Abstract
MET inhibition is effective in some MET-amplified esophagogastric cancer (EGC) patients, but understanding acquired and de novo resistance mechanisms will be critical to improving therapy. We identified KRAS mutation as a novel cause of acquired resistance in a patient after a two-year response to MET inhibitor. We also observed that 40–50% of MET-amplified EGC patients harbor co-amplification of HER2 and/or EGFR concurrently in the same tumor cells, which can drive de novo resistance. One patient with concurrent MET and HER2-amplification was refractory to HER2 blockade, but responded to combined MET/HER2 inhibition. We also found striking heterogeneity in MET-amplification between distinct metastatic lesions and primary tumors in individual EGC patients. In these patients, MET inhibition led to mixed responses and disease progression through outgrowth of non-MET-amplified clones, which could be monitored in circulating tumor DNA. Thus, receptor co-amplification and molecular heterogeneity may be key drivers of clinical resistance in MET-amplified EGC.
Keywords: MET, gastric cancer, esophageal cancer, drug resistance
INTRODUCTION
Cancers involving the esophagus, gastroesophageal junction, and stomach (collectively referred to as esophagogastric cancer, EGC) are the second leading cause of cancer death worldwide(1). While the identification and pharmacologic targeting of actionable molecular alterations have led to effective therapeutic strategies in many tumor types(2–4), targeted therapies have yet to substantially impact the treatment of EGC. The only FDA-approved therapy targeting a specific molecular alteration in EGC is trastuzumab, a monoclonal antibody against the Human Epidermal Growth Factor Receptor 2 (HER2, ERBB2), which is approved in combination with chemotherapy for the ~20% of gastric and gastroesophageal junction cancers with HER2-amplification or overexpression. However, the addition of trastuzumab to chemotherapy produced only a modest improvement in survival and a 12% absolute increase in response rate(5). Therefore, the development of new therapeutic approaches that can effectively exploit key molecular targets in EGC is of critical clinical importance.
MET gene amplification and MET protein overexpression are observed in ~5% and ~50% of EGC, respectively(6, 7). MET encodes a receptor tyrosine kinase (RTK), which is typically activated by its ligand, hepatocyte growth factor (HGF), and signals through downstream pathways involved in oncogenesis(8). Although clinical efforts to target the MET pathway in EGC patients harboring MET protein overexpression have yielded disappointing results(9, 10), there is evidence that MET-amplification (as opposed to protein overexpression only) may be a true oncogenic driver in EGC. MET-amplified EGC cell lines display exquisite sensitivity to MET inhibitors(11), and patients with MET-amplified EGC have experienced tumor shrinkage after treatment with small molecule MET inhibitors or monoclonal antibodies against MET(12, 13). Recently, a phase I clinical trial of the MET kinase inhibitor AMG337 in 13 patients with MET-amplified EGC demonstrated a 62% response rate(14).
Thus, MET-amplification may represent a promising therapeutic target for EGC, but a better understanding of the clinical mechanisms of resistance to MET inhibition will be vital to the successful therapeutic implementation of these agents. Here, through molecular analyses of tumor biopsies from MET-amplified EGC patients treated with MET kinase inhibitors, we define key mechanisms of acquired and de novo resistance and identify important characteristics of EGC biology that may have broad implications for the use of targeted therapies in this disease.
RESULTS
Clinical acquired resistance to MET kinase inhibition
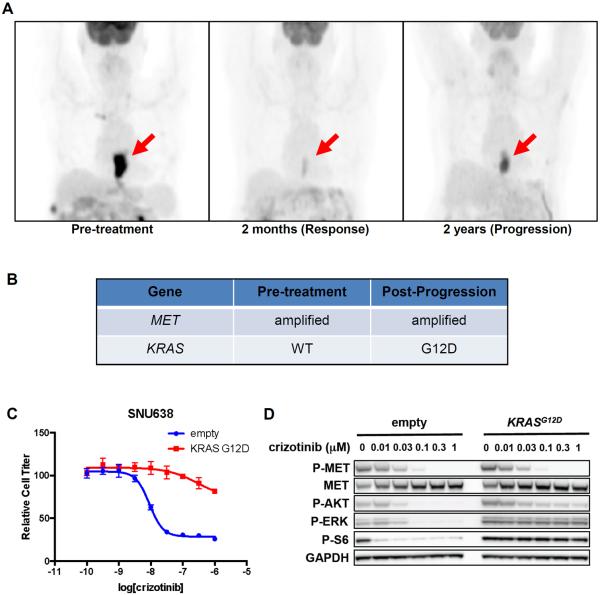
Robust and, in some cases, durable tumor responses to MET inhibition have been observed in some MET-amplified EGC patients(12–14), but as with all targeted therapies, the inevitable emergence of acquired resistance limits clinical benefit. Understanding the mechanisms of clinical acquired resistance to therapy may offer opportunities to overcome resistance and to prolong therapeutic response. Patient #1 was diagnosed with MET-amplified (4.3-fold) adenocarcinoma of the distal esophagus, and was treated with the experimental MET kinase inhibitor AMG337 (NCT01253707) following disease progression on first-line chemotherapy(14). AMG337 is a highly selective small molecule MET kinase inhibitor, which was found to bind only to MET in a competitive binding assay conducted against 402 human kinases(15). The patient experienced a profound tumor response lasting two years, until he developed difficulty swallowing and was found to have disease progression at the site of his primary tumor (Fig. 1A).
Fig. 1. KRAS mutation can drive clinical acquired resistance to MET inhibition.
(A) PET images of Patient #1 at baseline, initial response, and disease progression during AMG337 therapy. Red arrows indicate primary tumor. (B) Molecular analysis of pre-treatment and post-progression tumor biopsies. (C) SNU638 cells exogenously expressing KRASG12D or empty vector were treated with the indicated concentrations of crizotinib for 72h, and cell titer was determined. (D) SNU638 cells exogenously expressing KRASG12D or empty vector were treated with the indicated concentrations of crizotinib for 24h, and lysates were probed with the indicated antibodies.
A biopsy of his primary tumor was obtained upon disease progression, and molecular analysis was performed on this specimen as well as on the original biopsy obtained from the same site at diagnosis. Both biopsies harbored MET-amplification, but the post-progression biopsy also harbored a KRASG12D mutation that was not detected in the pre-treatment biopsy (Fig. 1B; Tables S1, S2). KRAS mutation or amplification can occur in ~10% or ~7% of treatment-naïve EGCs, respectively(6). The emergence of KRAS mutations has been observed as a cause of clinical acquired resistance to other RTK inhibitors, including anti-EGFR antibodies in colorectal cancer(16, 17). Similarly, exogenous expression of KRASG12D in a MET-addicted EGC cell line SNU638 conferred resistance to MET kinase inhibition with crizotinib (Fig. 1C). Although crizotinib is FDA-approved as an ALK inhibitor for ALK-rearranged lung cancer, it is also a highly potent inhibitor of MET, and responses to crizotinib have been observed in MET-amplified EGC patients(12). KRASG12D expression in SNU638 cells maintained signaling downstream of MET despite crizotinib treatment, as evidenced by sustained phosphorylation of ERK, ribosomal protein S6 (S6), and to a lesser degree AKT (Fig. 1D). These data support KRAS mutation as the cause of acquired resistance to MET kinase inhibition in this patient.
RTK co-amplification drives de novo resistance
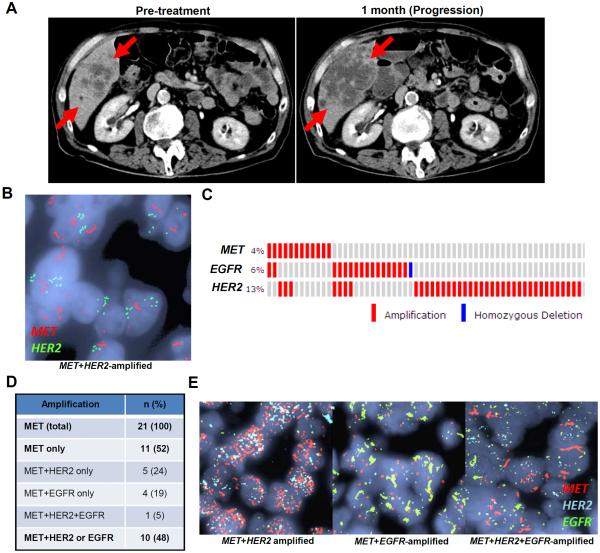
Despite the striking responses seen in some patients, not all patients with MET-amplified EGC respond to MET kinase inhibition. We sought to define the common mechanisms of de novo resistance to MET inhibition in order to guide potential therapeutic strategies to overcome resistance. Patient #2 was diagnosed with metastatic gastric adenocarcinoma, and standard HER2 testing revealed 7-fold HER2-amplification. The patient was initially treated with FOLFOX + trastuzumab for four months, followed by 5-FU and radiation, but developed new metastatic disease. Additional molecular analysis of the patient's original biopsy revealed 5-fold MET-amplification, and the patient was treated with AMG337(14). Unfortunately, the patient's disease progressed rapidly despite treatment (Fig. 2A), requiring discontinuation of therapy after less than one month. Remarkably, multi-color FISH analysis for MET and HER2 showed that co-amplification of these genes existed simultaneously in the same tumor cells (Fig. 2B). Thus, in this patient, co-amplification of two driver RTKs may have led to redundant activation of downstream signaling pathways, thereby driving resistance to MET kinase inhibition.
Fig. 2. Frequent co-amplification of RTKs in MET-amplified EGC.
(A) CT images of Patient #2 taken pre-treatment and upon disease progression one month after initiation of AMG337. Red arrows indicate tumor. (B) FISH images from Patient #2 showing co-amplification of MET and HER2 occurring in the same tumor cells. (C) Analysis of TCGA data showing co-occurrence of RTK amplification in EGC. (D) Multi-color FISH analysis of 21 MET-amplified EGC patients for co-amplification of HER2 and/or EGFR in the same tumor cells. (E) Representative multi-color FISH images from cases in (D).
To determine how commonly RTK co-amplification might exist as a potential driver of de novo resistance in MET-amplified EGC, we evaluated publicly available data from The Cancer Genome Atlas (TCGA) study of EGC(6) and identified 12 cases with MET-amplification. Interestingly, 5 of 12 (42%) MET-amplified EGCs harbored co-amplification of either HER2 or EGFR, based on sequencing analyses (Fig. 2C), suggesting that RTK co-amplification could be a common cause of de novo resistance. This finding is consistent with a recently published study of HER2-amplified EGC, which also showed a high frequency of co-occurring amplifications(18).
However, analysis of sequencing data alone cannot determine whether RTK co-amplification occurs in the same tumor cells, as in Patient #2, or if a tumor might harbor distinct subclonal populations of cells, each amplified for one specific RTK. Therefore, we identified 21 cases of MET-amplified EGC that had available tissue obtained at diagnosis, and analyzed each using a multi-color FISH assay allowing simultaneous assessment of MET, HER2, and EGFR copy number. Amplification of each gene was also confirmed by standard FISH. Consistent with the TCGA data, we found that 48% (10/21) of these MET-amplified EGCs harbored co-amplification of either HER2 and/or EGFR, but that co-amplification was present simultaneously within the same tumor cells (Figs. 2D–E). Remarkably, one case (5%) displayed amplification of all three RTKs within the same tumor cells. Collectively, these data suggest that RTK co-amplification is a common occurrence in MET-amplified EGC and has the potential to be an important driver of clinical resistance to MET inhibition.
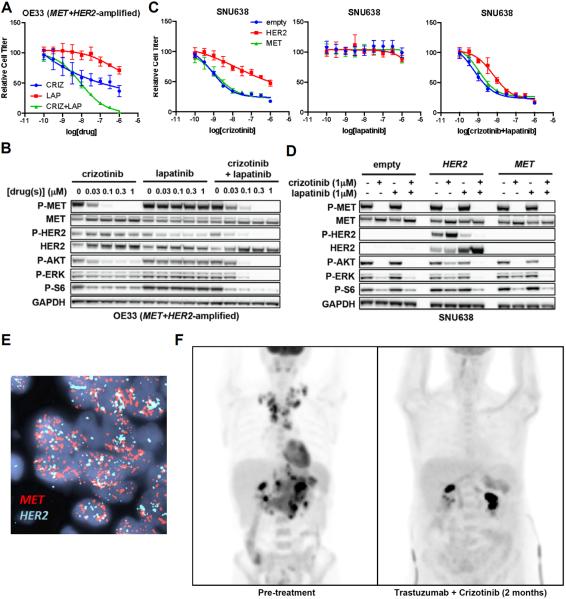
Since EGFR has previously been implicated in resistance to MET inhibition in EGC cells(19), we focused on modeling the effects of concomitant HER2 amplification as a driver of MET inhibitor resistance, especially since HER2 co-amplification was observed in 6/21 cases of MET-amplified EGC. We identified a cell line, OE33, that harbors concomitant amplification of MET and HER2. OE33 cells were resistant to MET kinase inhibition with crizotinib alone, or to HER2 inhibition with lapatinib alone (Fig. 3A). However, OE33 cells were highly sensitive to combined MET/HER2 inhibition with both drugs in combination. Consistent with these findings, the combination of crizotinib and lapatinib produced robust suppression of downstream signaling effectors, including phosphorylated ERK (P-ERK), P-AKT, and P-S6, whereas incomplete suppression was achieved with either drug alone (Fig. 3B). Similarly, we found that exogenous HER2 overexpression in MET-addicted SNU638 cells conferred resistance to MET kinase inhibition with crizotinib, but that combined MET/HER2 inhibition could overcome resistance (Fig. 3C). HER2 overexpression led to maintenance of P-ERK, P-AKT, and P-S6 in the presence of crizotinib alone, but the combination of crizotinib and lapatinib retained the ability to suppress phosphorylation of these downstream effectors (Fig. 3D). These data suggest that RTK co-amplification can lead to redundant activation of downstream effector pathways and that combined RTK inhibition may be required to restore sensitivity.
Fig. 3. Combined inhibition of co-amplified RTKs is required for response.
(A) OE33 cells were treated with the indicated concentrations of crizotinib alone, lapatinib alone, or both drugs in combination for 72h, and cell titer was determined. (B) Western blot of OE33 cells treated with the indicated concentrations of crizotinib, lapatinib, or both drugs in combination for 24h. (C) SNU638 cells exogenously expressing HER2, MET, or empty vector control were treated with the indicated concentrations of drug(s) for 72h, and cell titer was determined. (D) The same cells from (C) were treated with the indicated concentrations of drug for 24h, and western blotting was performed with the indicated antibodies. (E) FISH images showing >25-fold amplification of both MET and HER2 in the same tumor cells from Patient #3. (F) PET images from Patient #3 obtained pre-treatment and after 2 months of therapy with combined MET/HER2 inhibition.
Patient #3 presented with epigastric pain, bloating and early satiety. Endoscopy and biopsy revealed gastric adenocarcinoma with amplification of HER2. The patient was treated on a clinical trial for HER2-positive gastric cancer with capecitabine, oxaliplatin, bevacizumab, and the anti-HER2 monoclonal antibody trastuzumab; however, her disease progressed rapidly through treatment. More extensive molecular analysis of her biopsy revealed that her tumor harbored >25-fold amplification of both HER2 and MET present in the same tumor cells (Fig. 3E), likely explaining why her disease progressed despite trastuzumab-containing chemotherapy. Accordingly, the patient was treated with the combination of trastuzumab, crizotinib, and weekly paclitaxel (as described in Methods), which the patient tolerated well with some increased peripheral edema. Strikingly, a repeat PET-CT scan performed 2 months after initiation of therapy revealed near-complete resolution of her disease (Fig. 3F). This case suggests that co-amplification of MET and HER2 led to resistance to anti-HER2 therapy alone, but that, consistent with the in vitro results above, combined MET/HER2 inhibition led to a dramatic clinical response. Of note, the patient's tumor did not harbor rearrangements in ALK or ROS1, which are known to confer sensitivity to crizotinib in non-small cell lung cancer (Figure S1)(2, 4). Although a contribution of paclitaxel to the response cannot be excluded, this is less likely given that clinical evidence of response occurred rapidly, before substantial exposure to paclitaxel, and that the patient's disease had progressed rapidly through combination cytotoxic chemotherapy in the prior line of therapy. Based on these data, combined RTK inhibition may be an effective therapeutic strategy for EGCs harboring RTK co-amplification and warrants further clinical investigation.
Heterogeneity as a driver of resistance and mixed response in EGC
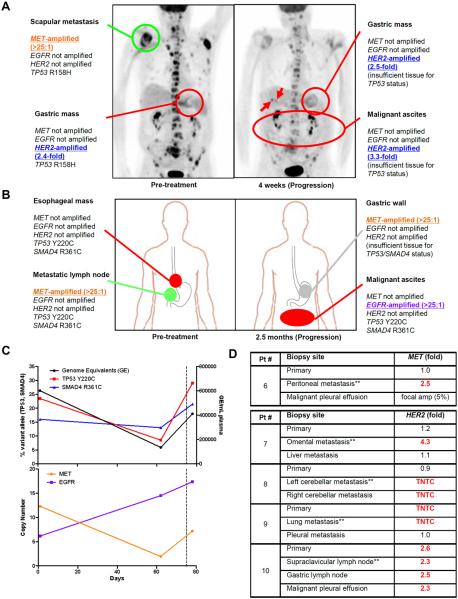
Patient #4 presented with shoulder pain and was found to have widespread bone metastases. Biopsy of a right scapular lesion established the diagnosis of adenocarcinoma, and endoscopy revealed a primary gastric cancer. Molecular analysis was performed on his scapular biopsy and revealed >25-fold MET-amplification. The patient was treated with AMG337, but after less than a month, he developed decreased oral intake and shortness of breath(14). He was found to have new ascites and pleural effusion, and endoscopy confirmed progression of his gastric mass. Imaging revealed the development of new liver metastases, but surprisingly also showed that his bone metastases had responded to therapy (Fig. 4A).
Fig. 4. Molecular heterogeneity can drive mixed response and treatment failure in EGC.
(A) PET images from Patient #4 obtained pre-treatment and upon disease progression after only 4 weeks of treatment with AMG337. The molecular profiles of individual biopsied lesions are shown. Red indicates progressing lesions, and green indicates responding lesions. Arrows demarcate new liver metastases. (B) Diagram of biopsied lesions from Patient #5 obtained pre-treatment and upon disease progression 2.5 months after initiation of AMG337. The molecular profiles of individual biopsied lesions are shown. Red indicates progressing lesions, green indicates responding lesions, and gray indicates response undetermined. (C) Serial ctDNA analysis of plasma from Patient #5 obtained pre-treatment, at the time of tumor response (2 months), and at disease progression (2.5 months). At each time point, plasma was analyzed for total genome equivalents of cell-free DNA, and for ctDNA levels of “truncal” mutations in TP53 and SMAD4, as well as MET and EGFR copy number. Dashed line represents timing of treatment discontinuation. (D) FISH analysis of multiple tumor biopsies obtained from distinct metastatic sites and primary tumors from individual patients with MET-amplified or HER2-amplified EGC. The biopsy upon which initial molecular testing was performed is indicated by (**). The fold-amplification is shown for each biopsy with biopsies harboring amplification shown in red. TNTC = amplification “too numerous to count.” The malignant pleural effusion from Patient #6 showed focal MET-amplification in only 5% of tumor cells.
To determine the cause of this mixed response to therapy, we performed molecular analyses on additional tumor biopsies obtained from this patient during the course of his clinical care. Prior to initiating therapy, a biopsy of his primary gastric tumor had also been obtained. However, since it is not standard clinical practice to perform molecular analysis on more than one biopsy from the same patient at diagnosis, this specimen had not undergone molecular testing, and only the scapular biopsy was initially analyzed. Surprisingly, retrospective analysis of his pre-treatment gastric biopsy showed no evidence of MET-amplification, but rather showed low-level HER2-amplification (Fig. 4A). Importantly, despite differences in gene amplification, both the scapular biopsy and the primary tumor harbored the same TP53 R158H mutation, indicating that tumor cells in each lesion shared a common clonal origin. Moreover, analysis of a repeat gastric mass biopsy and tumor cells from the patient's ascites, each obtained at the time of disease progression, displayed absence of MET-amplification and presence of low-level HER2-amplification, similar to the primary tumor. Overall, these data are consistent with a mixed response, in which AMG337 led to regression of the patient's MET-amplified bone metastases, but disease progression due to outgrowth of a non-MET-amplified, HER2-amplified clone. This case illustrates how heterogeneity in gene amplification between separate tumor sites in the same patient can lead to mixed response and treatment failure.
Similarly, Patient #5 was diagnosed with adenocarcinoma arising in the distal esophagus with evidence of infiltration into the gastric cardia. A biopsy of a gastrohepatic ligament lymph node confirmed metastatic disease, and molecular analysis of this biopsy revealed >25-fold MET-amplification. The patient was treated with AMG337(14) and experienced a marked reduction in the size of his metastatic lymph nodes, including the biopsied node, achieving a partial response after two months of therapy (Fig. S1). However, two weeks later, the patient developed difficulty swallowing and new ascites. Endoscopy was consistent with progression of his primary tumor. His primary tumor was not re-biopsied, but a biopsy was obtained from a region of tumor infiltration in the gastric cardia which, similar to his original lymph node biopsy, demonstrated >25-fold amplification of MET. However, retrospective analysis of the patient's original esophageal mass biopsy obtained prior to MET inhibitor therapy showed no evidence of MET-amplification (Fig. 4B). Tumor cells collected from the patient's ascites at the time of disease progression also showed no evidence of MET-amplification. Remarkably, these cells instead showed >25-fold EGFR-amplification, even though no evidence of EGFR-amplification was detected in the primary tumor. The same TP53 and SMAD4 mutations were detected in the initial metastatic lymph node biopsy, the initial esophageal mass biopsy, and in the ascites tumor cells at progression, suggesting that these were early “truncal” events present in a common tumor clone from which tumor cells at all three sites arose, despite the heterogeneity in gene amplification observed between them (Fig. 4B).
“Liquid biopsy” to assess cell-free circulating tumor DNA (ctDNA), which is shed into the bloodstream by tumor cells throughout the body, has the potential to capture the clonal heterogeneity of tumor cells residing in different tumor lesions within an individual patient(20, 21). Peripheral blood was collected at baseline and throughout MET inhibitor therapy, and ddPCR was used to monitor the levels of specific molecular alterations present in ctDNA (Fig. 4C). Levels of shared “truncal” mutations (in TP53 and SMAD4) decreased initially with treatment but rose upon disease progression, and largely paralleled the overall levels of cell-free DNA. Conversely, elevated MET copy number detectable in ctDNA decreased to near-normal levels during the first two months of therapy, indicating effective suppression of MET-amplified tumor clones, consistent with the response observed in the patient's nodal disease. However, ctDNA analysis showed that increased EGFR copy number was detectable prior to initiation of MET inhibitor, suggesting that EGFR-amplified clones were already present prior to treatment, but were likely present in different tumor lesions other than those biopsied pre-treatment. A marked increase in EGFR copy number detectable in ctDNA was observed throughout treatment, consistent with the outgrowth of resistant EGFR-amplified clones observed in the ascites upon progression. Collectively, these findings illustrate that extensive molecular heterogeneity in RTK amplification can arise between different EGC tumor lesions in the same patient. The selective pressure applied by targeted therapies can lead to outgrowth of resistant tumor clones, leading to lesion-specific mixed responses and treatment failure.
Accordingly, a biopsy of a single tumor lesion for molecular profiling, currently the standard diagnostic approach for selection of targeted therapy trials, may fail to capture the molecular heterogeneity of a patient's overall tumor burden, and may therefore be insufficient to guide therapy in EGC. To determine how commonly heterogeneity in RTK amplification might occur between primary tumors and different metastatic sites in the same patient, we identified additional patients with either MET-amplified or HER2-amplified EGC in whom tissue was available from the primary tumor and multiple metastatic sites in the same patient, but in which only a single metastatic lesion had undergone initial molecular testing to detect RTK amplification. We identified one such patient with MET-amplified EGC and four with HER2-amplified EGC. The MET-amplified patient and two of four HER2-amplified patients showed absence of MET or HER2-amplification (respectively) in the primary tumor, despite the known presence of amplification in the metastatic lesion initially tested (Figs. 4D, S2). Interestingly, a third HER2-amplified patient harbored HER2-amplification in the primary tumor as well as in the originally-tested metastatic biopsy, but not in a biopsy of a different metastasis.
Taken together, these data illustrate that heterogeneity and discordance in RTK amplification between the primary tumor and different metastatic lesions can occur commonly in individual EGC patients. Thus, detection of RTK amplification in a biopsy of a single metastatic lesion may not reliably predict the presence of this same “driver” RTK amplification in the primary tumor or at other metastatic sites. Interestingly, in a small cohort of MET-amplified EGC patients treated with AMG337(14), we found that those patients who benefited the most from therapy harbored MET-amplification (as the sole amplified RTK) in their primary tumor, several of whom remained on study for more than one year. Conversely, patients in whom MET-amplification was originally detected in a metastasis had shorter times to progression (range 0.8–2.5 months), as their primary tumors and other metastatic sites were ultimately found to lack MET-amplification (Table S3). While small patient numbers limit any definitive conclusions, these data suggest that it may be important to confirm the presence of RTK amplification in the primary tumor prior to initiation of targeted therapy in EGC. Overall, the potential for molecular heterogeneity appears to play a key role in drug resistance with several important diagnostic and therapeutic implications.
DISCUSSION
MET inhibition can lead to striking, and at times durable, responses in some patients with MET-amplified EGC, and represents a promising therapeutic strategy(12–14). The validity of MET as a therapeutic target in EGC has been questioned based on recent negative Phase III studies targeting this pathway in EGC(9, 10). However, these studies were not conducted specifically in the MET-amplified EGC population, and the majority of patients treated had MET protein overexpression only. One possible explanation is that the immunohistochemistry threshold used to define MET overexpression may not be stringent enough to identify those patients whose tumors are truly dependent on MET signaling. Alternatively, since MET-amplification involves a genetic change, it is perhaps more likely to represent a driving event upon which the tumor is dependent, compared to MET protein overexpression only, which may result as a consequence of other mechanisms within the tumor cell, making it less likely to be a driving molecular event. Thus, the therapeutic potential for MET inhibition may be limited to EGC patients with MET-amplification, and these clinical trials underscore how critical appropriate patient selection may be to the successful clinical implementation of MET inhibitors.
We evaluated the causes of acquired and de novo resistance to MET kinase inhibition in MET-amplified EGC to better understand the clinical factors influencing the efficacy of these agents. While the patients in this study were treated with a small molecule MET kinase inhibitor, tumor responses in MET-amplified EGC patients have also been observed with anti-MET antibodies(13), so it is likely that the resistance mechanisms we identified may be relevant to these agents, as well. We report here the first mechanism of clinical acquired resistance to be identified in a MET-amplified EGC patient, which followed a two-year response to a MET kinase inhibitor. Acquired resistance in this patient was driven by the emergence of a KRAS mutation, which can bypass pathway suppression by a MET inhibitor. A detailed understanding of additional mechanisms that can drive clinical acquired resistance in MET-amplified tumors may guide future therapeutic strategies to overcome resistance. We also sought to understand causes of de novo resistance to explain why some patients fail to respond to MET inhibition altogether. Our study revealed two unique characteristics of EGC that appear to play key roles in therapeutic resistance.
First, we found that RTK co-amplification is a common occurrence in MET-amplified EGC, with 40–50% of cases harboring co-amplification of either HER2 or EGFR. Our findings are consistent with a recent study showing frequent co-occurrence of oncogenic alterations by copy-number analysis in HER2-amplified EGC, providing a potential explanation for the limited benefit of trastuzumab and suggesting that this observation is not limited to the MET-amplified subtype(5, 18). However, by using multi-color FISH analysis, our study confirmed that co-amplification of RTKs actually occurs within the same tumor cells rather than in distinct subclones within the same tumor. It is unusual to observe frequent co-occurrence of functionally similar driver alterations in a single tumor, and most comprehensive genomic analyses of other tumor types have shown mutual exclusivity of molecular alterations that activate the same signaling pathways(22–24). Interestingly, RTK amplifications occur predominantly in EGCs belonging to the chromosomal instability (CIN) subtype, according to TCGA classification, which is characterized by frequent somatic copy number alterations(6). The propensity for copy number alterations in this subtype is one possible explanation for the frequency of RTK co-amplification observed. The potential for pathway crosstalk and the ability of MET to heterodimerize with HER family members, including EGFR and HER2(25), may be another potential explanation co-amplification. We present preclinical and clinical evidence that combined targeting of co-amplified RTKs is required for response through suppression of redundant pathway activation. Patients with co-amplification of MET and HER2 failed to respond to MET kinase inhibition (Patient #2, Fig. 2A) or HER2 inhibition (Patient #3), but combined MET/HER2 inhibition led to dramatic clinical response in one patient (Patient #3, Fig. 3F). Collectively, these observations suggest that comprehensive molecular analyses (as opposed to single biomarker assessment, as is the current diagnostic standard for HER2 testing) should be performed prior to selection of a targeted therapy strategy for EGC patients to identify potential co-occurring driver alterations that might preclude efficacy. Furthermore, these data suggest that RTK inhibitor combinations or dual-targeting agents directed against multiple RTKs may warrant clinical evaluation in EGC.
Additionally, we observed that dramatic heterogeneity in MET-amplification can occur between different metastatic lesions and the primary tumor in individual EGC patients, leading to mixed responses to MET kinase inhibition and treatment failure due to outgrowth of non-MET-amplified clones. We also found that similar heterogeneity can be observed in patients with HER2-amplified disease (Fig. 4D, Fig. S3), suggesting that this characteristic of EGC may have important implications beyond the MET-amplified subtype. The presence of RTK amplification in some metastatic lesions, but not in the primary tumor or in other metastases from the same patient suggests that perhaps RTK amplification is not an early or “founding” event in EGC development, but may arise at a later stage in specific tumor clones, possibly leading to increased aggressiveness or metastatic potential. Still, it is interesting that even though MET-amplification may have occurred late in the development of some EGCs (Patient #4 and #5, Figs. 4A–C)—as opposed to the “truncal” TP53 and/or SMAD4 mutations present in all tumor cells in these patients—tumor cells with MET-amplification still appear to be MET-dependent. This is evidenced by the tumor response to MET kinase inhibition observed at MET-amplified metastatic sites in each patient, and by the reduction in clonal abundance of MET copy gain observed in ctDNA from Patient #5 (Fig. 4C). Interestingly, our small cohort of MET-amplified patients suggested that patients with MET-amplification detected in their primary tumors derived greater clinical benefit from MET kinase inhibition, though small patient numbers limit a definitive correlation (Table S3). This suggests that testing of the primary tumor may identify patients in whom MET-amplification arose as an earlier event, and thus whose overall tumor burden may more likely be MET-dependent.
Due to the extensive intra-patient heterogeneity observed in EGC, molecular profiling of a single biopsied tumor lesion may be insufficient to guide targeted therapy in this disease. If multiple biopsy specimens are available for a given patient, molecular testing of each (including the primary tumor) may unveil the existence of molecular heterogeneity that could lead to treatment failure, as evidenced by the cases presented above. However, it is not feasible to biopsy every lesion in an individual patient. Thus, liquid biopsy approaches assessing ctDNA or circulating tumor cells may provide the means to capture the molecular heterogeneity present in multiple tumor lesions in an individual patient. Indeed, analysis of ctDNA from peripheral blood drawn from Patient #5 prior to therapy (Fig. 4C) was able to detect the presence of EGFR-amplification that eventually led to treatment failure, even though no evidence of EGFR-amplification was observed in two tumor biopsies obtained pre-treatment. Serial ctDNA analysis also allowed real-time monitoring of dynamic shifts in the abundance of distinct MET-amplified or EGFR-amplified clones that predicted treatment outcome. Therefore, incorporation of liquid biopsy approaches to capture intra-patient tumor heterogeneity may be valuable to future efforts to implement targeted therapy strategies in EGC.
In summary, we find that frequent RTK co-amplification and heterogeneity of RTK-amplification are key molecular features driving lack of benefit from MET inhibition (and perhaps other targeted therapies) in EGC patients. Thus, comprehensive molecular analysis to detect potential concurrent RTK gene amplifications should be performed prior to initiation of targeted therapy, and targeted therapy combinations may be needed. Furthermore, due to the potential for molecular heterogeneity within an individual EGC patient, concurrent molecular analysis of all available biopsy specimens, particularly the primary tumor, should be considered prior to initiation of therapy to identify patients in whom RTK-amplification is most likely to represent an early driving molecular event. These important characteristics of EGC biology may have important clinical implications for the development of targeted therapy strategies for this disease.
METHODS
Patient samples, cell lines, and reagents
Patient tumor and blood specimens were obtained from patients treated at the Massachusetts General Hospital under Institutional Review Board-approved studies. For patients with multiple blood or tumor samples analyzed, DNA fingerprinting by short tandem repeat (STR) analysis (BioSynthesis, Lewisville, TX) was performed to confirm that samples originated from the same patient. All patients provided written, informed consent, and studies were conducted in accordance with the Declaration of Helsinki. Patient #3 was treated with crizotinib 250mg orally twice daily, trastuzumab 6mg/kg intravenously every three weeks, and weekly paclitaxel 80mg/m2 (beginning the second week of treatment) off-label with informed consent.
OE33 and SNU638 cells were obtained from the Massachusetts General Hospital Center for Molecular Therapeutics, which performs routine cell line authentication testing by single-nucleotide polymorphism and STR analysis, and were passaged less than six months following receipt. Cells were grown in RPMI (GIBCO) with 10% FBS and assayed in DMEM/F12 with 5% FBS. Crizotinib and lapatinib (Selleck Chemicals) were dissolved in DMSO.
Fluoresence in situ hybridization (FISH)
Standard FISH for MET, HER2, and EGFR and break-apart FISH assays for the detection of ALK or ROS1 rearrangements(2, 4) were performed in the Massachusetts General Hospital Molecular Pathology Clinical Laboratory using CLIA-certified clinical assays. Multi-color FISH was performed using a probe mix containing equal parts of custom Kreatech MET (7q31) red, EGFR (7p11) green, and HER2/ERBB2 (17q12) blue FISH probes (Leica Biosystems).
Positron Emission Tomography (PET), Computed tomography (CT), and RECIST measurements
PET and spiral CT scans were obtained using standard procedures in the Department of Radiology at the Massachusetts General Hospital as part of the routine clinical care of these patients. Response Evaluation Criteria in Solid Tumors (RECIST) measurements were performed by radiologists in the Tumor Imaging Metrics Core at the Dana-Farber/Harvard Cancer Center using standard methods. PET or CT images corresponding to specific RECIST target lesions as defined by the Tumor Imaging Metrics Core were obtained for display.
Western blot analysis and antibodies
Western blotting was performed using standard methods. After treatment with indicated drugs, cells were washed with cold PBS and lysed in the following lysis buffer: 20 mM Tris pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 5 mM sodium pyrophosphate, 50 mM NaF, 10 nM β-glycerophosphate, 1 mM sodium vanadate, 0.5 mM DTT, 4 μg/mL leupeptin, 4 μg/mL pepstatin, 4 μg/mL aprotinin, 1 mM phenylmethylsulfonyl fluoride. Lysates were centrifuged at 16,000 × g for 5 min at 4°C. Protein concentrations were determined by BCA assay (Thermo Scientific). Proteins were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Hybond-P, Amersham). Immunoblotting was performed per antibody manufacturer's specifications. Antibodies for MET, P-MET (Y1234/1235) HER2, P-HER2 (Y1221/1222), P-AKT (S473), and P-S6 (S240/244), (Cell Signaling) were used at 1:1000 dilution; P-ERK (Cell Signaling) was used at 1:2000 dilution. GAPDH (Millipore) was used at 1:1000 dilution. Protein detection on Western blots was performed using SuperSignal chemiluminescence (Thermo Scientific).
Determination of relative cell titer
For in vitro viability assays, cells were plated onto parallel 96-well plates at 2,000 cells/well in RPMI (GIBCO) with 5% FBS. After 24 hours, serial dilutions of inhibitor were added to the wells. Plates were incubated for 3 days, and cell titer was measured by CellTiter-Glo assay (Promega). Relative cell titer was calculated as a percent of the value for cells without inhibitor treatment. Nonlinear regression curves were calculated and displayed using GraphPad Prism 5.
Plasmids
Wild-type MET (23889) and HER2 (23888) in pDONR223 were obtained from Addgene. cDNAs were transferred to the pLenti CMV Puro DEST (Addgene 17452) expression vector using LR Clonase II (Life Technologies).
Lentiviral infections
Transfection complexes containing the lentiviral expression vector and packaging plasmids psPAX2 and pVSVG were assembled using TransIT-LT1 Transfection Reagent (Mirus) and added to 293T cells for production of lentiviral supernatant. All infections were performed in the presence of polybrene (8μg/mL). Following addition of lentiviral supernatants, cells were centrifuged for 1 hour at 2500rpm. After 24 hours, growth medium containing 1μg/mL puromycin (Sigma) was added for 72 hours to select for a stable population of infected cells.
Plasma Collection
At least 10 mL of whole blood was collected by peripheral blood draw using EDTA as an anticoagulant. Plasma was separated within 4 hours through 2 different centrifugation steps (the first at room temperature for 10 minutes at 1,600 × g and the second at 3,000 × g for the same time and temperature), obtaining up to 3 mL of plasma. Plasma was stored at −80°C until ctDNA extraction.
ctDNA isolation and quantification of genome equivalents
ctDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN) according to the manufacturer's instructions. 6 μL of ctDNA were used as template for each reaction. All samples were analyzed in triplicate. PCR reactions were performed using 10 μL final volume containing 5 μL GoTaq qPCR Master Mix, 2X with CXR Reference Dye) (Promega) and LINE-1 [12.5 μmol] forward and reverse primers. DNA at known concentrations was also used to build the standard curve. Primer sequences are available upon request.
Droplet digital PCR analysis
Isolated circulating free DNA was amplified using ddPCR™ Supermix for Probes (Bio-Rad) using KRAS p.G12D (PrimePCR™ ddPCR™ Mutation Assay, Bio-Rad), TP53 p.R158H*, TP53 p.Y220C and SMAD4 p.R361C assays (custom designed) for point mutations, and ERBB2, MET, EGFR and EIF2C1 (reference) for gene copy number variations (PrimePCR™ ddPCR™ Copy Number Assay, Bio-Rad). ddPCR was then performed according to the manufacturer's protocol, and the results were reported as percentage or fractional abundance of mutant DNA alleles to total (mutant plus wild type) DNA alleles. 8 to 10 μL of DNA template was added to 10 μL of ddPCR™ Supermix for Probes (Bio-Rad) and 2 μL of the primer/probe mixture. This reaction mix was added to a DG8 cartridge together with 60μL of Droplet Generation Oil for Probes (Bio-Rad) and used for droplet generation. Droplets were then transferred to a 96 well plate (Eppendorf), and then thermal cycling was performed with the following conditions: 5 minutes at 95°C, 40 cycles of 94°C for 30s, 55°C for 1 minute followed by 98°C for 10 minutes (Ramp Rate 2°C/sec). Droplets were analyzed with the QX200™ Droplet Reader (Bio-Rad) for fluorescent measurement of FAM and HEX probes. Gating was performed based on positive and negative controls, and mutant populations were identified. The ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad) to obtain Fractional Abundance and Copy Number Variations of the mutant/amplified DNA alleles in the wildtype/normal background. The quantification of the target molecule was presented as number of total copies (mutant plus WT) per sample in each reaction. Fractional Abundance is calculated as follows: F.A. % = (Nmut/(Nmut+Nwt))*100), where Nmut is number of mutant events and Nwt is number of WT events per reaction. ddPCR analysis of normal control plasma DNA (from cell lines) and no DNA template controls were always included. Samples with low positive events were repeated at least twice in independent experiments to validate the obtained results.
Supplementary Material
Significance.
Co-amplification of driver oncogenes occurs frequently in EGC and can drive therapeutic resistance, supporting a role for comprehensive molecular analysis prior to targeted therapy. EGCs can also exhibit extensive heterogeneity in gene amplification between distinct tumor lesions within the same patient, suggesting that molecular profiling of a single-lesion biopsy may be insufficient to guide targeted therapy selection.
ACKNOWLEDGMENTS
We would like to acknowledge Darrell Borger, Nicholas Jessop, and members of the Massachusetts General Hospital Molecular Pathology Clinical Laboratory for performing FISH analyses, and Gataree Ngarmchamnanrith and the Amgen NCT01253707 study team.
GRANT SUPPORT This study is supported by grants from the NIH Gastrointestinal Cancer SPORE P50 CA127003, a Damon Runyon Clinical Investigator Award, NIH/NCI 1K08CA166510 (all to RBC); and a NIH/NCI Cancer Clinical Investigator Team Leadership Award supplement to P30CA006516 (to TSH).
Footnotes
Disclosure of Potential Conflicts of Interest: R.B. Corcoran is a consultant/advisory board member for Genentech, Merrimack Pharmaceuticals, and Avidity Nanomedicines.
REFERENCES
- 1.Howe HL, Wu X, Ries LA, Cokkinides V, Ahmed F, Jemal A, et al. Annual report to the nation on the status of cancer, 1975–2003, featuring cancer among U.S. Hispanic/Latino populations. Cancer. 2006;107:1711–42. doi: 10.1002/cncr.22193. [DOI] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. The New England journal of medicine. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. The New England journal of medicine. 2014;371:1963–71. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 6.Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janjigian YY, Tang LH, Coit DG, Kelsen DP, Francone TD, Weiser MR, et al. MET expression and amplification in patients with localized gastric cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20:1021–7. doi: 10.1158/1055-9965.EPI-10-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer metastasis reviews. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 9.Shah MA, Bang Y-J, Lordick F, Tabernero J, Chen M, Hack SP, et al. METGastric: A phase III study of onartuzumab plus mFOLFOX6 in patients with metastatic HER2-negative (HER2−) and MET-positive (MET+) adenocarcinoma of the stomach or gastroesophageal junction (GEC) J Clin Oncol 33. 2015;(suppl) abstr 4012. [Google Scholar]
- 10.Cunningham D, Al-Batran S-E, Davidenko I, Ilson DH, Murad AM, Tebbutt NC, et al. RILOMET-1: An international phase III multicenter, randomized, double-blind, placebo-controlled trial of rilotumumab plus epirubicin, cisplatin, and capecitabine (ECX) as first-line therapy in patients with advanced MET-positive gastric or gastroesophageal junction (G/GEJ) adenocarcinoma. J Clin Oncol 31. 2013;(suppl) abstr TPS4153. [Google Scholar]
- 11.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2316–21. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4803–10. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catenacci DV, Henderson L, Xiao SY, Patel P, Yauch RL, Hegde P, et al. Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer discovery. 2011;1:573–9. doi: 10.1158/2159-8290.CD-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak EL, LoRusso P, Hamid O, Janku F, Kittaneh M, Catenacci DVT, et al. Clinical activity of AMG 337, an oral MET kinase inhibitor, in adult patients (pts) with MET-amplified gastroesophageal junction (GEJ), gastric (G), or esophageal (E) cancer. J Clin Oncol 33. 2015;(suppl 3) abstr 1. [Google Scholar]
- 15.Hughes PE, Yang Y, Rex K, Zhang Y, Kaplan-Lefko PJ, Caenepeel S, et al. Abstract 728: AMG 337, a novel, potent and selective MET kinase inhibitor, has robust growth inhibitory activity in MET-dependent cancer models. Cancer research. 2014;74:728. doi: 10.1158/1535-7163.MCT-15-0871. [DOI] [PubMed] [Google Scholar]
- 16.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz LA, Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Fox C, Peng S, Pusung M, Pectasides E, Matthee E, et al. Preexisting oncogenic events impact trastuzumab sensitivity in ERBB2-amplified gastroesophageal adenocarcinoma. The Journal of clinical investigation. 2014;124:5145–58. doi: 10.1172/JCI75200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi J, McTigue MA, Rogers A, Lifshits E, Christensen JG, Janne PA, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer research. 2011;71:1081–91. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer discovery. 2014;4:650–61. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanizaki J, Okamoto I, Sakai K, Nakagawa K. Differential roles of trans-phosphorylated EGFR, HER2, HER3, and RET as heterodimerisation partners of MET in lung cancer with MET amplification. British journal of cancer. 2011;105:807–13. doi: 10.1038/bjc.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.