Abstract
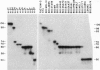
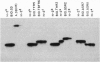
A hallmark of major histocompatibility complex (MHC) genes is their extraordinarily high level of polymorphism. Polymorphic residues on MHC molecules determine which peptide ligands they bind and present to effector T lymphocytes. Although the genetic mechanisms responsible for MHC polymorphism have been delineated, the timetable and the pathway of their diversification remain unclear. To trace MHC evolution, we have characterized a highly polymorphic microsatellite containing tandem repeats (TRs) of two tetranucleotide units, TGGA and GGCA, located at the 3' end of the second intron in the class II Eb gene of mouse. On the basis of length as well as sequence variations, 11 TR alleles were defined in 55 inbred mouse strains, which included MHC recombinant haplotypes and haplotypes derived from different subspecies of mouse. In this extensive sampling, a striking concordance was observed between the serologically identified class II proteins and the associated TR alleles. Examination of several strains carrying the same MHC haplotypes as well as strains carrying recombinant MHC haplotypes indicates that TR alleles are extremely stable. These observations suggest that TR polymorphism predates the separation of various subspecies of mouse. On the basis of sequence divergence, a genealogical tree has been constructed to depict evolution of the different TR alleles. Finally, evidence is presented that suggests this microsatellite polymorphism is generated by slipped-strand mispairing during DNA replication.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arden B., Klein J. Biochemical comparison of major histocompatibility complex molecules from different subspecies of Mus musculus: evidence for trans-specific evolution of alleles. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2342–2346. doi: 10.1073/pnas.79.7.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich A. B., Vu T. H., Jones P. P. Characterization of the molecular defects in the mouse E beta f and E beta q genes. Implications for the origin of MHC polymorphism. J Immunol. 1990 Mar 1;144(5):1957–1964. [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Boylan K. B., Ayres T. M., Popko B., Takahashi N., Hood L. E., Prusiner S. B. Repetitive DNA (TGGA)n 5' to the human myelin basic protein gene: a new form of oligonucleotide repetitive sequence showing length polymorphism. Genomics. 1990 Jan;6(1):16–22. doi: 10.1016/0888-7543(90)90443-x. [DOI] [PubMed] [Google Scholar]
- Buus S., Sette A., Grey H. M. The interaction between protein-derived immunogenic peptides and Ia. Immunol Rev. 1987 Aug;98:115–141. doi: 10.1111/j.1600-065x.1987.tb00522.x. [DOI] [PubMed] [Google Scholar]
- Coggins L. W., O'Prey M. DNA tertiary structures formed in vitro by misaligned hybridization of multiple tandem repeat sequences. Nucleic Acids Res. 1989 Sep 25;17(18):7417–7426. doi: 10.1093/nar/17.18.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein S. L., Arn J. S., Sachs D. H. Seven new MHC recombinant strains defining new H-2 haplotypes. Immunogenetics. 1990;32(2):142–145. doi: 10.1007/BF00210453. [DOI] [PubMed] [Google Scholar]
- Figueroa F., Günther E., Klein J. MHC polymorphism pre-dating speciation. Nature. 1988 Sep 15;335(6187):265–267. doi: 10.1038/335265a0. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Folsom V., Tonegawa S. Cell type-specific enhancer element associated with a mouse MHC gene, E beta. Nature. 1984 Aug 16;310(5978):594–597. doi: 10.1038/310594a0. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptfeld-Dolejsek V., Shreffler D. C. Antigenic properties of 36 new H-2 congenic strains and 4 independently derived strains, W10LT, DDD, BZH, and FM. Immunogenetics. 1989;30(2):132–136. doi: 10.1007/BF02421544. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Neumann R., Wilson V. Repeat unit sequence variation in minisatellites: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell. 1990 Feb 9;60(3):473–485. doi: 10.1016/0092-8674(90)90598-9. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Klein J., Figueroa F., David C. S. H-2 haplotypes, genes and antigens: second listing. II. The H-2 complex. Immunogenetics. 1983;17(6):553–596. doi: 10.1007/BF00366126. [DOI] [PubMed] [Google Scholar]
- Klein J., Figueroa F. Polymorphism of the mouse H-2 loci. Immunol Rev. 1981;60:23–57. doi: 10.1111/j.1600-065x.1981.tb00361.x. [DOI] [PubMed] [Google Scholar]
- Kobori J. A., Strauss E., Minard K., Hood L. Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex. Science. 1986 Oct 10;234(4773):173–179. doi: 10.1126/science.3018929. [DOI] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- Marshall J. T. Systematics of the genus Mus. Curr Top Microbiol Immunol. 1986;127:12–18. doi: 10.1007/978-3-642-71304-0_2. [DOI] [PubMed] [Google Scholar]
- McIntyre K. R., Seidman J. G. Nucleotide sequence of mutant I-A beta bm12 gene is evidence for genetic exchange between mouse immune response genes. Nature. 1984 Apr 5;308(5959):551–553. doi: 10.1038/308551a0. [DOI] [PubMed] [Google Scholar]
- Morral N., Nunes V., Casals T., Estivill X. CA/GT microsatellite alleles within the cystic fibrosis transmembrane conductance regulator (CFTR) gene are not generated by unequal crossingover. Genomics. 1991 Jul;10(3):692–698. doi: 10.1016/0888-7543(91)90454-m. [DOI] [PubMed] [Google Scholar]
- Nathenson S. G., Geliebter J., Pfaffenbach G. M., Zeff R. A. Murine major histocompatibility complex class-I mutants: molecular analysis and structure-function implications. Annu Rev Immunol. 1986;4:471–502. doi: 10.1146/annurev.iy.04.040186.002351. [DOI] [PubMed] [Google Scholar]
- Padgett K. A., Shreffler D. C., Saha B. K. Molecular mapping of murine I region recombinants. III. Crossing over at two discrete sites within the beta 1-beta 2 intron of the E beta gene. J Immunol. 1991 Oct 15;147(8):2764–2770. [PubMed] [Google Scholar]
- Pardue M. L., Lowenhaupt K., Rich A., Nordheim A. (dC-dA)n.(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J. 1987 Jun;6(6):1781–1789. doi: 10.1002/j.1460-2075.1987.tb02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess O., Kammerbauer C., Roewer L., Steimle V., Andreas A., Albert E., Nagai T., Epplen J. T. Hypervariability of intronic simple (gt)n(ga)m repeats in HLA-DRB genes. Immunogenetics. 1990;32(2):110–116. doi: 10.1007/BF00210448. [DOI] [PubMed] [Google Scholar]
- Sagai T., Sakaizumi M., Miyashita N., Bonhomme F., Petras M. L., Nielsen J. T., Shiroishi T., Moriwaki K. New evidence for trans-species evolution of the H-2 class I polymorphism. Immunogenetics. 1989;30(2):89–98. doi: 10.1007/BF02421536. [DOI] [PubMed] [Google Scholar]
- Stallings R. L., Ford A. F., Nelson D., Torney D. C., Hildebrand C. E., Moyzis R. K. Evolution and distribution of (GT)n repetitive sequences in mammalian genomes. Genomics. 1991 Jul;10(3):807–815. doi: 10.1016/0888-7543(91)90467-s. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Minard K., Horvath S., McNicholas J., Srelinger J., Wake C., Long E., Mach B., Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982 Nov 4;300(5887):35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989 Aug 25;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco D., Arnheim N. The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol. 1986 Nov;6(11):3934–3947. doi: 10.1128/mcb.6.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990 Jan 12;60(1):95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- Wakeland E. K., Boehme S., She J. X., Lu C. C., McIndoe R. A., Cheng I., Ye Y., Potts W. K. Ancestral polymorphisms of MHC class II genes: divergent allele advantage. Immunol Res. 1990;9(2):115–122. doi: 10.1007/BF02918202. [DOI] [PubMed] [Google Scholar]
- Wakeland E. K., Darby B. R. Recombination and mutation of class II histocompatibility genes in wild mice. J Immunol. 1983 Dec;131(6):3052–3057. [PubMed] [Google Scholar]
- Wakeland E. K., Price-LaFace M., Henson V., Peck A. B. Production of 35 H-2 homozygous strains from wild mice. Immunogenetics. 1987;26(1-2):115–119. doi: 10.1007/BF00345465. [DOI] [PubMed] [Google Scholar]
- Weber J. L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990 Aug;7(4):524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]
- Weller P., Jeffreys A. J., Wilson V., Blanchetot A. Organization of the human myoglobin gene. EMBO J. 1984 Feb;3(2):439–446. doi: 10.1002/j.1460-2075.1984.tb01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff R. K., Plaetke R., Jeffreys A. J., White R. Unequal crossingover between homologous chromosomes is not the major mechanism involved in the generation of new alleles at VNTR loci. Genomics. 1989 Aug;5(2):382–384. doi: 10.1016/0888-7543(89)90076-1. [DOI] [PubMed] [Google Scholar]
- Zimmerer E. J., Passmore H. C. Structural and genetic properties of the Eb recombinational hotspot in the mouse. Immunogenetics. 1991;33(2):132–140. doi: 10.1007/BF00210827. [DOI] [PubMed] [Google Scholar]