Abstract
The ETS factor FLI1 is a key modulator of lupus disease expression. Over-expressing FLI1 in healthy mice, results in the development of an autoimmune kidney disease similar to that observed in lupus. Lowering the global levels of FLI1 in two lupus strains (Fli1+/−) significantly improved kidney disease and prolonged survival. T cells from MRL/lpr Fli1+/− lupus mice have reduced activation and IL-4 production, Neuraminidase1 (Neu1) expression, and the levels of the glycosphingolipid (GSL) lactosylceramide (LacCer). Here we demonstrate that MRL/lpr Fli1+/− mice have significantly decreased renal Neu1 and LacCer levels. This corresponds with a significant decrease in the number of total CD3+ cells, as well as CD4+ and CD44+CD62L− T cell subsets in the kidney of MRL/lpr Fli1+/− mice compared to the Fli1+/+ nephritic mice. We further demonstrate that the percentage of CXCR3+ T cells and Cxcr3 message levels in T cells are significantly decreased and corresponds with a decrease in renal CXCR3+ cells and in Cxcl9 and Cxcl10 expression in the MRL/lpr Fli1+/− compared to the Fli1+/+ nephritic mice. Our results suggest that reducing the levels of FLI1 in MRL/lpr mice may be protective against development of nephritis in part through down-regulation of CXCR3, reducing renal T cell infiltration and GSL levels.
Introduction
Systemic lupus erythematosus (SLE or lupus), an autoimmune disease that can affect most organ systems, is characterized by autoantibody production and deposition of immune complexes in target organs and inflammation. Abnormalities in B cells and T cells contribute to loss of self-tolerance and production of autoantibodies. T cells are an important driver of disease and exhibit altered signaling, activation, gene expression and migration to affected organs and removal of T cells in lupus mouse strains decreased autoantibody production and nephritis and increased survival (1–4).
CD1 outbred mice globally over-expressing FLI1 (Friend leukemia virus integration 1) develop a lupus like disease, including autoreactive T cell expansion (5). Reduction of FLI1 levels either globally or only in hematopoietic cells in MRL/lpr and/or NZM2410 lupus strains significantly improved disease and survival (6, 7), suggesting specific effects of FLI1 in immune cells. Global reduction of FLI1 in MRL/lpr and/or NZM2410 lupus strains decreases renal and serum MCP-1 and IL-6 levels and renal inflammatory cell infiltration (6, 8–10). NZM2410 mice with reduced FLI1 levels (Fli1+/−) were shown to have decreased numbers of renal infiltrating inflammatory cells (11). Recent studies demonstrated that FLI1 regulates the expression of MCP-1, RANTES and IL-6 (9, 12, 13). Together, these studies suggest that globally reducing FLI1 levels in lupus mouse strains improves disease by decreasing renal chemokine production and thus, inflammatory infiltration.
We previously demonstrated that MRL/lpr T cells with reduced FLI1 levels exhibit significantly decrease activation (Ca2+ flux), IL-4 production and autoantibody production regardless of the FLI1 levels in the B cells (14). We further showed that the likely mechanism by which FLI1 exerts these effects in T cells is by regulating neuraminidase 1 (Neu1) expression and the levels of the glycosphingolipid (GSL) lactosylceramide (LacCer). These results indicate that FLI1 may have specific effects on T cell function that is independent of T cell extrinsic effects of FLI1 in MRL/lpr lupus mice. We also demonstrated that lupus mice and humans with nephritis have significantly elevated renal GSL metabolism characterized by elevated LacCer and Neu1 levels compared to their lupus counterparts without nephritis and unaffected controls (15).
Here we provide further understanding of the role FLI1 plays in disease expression with respect T cell function and effects on the kidney of MRL/lpr mice. Specifically, we demonstrate effects of FLI1 levels on CXCR3 expression in T cells and their infiltration of the kidney and on renal GSL metabolism. Together, our results suggest that intrinsic FLI1 levels in T cells play an important role in their migration to the kidney in part by regulating the expression of CXCR3 and renal chemokine expression. The decreased renal inflammatory infiltration and chemokine expression in by reducing FLI1 likely reduces GSL metabolism. A more solid understanding of the role and mechanisms by which FLI1 mediates T cell function and renal disease in lupus will be invaluable in identifying pathways and molecules that may serve as potential targets for therapeutic intervention, as well as provide better insight of disease progression in general.
Materials and Methods
Mice
All animal experiments and methods of euthanasia were approved by the Ralph H. Johnson VAMC Institutional Animal Care and Use Committee (IACUC). Mice were housed and maintained under pathogen-free conditions at the Ralph H. Johnson VAMC Animal Care Facility (Charleston, SC). MRL/lpr Fli1+/+ and Fli1+/− mice (6) and C57BL/6 Fli1+/+ and Fli1+/− mice (16) were obtained from matings between Fli1+/+ and Fli1+/− mice within each strain in our colony. Wild-type MRL/lpr and C57BL/6 Fli1+/+ mice obtained from Jackson Laboratories (Bar Harbor, ME) were used for matings every other generation to avoid genetic drift in the colonies. Age-matched animals of both genders were used in experiments.
Lactosylceramide quantification and neuraminidase activity assays
Lactosylceramide (LacCer) was measured quantitatively by the Lipidomics Core at MUSC using 2 mg of kidney homogenates as described previously (15). The AmplexRed NEU Assay kit (Invitrogen, Grand Island, NY) was used to measure NEU enzyme activity in 50 μg of kidney homogenate following the manufacturer’s instructions (15) and is presented in arbitrary units.
MALDI-FTICR imaging of kidney sections
Direct profiling of LacCer expression in kidney tissue sections was performed using matrix assisted laser desorption/ionization Fourier Transform Ioncyclotron Resonance imaging mass spectrometry (MALDI-FTICR) as we described previously (15).
Semi-quantitative RTPCR assays
RNA was prepared from isolated T cells or kidney using the RNeasy kit (Qiagen, Hilden, Germany) following manufacturer’s directions and cDNA generated using 0.5–1 μg RNA using the iScript cDNA Synthesis kit (BioRad, Hercules, CA). Real-time PCR was performed with the cDNA using the Lightcycler 480 SYBR Green I Master kit and Lightcycler 480 II (Roche, Indianapolis, IN). Primers used for real-time PCR include: Neu1 forward 5′-ACGATGTAGACACAGGGATAGTG-3′ and reverse 5′-GTCGTCCTTACTCCAAACCAAC-3′; Cxcr3 forward 5′-GGTTAGTGAACGTCAAGTGCT-3′ and reverse 5′-CCCCATAATCGTAGGGAGAGGT-3′; Cxcl9 forward 5′-GCCATGAAGTCCGCTGTTCT-3′ and reverse 5′-GGGTTCCTCGAACTCCACACT-3′; Cxcl10 forward 5′-GACGGTCCGCTGCAACTG-3′ and reverse 5′-GCTTCCCTATGGCCCTCATT-3′; βactin forward 5-AGATTACTGCTCTGGCTCCTAG-3′ and reverse 5′-CCTGCTTGCTGATCCACATC-3′. Relative message levels of Neu1, Cxcr3, Cxcl9 and Cxcl10 were calculated using the ΔΔCT method. Relative expression after normalizing to βactin or GAPDH was similar and the βactin-normalized values are presented. The ΔΔCT from one MRL/lpr Fli1+/+ mouse was set to one and all other mice compared to that mouse (“n”= the number of animals analyzed).
Immunofluorescence and Immunohistochemistry
Frozen and OTC embedded kidneys were sectioned at 5 microns. Sections were thawed at room temperature and fixed with acetone at −20°C for 10 min. Sections were fixed with 10% phosphate buffered formalin, washed with PBS + 0.05% Tween-20 (PBST) and blocked with 5% BSA in PBST. Sections were washed with PBST/1% BSA and incubated with a rabbit anti-LacCer (Biorbyt, San Francisco, CA) in PBST/5% BSA or rabbit anti-CXCR3 (Novus Biologicals, Littleton, CO). Anti-LacCer was detected with anti-rabbit FITC (Southern Biotechnology, Birmingham, AL) and nuclei detected by addition of DAPI. Anti-CXCR3 was detected with anti-rabbit HRP and nuclei detected by addition of Gill’s hematoxylin. Images of sections were taken using a Nikon Eclipse 80i microscope, DS camera and software (Nikon Instruments, Melville, NY) at the magnifications indicated.
T cell isolation
T cells were isolated using Pan T cell Isolation Kit (Miltenyi, Cologne, Germany) as described previously (14). This method removes the CD3+CD4−CD8−B220+ (double negative) T cell population that accumulates in the MRL/lpr strain. T cells were analyzed after isolation by flow cytometry and were 90–95% pure. Approximately 1–4% of the isolated T cells in the MRL/lpr strain were CD4−CD8−.
Flow cytometry
Kidneys were harvested from euthanized MRL/lpr (MRL Fli1+/+) or MRL/lpr Fli1+/− (MRL Fli1+/−) mice, rinsed in PBS, minced and digested with collagenase, type I (Calbiochem, La Jolla, CA). Digested kidneys were passed over a 40 μm cell strainer washed in cold PBS and resuspended in PBS, 1% FBS, 0.1% NaAzide (flow buffer). Primary T cells were isolated from spleen, as described above, and resuspended in flow buffer. Cells were blocked with Fc receptor block, labeled with fluorophore-conjugated antibodies and analyzed by flow cytometry. Fluorophore-conjugated antibodies included anti-mouse CD3, CD4, CD8, CD62L, and CD44 antibodies (BD Biosciences, San Jose, CA). For the kidney analyses, cells were gated on CD3 and then analyzed for CD4 and CD8 staining or CD44 and CD62L staining. Cell counts represent the total number of cells expressing a marker as calculated by multiplying the percentage of cells expressing a specific marker(s) by the number of total cells in the sample. Flow was initially performed on cells from perfused and non-perfused kidneys. There were no significant differences in the percentages of the T cell subsets obtained between perfused and non-perfused kidneys for CD3+ T cells; therefore, perfusion was not performed on subsequent analyses. Data from the perfused and non-perfused kidneys were averaged and are presented.
Western immunoblot
Whole cell extracts were prepared by incubating cells with RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% NaDeoxycholate, 0.1% SDS, 25 mM DTT, 10 mM EDTA, 1 mM PMSF) plus 1 μl each of protease and phosphatase inhibitor mixes (Sigma, St Louis, MO) per 100 μl buffer. After removing cell debris by centrifugation, protein concentration was determined using a micro BCA assay (Pierce/ThermoScientific, Rockford, IL). Extracts (20–50 μg per well) were separated on a Criterion TGX gel (Biorad, Hercules, CA), transferred to PVDF membrane and probed with rabbit anti-FLI1 (Santa Cruz Biotechnology, Dallas, TX) or rabbit anti-βactin (Cell Signaling Technology, Danvers, MA) antibodies. Anti-FLI1 was detected with an anti-rabbit biotin antibody (Pierce/ThermoScientific) followed by an AlexaFluor-647 streptavidin-conjugated antibody (Life Technologies, Grand Island, NY). Blots were stripped and reprobed with anti-βactin and detected with a goat anti-rabbit AlexaFluor-647 antibody (Life Technologies). Blots were scanned on an Odyssey Infrared Imaging system and software (LI-COR, Lincoln, NE).
Transfections
The murine MS1 endothelial cell and human Jurkat T cell lines were obtained from American Type Culture Collection (Manassas, VA) and maintained according to their recommendations. The mouse Neu1 promoter/reporter (P/R) construct containing the −435 to −11 region of the promoter (pGL3 mNeu1 −435) was described previously (14). The pSG hCXCR3 P/R construct (Switchgear Genomics/Active Motif, Carlsbad, CA) contains the human CXCR3 768 bp proximal promoter region driving Renilla expression. MS1 cells were seeded at 2×105 cells per well in a six-well plate in DMEM supplemented with 5% FBS the day before transfection. The pGL3 mNeu1 −435 or control (pGL3 Basic) constructs (1 μg) were transfected into the MS1 cells with increasing amounts of the pcDNA3 flag-tagged FLI1 (Flag FLI1) expression vector (17) using Fugene (Promega, Madison, WI). Jurkat cell transfections were performed as previously described (18) using 0.5 μg pSG hCXCR3 and increasing amounts of FLI1 expression vectors pSG5 FLI1 or pSG5 FLI1DBM (FLI1 DNA binding mutant) (12) or pcDNA3 Flag FLI1, as indicated in the figures. Cells were stimulated with 10 ng/ml PMA and 100 ng/ml ionomycin (PMA/ion) 4 hours after transfection and cells were harvested 24 hours after transfection. Total molar amount of DNA was kept constant from well to well by addition of empty pSG5 or pcDNA3. Transfection efficiency was normalized by addition of the Renilla expression vector pCMV-TK RL (Promega, Madison, WI) for the pGL3 mNeu1 P/R transfections or luciferase expression vector pGL3 Control for the pSG hCXCR3 P/R transfections. Transfections were performed three times with similar results. Representative results are presented.
Electrophoretic Mobility Shift Assay
EMSAs were performed essentially as described in (17). Nuclear extracts (NE) were prepared from Jurkat T cells stimulated with 10 ng/ml of PMA and 100 ng/ml of Ionomycin (Sigma, St Louis, MO) for 24 hours. Probes, annealed Cy5.5-labeled oligos containing putative conserved ETS binding sites of the hCXCR3 promoter (shown in Fig. 6E), were incubated with 10 μg of NE in binding buffer (10 mM HEPES, 10 mM Tris-HCl, 100 mM NaCl, 15 mM KCl, 0.5 mM EDTA, 1 mM DTT, 10% glycerol, 1 μg poly dI:dC). Specific binding was identified by pre-incubating the NE with 100X molar excess of unlabeled specific or non-specific (putative ETS site scrambled) annealed oligos or with 1 μg rabbit anti-FLI1, rabbit anti-ETS1 antibody or normal rabbit IgG antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Binding complexes were separated on a 4% polyacrylamide gel in 0.25X TBE running buffer (TBE: 0.1 M Tris (pH 7.2), 0.09 M boric acid, 0.001 MEDTA) and gels imaged on the Odyssey Infrared Imaging system (Li-Cor). Oligos were synthesized by Integrated DNA Technologies (Iowa City, IA). All EMSAs were performed at least twice with two independent preparations of NE. Representative gels are shown.
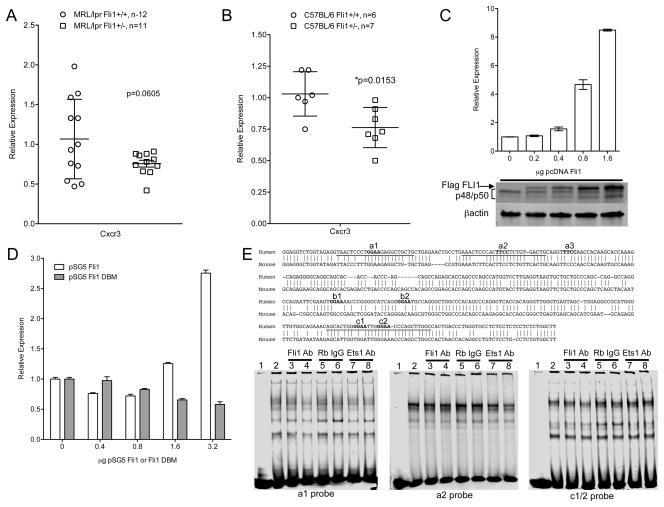
Fig. 6.
FLI1 regulates Cxcr3 expression in T cells. Cxcr3 message levels were measured by real-time PCR in reverse-transcribed RNA from negatively isolated splenic T cells of 16–18 week-old MRL/lpr (A) and C57BL/6 (B) Fli1+/+ and Fli1+/− mice. C) The Jurkat T cell line was transfected with a CXCR3 promoter/reporter vector containing the human CXCR3 (hCXCR3) promoter driving luciferase expression and increasing amounts of the FLI1 expression vector pcDNA Flag FLI1 (Flag FLI1). Cells were stimulated for 24 hours and whole cell extracts were analyzed for luciferase expression and subjected to western immunoblotting with antibodies to FLI1 and βactin (below graph). D) The Jurkat T cell line was transfected as indicated in (C) with the hCXCR3 promoter/reporter vector, but with increasing amounts of the FLI1 expression vector pSG5 FLI1 or the FLI1 DNA binding mutant (DBM) expression vector pSG5 FLI1 DBM. Transfections were performed at least three times with similar results. Representative experiments are presented. E) In vitro DNA binding assays were performed with the indicated hCXCR3 probes containing putative conserved ETS binding sites and nuclear extract from stimulated Jurkat T cells without or with anti-FLI1 or anti-ETS1 antibodies or normal IgG. Aligned sequences of the human and mouse CXCR3 promoters with locations of probes and putative ETS sites are shown above the EMSA blots.
Adoptive Transfer of MRL/lpr T cells
T cells were isolated as described above from spleens of three 7–8 week-old MRL/lpr Fli1+/+ and Fli1+/− mice, combined and resuspended in sterile 1X PBS. A total of 8×106 cells were transferred by tail vein injection into 7–8 week-old MRL/lpr Fli1+/+ and Fli1+/− mice. Three transfers were performed: Group 1, four Fli1+/+ mice received Fli1+/+ T cells; Group 2, four Fli1+/− mice received Fli1+/− T cells; and Group 3, eight Fli1+/− mice received Fli1+/+ T cells. Urine and serum were collected prior to transfer and 2, 4, 6, 8 and 12 weeks after transfer. Urine was collected over a period of 24 hours using metabolic cages. Recipient mice were euthanized 12 weeks after transfer (19–20 weeks of age) and kidneys were collected.
Statistics
Statistical analyses were performed using PRISM software (GraphPad Software Inc., LaJolla, CA) at a 95% confidence level. Real-time PCR differences were calculated using the unpaired t-test, two tailed with Welch’s correction adjusting for multiple comparisons as necessary. All other analyses were performed using two-way ANOVA adjusting for multiple comparisons (Sidak’s). Adjusted p values are presented and all error bars represent standard deviation.
Results
Elevated renal LacCer levels in glomeruli of MRL/lpr Fli1+/+ mice are decreased in MRL/lpr Fli1+/− mice
MRL/lpr Fli1+/− mice have FLI1 levels that are 50% of those in MRL/lpr Fli1+/+ mice and have significantly improved disease (6). We recently demonstrated that renal and urine lactosylceramide (LacCer) levels, Neu1 levels and/or NEU activity are elevated in MRL/lpr (Fli1+/+) mice (16–18 weeks of age) and human lupus patients with nephritis compared to non-nephritic lupus mice and patients, respectively, or compared to healthy controls (15). To determine if renal LacCer levels in MRL/lpr Fli1+/− mice, which have improved renal pathology and function, are reduced compared to Fli1+/+ mice, we measured LacCer levels in kidney homogenates from 16–18 week-old MRL/lpr Fli1+/+ and Fli1+/− mice. The Fli1+/− mice tended to have decreased levels of each of the major LacCer species expressed in the kidney (C16, C22, C24 and C24:1) and a significant reduction in total LacCer levels compared to the Fli1+/+ mice (Fig. 1A). LacCer levels across the tissue were assessed on renal sections by MALDI-FTICR imaging. As we demonstrated previously (15), LacCer species C16, C24 and C24:1 were observed to be higher across the tissue in the MRL/lpr Fli1+/+ kidneys compared to healthy C57BL/6 (B6) kidneys, while the LacCer levels in the Fli1+/− kidneys appear to have intermediate levels; higher than the B6 and lower than the Fli1+/+ (Fig. 1B and data not shown).
Fig. 1.
The level of Lactosylceramide is significantly reduced in MRL/lpr Fli1+/− kidneys. A) Individual species and total levels of the major LacCer species in the kidney were measured in renal cortices from 16–18 week-old MRL/lpr Fli1+/+ and Fli1+/− mice. B) MALDI-FITCR images of C16-LacCer expression in renal sections from 16–18 week-old C57BL/6 and MRL/lpr Fli1+/+ and Fli1+/− mice. Sections from three mice of each strain were analyzed with representative images presented. C) Immunofluorescence for LacCer in renal sections from 16–18 week-old MRL/lpr Fli1+/+ mice. Sections from three mice were analyzed with representative images presented.
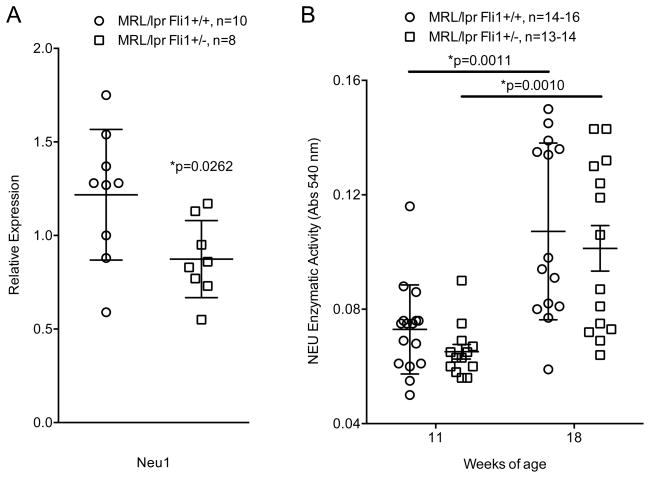
In addition to changes in LacCer, the kidneys of MRL/lpr Fli1+/− mice had significantly decreased Neu1 expression compared to Fli1+/+ mice at 16–18 weeks of age (Fig. 2A). However, a significant decrease in Neu1 expression did not result in a significant decrease in NEU activity at either 11 weeks (pre/early disease) or 18 weeks (diseased) of age (Fig. 2B), which may be due to activity of the other NEUs (NEU2, 3 and 4) that may be unaffected by FLI1. Since FLI1 can regulate Neu1 transcription in T cells (14), it may similarly regulate Neu1 in the kidney. Endothelial cells are the only renal cell type shown to express FLI1 in the kidney (8, 19). Therefore, we transfected the mouse endothelial cell line, MS1, with a Neu1 promoter/reporter construct and increasing amounts of a FLI1 expression vector. Unlike in T cells, FLI1 had no effect on Neu1 promoter activity in MS1 endothelial cells (data not shown). Together, these results indicate that globally reducing FLI1 levels in MRL/lpr mice decreases renal Neu1 and LacCer levels, but likely is not due to FLI1 regulation of Neu1 expression in the kidney. Intense LacCer staining is observed in the mesangial region of human lupus renal biopsies (12) and we here we demonstrate specific LacCer staining in the glomeruli of MRL/lpr mice (Fig. 1C). T cell infiltration was shown to occur predominantly in the glomerular region in MRL/lpr kidneys (20). Since MRL/lpr Fli1+/− mice exhibit reduced immune cell infiltration of the kidney including T cells (10), reduced T cell infiltration may be one mechanism by which reducing FLI1 levels impacts GSL metabolism in the kidney.
Fig. 2.
Neu1 expression is significantly reduced in MRL/lpr Fli1+/− kidneys. A) Neu1 expression measured by real-time PCR in reverse-transcribed RNA isolated from renal cortices of 18 week-old MRL/lpr Fli1+/+ and Fli1+/− mice. B) NEU activity measured in extracts from renal cortices of 11 and 18 week-old MRL/lpr Fli1+/+ and Fli1+/− mice.
CD4+ and activated T cells are significantly reduced in MRL/lpr Fli1+/− kidneys
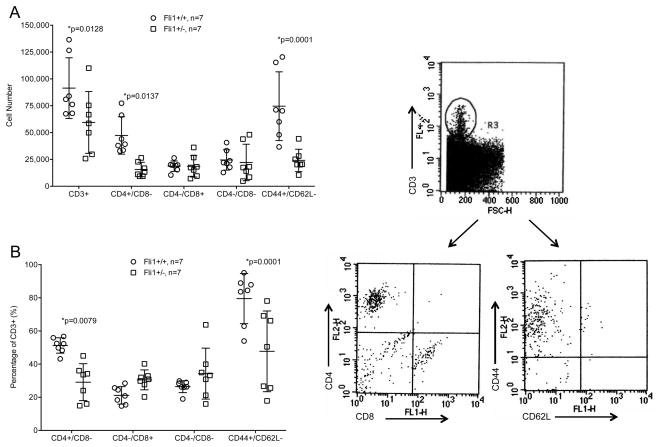
To further examine the effects of FLI1 on T cell infiltration in the kidneys, we quantified T cell subsets in the kidneys of MRL/lpr Fli1+/+ and Fli1+/− mice by flow cytometry. Antibodies to CD3, CD4, CD8, CD44 and CD62L were used to stain single cell suspensions of kidneys from 14–16 week-old MRL/lpr mice. The overall numbers of T cells (all CD3+ cells) were significantly decreased, as were the numbers and percentage of CD4+ T cells (Fig. 3A and 3B). The percentage of CD3+ cells that were CD8+ or CD4−/CD8− double negative (DN) T cells were slightly increased in the Fli1+/− mice compared to the Fli1+/+ mice, but there were no differences in their overall numbers. The decrease in the percentage of CD4+ T cells and increase in percentage of CD8+ T cells in the Fli1+/− kidneys is likely due to the decrease in the numbers of CD4+ T cells and not due to an increase in the number of CD8+ T cells. Moreover, both the number and percentage of activated CD3+ cells (CD44+/CD62L−; Fig. 3A and B) in the kidneys were significantly decreased in the Fli1+/− compared to the Fli1+/+ mice. These results indicate that reducing FLI1 levels in MRL/lpr mice specifically decreases the renal infiltration of CD4+ T cells, as well as decreases the number of activated T cells in the kidney.
Fig. 3.
Renal infiltrating T cells are significantly decreased in MRL/lpr Fli1+/− mice. CD3+ T cells and indicated subsets gated on CD3+ cells, from kidneys of 14–16 week-old MRL/lpr Fli1+/+ and Fli1+/− mice were quantified by flow cytometry. Cell numbers (A) and percentage of CD3+ gated cells (B) are presented. Example of the gating strategy is shown to the right of the graphs.
CXCR3 expression in T cells and CXCR3 ligands in the kidney are significantly reduced in MRL/lpr Fli1+/−
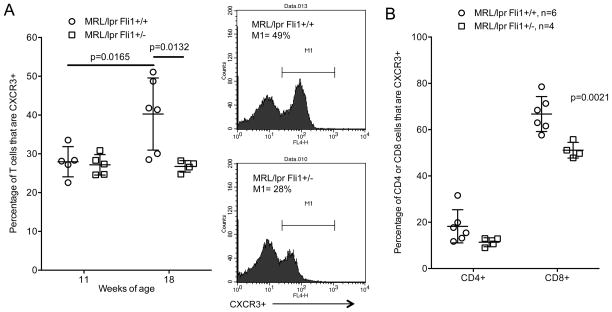
We next analyzed if FLI1 regulates expression of chemokine receptors as a possible mechanism by which FLI1 impacts T cell migration to the kidney. Flow analysis was performed on spleen cells isolated from 18 week-old MRL/lpr mice to examine expression of various chemokine receptors. Of the chemokine receptors analyzed (CCR4, CCR6, CCR7, CXCR3, CXCR4, CXCR5), CXCR3 was the most abundant on both CD4+ and CD8+ populations (data not shown). We then performed flow cytometry on T cells negatively isolated from spleens of 11 and 18 week-old Fli1+/+ and Fli1+/− mice. Between 11 and 18 weeks of age the percentage of CXCR3+ T cells significantly increased (p=0.0165) in the Fli1+/+ mice while no increase was observed in the Fli1+/− mice (Fig. 4A). The percentage of CXCR3+ T cells in the Fli1+/+ mice was significantly higher compared to Fli1+/− mice at 18 weeks of age (p=0.0132). Differences in the percentage of CXCR3+ T cells between Fli1+/+ and Fli1+/− mice at 18 weeks of age were observed in both the CD4+ and CD8+ subsets with the differences in CD8+ cells being significant (Fig. 4B).
Fig. 4.
The percentages of CXCR3+ T cells are significantly decreased in 18 week-old MRL/lpr Fli1+/− mice. A) Percentages of CXCR3+ cells were quantified by flow cytometry in T cells negatively isolated from spleens of 11 and 18 week-old MRL/lpr Fli1+/+ and Fli1+/− mice (n=4–6 for each group. Example of flow data is shown to the right of the graph. B) Percentages of CD4+ and CD8+ T cells that are CXCR3+ in the 18 week-old samples analyzed in (A).
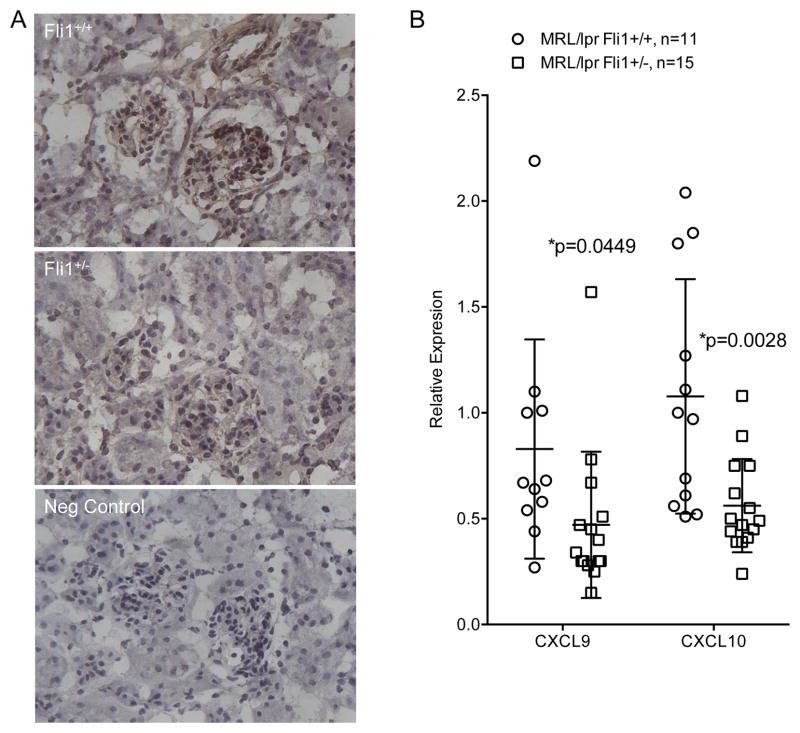
We then analyzed renal sections and observed reduced CXCR3 staining in the MRL/lpr Fli1+/− compared to Fli1+/+ mice (Fig. 5A). Of the CXCR3 ligands CXCL9 (MIG), CXCL10 (IP-10) and CXCL11 (I-TAC), mRNA levels of Cxcl9 and Cxcl10 are more highly expressed than Cxcl11 in the kidneys of MRL/lpr mice (21, 22). Measures of renal message levels showed a significant decrease of Cxcl9 and Cxcl10 in the Fli1+/− compared to Fli1+/+ mice at 18 weeks of age (Fig. 5B). Together, these results suggest that decreased T cell infiltration of the kidney in MRL/lpr Fli1+/− mice may be due in part to decreased CXCR3 expression in T cells and decreased renal expression of its ligands Cxcl9 and Cxcl10.
Fig. 5.
Renal CXCR3+ cells are reduced and expression of CXCR3 ligands Cxcl9 and Cxcl10 is significantly decreased in MRL/lpr Fli1+/− mice. A) Renal sections from 16–18 week-old MRL/lpr Fli1+/+ and Fli1+/− mice were analyzed for CXCR3+ cells by immunohistochemistry. Negative control (Neg Control) is secondary only. All images were taken at 40x. Sections are representative of three mice analyzed from each strain. B) Cxcl9 and Cxcl10 message levels were measured by real-time PCR in reverse transcribed RNA isolated from renal cortices of 18 week-old MRL/lpr Fli1+/+ and Fli1+/− mice.
FLI1 regulates CXCR3 expression in T cells
Next, we determined if FLI1 regulates CXCR3 in T cells. Cxcr3 message levels in T cells from the 16–18 week-old mice were decreased in MRL/lpr Fli1+/− T cells compared to the Fli1+/+ T cells with the differences approaching significance (Fig. 6A). Importantly, T cells with reduced levels of FLI1 from healthy, non-autoimmune prone (C57BL/6 Fli1+/−) mice had significantly decreased Cxcr3 message levels compared to T cells from Fli1+/+ mice (p=0.0153; Fig. 6B), indicating that FLI1 may regulate Cxcr3 transcription. We then transfected the Jurkat T cell line with a human CXCR3 promoter/reporter (hCXCR3) (P/R) construct and a FLI1 expression vector (pcDNA Flag FLI1). Both isoforms of FLI1 are expressed endogenously in Jurkat cells (p50 and p48) and the transfected Flag FLI1 migrates slower than the endogenous FLI1 due to the presence of the Flag epitope (Fig. 6C, blot). Exogenous FLI1 resulted in an increase in expression of the endogenous protein, which was expected based on prior observations that FLI1 regulates its own expression (5, 17, 18). hCXCR3 promoter activity increased in response to Flag FLI1 in a dose-dependent manner (Fig. 6C). This effect of FLI1 may be the result of direct binding to the hCXCR3 promoter as transfection with an expression vector for a FLI1 DNA binding mutant (pSG5 FLI1 DBM) failed to activate the hCXCR3 promoter compared to wild-type FLI1 (pSG5 FLI1) (Fig. 6D). The pSG5 (SV40 driven) vector does not express as highly as the pcDNA (CMV driven) vector, accounting for the relative fold differences in hCXCR3 promoter activation by wild-type FLI1 in Fig. 6C and Fig. 6D (compare 1.6 μg of pcDNA Flag FLI1 to 1.6 μg of pSG5 FLI1).
To further determine if FLI1 binds directly to the hCXCR3 promoter, electrophoretic mobility shift assays (EMSA) were performed. Putative ETS binding sites, identified by the core binding sequence (GGAA) in the human CXCR3 proximal promoter sequence, were chosen for EMSA analyses based on conservation with the mouse sequence. Three highly conserved putative ETS binding sites were identified (a1, a2 and c2) (Fig. 6E). Site a2 has the highest homology with the canonical FLI1 binding site ACCGGAA(G/A)(T/C) (23). Probes encompassing a1, a2 and c2 (underlined in the sequence in Fig. 6E) were incubated with nuclear extract (NE) prepared from stimulated Jurkat T cells. Specific binding was demonstrated by addition of excess cold-specific sequences compared to cold non-specific (scrambled) sequences (data not shown). Although we identified bands that were a result of specific binding, neither FLI1 nor ETS1 were observed to bind these probes following addition of anti-FLI1, anti-ETS1 or normal rabbit IgG antibodies (Fig. 6E). Over-expressed FLI1 in NE from Flag FLI1-transfected Jurkat cells (as in Fig. 6C) also failed to bind to these probes (data not shown). These results suggest that although FLI1 requires its DNA binding domain to fully activate the hCXR3 promoter, FLI1 may regulate hCXCR3 from a less conserved and/or non-canonical FLI1/ETS binding site or by indirect regulation.
FLI1 levels in transferred T cells impact renal T cell infiltration in recipients
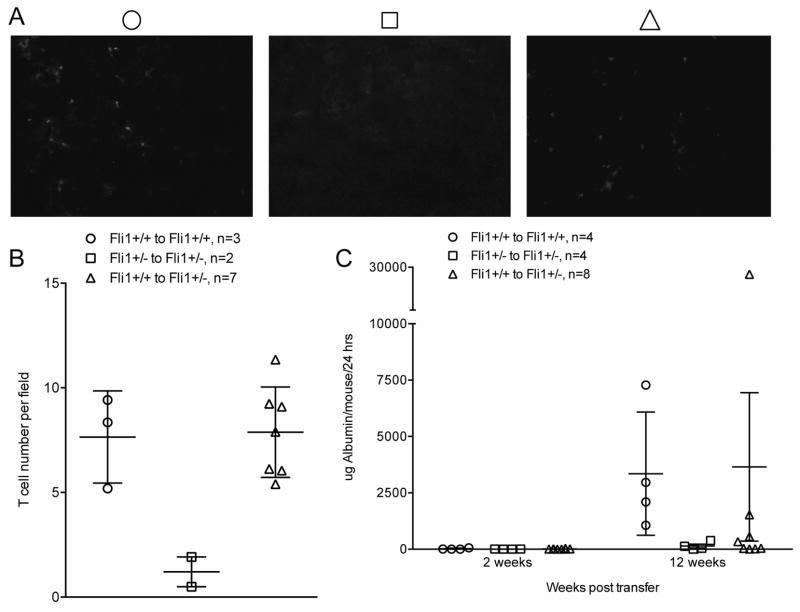
The contribution of FLI1 levels in MRL/lpr T cells to T cell migration to the kidney and progression of nephritis was analyzed in an adoptive transfer experiment. To avoid extrinsic effects of disease on T cell function prior to transfer, CD3+ T cells were negatively isolated from spleens of MRL/lpr Fli1+/+ and Fli1+/− mice at 7–8 weeks of age (non-nephritic) and transferred into 7–8 week-old MRL/lpr Fli1+/+ and Fli1+/− mice. Control groups included Fli1+/+ mice receiving Fli1+/+ T cells and Fli1+/− mice receiving Fli1+/− T cells, with the experimental group being Fli1+/− mice that received Fli1+/+ T cells. Urine was collected prior to transfer and every two weeks after transfer for a total of 12 weeks. Kidneys were examined at the end point. All of the Fli1+/+ mice that received Fli1+/+ T cells had 5 to 10 times more renal CD4+ T cells compared to the Fli1+/− mice that received Fli1+/− T cells (Fig. 7A & B). Interestingly, the number of CD4+ T cells in the kidneys of all the Fli1+/− mice that received Fli1+/+ T cells reflected the T cell numbers in the Fli1+/+ to Fli1+/+ transfer (Fig. 7A & B), suggesting that intrinsic levels of FLI1 in T cells impact T cell migration to the kidney independent of extrinsic levels of FLI1.
Fig. 7.
Adoptive transfer of MRL/lpr Fli1+/+ T cells to MRL/lpr Fli1+/− mice results in increased numbers of renal CD4+ T cells. T cells were negatively isolated from the spleens of MRL/lpr Fli1+/+ and Fli1+/− mice and transferred into 8 week-old MRL/lpr Fli1+/+ or Fli1+/− mice. Renal sections in recipient mice were analyzed 12 weeks after T cell transfer (20 weeks of age) for CD4+ cells by immunofluorescence. Representative images are presented in (A) and average T cell numbers per field of all mice are presented graphically in (B). C) 24-hour urine samples were collected from recipients prior to and every two weeks after T cell transfer and albumin levels as a measure of proteinuria were quantified. Averaged data for recipients within each transfer group at the 2- and 12-week post-transfer time points are presented.
No differences were observed in IgG or C3 deposition between any of the transfer groups (data not shown). This was not unexpected as significant differences in renal IgG and C3 deposition were not observed between MRL/lpr Fli1+/+ and Fli1+/− mice previously (6). We measured proteinuria to determine if the transfer of Fli1+/+ T cells had an effect on renal disease in Fli1+/− mice. Although proteinuria steadily increased over the 12 weeks of the study in the Fli1+/+ mice that received Fli1+/+ T cells and remained nearly undetectable in the Fli1+/− mice that received Fli1+/− T cells as expected, only two of the recipients in the experimental group had an increase similar to those observed in the Fli1+/+ to Fli1+/+ group (Fig. 7C). No significant differences in renal pathology between the Fli1+/− that received Fli1+/+ T cells and the control Fli1+/− group that received Fli1+/− T cells were observed (data not shown). Together these results suggest that intrinsic FLI1 levels are important in promoting T cell migration to the kidney in MRL/lpr mice. However, a onetime transfer of Fli1+/+ T cells likely was not sufficient to exacerbate nephritis in Fli1+/− mice or T cell extrinsic effects of FLI1 also are important in disease progression.
Discussion
MRL/lpr mice that have reduced levels of FLI1 (Fli1+/−) have improved disease and survive significantly longer than Fli1+/+ mice (6). FLI1 expression is highly expressed in the adult spleen, including mature T and B cells. There are no differences in lymphadenopathy or the percentages of CD4+, CD8+ or memory/activated (CD44+CD62L−) T cells in the spleens of nephritic MRL/lpr Fli1+/+ mice compared to MRL/lpr Fli1+/− mice, yet improved renal disease in the Fli1+/− mice is characterized by decreased infiltration of immune cells in the kidney (6). Moreover, MRL/lpr Fli1+/− T cells show reduced migration in vitro compared to wild-type MRL/lpr (Fli1+/+) mice (10). We show here that MRL/lpr Fli1+/− mice have significantly decreased numbers of CD3+ cells, specifically CD4+ and activated/memory (CD44+CD62L−) CD3+ subsets, in the kidney compared to MRL/lpr Fli1+/+ mice. Previously, we demonstrated that Fli1 levels in MRL/lpr T cells do not have an effect on apoptosis (14). Together these results support a role for FLI1 in T cell activation/migration rather than on T cell development, survival and/or differentiation.
FLI1 is expressed in kidney, but at 14-fold lower levels compared to spleen at the message level (24, 25) and is only detected in endothelial cells of the kidney (8, 19). MRL/lpr Fli1+/+ immune cells, including T cells, do not readily migrate to the kidney when transferred into MRL/lpr Fli1+/− mice (10). Chemokines Ccl2 (Mcp-1), Ccl3 (Mip-1α), Ccl4 (Mip-1β) and Ccl5 (Rantes) are significantly decreased at the message level in the kidneys of MRL/lpr Fli1+/− compared to Fli1+/+ mice and FLI1 was demonstrated to regulate the promoter activity of several different cytokine genes in endothelial cells (8, 9, 12, 13). Here we demonstrated that the expression Cxcl9 and Cxcl10, CXCR3 ligands, also are significantly decreased in the kidneys of MRL/lpr Fli1+/− compared to the Fli1+/+ mice. These results suggest that FLI1 regulation of chemokine levels in renal endothelial cells mediate immune cell infiltration to the kidney. However, MRL/lpr Fli1+/− immune cells show decreased migration in vitro towards CCL2 and CCL5 (10), suggesting that intrinsic FLI1 levels also impact immune cell migration. The mechanism(s) responsible for this effect of FLI1 are unknown as the percentages of CD3+ cells expressing receptors for CCL2 and CCL5 were not significantly decreased in the Fli1+/− mice (10).
To identify possible mechanisms by which FLI1 intrinsically mediates T cell migration, we analyzed effects of FLI1 levels on chemokine receptors. We demonstrated that the percentage of CXCR3-expressing CD3+ T cells in the spleen of MRL/lpr mice significantly increases from 29% at 11 weeks of age to approximately 40% at 18 weeks of age. This increase was not observed in MRL/lpr Fli1+/− mice. The significantly reduced numbers of CD3+CD4+ and CD3+CD44+CD62L− T cells in the kidney coincided with significantly reduced numbers of CXCR3+ T cells in the spleen and fewer CXCR3+ cells infiltrating the kidney in the Fli1+/− mice. The percentages of both CD4+CXCR3+ and CD8+CXCR3+ T cells were decreased in the spleens of Fli1+/− compared to the Fli1+/+ mice, but only the percentage of CD8+CXCR3+ T cells were significantly decreased. The greater percentage (and number) of CD4+ than CD8+ cells in the kidney may account for the fewer CD4+CXCR3+ cells (~18%) compared to CD8+CXCR3+ cells (~67%) in the spleen of Fli1+/+ mice if CD4+CXCR3+ cells migrate more readily to the kidney than CD8+CXCR3+ cells. This also may explain the lack of a significant decrease in CD4+CXCR3+ cells in the spleens of Fli1+/− compared to Fli1+/+ mice. Future analyses to measure the number of CD4+ versus CD8+ that are CXCR3+ in the kidney should address this question. Alternatively, additional FLI1-regulated chemokine receptors or pathways may play a role in the migration of CD8+ versus CD4+ T cells to the kidney in MRL/lpr mice.
CXCR3 is important for T cell migration (26–28) and is implicated as an important molecule in the progression of lupus nephritis. The CXCR3 ligand CXCL10 is one of the first chemokines expressed in the kidney of MRL/lpr lupus mice (29) and genetically deleting Cxcr3 reduced kidney T cell infiltration in MRL/lpr mice (22). CXCL10 serum levels are elevated in lupus patients, renal infiltrating CD4+CXCR3+ T cells are abundant, and Cxcr3 mRNA is detected in the urine of lupus patients with nephritis (30, 31). CXCR3 clearly plays an important role in mouse and human lupus and our results indicate that FLI1 plays a role in regulating CXCR3 expression in T cells. CXCR3 ligands are potent stimulators of Ca2+ flux in activated naïve CD4+ and CD8+ T cells (26). We previously demonstrated that MRL/lpr Fli1+/− T cells had significantly decreased activation as measured by Ca2+ flux when stimulated through the TCR (14). As CXCR3 and CD3e are spatially associated in the plasma membrane and exhibit signaling synergy (32, 33), it is possible that FLI1 plays a role in disrupting TCR signaling and Ca2+ flux by down-regulating Cxcr3 transcription in T cells to reduce T cell activation and migration in response to CXCR3 ligands.
Further studies presented here indicate that FLI1 may directly regulate Cxcr3 transcription. Cxcr3 message levels in Fli1+/− T cells were significantly decreased compared to Fli1+/+ T cells and hCXCR3 promoter activity in Jurkat T cells was dose-dependently increased by FLI1 and required its DNA binding domain. However, in vitro binding analyses of the most highly conserved ETS binding sites did not appear to bind FLI1. It is possible that FLI1 regulates Cxcr3 promoter activity through a non-conserved (such as a3, b1 or b2 shown in Fig. 6E sequence) and/or non-canonical ETS binding site that remains to be identified. The putative ETS sites in hCXCR3 tested for FLI1 binding were chosen based on numerous studies, including some from our laboratory, demonstrating that FLI1 binds and acts through conserved canonical ETS sites that are more proximal to the transcription start site (9, 12, 17, 34–41). However, we recently demonstrated that FLI1 also can regulate promoters through more distal sites (12, 13). Alternatively, FLI1 may indirectly regulate Cxcr3 transcription. We believe this is less likely since FLI1 activation of the hCXCR3 promoter required its DNA binding domain. Additional studies to identify the Cxcr3 promoter region(s) and specific sequences that are required for FLI1 activation are ongoing.
Previous adoptive transfer studies demonstrated that migration of MRL/lpr CD3+ cells to the kidney was reduced when either the donor CD3+ cells were Fli1+/− or the recipient animal was Fli1+/− (10). However, this study transferred total spleen cells from 18–24 week-old MRL/lpr (active disease state for Fli1+/+ mice) and examined infiltration 18 hours after transfer. Thus, extrinsic effects of disease on the T cells prior to transfer or effects of the other spleen cell types on the co-transferred T cells could have influenced their migration, and long-term effects were not examined. The adoptive transfer approach in this study was designed to avoid extrinsic effects of overt disease on the T cells prior to transfer by transferring T cells from pre-nephritic 7–8 week-old donors into age-matched recipients and to examine effects over a longer period (12 weeks). This experiment demonstrated that the number of T cells present in the kidney of Fli1+/− mice that received Fli1+/+ T cells was similar to Fli1+/+ mice that received Fli1+/+ T cells, further supporting an intrinsic role of FLI1 on lupus T cell migration to the kidney. Only two of the Fli1+/− recipients of Fli1+/+ T cells had significant increases in proteinuria compared to Fli1+/− recipients of Fli1+/− T cells. It is likely that a significant effect on proteinuria may not result from a one-time transfer. Alternatively, the continued presence of the recipient Fli1+/− T cells and/or the reduced chemokine expression in the kidney may provide a protective effect against the more pathogenic Fli1+/+ T cells.
We recently demonstrated that MRL/lpr mice and human lupus patients with nephritis have significantly altered renal GSL levels compared to lupus counterparts without nephritis and healthy controls, including increased levels of renal and/or urine LacCer and Neu1 (15). Interestingly, we demonstrated in this study that MRL/lpr Fli1+/− mice have significantly decreased total LacCer levels and Neu1 expression levels compared to Fli1+/+ mice. FLI1 had no effect on Neu1 promoter activity in an endothelial cell line, suggesting the decrease in Neu1 expression in the Fli1+/− kidney is not likely due to FLI1 regulation of Neu1 in renal endothelial cells. NEU activity tended to be lower, but was not significantly decreased in the Fli1+/− kidney. There are three other NEUs that are expressed in the kidney (Neu2, 3 and 4) and FLI1 may not have an effect on the expression of the other NEUs. Therefore, the lack of a significant decrease in renal NEU activity, despite a significant decrease in Neu1 message, may be due to continued activity and/or compensation by the other NEUs. Renal LacCer elevation is observed at roughly the same time as major inflammatory infiltration in the MRL/lpr kidney (15). Based on our previous studies demonstrating that Fli1+/− T cells have decreased levels of LacCer and Neu1 and that FLI1 can regulate Neu1 promoter activity in T cells (14), the decrease in renal infiltrating T cells in the Fli1+/− mice may explain, in part, the decrease in renal LacCer and Neu1 levels. Alternatively, or additionally, the decrease in renal GSL metabolism may be due to decreased local LacCer and Neu1 expression in response to decreased immune cell infiltration and cytokine expression. Studies aimed at identifying if LacCer levels are decreased locally in renal cells and/or is due to decreased numbers of infiltrating Neu1/LacCer-expressing T cells in Fli1+/− are currently being pursued.
In summary, previous studies and the data presented here suggest that FLI1 regulation of renal chemokine expression and T cell chemokine receptor expression both play an important role in reducing the number of renal infiltrating T cells and other inflammatory cells to protect against and/or slow disease progression. FLI1 likely mediates disease expression, in part, through direct regulation of CXCR3 to reduce T cell activation and migration and in part through down-regulation of CXCR3 ligands Cxcl9 and Cxcl10 in the kidney. Additionally, reduced renal GSL metabolism in MRL/lpr Fli1+/− mice may be due to reduced numbers of T cells expressing LacCer/Neu1 and/or to reduced local LacCer/Neu1 expression in response to reduced T cell infiltration. Together, these results support a role for FLI1 in mediating both intrinsic (Cxcr3 expression) and extrinsic (renal Cxcl9 and Cxcl10 expression) effects on T cell migration and disease expression (GSL metabolism). Further elucidation of the T cell intrinsic and extrinsic mechanisms will be important in fully understanding how FLI1 mediates disease progression and identifying potential molecules and/or pathways as therapeutic targets.
Acknowledgments
Funding: Support for these studies was provided in part by the Veteran’s Administration Merit Review grant BX000115, NIAMS AR053376, and pilot award from the SC Lipidomics and Pathobiology COBRE P20 RR017677 to TKN; NCI R01CA135087 and Department of Defense W81XWH-10-1-0136 to RRD; and the Lipidomics Shared Resource of the Hollings Cancer Center at the Medical University of South Carolina supported by a Cancer Center Support Grant (P30 CA138313) and the Lipidomics Core in the SC Lipidomics and Pathobiology COBRE (P20 RR017677). The contents of this publication do not represent the views of the Department of Veterans Affairs or the United States Government or any of the other funding entities. The funders had no role in study design, data collection and analyses, decision to publish, or preparation of the manuscript.
The authors thank Dr. John Zhang for assistance with tail vein injections and Zainab Amani for assistance in quantifying cell numbers in stained sections in the adoptive transfer experiment.
References
- 1.Seaman WE, Wofsy D, Greenspan JS, Ledbetter JA. Treatment of autoimmune MRL/Ipr mice with monoclonal antibody to Thy-1.2: a single injection has sustained effects on lymphoproliferation and renal disease. J Immunol. 1983;130:1713–1718. [PubMed] [Google Scholar]
- 2.Santoro TJ, Portanova JP, Kotzin BL. The contribution of L3T4+ T cells to lymphoproliferation and autoantibody production in MRL-lpr/lpr mice. The Journal of experimental medicine. 1988;167:1713–1718. doi: 10.1084/jem.167.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jevnikar AM, Grusby MJ, Glimcher LH. Prevention of nephritis in major histocompatibility complex class II-deficient MRL-lpr mice. The Journal of experimental medicine. 1994;179:1137–1143. doi: 10.1084/jem.179.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng SL, Madaio MP, Hughes DP, Crispe IN, Owen MJ, Wen L, Hayday AC, Craft J. Murine lupus in the absence of alpha beta T cells. J Immunol. 1996;156:4041–4049. [PubMed] [Google Scholar]
- 5.Zhang L, Eddy A, Teng YT, Fritzler M, Kluppel M, Melet F, Bernstein A. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;15:6961–6970. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang XK, Gallant S, Molano I, Moussa OM, Ruiz P, Spyropoulos DD, Watson DK, Gilkeson G. Decreased expression of the Ets family transcription factor Fli-1 markedly prolongs survival and significantly reduces renal disease in MRL/lpr mice. J Immunol. 2004;173:6481–6489. doi: 10.4049/jimmunol.173.10.6481. [DOI] [PubMed] [Google Scholar]
- 7.Mathenia J, Reyes-Cortes E, Williams S, Molano I, Ruiz P, Watson DK, Gilkeson GS, Zhang XK. Impact of Fli-1 transcription factor on autoantibody and lupus nephritis in NZM2410 mice. Clinical and experimental immunology. 2010;162:362–371. doi: 10.1111/j.1365-2249.2010.04245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki E, Karam E, Williams S, Watson DK, Gilkeson G, Zhang XK. Fli-1 transcription factor affects glomerulonephritis development by regulating expression of monocyte chemoattractant protein-1 in endothelial cells in the kidney. Clin Immunol. 2012;145:201–208. doi: 10.1016/j.clim.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato S, Lennard Richard M, Brandon D, Jones Buie JN, Oates JC, Gilkeson GS, Zhang XK. A critical role of the transcription factor fli-1 in murine lupus development by regulation of interleukin-6 expression. Arthritis & rheumatology. 2014;66:3436–3444. doi: 10.1002/art.38818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato S, Zhang XK. The Friend leukaemia virus integration 1 (Fli-1) transcription factor affects lupus nephritis development by regulating inflammatory cell infiltration into the kidney. Clinical and experimental immunology. 2014;177:102–109. doi: 10.1111/cei.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagafuku M, Okuyama K, Onimaru Y, Suzuki A, Odagiri Y, Yamashita T, Iwasaki K, Fujiwara M, Takayanagi M, Ohno I, Inokuchi J. CD4 and CD8 T cells require different membrane gangliosides for activation. Proc Natl Acad Sci U S A. 2012;109:E336–342. doi: 10.1073/pnas.1114965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennard Richard ML, Nowling TK, Brandon D, Watson DK, Zhang XK. Fli-1 controls transcription from the MCP-1 gene promoter, which may provide a novel mechanism for chemokine and cytokine activation. Mol Immunol. 2015;63:566–573. doi: 10.1016/j.molimm.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lennard Richard ML, Sato S, Suzuki E, Williams S, Nowling TK, Zhang XK. The Fli-1 transcription factor regulates the expression of CCL5/RANTES. J Immunol. 2014;193:2661–2668. doi: 10.4049/jimmunol.1302779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard EM, Thiyagarajan T, Bunni MA, Basher F, Roddy PO, Siskind LJ, Nietert PJ, Nowling TK. Reducing FLI1 levels in the MRL/lpr lupus mouse model impacts T cell function by modulating glycosphingolipid metabolism. PloS one. 2013;8:e75175. doi: 10.1371/journal.pone.0075175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowling TK, Mather AR, Thiyagarajan T, Hernandez-Corbacho MJ, Powers TW, Jones EE, Snider AJ, Oates JC, Drake RR, Siskind LJ. Renal Glycosphingolipid Metabolism Is Dysfunctional in Lupus Nephritis. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2014050508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowling TK, Fulton JD, Chike-Harris K, Gilkeson GS. Ets factors and a newly identified polymorphism regulate Fli1 promoter activity in lymphocytes. Mol Immunol. 2008;45:1–12. doi: 10.1016/j.molimm.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svenson JL, Chike-Harris K, Amria MY, Nowling TK. The mouse and human Fli1 genes are similarly regulated by Ets factors in T cells. Genes and immunity. 2009;11:161–172. doi: 10.1038/gene.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem. 2006;54:385–395. doi: 10.1369/jhc.4A6514.2005. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Ito S, Chino Y, Iwanami K, Yasukochi T, Goto D, Matsumoto I, Hayashi T, Uchida K, Sumida T. Use of laser microdissection in the analysis of renal-infiltrating T cells in MRL/lpr mice. Modern rheumatology/the Japan Rheumatism Association. 2008;18:385–393. doi: 10.1007/s10165-008-0074-8. [DOI] [PubMed] [Google Scholar]
- 21.Teramoto K, Negoro N, Kitamoto K, Iwai T, Iwao H, Okamura M, Miura K. Microarray analysis of glomerular gene expression in murine lupus nephritis. Journal of pharmacological sciences. 2008;106:56–67. doi: 10.1254/jphs.fp0071337. [DOI] [PubMed] [Google Scholar]
- 22.Steinmetz OM, Turner JE, Paust HJ, Lindner M, Peters A, Heiss K, Velden J, Hopfer H, Fehr S, Krieger T, Meyer-Schwesinger C, Meyer TN, Helmchen U, Mittrucker HW, Stahl RA, Panzer U. CXCR3 mediates renal Th1 and Th17 immune response in murine lupus nephritis. J Immunol. 2009;183:4693–4704. doi: 10.4049/jimmunol.0802626. [DOI] [PubMed] [Google Scholar]
- 23.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- 25.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van Nimwegen E, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegner J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabin RL, Park MK, Liao F, Swofford R, Stephany D, Farber JM. Chemokine receptor responses on T cells are achieved through regulation of both receptor expression and signaling. J Immunol. 1999;162:3840–3850. [PubMed] [Google Scholar]
- 27.Lord GM, Rao RM, Choe H, Sullivan BM, Lichtman AH, Luscinskas FW, Glimcher LH. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood. 2005;106:3432–3439. doi: 10.1182/blood-2005-04-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taqueti VR, Grabie N, Colvin R, Pang H, Jarolim P, Luster AD, Glimcher LH, Lichtman AH. T-bet controls pathogenicity of CTLs in the heart by separable effects on migration and effector activity. J Immunol. 2006;177:5890–5901. doi: 10.4049/jimmunol.177.9.5890. [DOI] [PubMed] [Google Scholar]
- 29.Perez de Lema G, Maier H, Nieto E, Vielhauer V, Luckow B, Mampaso F, Schlondorff D. Chemokine expression precedes inflammatory cell infiltration and chemokine receptor and cytokine expression during the initiation of murine lupus nephritis. J Am Soc Nephrol. 2001;12:1369–1382. doi: 10.1681/ASN.V1271369. [DOI] [PubMed] [Google Scholar]
- 30.Narumi S, Takeuchi T, Kobayashi Y, Konishi K. Serum levels of ifn-inducible PROTEIN-10 relating to the activity of systemic lupus erythematosus. Cytokine. 2000;12:1561–1565. doi: 10.1006/cyto.2000.0757. [DOI] [PubMed] [Google Scholar]
- 31.Enghard P, Humrich JY, Rudolph B, Rosenberger S, Biesen R, Kuhn A, Manz R, Hiepe F, Radbruch A, Burmester GR, Riemekasten G. CXCR3+CD4+ T cells are enriched in inflamed kidneys and urine and provide a new biomarker for acute nephritis flares in systemic lupus erythematosus patients. Arthritis and rheumatism. 2009;60:199–206. doi: 10.1002/art.24136. [DOI] [PubMed] [Google Scholar]
- 32.Dar WA, Knechtle SJ. CXCR3-mediated T-cell chemotaxis involves ZAP-70 and is regulated by signalling through the T-cell receptor. Immunology. 2007;120:467–485. doi: 10.1111/j.1365-2567.2006.02534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton P, O’Boyle G, Jenkins Y, Ali S, Kirby JA. T cell extravasation: demonstration of synergy between activation of CXCR3 and the T cell receptor. Mol Immunol. 2009;47:485–492. doi: 10.1016/j.molimm.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gosiengfiao Y, Horvat R, Thompson A. Transcription factors GATA-1 and Fli-1 regulate human HOXA10 expression in megakaryocytic cells. DNA and cell biology. 2007;26:577–587. doi: 10.1089/dna.2007.0575. [DOI] [PubMed] [Google Scholar]
- 35.Tamir A, Howard J, Higgins RR, Li YJ, Berger L, Zacksenhaus E, Reis M, Ben-David Y. Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: evidence for direct transcriptional repression of the Rb gene during differentiation. Mol Cell Biol. 1999;19:4452–4464. doi: 10.1128/mcb.19.6.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodge DR, Xiao W, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001;276:39508–39511. doi: 10.1074/jbc.C100343200. [DOI] [PubMed] [Google Scholar]
- 37.Jinnin M, Ihn H, Mimura Y, Asano Y, Yamane K, Tamaki K. Matrix metalloproteinase-1 up-regulation by hepatocyte growth factor in human dermal fibroblasts via ERK signaling pathway involves Ets1 and Fli1. Nucleic Acids Res. 2005;33:3540–3549. doi: 10.1093/nar/gki648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lakhanpal GK, Vecchiarelli-Federico LM, Li YJ, Cui JW, Bailey ML, Spaner DE, Dumont DJ, Barber DL, Ben-David Y. The inositol phosphatase SHIP-1 is negatively regulated by Fli-1 and its loss accelerates leukemogenesis. Blood. 2011;116:428–436. doi: 10.1182/blood-2009-10-250217. [DOI] [PubMed] [Google Scholar]
- 39.Landry JR, Kinston S, Knezevic K, Donaldson IJ, Green AR, Gottgens B. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106:2680–2687. doi: 10.1182/blood-2004-12-4755. [DOI] [PubMed] [Google Scholar]
- 40.Lesault I, Quang CT, Frampton J, Ghysdael J. Direct regulation of BCL-2 by FLI-1 is involved in the survival of FLI-1-transformed erythroblasts. Embo J. 2002;21:694–703. doi: 10.1093/emboj/21.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puri S, Rodova M, Islam MR, Magenheimer BS, Maser RL, Calvet JP. Ets factors regulate the polycystic kidney disease-1 promoter. Biochem Biophys Res Commun. 2006;342:1005–1013. doi: 10.1016/j.bbrc.2006.02.045. [DOI] [PubMed] [Google Scholar]