Abstract
Epigenetic regulators have emerged as critical factors governing the biology of cancer. Here, in the context of melanoma, we show that RNF2 is prognostic, exhibiting progression-correlated expression in human melanocytic neoplasms. Through a series of complementary gain-of-function and loss-of-function studies in mouse and human systems, we establish that RNF2 is oncogenic and pro-metastatic. Mechanistically, RNF2-mediated invasive behavior is dependent on its ability to mono-ubiquitinate H2AK119 at the promoter of LTBP2, resulting in silencing of this negative regulator of TGFβ signaling. In contrast, RNF2's oncogenic activity does not require its catalytic activity nor does it derive from its canonical gene repression function. Instead, RNF2 drives proliferation through direct transcriptional up-regulation of the cell cycle regulator CCND2. We further show that MEK1 mediated phosphorylation of RNF2 promotes recruitment of activating histone modifiers UTX and p300 to a subset of poised promoters, which activates gene expression. In summary, RNF2 regulates distinct biological processes in the genesis and progression of melanoma via different molecular mechanisms.
Keywords: RNF2, Polycomb, Melanoma, Metastasis, Epigenome, MEK1, Proliferation, Invasion
INTRODUCTION
Epigenetic factors offer important new targets for cancer therapy given their crucial role in the regulation of major cancer-relevant transcriptional programs and their potential reversibility (1). Significant effort has been directed towards identifying key epigenetic regulators in certain cancer contexts and elucidating the specific mechanisms, cell biological processes and surrogate transcriptional networks governed by these factors. However, we have limited understanding of the roles of epigenetic regulators in melanoma progression.
Melanoma is an aggressive cancer with escalating incidence worldwide (2). Melanoma deaths stem primarily from widespread metastatic disease (2), though genetic determinants and molecular mechanisms driving this disease remain poorly understood. Recent integrated genomic and functional screening efforts have identified pro-invasive determinants of melanoma metastasis with potential prognostic significance (3). The list of 18 prognostic determinants that emerged from this screen was identified based on evidence of pro-invasive and oncogenic capabilities in vitro and in vivo, in addition to genomic and expression alterations in human melanomas. On this list of 18, four were known epigenetic regulators: ASF1B (4), HMGB1 (5), RNF2 (6) and UCHL5 (7).
In this study, we focus on RNF2, a component of the polycomb repressor complex-1. RNF2 catalyzes mono-ubiquitination of lysine 119 of histone H2A (H2AK119Ub) (6, 8) and is overexpressed in gastrointestinal tumors, lymphomas and pancreatic cancers (9, 10). However, it is not known whether RNF2 overexpression is relevant functionally and, if so, what mechanisms, biological functions or transcriptional networks are governed by RNF2 in a cancer context. Here we elucidate the functional and biological roles of RNF2 in melanoma.
RESULTS
RNF2 is a prognostic metastasis oncogene in human melanoma
RNF2 was previously identified as a candidate pro-metastasis oncogene (3). Here, we set out to validate its pro-metastatic and oncogenic activities and discern functional mechanisms. Metastatic function was assessed in multiple melanoma cell models, including two primary immortalized human melanocyte lines constitutively expressing TERT, p53DD, CDK4R24C and either BRAFV600E or NRASG12D mutant proteins (11) (HMEL-BRAFV600E and pMEL-NRASG12D) and two established human melanoma cell lines WM115 and 1205Lu. Lentiviral transduction and overexpression of wild type RNF2 (hereafter RNF2WT) (Supplementary Figure 1A) promoted invasion in a Boyden chamber matrigel invasion assay in HMEL-BRAFV600E, WM115 and 1205Lu cells (Figure 1A and Supplementary Figure 1B). Similarly, RNF2WT enhanced metastatic ability as measured by spontaneous distant metastasis (lung/liver/lymph node) in nude mice with tumor burden of 1.5cm following intradermal injection of transduced WM115, 1205Lu and pMEL-NRASG12D cells (Figure 1B).
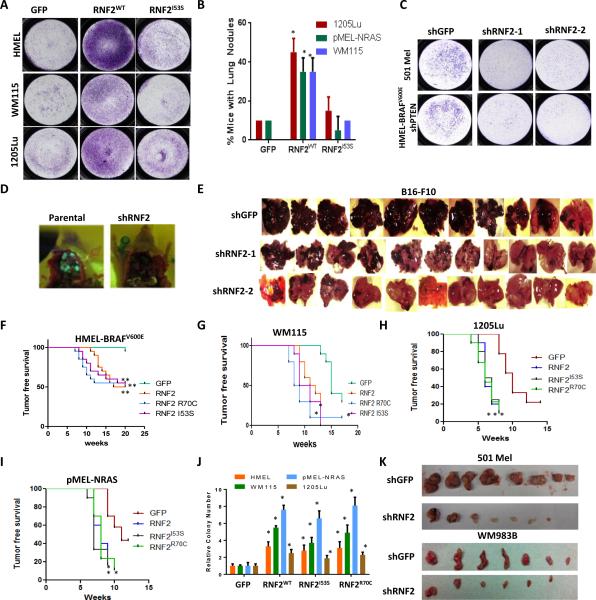
Figure 1. RNF2 overexpression promotes invasion and metastasis in catalytic activity dependent manner.
(A) RNF2 overexpression promotes invasion in multiple melanocytic and melanoma-derived cell lines in catalytic activity dependent manner. GFP, RNF2WT or RNF2I53S were overexpressed by lentiviral transduction in HMELBRAFV600E (primary melanocytes), WM115 and 1205Lu cells and invasion capacity measured using Boyden Chamber matrigel invasion assay. Representative image of invasive cells is shown. pMEL-NRASG12D cells were not tested in invasion assay due to high background. (B) RNF2 overexpression promotes metastasis. Percentage of mice with lung nodules (at the time of euthanasia due to tumor burden) is shown in the graph. HMEL-BRAFV600E cells were not used in the metastasis assay due to high latency. (* denotes significant change t-test p < 0.05). (C) 501Mel and HMEL-BRAF-shPTEN cells with stably integrated shGFP, shRNF2-1, shRNF2-2 were subjected to Boyden chamber matrigel invasion assay. Representative images of invaded cells are shown. (D) Representative image showing lung seeding of HMEL-BRAFV600E-shPTEN cells alone or with shRNF2. Cells are labeled with GFP and hence the lung seeding noted by green nodules in the lung. (E) B16-F10 mouse cells with stably integrated shGFP, shRNF2-1 or shRNF2-2 were injected intravenously in C57BL/6 mice. Mice were sacrificed after 16 days and lung seeding noted by color of black melanocytes in lung. (F-I) Kaplan-Meier curve showing tumor free survival of mice following intradermal injection of (F) HMEL-BRAFV600E cells, (G) WM115 cells, (H) 1205Lu cells and (I) pMEL-NRASG12D overexpressing GFP, RNF2 wild type or catalytic mutant derivative (R70C or I53S). Mantel-Cox p values for graph comparisons between GFP and individual RNF2 derivatives are as follows: HMEL-BRAFV600E = p<0.005; WM115 = p < 0.01; 1205Lu = p< 0.01; pMEL-NRASG12D = p<0.01. Asterisk (*) represents p < 0.01 and double asterisk (**) represents p < 0.005. (J) Graph showing relative colony number from a soft-agar colony formation assay in HMEL-BRAFV600E, pMEL-NRASG12D, WM115 cells and 1205Lu cells overexpressing GFP, RNF2 wild type or catalytic mutant derivative (R70C or I53S). Asterisk (*) denotes significant change t-test p < 0.05. (K) 501Mel or WM983B cells stably expressing shGFP or shRNF2 were subjected to tumor formation assay by intradermal injection in immunodeficient mice. Image shows subcutaneous tumors after 8 weeks post injection.
To complement this approach, loss-of-function studies in the highly invasive human melanoma cell lines 501Mel (harboring high levels of RNF2, Supplementary Figure 1C) and engineered HMEL-BRAFV600E melanocyte with stable shRNA targeting PTEN (HMEL-BRAFV600E-shPTEN) showed significant reduction in invasive potential in vitro upon RNF2 knockdown with two independent shRNAs (Figure 1C and Supplementary Figures 1D-E). Since pro-invasive properties are critical for seeding to distant organs during metastasis (12), we tested if RNF2 was required for seeding to distant organs. Indeed, RNF2 silencing in HMEL-BRAFV600E-shPTEN cells reduced lung seeding potential (Figure 1D and Supplementary Figure 1F). Furthermore, in an immunocompetent C57BL/6 host, knockdown of RNF2 in highly invasive B16-F10 cells similarly reduced lung seeding (Figure 1E and Supplementary Figure 1G).
Next, to explore RNF2's role as an oncogene, we assessed tumor formation following intradermal injection of RNF2WT overexpressing HMEL-BRAFV600E and pMEL-NRASG12D melanocytes as well as WM115 and 1205Lu melanoma cells. RNF2WT significantly increased tumorigenic potential compared to control (Figures 1F-I and Supplementary Figures 2A-D) in all four cell-lines tested. Similar activity of RNF2WT was observed in cell-based soft agar colony formation assay, a surrogate for tumorigenesis (Figure 1J). Reciprocally, shRNA-mediated knockdown of RNF2 in highly tumorigenic 501Mel and WM983B cells, which express high levels of RNF2 (Supplementary Figure 1C), resulted in significant reduction in tumor burden (Figure 1K and Supplementary Figures 2E-G). Consistently, proliferation defect was seen in 501Mel, HMEL-BRAFV600E-shPTEN and B16-F10 cells upon RNF2 knockdown (Supplementary Figures 2H-J).
To substantiate the relevance of RNF2 in human melanoma, we verified that RNF2 expression correlates with disease progression at the mRNA and protein levels. Specifically, as summarized in Supplementary Figure 3A, RNF2 mRNA expression was elevated in primary melanoma tissue compared to skin and nevi (13) and, in an independent cohort, was significantly higher in metastatic lesions when compared to localized primary tumors (Supplementary Figure 3B). Correspondingly, TMA (Tissue Microarray) analysis verified progression-correlated expression across 480 cores derived from 170 patients (132 benign nevi cores from 36 patients), 196 primary melanoma cores derived from 59 patients, 60 lymph node metastasis cores derived from 29 patients, and 92 visceral metastasis cores derived from 46 patients (Figure 2A and Supplementary Figure 3C). Overall, RNF2 expression was low in normal skin cells, including melanocytes, and progressively increased from nevi to primary to lymph node metastases.
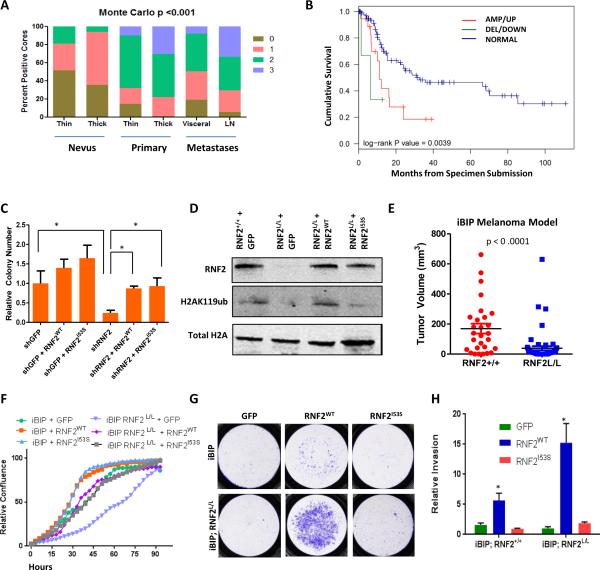
Figure 2. RNF2 promotes tumorigenesis in catalytic activity independent manner.
(A) Bar plot showing distribution of RNF2 immunoreactive intensity counts (0,1,2,3) in nevi (thin and thick), primary (thin and thick) and metastasis (visceral and lymph node). (B) Kaplan-Meier curve showing cumulative survival of three groups of patients defined by copy number change and expression in a TCGA cohort with available survival data (108): amplified/upregulated (AMP/UP, 12/18, Red), deleted/downregulated (DEL/DOWN, 2/4, Green) and no copy number/expression change (‘Normal’, 44/104, Blue). (C) Graph shows relative number of soft agar colonies in WM983B cells rescues with GFP, RNF2 wild type or catalytic mutant derivative (R70C or I53S) (* denotes significant change t-test p < 0.05). (D) Western blot showing levels of Rnf2, H2AK119ub and total H2A in iBIP mice tumor cells with (Rnf2+/+) or without RNF2 (Rnf2L/L) overexpressing GFP, RNF2WT and RNF2I53S. (E) Scatter plot showing ear tumor volume in iBIP mice with iBIP;RNF2+/+ or iBIP;RNF2L/L genotype after doxycycline (2mg/ml) administration and treatment with 4-hydroxytamoxifen (1μM). t-test p<0.0001. (F-H) (F) Proliferation assay, (G) Invasion assay images and (H) invasion assay quantitation in iBIP mice tumor cells with (Rnf2+/+) or without RNF2 (Rnf2L/L) overexpressing GFP, RNF2WT and RNF2I53S. Asterisk denotes significant change t-test p < 0.05.
Leveraging the clinically annotated multi-dimensional dataset on melanoma generated by The Cancer Genome Atlas (TCGA) Network (14 2013-04-06), we investigated the relationship between RNF2 copy number and expression correlation with cumulative overall survival. Of the 268 samples with copy number and expression data, we found copy number gains of RNF2 in 42 (15.7%, defined by segmented copy number value greater than 0.5), copy number loss in 6 samples (2.2%, defined by copy number value less than 0.5) and overexpression of RNF2 in 13 of 268 tumors (4.9%, defined by normalized expression z scores greater than 2). Overall, 44 tumors showed copy number gain or overexpression of RNF2 with overlap of 11 samples (p = 2.5e-8, fisher's exact test), whereas 218 tumors showed neither copy number change nor expression difference (hereafter referred to as “RNF2 normal”). Further we found that amplification/overexpression of RNF2 significantly co-occurred with NRAS mutations (Odds ratio = 3.2, p = 0.00077) and was significantly mutually exclusive with BRAF mutations (Odds Ratio=0.37, P=0.0046). Survival intervals from date of specimen submission to patients’ death or last follow-up were available in 154 cases. Among these 154 cases, we found that, indeed, elevated RNF2 levels were associated with poorer overall survival (log-rank P value < 0.0039, Figure 2B), confirming the prognostic significance of RNF2 in melanoma.
RNF2 has both catalytic dependent and independent activities
Given RNF2's known transcriptional repressor and catalytic activities, we sought to determine whether RNF2's catalytic activity is required for its pro-invasion and protumorigenic phenotypes. Mutant forms of RNF2: RNF2I53S and RNF2R70C, shown previously to lack catalytic activity (15, 16), were engineered. We found that as expected, these mutants showed diminished invasion and metastasis activity compared to RNF2WT (Figure 1A-B and Supplementary Figures 1A-B). However, to our surprise, both RNF2I53S and RNF2R70C mutants retained the capacity to enhance proliferation and anchorage independent growth in vitro, and tumorigenicity in vivo, at levels comparable to RNF2WT in all 4 melanoma/melanocytic cell models (Figures 1F-J and Supplementary Figures 2A-D, 3D). This observation suggested that RNF2's pro-tumorigenic potential does not require its catalytic activity. To verify this, we performed rescue experiments with vectors encoding the open reading frames of wild type and catalytic mutants of RNF2 in WM983B cells wherein RNF2 was silenced with a 3’ UTR directed shRNA. Consistent with the overexpression data in HMEL-BRAFV600E cells, RNF2WT and RNF2I53S expression were similarly able to restore soft agar colony formation ability (Figure 2C and Supplementary Figure 3E).
To address the possible confounding effect of endogenous RNF2 expression in the above study, we engineered a mouse line bearing a conditional RNF2 knockout allele with LoxP sites flanking exon 2 (Supplementary Figure 3F), where Cre-mediated recombination results in loss of RNF2 protein expression (Figure 2D). The RNF2L/L allele was introduced into an inducible melanoma model called iBIP [inducible Braf Ink/Arf Pten] which harbors the following alleles: Ink4a/Arf−/−, Tyr-CreERT2, Rosa26-LoxP-Stop-LoxP-Rtta, TetO-BrafV600E, and PtenL/L (17). The iBIP mouse model allows temporal and spatial control of tumor development and growth through melanocyte-specific, doxycycline-dependent BrafV600E activation, restricted to the same cells as those undergoing 4-hydroxytamoxifen (OHT)-dependent Pten deletion in the Ink4a/Arf germline knockout background. Comparison of melanoma tumor burden following topical 4-OHT application in littermate iBIP;RNF2+/+ and iBIP;RNF2L/L mice showed that RNF2 deficiency was associated with significant reduction in tumor burden at 14 weeks and improved survival (Figure 2E and Supplementary Figure 3G).
Using this genetic system in which RNF2 can be rendered homozygous null, we re-assessed the differential requirement of RNF2 catalytic activity in cellular proliferation and invasion. Specifically, melanoma cells derived from iBIP-RNF2L/L animals were transduced with lentivirus encoding RNF2WT and RNF2I53S (Figure 2D) and assayed for proliferation and invasion along with the levels of H2AK119 mark. Consistent with studies above, RNF2 catalytic activity was dispensable for proliferation enhancement yet required for invasion (Figure 2F-H). Taken together, these in vitro and in vivo functional assays suggested that, unlike its metastatic function, RNF2's oncogenic potential is not dependent on its catalytic activity.
RNF2 promotes TGFβ signaling via down-regulation of LTBP2
To explore the mechanistic basis of RNF2's cancer relevant activities, transcriptome profiling (Supplementary Figure 4A) and ChIP-seq (Chromatin Immunoprecipitation followed by deep sequencing, performed using V5-antibody) occupancy profiling were performed in HMEL-BRAFV600E melanocytes with enforced expression of RNF2WT (hereafter HMEL-BRAFV600E-RNF2WT). These RNF2 ChIP-Sequencing studies were also conducted in primary tumor cells derived from HMEL-BRAFV600E-RNF2WT melanocytes. ChIP-Sequencing data analysis showed RNF2-occupied loci exhibited significantly higher enrichment of RNF2 compared to input (Supplementary Figure 4B) and were evolutionary conserved among 44-species (Supplementary Figure 4C). Analyses of the distribution of RNF2 occupancy sites in relation to transcription start sites (TSS) revealed 3,465 genes in +/−5Kb vicinity of the RNF2 occupied loci in HMEL-BRAFV600E-RNF2WT melanocytes (Supplementary Figures 4D-E and Supplementary Table 1). Overlap of expression and occupancy datasets showed that 363 genes, whose promoters were occupied by RNF2 in HMEL-BRAFV600E-RNF2WT melanocytes, exhibited altered gene expression upon RNF2WT overexpression compared to GFP in HMEL-BRAFV600E cells (Figure 3A and Supplementary Table 2). While 47% of these genes with RNF2 occupancy were found to have decreased expression (compared to GFP) consistent with the classical repressive function of the RNF2-polycomb complex, it is worth noting that 53% of RNF2-occupied genes showed increased expression pointing to a likely role for RNF2 in transcriptional activation (see discussion) (Figure 3A).
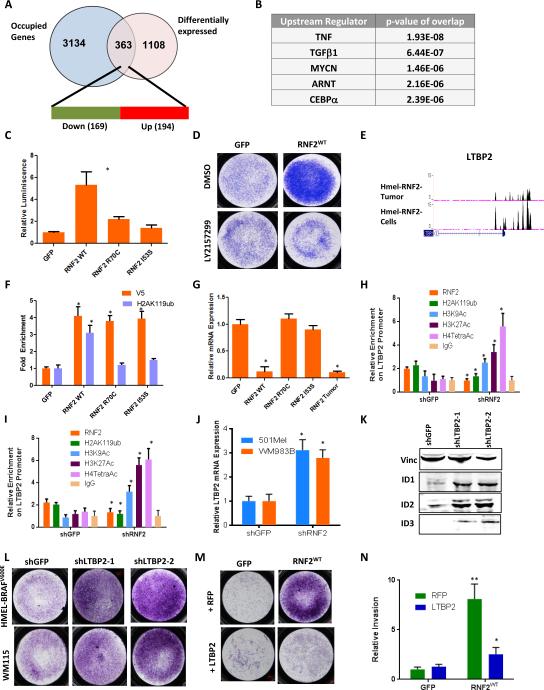
Figure 3. RNF2 promotes TGFβ signaling.
(A) Overlap of genes with corresponding promoters occupied by RNF2 (using ChIP-Seq data) and differentially expressed genes. 363 genes show overlap of which 169 (47%, green) are downregulated and 194 (53%, red) are upregulated. (B) Top 5 pathways from upstream regulating factor enrichment by IPA (Ingenuity Pathway Analysis). Note that TGFβ target genes were one of the most significantly deregulated and occupied genes. (C) Luciferase assay showing increased TGFβ responsive promoter activity in HEK293 cells with overexpression of RNF2WT, but not with RNF2R70C and RNF2I53S. (D) Representative image from a Boyden chamber matrigel invasion experiment in HMEL-BRAFV600E cells overexpressing GFP or RNF2WT and treated with DMSO or LY2157299 (TGFβRI Inhibitor). Invaded cells stained with Crystal Violet are shown. (E) Occupancy of RNF2 on the LTBP2 promoter. Two ChIP-Seq tracks are shown: top: HMEL-BRAFV600E-RNF2WT tumor cells; bottom: HMELBRAFV600E-RNF2WT cells. (F-G) (F) qPCR validation showing enrichment of V5-RNF2 (V5 antibody) and H2AK119ub on LTBP2 promoter and (G) mRNA expression of LTBP2 in HMEL-BRAFV600E cells overexpressing GFP, RNF2 wild type or catalytic mutant (R70C and I53S) derivative. (H-I) Graph shows relative occupancy enrichment of RNF2 (endogenous), H2AK119ub, H3K9Ac, H3K27Ac, H4TetraAc and IgG on LTBP2 promoter as obtained by ChIP-qPCR in shGFP or shRNF2 infected 501Mel (H) and WM983B (I) cells. (J) Relative mRNA expression of LTBP2 in shGFP or shRNF2 infected 501Mel or WM983B cells. (K) Western blot showing protein levels of TGFβ target genes ID1, ID2 and ID3 in HMEL-BRAFV600E cells with knockdown of LTBP2 using two shRNAs. (L-M) Representative image of invaded cells from a triplicate Boyden chamber matrigel invasion experiment in (L) HMEL-BRAFV600E or WM115 cells with knockdown of LTBP2 using two shRNAs and (M) HMEL-BRAFV600E cells overexpressing GFP or RNF2WT along with either RFP or LTBP2. (N) Graph showing quantitation of experiment shown in panel (M). Across all panels “*” denotes significant change t-test p < 0.05 and “**” represents p value <0.01.
Pathway enrichment analysis of RNF2-occupied genes with increased expression showed enrichment in proliferation pathways, in addition to nucleotide synthesis and hypoxia pathways (Supplementary Figure 4F, top 5 pathways shown), while RNF2-occupied genes with decreased expression are associated with regulation of transcription and nucleotide synthesis (Supplementary Figure 4G, top 5 pathways shown). Among the RNF2-occupied genes exhibiting the most robust alterations in expression were those linked to TGFβ signaling (Figure 3B), in line with the known role of TGFβ in invasion and metastasis (18). Thus, we next sought to determine whether RNF2 could modulate TGFβ pathway activation. First, we showed that indeed, overexpression of RNF2WT, but not RNF2I53S, enhanced luciferase reporter activity driven by a generic TGFβ-responsive promoter in HEK293 cells (Figure 3C and Supplementary Figure 4H) and drove increased expression of TGFβ target genes (ID1, ID2 and ID3) in HMEL-BRAFV600E melanocytes (Supplementary Figure 4I). Consistent with a functional role of RNF2-driven TGFβ pathway activation in invasion, treatment of RNF2 overexpressing cells with an inhibitor of TGFβ pathway (LY2157299, (19)) resulted in reduced invasion in Boyden chamber matrigel invasion assays (Figure 3D).
To identify candidate direct targets of RNF2 that govern TGFβ pathway activation, gene expression and promoter occupancy profiles were overlaid to define 363 genes (Figure 3A); among which one of the most significantly changed genes was LTBP2 (Figure 3E and Supplementary Table 2), a member of the Latent TGFβ binding family of proteins that resides in the extracellular matrix and regulates bioavailability of TGFβ ligand (20) to positively or negatively influence TGFβ signaling (21). This finding gains added significance since LTBP2 is down-regulated upon RNF2 overexpression and has been shown previously to inhibit the migration capacity of human melanoma cells (22). Thus, we next performed ChIP-qPCR to examine the LTBP2 promoter for occupancy by RNF2 in accordance with histone H2AK119ub modification. As shown in Figure 3F, while the LTBP2 promoter was occupied by RNF2 in RNF2WT, RNF2R70C and RNF2I53S expressing HMEL-BRAFV600E melanocytes, the H2AK119ub mark was only observed in RNF2WT, not RNF2R70C or RNF2I53S, expressing cells. In other words, the catalytic dead RNF2 was defective in catalyzing H2AK119ub, and RNF2 enzymatic activity is not required for RNF2 binding at the promoter of LTBP2. Consistent with RNF2 catalytic activity dependent repression, quantitative RT-PCR confirmed down-regulation of LTBP2 mRNA only in RNF2WT, but not RNF2R70C or RNF2I53S, expressing cells (Figure 3G). This was also validated in human melanoma cell lines 501Mel and WM983B where we noted RNF2 occupancy in parental cells and loss of H2AK119ub signal in 501Mel and WM983B cells upon RNF2 knockdown (Figures 3H-I). Consistently, activating histone acetylation marks were enriched on LTBP2 promoter (Figures 3H-I) and its mRNA expression was increased upon RNF2 knockdown (Figure 3J). Additionally, LTBP2 mediated modulation of TGFβ signaling is supported by the correlation of LTBP2 knockdown with up-regulation of TGFβ targets genes ID1, ID2, and ID3 (Figure 3K and Supplementary Figure 4J), as well as with enhanced invasion activity in vitro (Figure 3L). Finally, the functional epistatic link between RNF2 and LTBP2 is supported by the demonstration that LTBP2 overexpression partially inhibited the invasive activity of RNF2WT overexpressing melanocytes (Figure 3M-N).
RNF2 promotes tumorigenesis through up-regulation of CyclinD2
As noted above, many genes proximal to RNF2 occupancy sites in HMEL-RNF2WT melanocytes showed increased expression (Figure 3A). Indeed, the most significantly up-regulated and occupied gene was CCND2, which encodes the cell cycle regulator CyclinD2 (Figure 4A and Supplementary Table 2). ChIP-qPCR confirmed RNF2 occupancy at the CCND2 promoter in HMEL-BRAFV600E cells overexpressing RNF2WT, RNF2R70C and RNF2I53S (Figure 4B). Additionally, CCND2 expression was induced by ectopic RNF2 (wildtype or catalytic dead) and remained high in HMEL-BRAFV600E cells overexpressing RNF2WT, RNF2R70C and RNF2I53S (Figure 4C), suggesting that transcriptional activation did not require catalytic activity and histone H2AK119 ubiquitination. Indeed, no enrichment of the H2AK119ub mark was detected on the CCND2 promoter (Figure 4B), which instead possessed activating chromatin modifications, including H3K9Ac, H3K27Ac, H4TetraAc and H3K4me3, in HMEL-BRAFV600E cells overexpressing both wild type and catalytic mutants of RNF2 (Figure 4B). Accordingly, RNF2 knockdown caused repression of CCND2 expression (Figure 4D) and removal of activation marks in WM983B and 501Mel cells (Figure 4E-F).
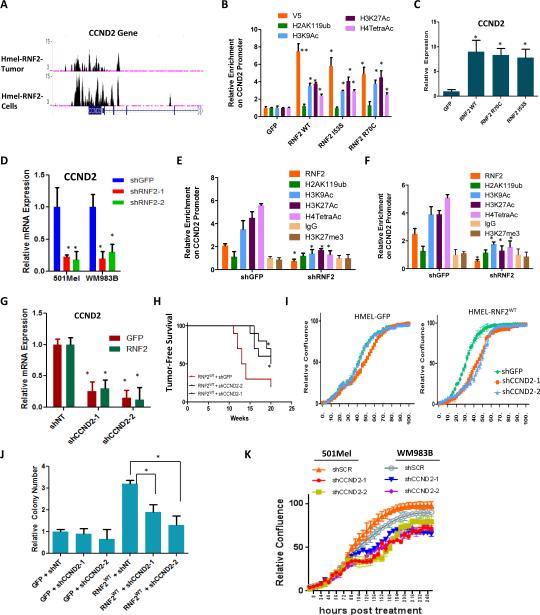
Figure 4. Oncogenic activity of RNF2 depends on upregulation of CCND2.
(A) Occupancy of RNF2 on the promoter of CCND2. Two ChIP-Seq tracks are shown: top: HMEL-BRAFV600E-RNF2WT tumor cells; bottom: HMEL-BRAFV600E-RNF2WT cells. (B) Graph shows relative occupancy enrichment of V5-RNF2 (using V5 antibody), H2AK119ub, H3K9Ac, H3K27Ac and H4TetraAc on CCND2 promoter as obtained by ChIP-qPCR in GFP or RNF2WT or RNF2I53S or RNF2R70C overexpressing HMELBRAFV600E cells. (C) Graph shows relative CCND2 expression in HMEL-BRAFV600E cells overexpressing GFP, RNF2 wild type or catalytic mutant derivative (R70C or I53S). Values were normalized to GFP cells as 1. (D) Graph showing mRNA expression levels of CCND2 in 501Mel and WM983B cells with RNF2 knockdown. (E-F) Graph shows relative occupancy enrichment of RNF2(endogenous), H2AK119ub, H3K9Ac, H3K27Ac, H4TetraAc, H3K27me3 and IgG on CCND2 promoter as obtained by ChIP-qPCR in shGFP or shRNF2 infected 501Mel (E) and WM983B (F) cells. (G) Graph shows mRNA expression of CCND2 in HMEL-BRAFV600E cells with GFP or RNF2WT overexpression with two stably integrated CCND2 shRNAs. (H-J) Assays for tumorigenicity in RNF2WT overexpressing HMEL-BRAFV600E cells with CCND2 knockdown (two shRNAs). (H) Kaplan-Meier curve showing tumor free survival (Mantel Cox p < 0.05), (I) relative cell density from in vitro proliferation assay and (J) soft agar colony counts (K) Proliferation curves for 501Mel and WM983B cells infected with shRNAs for GFP (shGFP) or CCND2 (shCCND2-1 and shCCND2-2). Across all panels “*” denotes significant change t-test p < 0.05 and “**” represents p value <0.01.
To assess the potential role of RNF2-directed CCND2 up-regulation in promoting increased proliferation and tumorigenesis, shRNA-mediated knockdown of CCND2 was performed in HMEL-BRAFV600E-RNF2WT overexpressing cells (Figure 4G). As shown in Figure 4H-J, in vivo tumor formation (Figure 4H), enhancement in 2D proliferative capacity (Figure 4I) and 3D anchorage independent growth (Figure 4J) conferred by RNF2WT overexpression were partially reversed upon CCND2 knockdown, suggesting that CCND2 contributes to pro-oncogenic activities of RNF2. Consistently, knockdown of CCND2 reduced proliferative capacity of 501Mel and WM983B cells, which express high levels of RNF2 (Figures 4K).
Pre-existing chromatin promoter states determine the genes activated by RNF2
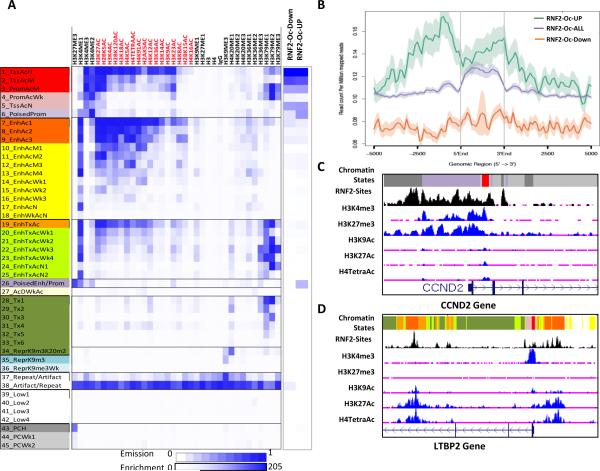
Next we sought to understand how RNF2 might promote gene activation contrary to its known role in gene repression. We considered the possibility that the transcriptional fate of genes regulated by RNF2 might depend on the chromatin states that pre-existed on their promoters before upregulation of RNF2. To identify these preexisting chromatin states of RNF2 regulated genes in the melanocytes before RNF2 overexpression, we performed epigenomic analyses for 35 histone marks in the HMEL-BRAFV600E cell system that was used in the RNF2 gain-of-function experiments (Rai et al, unpublished). There, we modeled histone modification profiles as 45 defined chromatin states using the ChromHMM modeling method (23) (See Methods) (Figure 5A) which captures important biological states such as poised or bivalent promoter/enhancer states (State 26 and State 6) (24). Each of these chromatin states were annotated based on the enrichment of different histone marks as well as the enrichment of known genomic elements (Figure 5A, Supplementary Figures 5A-B and Supplementary Table 3). We overlapped RNF2 binding sites to these chromatin states and found that, although all RNF2 binding sites in the genome overlapped with a number of states, the sites that were associated with genes showing altered expression were limited to promoter and poised states (Figure 5A). Interestingly, we noted that promoters of the genes upregulated by RNF2, including CCND2, were specifically enriched in State 26 while RNF2-downregulated promoters were markedly absent (Figure 5A). These downregulated promoters displayed only active promoter states (States1-3, 5). Therefore, we compared the cumulative presence of H3K27me3 marks on all, upregulated and downregulated RNF2-bound promoters. As shown in Figure 5B, H3K27me3 was significantly enriched at promoters that are up-regulated by RNF2, compared to promoters of genes destined for repression by RNF2 which lack enrichment of this mark. Consistent with this, a UCSC genome browser view of the CCND2 promoter showed prominent peaks of H3K27me3 and H3K4me3, characteristic of State 26 (Poised Enhancer/Promoter) and State 6 (Poised Promoter), around RNF2 binding sites immediately upstream of the TSS (Figure 5C). In contrast, analysis of the LTBP2 promoter, which is repressed when RNF2 is expressed, showed enrichment of active promoter states (States 1, 2) and active promoter marks (H3K4me3, H3K9Ac) as well as enhancer states (States 7,8,9) and enhancer (H3K27Ac) marks (Figure 5D). These data suggest that genes activated by RNF2 may be marked, or poised, by repression-associated mark H3K27me3 prior to RNF2-mediated activation and gain of histone acetylation marks.
Figure 5. RNF2 activated genes harbor H3K27me3 poised chromatin state.
(A) Overlap of RNF2 binding sites, upregulated and downregulated promoters with 45-State model predicted by ChromHMM of occupancy of 35-histone marks in HMEL-BRAFV600E cells (data described in Rai et al unpublished). X-axis shows histone modification antibodies used for modeling; Y-axis shows chromatin states and description of each state (also in Supplementary Table 3). Blue is enrichment. Scale is shown on the bottom. (B) Graph showing enrichment of H3K27me3 on all genes (RNF2-Oc-ALL), upregulated genes (RNF2-Oc-UP) and downregulated (RNF2-Oc-Down) containing RNF2 binding sites in their promoters. 5’ end, 3’ end and the distance from TSS is shown on the x-axis. Shadow represents standard error of mean. (C-D) UCSC genome browser view of CCND2 promoter (C) and LTBP2 promoter (D) showing chromatin state enrichment as well as RNF2 binding sites.
RNF2 recruits UTX and p300 to the CCND2 promoter
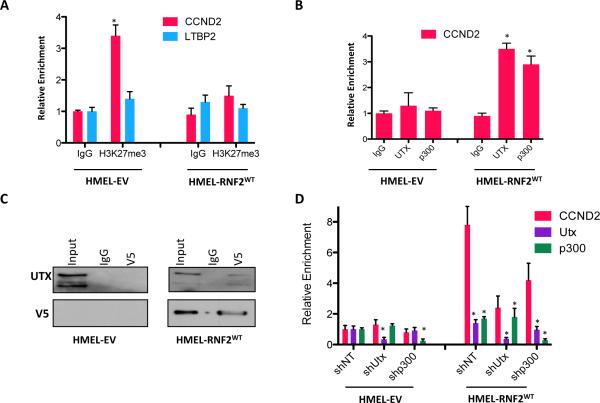
Recently, MLL2, UTX and p400 were identified as RNF2 associated proteins in mouse ES cells, which co-migrate on a sucrose gradient separately from RNF2-containing PRC1 components (25). This observation suggests that a fraction of RNF2 molecules may exist in an activating complex with MLL2, UTX or p400. Therefore, we hypothesized that a sub-fraction of RNF2 may preferentially recruit activating factors to the H3K27me3 containing poised promoters. To investigate this, we first tested whether RNF2 overexpression led to H3K27me3 loss on activated promoters. Indeed, RNF2 overexpression led to loss of H3K27me3 occupancy as well as gain of histone acetylation marks (H3K9Ac, H3K27Ac and H4TetraAc) on CCND2 promoter (Figure 6A and Figure 4B). These histone modification events upon RNF2 overexpression were consistent with RNF2's suggested interaction with UTX, an H3K27 demethylase and p300, a histone acetyltransferase (25). Indeed, ChIP-qPCR showed that UTX and p300 were enriched on CCND2 promoter after RNF2 overexpression (Figure 6B). Consistent with these observations, we noted interactions between RNF2 and UTX by coimmunoprecipitation in HMEL-BRAFV600E-RNF2WT cells (Figure 6C). Finally, we tested whether UTX and p300 recruitment by RNF2 had any impact on transcriptional activation of CCND2 promoter. Downregulation of UTX or p300 individually by shRNAs significantly reduced CCND2 expression in RNF2 overexpressing cells but not in control cells (Figure 6D). Together these observations suggest that recruitment of UTX and p300 to CCND2 promoter by RNF2 is critical for creating activating chromatin environment as well as transcriptional activation.
Figure 6. RNF2 recruits and requires UTX and p300 for CCND2 activation.
(A) Relative occupancy of H3K27me3 on CCND2 and LTBP2 promoters in HMELBRAFV600E-EV and HMEL-BRAFV600E-RNF2WT cells. (B) Relative occupancy of UTX and p300 on CCND2 promoter. (C) Western blot showing co-immunoprecipitation of UTX upon immunoprecipitation of RNF2 (using anti-V5) from HMEL-BRAFV600E-EV and HMEL-BRAFV600E-RNF2WT cells. (D) Relative expression of CCND2, UTX and p300 in HMEL-BRAFV600E-EV and HMEL-BRAFV600E-RNF2WT cells upon control (shNT), Utx (shUtx) and p300 (shp300) knockdown. Asterisk denotes significant change t-test p < 0.05.
MEK mediated phosphorylation of RNF2
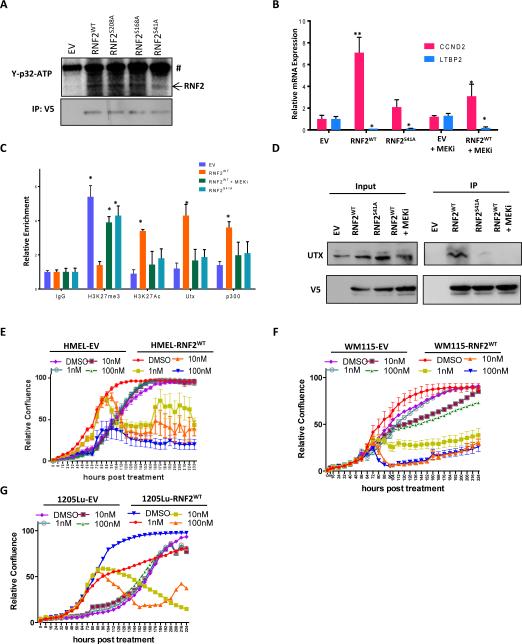
To understand how RNF2 might act as both an activator and a repressor in the same cell, we asked whether a particular modified form of RNF2 is important for gene activation. It was previously shown that RNF2 is phosphorylated in a MEK dependent manner and this phosphorylation may be associated with histone acetylation events (26). Since RNF2 overexpression was studied in the context of activated MAPK signaling (due to BRAFV600E mutation), which is known to activate MEK, we asked whether phosphorylation of RNF2 by MEK may be important for its role in gene activation in the context of melanoma. We first verified that MEK1 is indeed able to phosphorylate RNF2 using an in vitro kinase assay (Figure 7A). Moreover, serine 41 (26) to alanine mutant derivative of RNF2 showed significantly reduced phosphorylation compared to wild type whereas S168A and S208A mutant derivatives were phosphorylated to the same extent as wild type RNF2. Further, treatment of RNF2WT overexpressing HMEL-BRAFV600E cells with MEK inhibitor Trametinib led to a significant reduction in CCND2 gene activation by RNF2WT; whereas, LTBP2 expression remained unchanged and overexpression of RNF2S41A failed to activate CCND2 promoter (Figure 7B). Consistently, RNF2 induced H3K27me3 demethylation, H3K27Ac accumulation and UTX/p300 recruitment at the CCND2 promoter, which were abrogated by MEK inhibition in RNF2WT overexpressing cells (Figure 7C). In parallel, S41A mutant was inefficient in promoting H3K27me3 demethylation, inducing H3K27Ac and UTX/p300 recruitment (Figure 7C). Consistently, MEK inhibition and S41A mutant drastically reduced interaction between RNF2 and UTX (Figure 7D). Finally, we showed that MEK inhibition selectively reduces increased proliferation conferred by RNF2 overexpression in HMEL-BRAFV600E, WM115 and 1205Lu cells (Figure 7E-G), suggesting a therapeutic strategy to suppress RNF2 mediated tumorigenesis.
Figure 7. MEK dependent phosphorylation of RNF2 at Serine 41 is required for recruitment of UTX and p300 to CCND2 promoter.
(A) Western blot image showing γ-p32-ATP signal from in vitro kinase assay (upper panel) performed using purified MEK1 kinase and immunoprecipitated GFP/RNF2WT/RNF2S41A proteins from HEK293 cells (loading control in bottom panel) as substrate. # denotes non-specific band. (B) Relative mRNA expression of CCND2 and LTBP2 upon MEKi (Trametinib, 5nM) in HMEL-BRAFV600E-EV and HMEL-BRAFV600E-RNF2WT cells or HMEL-BRAFV600E-RNF2S41A cells. (C) Relative occupancy of UTX and p300 on CCND2 promoter in untreated or MEKi (Trametinib, 5nM) treated HMEL-BRAFV600E-EV and HMEL-BRAFV600E-RNF2WT cells or HMEL-BRAFV600E-RNF2S41A cells. (D) Co-immunoprecipitation of UTX with RNF2 (anti-V5) in untreated or MEKi (Trametinib, 5nM) treated HMEL-BRAFV600E-EV and HMEL-BRAFV600E-RNF2WT cells or HMEL-BRAFV600E-RNF2S41A cells. (E-G) Proliferation curves for EV and RNF2WT expressing HMEL-BRAFV600E, WM115 and 1205Lu cells after treatment with DMSO or MEKi (Trametinib at 1nM, 10nM and 100nM). Across all panels “*” denotes significant change t-test p < 0.05 and “**” represents p value <0.01.
Together, these data support a model wherein MEK mediated RNF2 phosphorylation may induce its interaction with histone modifiers, such as UTX and p300, and their recruitment to poised H3K27me3 containing promoter. This recruitment, and subsequent loss of H3K27me3 with gain of activating histone marks, selectively creates an activating environment on gene promoters that exist in a poised state.
DISCUSSION
In this study, we elucidated distinct molecular mechanisms by which RNF2 regulates proliferation and invasion, highlighting the complex and multi-faceted action of epigenetic regulators. Molecularly, the intersection of RNF2 chromatin binding and gene expression analyses identified RNF2-occupied repressed and active promoters. Biologically, a series of reinforcing functional assays utilizing both somatic and genetically engineered germline model systems demonstrated that RNF2's catalytic activity is dispensable for CCND2 activation, which drives proliferation, but is required for suppression of LTBP2 and activation of TGFβ signaling for invasion and metastasis.
Although it has been suggested that RNF2 may promote gene repression by chromatin compaction independently of its catalytic activity (27), this is the first report of RNF2's role in gene activation independent of its E3 ubiquitin ligase activity. In this regard, we find that ~53% of genes with RNF2-occupied promoters were upregulated in RNF2 overexpressing melanocytes. As with other transcription factors, an intriguing question is how RNF2 might act as both an activator and repressor in the same cell type. A subset of genes activated by RNF2 in this study have poised promoters strongly enriched in H3K27me3 mark as well as showing weak enrichment of activating histone acetylation and methylation marks. We provide mechanistic insights that MEK1 dependent phosphorylation of RNF2 may promote its binding to activating chromatin modifiers such as UTX and p300, which in turn remove H3K27me3 and acetylate the promoter respectively to open chromatin for gene activation. MEK-dependent phosphorylation of RNF2 provides precedence for a mechanism that signaling proteins may utilize the same molecule to effect gene-specific outcomes in a context-dependent manner. Finally, our data also suggest that MEK inhibitors could be used to block RNF2's pro-tumorigenic function and therefore could be potentially beneficial in clinic to suppress growth of RNF2 amplified tumors.
TGFβ signaling has been shown to be critical for induction of pro-invasion and migration genes such as MMPs, N-Cadherin, Vimentin and Fibronectin. Here we identified RNF2 as an important epigenetic regulator of TGFβ signaling. Promoter occupancy and expression analyses in this study revealed that RNF2 can directly bind to the LTBP2 promoter to create a repressive environment through H2AK119 ubiquitination and consequent gene silencing. Although LTBP proteins have been reported to both negatively and positively regulate TGFβ signaling (20), our study suggests that, in melanoma, LTBP2 acts as a negative regulator of TGFβ signaling in invasion. We noted that apart from LTBP2, mRNA expression of EMT transcription factor ZEB2 was also increased in RNF2 expressing cells and may also contribute to pro-metastatic phenotype conferred by RNF2. Moreover, we provide strong evidence for requirement of its E3-ubiquitin ligase activity in the promotion of invasive and metastatic properties by RNF2. This, against the backdrop of the well-known opposing effect of TGFβ signaling, raises the possibility that inhibition of RNF2 catalytic activity offers a new therapeutic intervention to target the metastatic activity of TGFβ in metastatic melanoma.
An important question is whether pro-metastatic and pro-tumorigenic activities of RNF2 are completely independent of each other. Although we provide evidence that proinvasive/metastatic function is dependent on RNF2's catalytic activity and protumorigenic role is independent of it, our data do not completely rule out the possibility that RNF2's role in proliferation also contributes to its pro-metastatic phenotype.
Taken together, our findings provide strong evidence that epigenetic regulators, such as RNF2, directly and functionally control powerful gene networks that are vital in multiple cancer processes.
METHODS
Cell Culture, Proliferation Assays, Soft Agar Colony formation assay and Boyden Chamber Invasion Assay
Cells were grown in standard tissue culture conditions (5% CO2, 37°C). HMEL-BRAFV600E cells were kind gift of Dr. David Fisher. 1205Lu, WM115, 501Mel, WM983B cells were obtained either from ATCC or Coriell and maintained according to manufacturer's instructions. Cell lines were authenticated by STR (short tandem repeat) profiling and tested every 2 months for mycoplasma contamination. Cell proliferation assays were performed using Incucyte instrument (Essen Bioscience). The instrument captures bright field images every two hours and calculates cell density based on the area occupied by cells compared to total area. Soft agar colony formation assay was performed as described earlier (3). Briefly, two layers of soft-agar (bottom layer 0.8% and top layer 0.5%) mixed with DMEM growth medium and fetal bovine serum were prepared. Two thousand cells were mixed in the top agar layer during plating and colony formation was monitored. Upon appropriate size of colonies, the colonies were stained with p-iodonitro tetrazolium violet, pictures taken and counted manually or with ImageJ software. Boyden chamber matrigel invasion assay was performed as described earlier (3). Briefly, chambers were brought to room chamber and hydrated in serum free media. One hundred thousand cells were seeded inside the chamber in serum-free media and assayed for ability to move to the bottom of the chamber in response to 10% serum containing media present in the well after 24-48 hrs.
Mice injections and tumor studies
Four to six weeks old NCR-NUDE mice were purchased from Taconic and injected intradermally with 1 million cells. Tumor volume was measured at designated time points. Mice were euthanized and tumor harvested when tumor size reached 1.5cm. Mice were maintained in either the animal facility at Harvard Center for Comparative Medicine or in animal facility at MD Anderson Cancer Center. All animal experiments were approved by an IACUC review board.
Tissue Microarray and immunohistochemistry
Tissue microarray for melanoma progression has been previously described (28). RNF2 immunohistochemistry was performed using Prestige rabbit polyclonal antibody (Sigma). TMA slides were heated at 65°C for 1hr, deparaffinized in xylene, and rehydrated. Antigen retrieval was performed by boiling at 115°C for 10mins and then at 95°C for 30sec. After cooling, slides were incubated in 3% H2O2 for 20mins, washed in PBS and blocked in goat serum. Following incubation with primary antibody (1:200) overnight, slides were incubated in secondary antibody for 1hr at 37°C. Slides were then washed and incubated in ABC elite reagent (Vector labs) and developed using ImmuPACT (Novagen). Manual blinded scoring of the TMA core intensity was performed by two independent pathologists.
Chromatin Immunoprecipitation and next-generation sequencing (ChIP-Sequencing)
Chromatin immunoprecipitation was performed as described earlier (29). Library preparation was done using NEB reagents as described earlier (29). Sequencing was performed in Hiseq2000 (Illumina). Data analysis was performed as described in the Supplementary Methods.
RNA isolation, quantitative PCR and Microarray
RNA was isolated using RNeasy Kit (Qiagen) per manufacturer's instructions. cDNA was prepared using SuperScript III (Life Technologies) using manufacturer's instructions. qPCR were performed using SybrGreener (Invitrogen) and Stratagene instrument. Microarray experiments were performed in MD Anderson Center for ncRNA Sequencing core facility. Microarray data was analyzed using LIMMA bioconductor package. Details of analysis are in Supplementary Methods. All genomic data sets are publicly available at NCBI's GEO database (GSE51928, GSE51929 and GSE51930).
Survival analysis in TCGA data
TCGA melanoma data (2013_04_06 stddata run) were retrieved from Genome Data Analysis Center of TCGA. Survival intervals from date of specimen submission to patients’ death/last follow-up were available in 154 cases. Statistical significance of survival differences was estimated by Kaplan-Meier curves and log rank test in R.
Protein isolation and Western blotting
Proteins were made using RIPA buffer (Boston BioProducts) and complete mini protease inhibitor cocktail (Roche). Western blotting was performed by standard procedure using Invitrogen or Bio-Rad precast 4-12% gels. Antibodies used were anti-V5 (Invitrogen), anti-vinculin (Sigma), anti-H2AK119ub (Millipore), anti-RNF2 (Sigma), anti-ID1 (SCBT), anti-ID2 (SCBT), anti-ID3 (SCBT). Secondary antibodies used were from LICOR. Blots were developed using LICOR Odyssey imager.
Mouse Models
Generation and characterization of iBIP mice and RNF2L/L mice are described in Supplementary Methods.
ChromHMM Analysis
We used ChromHMM (23) with default parameters to derive genome-wide chromatin state maps for all cell types as described in our unpublished study (Rai et al. unpublished). We binarized the input data with ChromHMM's BinarizeBed method using a p-value cutoff of 1e−4. We considered chromatin state models learned jointly on all chromatin marks at every increment of 5 states from 10 to 120 states. We chose a model with 45 states for our main analysis to balance interpretability and capturing informative state distinctions for the analysis here. In particular the model with 45 states was the model with the minimum number of states that was able to separate bivalent and poised states from active states.
Supplementary Material
SIGNIFICANCE.
Role of epigenetic regulators in cancer progression is being increasingly appreciated. We show novel roles for RNF2 in melanoma tumorigenesis and metastasis albeit via different mechanisms. Our findings support notion that epigenetic regulators, such as RNF2, directly and functionally control powerful gene networks that are vital in multiple cancer processes.
Acknowledgments
Grant Support
The work described in this manuscript was supported by grants from the National Institutes of Health (5U01 CA141508 and U01 CA168394 to LC; R01ES024995 and U01 HG007912 to JE), National Cancer Institute (1K99CA160578-01 to KR), Cancer Prevention and Research Institute of Texas (R1204 to LC) and National Science Foundation (1254200 to JE). Authors were supported by various fellowships: KR (Charles A. King post-doctoral fellowship), JE (Alfred P. Sloan fellowship) and PF (California Institute for Regenerative Medicine Training Grant TG2-01169 and UCLA Training Program).
Footnotes
Conflict of Interest Statement: Authors declare no conflict of interest.
REFERENCES
- 1.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leong SP, Gershenwald JE, Soong SJ, Schadendorf D, Tarhini AA, Agarwala S, et al. Cutaneous melanoma: a model to study cancer metastasis. Journal of surgical oncology. 2011;103:538–49. doi: 10.1002/jso.21816. [DOI] [PubMed] [Google Scholar]
- 3.Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–60. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 5.Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochemical Society transactions. 2012;40:341–6. doi: 10.1042/BST20120014. [DOI] [PubMed] [Google Scholar]
- 6.Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. The International journal of developmental biology. 2009;53:355–70. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- 7.Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends in biochemical sciences. 2009;34:71–7. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 8.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Romero C, Rooman I, Skoudy A, Guerra C, Molero X, Gonzalez A, et al. The epigenetic regulators Bmi1 and Ring1B are differentially regulated in pancreatitis and pancreatic ductal adenocarcinoma. The Journal of pathology. 2009;219:205–13. doi: 10.1002/path.2585. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Beato M, Sanchez E, Gonzalez-Carrero J, Morente M, Diez A, Sanchez-Verde L, et al. Variability in the expression of polycomb proteins in different normal and tumoral tissues. A pilot study using tissue microarrays. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2006;19:684–94. doi: 10.1038/modpathol.3800577. [DOI] [PubMed] [Google Scholar]
- 11.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nature reviews Cancer. 2009;9:274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 13.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 14.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–74. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Molecular cell. 2006;24:701–11. doi: 10.1016/j.molcel.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 17.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. The Journal of clinical investigation. 2015;125:1459–70. doi: 10.1172/JCI78954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meulmeester E, Ten Dijke P. The dynamic roles of TGF-beta in cancer. The Journal of pathology. 2011;223:205–18. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 19.Bueno L, de Alwis DP, Pitou C, Yingling J, Lahn M, Glatt S, et al. Semi-mechanistic modelling of the tumour growth inhibitory effects of LY2157299, a new type I receptor TGF-beta kinase antagonist, in mice. European journal of cancer. 2008;44:142–50. doi: 10.1016/j.ejca.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Todorovic V, Rifkin DB. LTBPs, more than just an escort service. Journal of cellular biochemistry. 2012;113:410–8. doi: 10.1002/jcb.23385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyytiainen M, Penttinen C, Keski-Oja J. Latent TGF-beta binding proteins: extracellular matrix association and roles in TGF-beta activation. Critical reviews in clinical laboratory sciences. 2004;41:233–64. doi: 10.1080/10408360490460933. [DOI] [PubMed] [Google Scholar]
- 22.Vehvilainen P, Hyytiainen M, Keski-Oja J. Latent transforming growth factor-beta-binding protein 2 is an adhesion protein for melanoma cells. The Journal of biological chemistry. 2003;278:24705–13. doi: 10.1074/jbc.M212953200. [DOI] [PubMed] [Google Scholar]
- 23.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 25.Illingworth RS, Botting CH, Grimes GR, Bickmore WA, Eskeland R. PRC1 and PRC2 are not required for targeting of H2A.Z to developmental genes in embryonic stem cells. PloS one. 2012;7:e34848. doi: 10.1371/journal.pone.0034848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao PS, Satelli A, Zhang S, Srivastava SK, Srivenugopal KS, Rao US. RNF2 is the target for phosphorylation by the p38 MAPK and ERK signaling pathways. Proteomics. 2009;9:2776–87. doi: 10.1002/pmic.200800847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Molecular cell. 2010;38:452–64. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin Y, Ekmekcioglu S, Liu P, Duncan LM, Lizee G, Poindexter N, et al. Constitutive aberrant endogenous interleukin-1 facilitates inflammation and growth in human melanoma. Molecular cancer research : MCR. 2011;9:1537–50. doi: 10.1158/1541-7786.MCR-11-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Molecular cell. 2012;47:810–22. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.