevant to mitochondrial and aging-related disorders, where mutant mtDNA accumulates to high levels in segmental regions along the length of a fiber (Barron et al., 2005; Elson et al., 2002; Wanagat et al., 2001). The dysfunctional region is associated with local, clonal expansion of mutant mtDNA, and appears as OXPHOS-deficient, “ragged red fibers” on transverse sections of muscle tissue. Our discovery of mitochondrial domains provides a rationale for why these mtDNA defects are limited to discrete longitudinal domains. With clonal expansion of mutated mtDNA, the loss of OXPHOS activity would lower local mitochondrial fusion rates, thereby providing a mechanism to promote regionalization of the defect and prevent its spread throughout the fiber (Figure 7). We propose that by linking OXPHOS activity of the mitochondrion to fusion rates, the myofiber can promote health under normal conditions and restrict the spread of mtDNA defects during disease and aging.
Figure 7. Model: the linking of OXPHOS activity and mitochondrial fusion promotes organellar health and restricts spread of mtDNA defects in myofibers.
(Left) In wildtype myofibers, oxidative activity promotes fusion of mitochondria across the sarcomere, thereby promoting some homogenization of the organellar population along the longitudinal axis. Z-lines are depicted in cyan. (Right) In the presence of mutant mtDNA, clonal expansion of the genetic defect (due to unknown mechanisms) results in decreased OXPHOS activity. Decreased OXPHOS at the site of the defect is proposed to lower local fusion rates, thereby promoting compartmentalization and limiting spread of the mutant mtDNA genome throughout the fiber.
EXPERIMENTAL PROCEDURES
Mouse breeding
All mouse experiments were approved by the Caltech Institutional Animal Care and Use Committee. Mice with conditional or ubiquitous expression of mito-Dendra2 were described previously (Pham et al., 2012). To label type I fibers and visualize mitochondria, transgenic mice expressing CFP under control of the MyH7 promoter (Chakkalakal et al., 2012b) were crossed to mice with the ubiquitous mito-Dendra2 allele (MyH7-CFP; mito-Dendra2). Similarly, type IIA fibers were visualized using mice expressing DsRed under control of the MyH2 promoter (Chakkalakal et al., 2012b) (MyH2-DsRed; mito-Dendra2). To label type IIX/IIB fibers, we utilized the Pvalb-Cre allele (Hippenmeyer et al., 2005), which is selectively active in IIX/IIB fibers (Berchtold et al., 2000; Chakkalakal et al., 2012b). Pvalb-Cre; mito-Dendra2cond mice allowed visualization of mitochondria specifically in IIX/IIB fibers. To address the role of mitofusins in type IIA fibers, we used previously described conditional and excised knockout alleles of Mfn1 and Mfn2, in combination with the MLC-Cre allele (Bothe et al., 2000; Chen et al., 2010; Mourkioti et al., 2008).
For voluntary exercise experiments, we generated MyH2-DsRed; Pvalb-Cre; mito-Dendra2cond mice, which allows us to label and distinguish IIA and IIX/IIB fibers. At 6 weeks of age, mice were housed in isolation with (‘exercised’) or without (‘sedentary’) running wheels. Mice were euthanized at 10 weeks of age and muscles were harvested for analysis (see below).
For stochastic labeling of myonuclei, we took advantage of the Pax7-CreERT2 allele, in which tamoxifen-dependent Cre recombinase is expressed in the Pax7-positive satellite cells (Lepper et al., 2009). In the absence of tamoxifen, leaky activity of the Cre recombinase allows for low-level excision of floxed alleles (Haldar et al., 2009; Kemp et al., 2004; Liu et al., 2010). The Pax7-CreERT2 allele was combined with conditional expression alleles for mito-Dendra2 (Pham et al., 2012), cytosolic YFP (Srinivas et al., 2001), or nuclear-targeted GFP-βgalactosidase fusion protein (Stoller et al., 2008), allowing visualization of mitochondrial, cytosolic and nuclear domains respectively. All conditional expression alleles are integrated into the ROSA26 locus.
To assess the effect of mitochondrial fusion on the size of domains, we utilized previously described (Chen et al., 2003; Chen et al., 2007) conditional and excised knockout alleles of Mitofusin 1 and Mitofusin 2. Complete removal of Mfn1 or Mfn2 is embryonic lethal (Chen et al., 2003), so we instead removed these proteins from stochastically labeled myonuclei using the Pax7-CreERT2 allele. As the Pax7 and Mfn2 loci are both on chromosome 4, we isolated animals with a Pax7-CreERT2 Mfn2 − chromosome by screening for homologous recombination events between the alleles. Experimental and control littermates were then bred by crossing Pax7-CreERT2 Mfn2 −/+; Mfn1 −/+ mice to mito-Dendra2cond; Mfn1loxP/loxP; Mfn2loxP/loxP mice. To assess the effect of mitochondrial fission on the size of domains, we utilized Mff mutant mice (unpublished data) carrying a gene-trap allele that abolishes Mff expression (Loson et al., 2013). Experimental and control littermates were bred by crossing Pax7-CreERT2; Mff −/+ mice to mito-Dendra2cond; Mff −/+ mice.
Muscle preparation and imaging
For analysis of mitochondrial morphology or domain measurements, skeletal muscle was dissected and fixed in formalin for 3–4 hours at room temperature, followed by overnight at 4°C. Muscles were rinsed with PBS and teased apart using fine forceps. Samples were mounted on glass slides with Cytoseal XYL mounting media (Thermo Scientific). Images were acquired on a Zeiss LSM 710 confocal microscope using Plan-Apochromat objectives (63x, 40x or 20x). For mitochondrial morphology measurements, optical slices at 0.7 μm thickness were acquired, and Z-stacks were oversampled at 0.35 μm. For domain measurements, optical slices of 2.0 μm thickness were acquired.
For photoconversion or fusion experiments, acutely dissected whole EDL or soleus muscle was transferred to culturing media [DMEM (Sigma D5030) supplemented with 1x penicillin-streptomycin, 10 mM D-glucose, 2 mM L-glutamine, 10% dialyzed FBS, 25 mM HEPES, pH 7.4] in a coverglass bottom dish. Muscles were stabilized with a slice anchor (Warner Instruments) and imaged on a heated stage at 37°C. For photoconversion of Dendra2, a region of interest was irradiated with the 405 nm laser. To follow fusion, Z-stacks were acquired for 30 minutes immediately after photoconversion.
Oxygen consumption rates were measured in freshly dissected EDL muscles using a two chamber Clark oxygen electrode system (Hansatech Instruments), allowing simultaneous measurements of two muscles from the same animal. Chambers were equilibrated with the indicated culturing media, and a single EDL muscle was added to each chamber. After closing the chamber and allowing for equilibration of oxygen levels, basal respiration was measured as the average oxygen consumption rate (OCR) over 3 minutes. For acetoacetate-containing media, glucose was replaced with acetoacetate (10 mM), which was previously shown to increase OCR in cultured cells (Mishra et al., 2014). Respiratory parameters were measured by the sequential addition of oligomycin (1 μM, Sigma), CCCP (2 μM, Sigma), and antimycin A (1 μM, Sigma). The cellular respiratory control ratio (RCR) was calculated as the ratio of maximal OCR (CCCP-stimulated) to oligomycin-inhibited OCR.
Membrane potential measurements were performed by incubating EDL muscles in media supplemented with 200 nM TMRM (Molecular Probes) for 30 min at 37°C. Muscles were then switched to non-TMRM media and imaged. Relative fluorescence was quantified from pixels of mitochondrial signal using ImageJ.
For ex vivo analysis of mitochondrial domains, single fibers were isolated by enzymatic digestion. EDL muscles were incubated at 37°C in a shaker (90 rpm for 45 minutes) in DMEM (Invitrogen 11995) supplemented with collagenase type IV (Sigma). Fibers were then manually dissociated in DMEM containing 10% horse serum using mild trituration through decreasing bore sizes of pipet tips pre-coated with 10% horse serum. Individual fibers were transferred to Matrigel (BD Bioscience) coated dishes, and cultured for 72 hours at 37°C, 5% CO2. Individual mitochondrial domains were identified and imaged every 12 hours.
For analysis of nuclear GFP expression, single myofibers were isolated as above. Individual fibers were then transferred to 8-well Permanox slides (Nunc Lab-Tek) pre-coated with Matrigel. After settling, fibers were fixed for 20 minutes at 37°C with formalin, permeabilized for 10min at −20°C with acetone, and then blocked in PBS containing 10% FBS, 1% BSA, 2% Triton X-100. GFP was detected with AlexaFluor555-conjugated αGFP antibody (Molecular Probes) and myonuclei were stained with DAPI (Molecular Probes). Nuclear activation was quantified by comparing the number of GFP-positive nuclei versus total number of nuclei.
For analysis of protein levels, muscle lysates were prepared via homogenization and clarification in lysis buffer (50 mM Tris, 2% SDS, pH 6.8) supplemented with HALT protease inhibitors, and analyzed by Western blot. The following antibodies were used for detection: Mfn1 (Chen et al., 2003), Mfn2 (Cell Signaling), Mff (gift from A. van der Bliek), Hsp60 (Santa Cruz Biotech), and Tom20 (Santa Cruz Biotech).
Analysis of mitochondrial domains
Mitochondrial domains were imaged as described above. Single optical sections were taken near the center of individual fibers. Curved fibers were digitally straightened, and fluorescence intensities along the longitudinal axis were calculated in ImageJ. Raw intensities were filtered using a moving average, and domain size was calculated as the peak width at half-maximal intensity using custom scripts in Matlab (Mathworks). Only single, well-separated peaks were analyzed, and the intensity at both tails was required to be below 30% of the maximal intensity.
Supplementary Material
Acknowledgments
We thank Alexander van der Bliek for providing the α-Mff antibody. We are grateful to Hsiuchen Chen for help with experiments using Mfn1/2 and Mff mutant mice. This work was supported by grants from the NIH (GM062967) and the Muscular Dystrophy Association. P.M. was supported by a Baxter Senior Postdoctoral fellowship. G.V. was supported by grants from the NIH/NRSA (GM0076162T32) and the California Institute for Regenerative Medicine (Stem Cell Bridges Program, TB1-01176). We thank members of the Chan lab for helpful discussions and comments on the manuscript.
Footnotes
Author Contributions
P.M., G.V., A.P., and D.C. conceived and designed experiments. P.M. and A.P. performed experiments relating to mitochondrial morphology. G.V. and A.P. performed experiments relating to mitochondrial domains. P.M. and D.C. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agbulut O, Noirez P, Beaumont F, Butler-Browne G. Myosin heavy chain isoforms in postnatal muscle development of mice. Biology of the cell / under the auspices of the European Cell Biology Organization. 2003;95:399–406. doi: 10.1016/s0248-4900(03)00087-x. [DOI] [PubMed] [Google Scholar]
- Allen DL, Harrison BC, Maass A, Bell ML, Byrnes WC, Leinwand LA. Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. Journal of applied physiology. 2001;90:1900–1908. doi: 10.1152/jappl.2001.90.5.1900. [DOI] [PubMed] [Google Scholar]
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, Campos Y, Rivera H, de la Aleja JG, Carroccia R, Iommarini L, Labauge P, Figarella-Branger D, Marcorelles P, Furby A, Beauvais K, Letournel F, Liguori R, La Morgia C, Montagna P, Liguori M, Zanna C, Rugolo M, Cossarizza A, Wissinger B, Verny C, Schwarzenbacher R, Martin MA, Arenas J, Ayuso C, Garesse R, Lenaers G, Bonneau D, Carelli V. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain : a journal of neurology. 2008;131:338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- Barron MJ, Chinnery PF, Howel D, Blakely EL, Schaefer AM, Taylor RW, Turnbull DM. Cytochrome c oxidase deficient muscle fibres: substantial variation in their proportions within skeletal muscles from patients with mitochondrial myopathy. Neuromuscular disorders : NMD. 2005;15:768–774. doi: 10.1016/j.nmd.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiological reviews. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- Blaveri K, Heslop L, Yu DS, Rosenblatt JD, Gross JG, Partridge TA, Morgan JE. Patterns of repair of dystrophic mouse muscle: studies on isolated fibers. Developmental dynamics : an official publication of the American Association of Anatomists. 1999;216:244–256. doi: 10.1002/(SICI)1097-0177(199911)216:3<244::AID-DVDY3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bothe GW, Haspel JA, Smith CL, Wiener HH, Burden SJ. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis. 2000;26:165–166. [PubMed] [Google Scholar]
- Casas M, Figueroa R, Jorquera G, Escobar M, Molgo J, Jaimovich E. IP(3)-dependent, post-tetanic calcium transients induced by electrostimulation of adult skeletal muscle fibers. The Journal of general physiology. 2010;136:455–467. doi: 10.1085/jgp.200910397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012a;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Kuang S, Buffelli M, Lichtman JW, Sanes JR. Mouse transgenic lines that selectively label Type I, Type IIA, and Types IIX+B skeletal muscle fibers. Genesis. 2012b;50:50–58. doi: 10.1002/dvg.20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. The Journal of cell biology. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Zammit PS, Ruiz AP, Morgan JE, Partridge TA. A population of myogenic stem cells that survives skeletal muscle aging. Stem cells. 2007;25:885–894. doi: 10.1634/stemcells.2006-0372. [DOI] [PubMed] [Google Scholar]
- Eisner V, Lenaers G, Hajnoczky G. Mitochondrial fusion is frequent in skeletal muscle and supports excitation-contraction coupling. The Journal of cell biology. 2014;205:179–195. doi: 10.1083/jcb.201312066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson JL, Samuels DC, Johnson MA, Turnbull DM, Chinnery PF. The length of cytochrome c oxidase-negative segments in muscle fibres in patients with mtDNA myopathy. Neuromuscular disorders : NMD. 2002;12:858–864. doi: 10.1016/s0960-8966(02)00047-0. [DOI] [PubMed] [Google Scholar]
- Fry CS, Lee JD, Mula J, Kirby TJ, Jackson JR, Liu F, Yang L, Mendias CL, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nature medicine. 2015;21:76–80. doi: 10.1038/nm.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka H, Tandler B, Haldar SM, Jain MK, Hoppel CL. String mitochondria in mouse soleus muscle. Microscopy research and technique. 2013;76:237–241. doi: 10.1002/jemt.22158. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nature cell biology. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar M, Hedberg ML, Hockin MF, Capecchi MR. A CreER-based random induction strategy for modeling translocation-associated sarcomas in mice. Cancer research. 2009;69:3657–3664. doi: 10.1158/0008-5472.CAN-08-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall ZW, Ralston E. Nuclear domains in muscle cells. Cell. 1989;59:771–772. doi: 10.1016/0092-8674(89)90597-7. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS biology. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Amati-Bonneau P, Blakely EL, Stewart JD, He L, Schaefer AM, Griffiths PG, Ahlqvist K, Suomalainen A, Reynier P, McFarland R, Turnbull DM, Chinnery PF, Taylor RW. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain : a journal of neurology. 2008;131:329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- Keefe AC, Lawson JA, Flygare SD, Fox ZD, Colasanto MP, Mathew SJ, Yandell M, Kardon G. Muscle stem cells contribute to myofibres in sedentary adult mice. Nature communications. 2015;6:7087. doi: 10.1038/ncomms8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keire P, Shearer A, Shefer G, Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods in molecular biology. 2013;946:431–468. doi: 10.1007/978-1-62703-128-8_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp R, Ireland H, Clayton E, Houghton C, Howard L, Winton DJ. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic acids research. 2004;32:e92. doi: 10.1093/nar/gnh090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita I, Vilquin JT, Asselin I, Chamberlain J, Tremblay JP. Transplantation of myoblasts from a transgenic mouse overexpressing dystrophin prduced only a relatively small increase of dystrophin-positive membrane. Muscle & nerve. 1998;21:91–103. doi: 10.1002/(sici)1097-4598(199801)21:1<91::aid-mus12>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Legros F, Lombes A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Suckale J, Masjkur J, Magro MG, Steffen A, Anastassiadis K, Solimena M. Tamoxifen-independent recombination in the RIP-CreER mouse. PloS one. 2010;5:e13533. doi: 10.1371/journal.pone.0013533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Molecular biology of the cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattenberger Y, James DI, Martinou JC. Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS Lett. 2003;538:53–59. doi: 10.1016/s0014-5793(03)00124-8. [DOI] [PubMed] [Google Scholar]
- Mishra P, Carelli V, Manfredi G, Chan DC. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell metabolism. 2014;19:630–641. doi: 10.1016/j.cmet.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nature reviews Molecular cell biology. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourkioti F, Slonimsky E, Huth M, Berno V, Rosenthal N. Analysis of CRE-mediated recombination driven by myosin light chain 1/3 regulatory elements in embryonic and adult skeletal muscle: a tool to study fiber specification. Genesis. 2008;46:424–430. doi: 10.1002/dvg.20419. [DOI] [PubMed] [Google Scholar]
- Ogata T, Yamasaki Y. Ultra-high-resolution scanning electron microscopy of mitochondria and sarcoplasmic reticulum arrangement in human red, white, and intermediate muscle fibers. The Anatomical record. 1997;248:214–223. doi: 10.1002/(SICI)1097-0185(199706)248:2<214::AID-AR8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Pavlath GK, Rich K, Webster SG, Blau HM. Localization of muscle gene products in nuclear domains. Nature. 1989;337:570–573. doi: 10.1038/337570a0. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Myosin isoforms, muscle fiber types, and transitions. Microscopy research and technique. 2000;50:500–509. doi: 10.1002/1097-0029(20000915)50:6<500::AID-JEMT7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Pham AH, McCaffery JM, Chan DC. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 2012;50:833–843. doi: 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redenbach DM, Ovalle WK, Bressler BH. Effect of neonatal denervation on the distribution of fiber types in a mouse fast-twitch skeletal muscle. Histochemistry. 1988;89:333–342. doi: 10.1007/BF00500634. [DOI] [PubMed] [Google Scholar]
- Rouzier C, Bannwarth S, Chaussenot A, Chevrollier A, Verschueren A, Bonello-Palot N, Fragaki K, Cano A, Pouget J, Pellissier JF, Procaccio V, Chabrol B, Paquis-Flucklinger V. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain : a journal of neurology. 2012;135:23–34. doi: 10.1093/brain/awr323. [DOI] [PubMed] [Google Scholar]
- Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Developmental biology. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge EA, Karpati G, Hastings KE. Deletion mutants are functionally dominant over wild-type mitochondrial genomes in skeletal muscle fiber segments in mitochondrial disease. Cell. 1990;62:43–49. doi: 10.1016/0092-8674(90)90238-a. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoller JZ, Degenhardt KR, Huang L, Zhou DD, Lu MM, Epstein JA. Cre reporter mouse expressing a nuclear localized fusion of GFP and beta-galactosidase reveals new derivatives of Pax3-expressing precursors. Genesis. 2008;46:200–204. doi: 10.1002/dvg.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, Ehses S, Krause F, Chan DC, Alexander C, Bauer C, Youle R, Langer T, Martinou JC. SLP-2 is required for stress-induced mitochondrial hyperfusion. The EMBO journal. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.