Abstract
Tumor-associated myeloid cells, including dendritic cells (DCs) and macrophages, are immune suppressive. This study demonstrates a novel mechanism involving FOXO3 and NF-κB RelA that controls myeloid cell signaling and impacts their immune-suppressive nature. We find that FOXO3 binds NF-κB RelA in the cytosol, impacting both proteins by preventing FOXO3 degradation and preventing NF-κB RelA nuclear translocation. The location of protein–protein interaction was determined to be near the FOXO3 transactivation domain. In turn, NF-κB RelA activation was restored upon deletion of the same sequence in FOXO3 containing the DNA binding domain. We have identified for the first time, to our knowledge, a direct protein–protein interaction between FOXO3 and NF-κB RelA in tumor-associated DCs. These detailed biochemical interactions provide the foundation for future studies to use the FOXO3–NF-κB RelA interaction as a target to enhance tumor-associated DC function to support or enhance antitumor immunity.
Introduction
The tumor microenvironment (TME) is highly immunosuppressive due to a combination of factors that include tumor-produced cytokines and growth factors, as well as suppressive infiltrating immune cells. It has now become clear that many of the suppressive mechanisms within the TME are regulated by key transcription factors. Although there are many, among the most influential in regulating immune responses include NF-κB, p53, β-catenin, and Forkhead Box family members such as FOXO3 (1–4). When these transcription factors are overexpressed, they promote tumor growth and survival, as well as dysregulate immune cell function, impeding effective antitumor immunity (3, 5, 6). The functions of each of these transcription factors are known to promote survival of tumor cells (1). However, less is known regarding their role in regulating antitumor immunity, especially cross talk between the signaling pathways that can control immune cell functions.
FOXO3 has been described to play roles in the development and function of suppressive immune cells in infections and situations of immune stress, including the TME (7–11). In particular, it was demonstrated that FOXO3 is essential for FOXP3 induction in the development of T regulatory cells, central memory CD8+ T cells, and the suppressive function of myeloid cells including tumor-associated dendritic cells (TA-DCs) (10–12) and monocytes (9). Lee et al. (9) demonstrated that FOXO3A binds the promoter region of TGF-β1 to regulate transcription, which further enhanced cytokine production, but the mechanism by which FOXO3 regulates other functions in myeloid cells remains unclear. In this study, we aimed to determine the mechanisms regulated by FOXO3 influencing the TA-DC role in promoting or suppressing antitumor immunity. Intriguingly, FOXO3 was found to greatly impact or regulate cellular function from the cytoplasm rather than by regulating transcription of genes in the nucleus. These data demonstrate for the first time, to our knowledge, that FOXO3 participates in regulation of cellular functions through additional mechanisms and may be a critical target to regulate antitumor immunity and immune tolerance.
Materials and Methods
Experimental mice
Three different tumor models were selected to analyze FOXO3 protein expression in TA-DCs. The first model is the transgenic adenocarcinoma of the mouse prostate (TRAMP), spontaneous prostate tumor–bearing mice. TRAMP mice were used at 16 wk of age when 100% of the mice display hyperplasia and/or prostate intraepithelial neoplasia; although this is not fully developed carcinoma, the TME is already highly immunosuppressive (13). The second tumor model used was B16 melanoma. The use of B16 allowed us to use FOXO3−/− mice, which were not available on the TRAMP model. B16 were injected s.c. into the left hind flank of shaved C57Bl6, FOXO3−/−, or FOXO3+/− mice [kindly provided by Dr. Stephen Hedrick, University of California San Diego (7) and Dr. Karen Arden, Ludwig Institute]. The third model used 4T-1 breast tumors (ATCC) that were injected into the mammary fat pad of BALB/c mice. Use of the 4T-1 model allowed us to compare our results in TA-DCs across three different models to confirm that findings were common among TA-DCs and were not associated with one particular tumor model. All TA-DCs were purified, as described later, by magnetic beads from the tumor. As sources of control DC populations (Ctl DCs), C57BL/6 or BALB/c mice were used to generate bone marrow–derived DCs (BM-DCs) or DCs were purified from spleens and are indicated in each figure. To test T cell responses to wild type (WT) or FOXO3−/− DCs, we isolated TRP-2 Ag-specific T cells (obtained from the National Cancer Institute at Frederick) from the lymph nodes of TRP-2 transgenic mice (14).
Mice were housed under specific pathogen-free conditions and were treated in accordance with National Institutes of Health guidelines under protocols approved by the Animal Care and Use Committee of Loyola University Chicago (Maywood, IL).
Cell lines and transfection
To extensively test localization of FOXO3 and NF-κB proteins and biochemical interactions, we transduced cell lines to overexpress both proteins. The murine DC cell line DC2.4 was kindly provided by Dr. Ken Rock, University of Massachusetts Medical School, and macrophage cell line RAW264 was kindly provided by Dr. Elizabeth Kovacs, Loyola University. Given the difficulty in transducing myeloid cells, including myeloid cell lines, mechanisms of protein interactions were also tested with cell lines with higher transduction efficiencies, including the prostate tumor cell line TRAMP-C2 (ATCC) and human HEK 293T embryonic kidney cell line (kindly provided by Dr. Michael Nishimura, Loyola University Chicago). Cell lines were transfected with human FOXO3 in combination with human NF-κB RelA or human NF-κB–RelA–luciferase reporter constructs (kindly provided by Dr. Nancy Colburn, National Cancer Institute at Frederick). Transductions were performed using the FuGENE transfection reagent (Promega). At 48 h after transduction, cells were treated with 20 ng/ml recombinant (mouse or human) IL-6 (Peprotech) and tested protein expression and/or luciferase activity using the luciferase detection kit from Promega. FOXO3 mutant constructs were kindly provided by Dr. Mitsu Ikura, University of Toronto. FOXO3 mutants include: FL, full length (1–673); M1, mutant 1 with deletion in N terminus (151–673); M2, mutant 2 with deletion in N terminus and Forkhead domain (244–673); M3, mutant 3 with deletions in N and C termini (151–650); M4, mutant 4 with deletions in N and C termini including the transactivation domain (151–600); M5, mutant 5 with N and C termini and additional deletion 492–608; and M6, mutant 6 with N and C termini and additional deletion 398–608. Constructs are illustrated in Wang et al. (4).
Cell isolations
Single-cell suspensions were generated from tumors by excising tumors and placing in digest buffer that consisted of Collagenase D (Roche) and DNAse I (Roche). Cell digests were incubated for 20 min at 37°C with rotation. Single-cell suspensions were generated from spleen by pressing spleens between frosted slides and passing through a 70-μm filter (BD Biosciences). RBCs were removed by ACK lysis buffer. Single-cell suspensions were washed three times with PBS + 2% FBS. TA-DCs and Ctl DCs from spleen were isolated from single-cell suspensions using the Miltenyi AutoMACS cell separation system and Pan-DC magnetic beads (Miltenyi Biotech) according to the manufacturer’s instructions. Cell separations consistently yielded purity of >85% CD11c+/CD317+ cells (15). Human DCs were generated from peripheral blood monocytes by incubation in vitro for 9 d with rIL-4 and GM-CSF. Similarly, bone marrow Ctl DCs were derived from mouse bone marrow by a 9-d in vitro culture with recombinant mouse GM-CSF (Peprotech). TRP-2 Ag-specific T cells were isolated from lymph nodes of transgenic mice by harvesting inguinal, mesenteric, brachial, and axially nodes, and gently rolling nodes between frosted slides.
Western blotting and immunoprecipitation
Whole-cell lysates were generated from purified TA-DCs using lauryl-maltoside (Sigma Aldrich) immunoprecipitation buffer supplemented with one protease inhibitor tablet (Roche). Lysates were either used for a direct Western blot or immunoprecipitated for FOXO3 or NF-κB proteins as indicated. For immunoprecipitation, Abs were incubated with protein G-Sepharose beads (Ambion) for 1 h before adding cell lysates. Lysates were incubated with Ab-coated beads for 4 h at 4°C with rotation. Precipitated proteins were tested by Western blot analysis for coprecipitation. To confirm cellular localization of protein, we performed cytoplasmic or nuclear fractionation on lysates with the Paris protein fractionation kit (Ambion) per manufacturer’s instructions. Proteins were run by electrophoresis on a 12% polyacrylamide gel and transferred to a nitrocellulose membrane using the Rapid Transfer semidry transfer box (Bio-Rad). Membranes were blotted with anti-FOXO3 (R&D Systems, Cell Signaling), anti–phospho-FOXO3 (Cell Signaling), anti–NF-κB RelA, RelB, or c-rel (Cell Signaling), β-ACTIN (Sigma-Aldrich), LAMIN-B1 (Abcam), and anti-IκBα and anti-IKKα (Cell Signaling).
Cycloheximide
TA-DCs were purified from tumors as described earlier and treated with 50 μg/ml cycloheximide (Cell Signaling) for 0–24 h in a 24-well dish. Phosphorylated FOXO3 found in the cytoplasm can be rapidly degraded; therefore, the initial time points 0, 5, 10, 15, and 30 min were the primary focus of Western blot analysis. STAT3 degradation was observed to degrade in response to cycloheximide treatment and was therefore used as a control that protein degradation did occur upon treatment.
Chromatin immunoprecipitation
TA-DCs were purified from B16 tumors and splenic DCs were used as Ctl DCs isolated from aged matched mice. Chromatin immunoprecipitation (ChIP) was performed according to the SimpleChIP Plus (Cell Signaling) kit instructions. In brief, 4 × 106 cells were cross-linked in a 1% formaldehyde solution diluted in RPMI 1640 for 10 min at room temperature. Cells were washed twice in ice-cold PBS and resuspended in lysis buffer at 4°C with rotation for 10 min. Samples were then incubated with micrococcal nuclease to aid DNA fragmentation and placed at 37°C for 20 min. After micrococcal digestion, samples were sonicated to break nuclear membranes and release chromatin fragments. A small aliquot of chromatin fragments was RNase and proteinase k treated, and final chromatin concentration was determined. The remainder was then used in the ChIP assays. A total of 10 μg cross-linked chromatin was incubated with anti-FOXO3 Ab (Cell Signaling) or rabbit IgG as a control at 4°C overnight with rotation. Then ChIP grade agarose beads were added to the chromatin solution and incubated for 2 h at 4°C with rotation. Beads were washed three times in a low-salt wash and once in a high-salt wash to remove nonspecific binding. After washing, chromatin was eluted from the beads and protein was removed via proteinase K treatment at 65°C overnight. DNA was then purified using spin columns and was quantified via PCR using primers to a known DNA binding site of FOXO3 in the androgen receptor promoter (Forward 5′-TCTCCCTTCTGCTTGTCCTGGT-3′, Reverse 5′-TAGGCTCCAAAGCAGAAGCGAT-3′).
Flow cytometry, Amnis Image Stream, and immunofluorescent microscopy
TRP2 Ag-specific cells were tested for proliferation by labeling with CFSE, and incubated with BM-DCs from WT or FOXO3−/− mice pulsed with TRP2 peptide (SVYDFFVWL). Peptides were purchased from New England Peptide. IFN-γ production was tested after a 48-h coculture. In brief, TRP-2 cells from DC cocultures were treated with GolgiPlug and stimulated again with WT or FOXO3−/− for an additional 6 h. T cells were fixed and permeabilized with eBioscience buffers and stained for IFN-γ (eBioscience). TRP-2 T cells were analyzed on the Fortessa, and gates were set for CD3+/CD8+ cells.
Fcγ receptors were blocked in cell suspensions with anti-FcγRIIb Ab and incubated with the indicated Abs purchased from BD Pharmingen (CD11c), eBioscience (CD317), or Cell Signaling (FOXO3, RelA) for flow cytometry on the Fortessa (BD Pharmingen). The purification of DCs from tumors was tested by flow cytometry before and after magnetic bead isolation. DCs represented ∼8% of total CD45+ cells within the tumor preisolation. DCs were enriched to >85% of total CD45+ cells. For further characterization of TA-DC phenotype, expression of ZBTB46 and Clec9 were assessed, along with expression of costimulatory molecules. The majority of DCs were Clec9− and ZBTB46− (45–48%). However, there was a 10% increase in frequency of double-positive cells from the tumor compared with DCs isolated from spleen or from normal prostates, and no differences in expression of CD80 or CD86 were detected.
Cells were stained for microscopy after fixation with ice-cold 100% MeOH. Fixed cells were incubated with mouse anti-FOXO3 (R&D Systems) or rabbit anti–NF-κB RelA (Cell Signaling) at a 1:1000 dilution followed by staining with a 1:500 dilution, Alexa Flour 488 anti-mouse (Cell Signaling) or Alexa Flour 647 anti-rabbit secondary fluorescent Abs. Cells were fixed to positively charged slides by cytospin at 300 rpm for 5 min. One drop of Vectashield mounting media with DAPI (purchased from Vector Labs) was added to each slide and coverslipped. Cells were analyzed on an inverted fluorescent EVOS microscope at ×40 and ×100 magnification. For Amnis Image Stream analysis, cells were fixed and permeabilized in 100% ice-cold MeOH and then treated for 30 min with a rabbit anti-FOXO3 primary Ab (Cell Signaling) followed by FITC-conjugated anti-rabbit secondary and PE-conjugated NF-κB Abs (eBioscience) or appropriate isotype controls. DAPI (BioLegend) was added to all cells 15 min before analysis for staining of cell nuclei. Cells were acquired on the Amnis Image Stream X machine at ×60 magnification, using the 405, 488, and 785 lasers and bright field. Analysis was performed on the IDEAS software, using the colocalization and nuclear localization analysis wizards. Templates of software settings were created in IDEAS to standardize analysis.
Real-time PCR
RNA was isolated from TA-DCs purified from tumors or Ctl DCs by RNeasy Spin Columns (Qiagen) per manufacturer’s instructions. RNA quality was determined by analysis on an Agilent Bioanalyzer 2000. Primers, IL-12, IL-15, actin, and GAPDH for RT-PCR were purchased (SA Biosciences, Qiagen) and used per manufacturer’s instructions in combination with SYBR Green Master Mix (Qiagen). Samples were analyzed on a Bio-Rad iCycler RT-PCR machine and a QuantStudio6 (ABI). Cycle threshold (CT) values were determined by the Applied Biosystems 7300 SDS software. Data were analyzed by the ΔΔCT method or uploaded to the SABiosciences RT2 Profiler PCR Array Data Analysis Web-based software, version 3.5. The ΔΔCT uses the CT values of the gene of interest and housekeeping gene (HKG). ΔCT is determined by subtracting the CT(HKG) from the CT(gene of interest); then the ΔΔCT is determined by subtracting the ΔCT(control) from the ΔCT(experimental). Fold change is equal to 2(−ΔΔCt). Actin was used as the HKG and untreated FOXO+/− as the control group.
Small interfering RNA
DCs were isolated from tumors and cultured with commercial predesigned and validated siRNAs to Foxo3, NF-κB RelA, or scrambled negative control small interfering RNAs (siRNAs; purchased from Life Technologies) for 48 h per manufacturer’s instructions. Efficiency of knockdown was determined by RT-PCR and Western blot analysis.
In vitro FOXO3/RelA binding assay
Recombinant RelA and FOXO3, both produced in HEK 293 cells and purified via C-terminal DDK affinity tags, were purchased from OriGene. A DNA oligonucleotide incorporating the FOXO3 consensus sequence and fluorescently labeled at the 5′ end (5′-6-FAM CGCATCCTATGTAAACAACTCGAGTC-3′), as well as its complement, was purchased from Integrated DNA Technologies. Binding of FOXO3 to DNA in the presence and absence of RelA was assayed via fluorescence anisotropy (16, 17) using a Beacon 2000 spectrophotometer, exciting at 490 nm and detecting at 530 nm. All samples were buffer exchanged into 20 mM sodium phosphate (pH 7.4), 75 mM NaCl and degassed for 30 min before use. To form a DNA duplex, we annealed the labeled oligo and its complement by heating for 10 min to 95°C, then slowly cooling. Duplex DNA concentrations were 50 nM in all samples. The concentration of FOXO3 when present was 100 nM. RelA was added to FOXO3/DNA solutions at concentrations of 1 μM (10:1 RelA/FOXO3 ratio), 500 nM (5:1 RelA/FOXO3 ratio), 250 nM (2.5:1 RelA/FOXO3 ratio), and 50 nM (0.5:1 RelA/FOXO3 ratio). The concentration of RelA in the RelA/DNA control was 1 μM. All samples were formed by mixing and incubating on ice for 1 h. Measurements were taken at 25°C, reporting the averages of 40 consecutive measurements taken at 15-s intervals after thermal equilibration. Sample volumes were 100 μl.
ELISA
Cell lysates were generated from WT or FOXO3−/− DCs and fractionated into cytoplasmic and nuclear sections as described earlier. Nuclear lysates were added to the PathScan Total NF-κB p65 Sandwich ELISA kit (Cell Signaling), and the assay was carried out per manufacturer’s instructions.
Statistics
Statistical analyses for differences between group means were performed by unpaired (two-tailed) Student t test. Data are presented as mean ± SD, and a p value <0.05 was considered statistically significant.
Results
FOXO3 is localized to the cytoplasm of TA-DCs
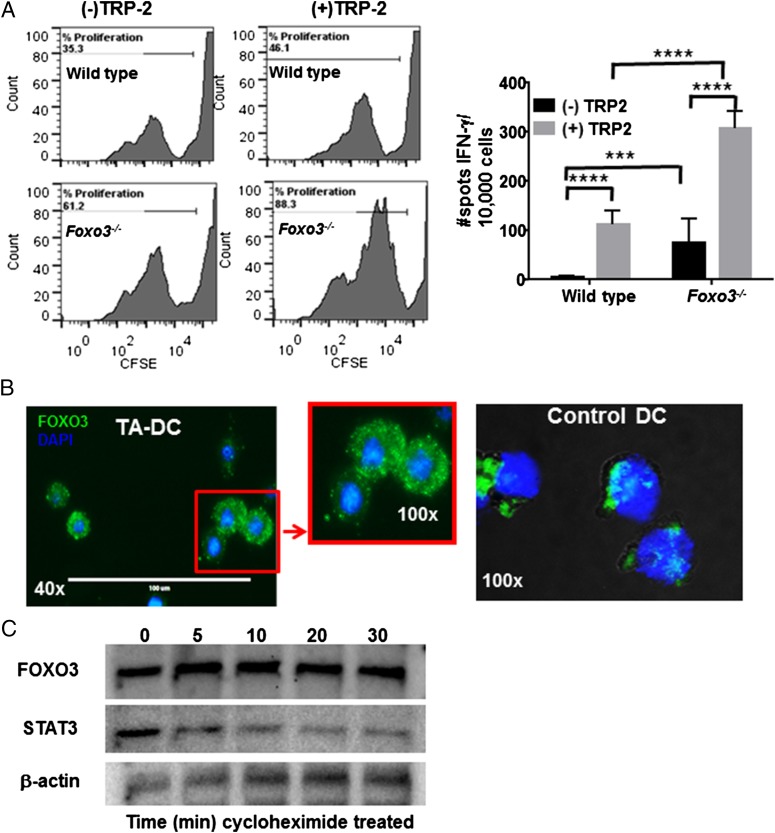
Previously, DCs were found to impact T cell function through a mechanism involving FOXO3. In those studies, it was determined that FOXO3 reduced proimmune stimulatory functions such as cytokine production (TNF-α and IL-6) and costimulatory molecule expression (7). In addition, it was observed that FOXO3 also increased tolerogenic functions such as IDO expression and TGF-β production (7, 18). We further confirmed that DCs deficient in Foxo3 induced increased proliferation and IFN-γ production in tumor Ag-specific CD8+ T cells (Fig. 1A). Therefore, to further our understanding of the role of this important transcription factor in DCs and its role in immune suppression, the cellular localization of FOXO3 was examined in DCs by immunofluorescence. Abundant levels of FOXO3 were found to be present in the cytosol in TA-DCs, whereas minimal levels of the protein were detected in the nucleus, indicated by nuclear DAPI staining (Fig. 1B). Much lower concentrations of FOXO3 were intermittently detected in control non–tumor-associated DC (Ctl DC) populations, which were isolated from normal prostate tissue, the spleen, or generated from bone marrow by in vitro culture with GM-CSF. FOXO3 expression was tested on both activated and nonactivated Ctl DC populations; regardless of the activation state of the Ctl DCs (activated by LPS, CpG, or anti-CD40), minimal FOXO3 protein was detected. The FOXO3 that was observed was found in both the cytoplasm and the nucleus (Fig. 1C). These data, together with reports demonstrating the impact of FOXO3 on DC function (7), suggest that in addition to transcriptional regulation of target genes, FOXO3 can also play a role in regulating cell signaling that may impact function from its location in the cytoplasm of TA-DCs.
FIGURE 1.
FOXO3 decreases DC immune-stimulating potential and is mostly expressed in the cytoplasm when upregulated. (A) DC stimulation of TRP2 Ag-specific T cells was tested in the presence or absence of FOXO3. (B) DCs (TA-DCs and Ctl DC) were stained for FOXO3 (green) and assessed by immunofluorescence microscopy to visualize cellular localization. DAPI was used to detect nuclear staining. Results are representative of four experiments. (C) TA-DCs were treated with cycloheximide for 0–30 min before Western blotting for FOXO3 or STAT3 as a control. ***p < 0.001, ****p < 0.0001.
To determine the temporal stability of cytoplasmic FOXO3, we isolated TA-DCs from murine tumors and treated them in vitro with cycloheximide to inhibit protein synthesis. Multiple tumor types including TRAMP, B16 melanoma, and 4T-1 breast tumors were used in different experiments to ensure the mechanisms identified were not an artifact of one particular murine tumor model. Under normal conditions, FOXO3 is a rapidly degraded protein when found in the cytosol (reviewed in Ref. 19); however, cycloheximide-treated TA-DCs maintained elevated levels of FOXO3 expression for 6 h posttreatment, whereas other proteins, such as STAT3, were degraded within 30 min, indicating that cytoplasmic FOXO3 in TA-DCs is comparatively stable (Fig. 1C). To further confirm that FOXO3 has different functional capabilities in TA-DCs compared with Ctl DCs, we used a ChIP assay to assess FOXO3 bound to known target DNA sequences (20). As anticipated by our findings that FOXO3 was primarily in the cytoplasm of TA-DCs, ChIP analysis confirmed that minimal FOXO3 in TA-DCs was bound to DNA consensus sequences under denaturing conditions (Supplemental Fig. 1A). As determined by RT-PCR, the quantity of FOXO3 bound to DNA was much lower in TA-DCs compared with Ctl DCs when the same concentration of FOXO3 was assessed from each sample (Supplemental Fig. 1B). It should be taken into consideration that target chromatin regions may be inactivated in TA-DCs, but not Ctl DCs. However, these data may also suggest that modifications directly on FOXO3 have occurred in the TME and/or the possibility that FOXO3 interacts with other proteins in the cytosol of TA-DCs that prevents nuclear mobilization and binding to target DNA sequences.
Cell type influences FOXO3 protein localization
A previous study by Karube et al. (21) demonstrated that cellular localization of FOXO3 was different in NK cells compared with other cell lines transfected to overexpress FOXO3. Because FOXO3 is a transcription factor and thought to function primarily in the nucleus, we tested whether dominant cytoplasmic expression of FOXO3 was unique to TA-DC or at least cells of the myeloid linage. We used an Amnis Image stream, which combines the power of confocal microscopy and flow cytometry, to visualize and quantify staining for FOXO3 in the cytoplasm or nucleus of various cells. Endogenous FOXO3 was detected in human peripheral blood monocyte-derived DCs (monocytes cultured with GM-CSF and IL-4 for 9 d) and murine BM-DCs (cultured with GM-CSF for 9 d) and was found to exist primarily in the cytoplasm. For further analysis of protein localization, cell lines were transfected to overexpress FOXO3. In addition to cell type, expression level was a factor considered in the cellular localization of the protein. Cells examined included the DC line, DC2.4, a prostate tumor cell line, TRAMPC2, RAW264 macrophages, and a human embryonic kidney cell line, HEK 293T (Supplemental Fig. 2). Staining of FOXO3 was not specifically exclusive for cytoplasmic expression in DCs, but the myeloid-derived cell types (DCs and macrophages) did express much more FOXO3 in the cytoplasm than in the nucleus compared with the other cell types. Transfection of TRAMP-C2 or HEK 293T resulted in 61 and 75%, respectively, expression of nuclear FOXO3, as demonstrated by the teal color obtained from the blending of green-labeled FOXO3 with the blue-labeled DAPI of the nucleus (Supplemental Fig. 2). These data suggest that cellular localization of the FOXO3 transcription factor is influenced by both cell type and amount of FOXO3 expressed. It is unclear whether the localization is solely due to a threshold effect of the amount of protein present because we were unsuccessful in efficiently transducing DCs to overexpress FOXO3. However, the observation that FOXO3 expressed in TA-DCs is exclusively expressed in the cytosol may indicate modifications induced by the TME.
FOXO3 colocalizes with NF-κB RelA in TA-DCs
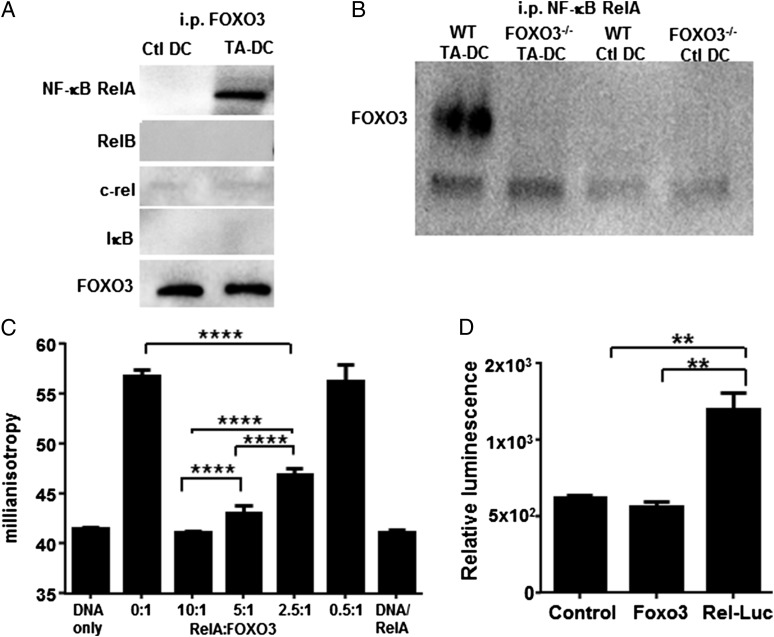
Given the apparent lack of transcription factor activity of FOXO3 in TA-DCs, we next tested the hypothesis that similar to other transcription factors deregulated in tumors, such as p53 (reviewed in Ref. 22), FOXO3 could impact functional activity by interfering with proinflammatory signaling pathways. One critical pathway to proinflammatory functional activation is NF-κB. NF-κB is also capable of controlling IL-6 and TNF-α production, both of which were previously reported to increase upon deletion of FOXO3 (7). To determine the cellular localization of NF-κB compared with FOXO3, we used Amnis Image Stream flow cytometry for both visual and quantitation analysis. Similar to FOXO3, NF-κB was detected primarily in the cytoplasmic regions of TA-DCs. Colocalization analysis revealed that between 40 and 80% of all cell types tested were double positive (data not shown). To test for the existence of a protein–protein interaction, we immunoprecipitated FOXO3 or NF-κB RelA from lysates generated from DCs isolated from normal prostate (Ctl DC) or TA-DCs isolated from TRAMP prostate tumors, followed by Western blotting for the detection of both proteins. Equal concentrations of precipitated FOXO3 were loaded into gels for analysis as demonstrated by the blot for FOXO3 (Fig. 2A). Analysis revealed coprecipitation of NF-κB RelA with FOXO3 and coprecipitation of FOXO3 with NF-κB RelA from TA-DC but not Ctl DC lysates (Fig. 2A, 2B). Further analysis of immunoprecipitated FOXO3 revealed that the interaction was specific to the NF-κB RelA species, because RelB, c-rel, and IκB were not detectable when tested from equivalent protein containing TA-DC lysates (Fig. 2A). Our data do not preclude the possibility that FOXO3 may interact with other NF-κB family members under different conditions. In fact, it was previously shown in a model of inflammation that FOXO3 inhibited NF-κB activity, and the mechanism was found to include interaction with IκB (23). Our results demonstrate IκB does not coprecipitate with FOXO3 in TA-DCs (Fig. 2A), which suggests there is a different mechanism by which FOXO3 can control NF-κB signaling in the TME and in DCs. These data identify a direct interaction between NF-κB RelA and FOXO3 proteins in TA-DCs.
FIGURE 2.
TA-DC FOXO3 forms a protein complex with NF-κB RelA. (A) FOXO3 or (B) NF-κB RelA was immunoprecipitated from WT DC or TA-DC lysates. The immunoprecipitated proteins were then analyzed by Western blot for FOXO3 and NF-κB family member proteins. (C) Fluorescence anisotropy indicates that RelA interacts directly with FOXO3. (D) NF-κB RelA responses were decreased in DC2.4 cells transfected to overexpress FOXO3 as measured by RelA luciferase reporters. Results are displayed as one independent experiment and are representative of three trials. **p < 0.01, ****p < 0.001.
We next sought to determine whether phosphorylation of FOXO3 that may occur in the TME could impact the protein’s ability to bind to RelA. TA-DCs were purified from murine tumors, lysates were generated and incubated with Sepharose beads coated with various species of FOXO3 including phospho-Ser318, phospho-Ser253, and phospho-Ser294, all of which are regulated by Akt signaling and are prime candidates for phosphorylation in the TME. RelA was also precipitated as a positive control, and lysates were incubated with unconjugated protein G-Sepharose beads or with an IgG isotype control as negative controls for nonspecific binding. Bands resulting from the IgG H and L chains were compared as a source of loading control. The results demonstrated that RelA did coprecipitate with each species of FOXO3 tested (Supplemental Fig. 3). These results are not all inclusive for all types of posttranslational modifications possible of FOXO3, but do reveal that phosphorylation through the Akt pathway does not preclude interaction with NF-κB RelA.
To further confirm the FOXO3–NF-κB protein–protein interaction, we first attempted to use EMSAs. However, because of the low frequency of TA-DCs in the tumor and minimal amount of protein that can be isolated from TA-DCs, we experienced significant technical difficulties with this assay. We were successful, however, using an in vitro binding assay with recombinant proteins. The objective of this assay was to determine whether the presence of one protein functionally impacted the other. To this end, we used the protein’s ability to bind specific DNA and tested whether adding in increasing concentrations of NF-κB RelA protein altered the ability of FOXO3 to bind to its target DNA sequence. Binding was assessed via fluorescence anisotropy (closely related to fluorescence polarization), measuring the binding of FOXO3 to a DNA oligonucleotide fluorescently labeled at the 5′ end. As fluorescence anisotropy reports on molecular tumbling, with protein bound, a fluorescently labeled oligonucleotide will tumble more slowly, yielding an anisotropy value higher than that for free DNA (16, 24). In the direct binding experiment, a 50 nM solution of DNA alone had a relatively low steady-state anisotropy near 40 mA. Addition of 100 nM FOXO3 resulted in a significant increase in anisotropy, consistent with formation of a FOXO3–DNA complex and reflecting the 0.3-μM affinity FOXO3 has for its consensus sequence; this group serves as the positive control (25) (Fig. 2C). Coincubation of this complex with increasing amounts of RelA, from 50 nM to 1 μM (reflecting RelA/FOXO3 ratios ranging from 0.5:1 to 10:1), resulted in consistently lower anisotropy values, indicating less FOXO3 is available to bind DNA (Fig. 2C). The anisotropy of a solution of 1 μM RelA mixed with 50 nM DNA was indistinguishable from that of DNA alone. These results demonstrate for the first time, to our knowledge, that RelA interacts directly with FOXO3 and this protein–protein interaction. These important findings show that, in their purest form, without any other cellular factors present, FOXO3 and NF-κB RelA proteins directly interact and that interaction is capable of impacting the functionality of FOXO3 as a transcription factor.
NF-κB RelA has critical roles in regulating the transcription of genes associated with proinflammatory cytokine production (22) and promotion of antitumor immunity. Therefore, we next sought to determine the impact of FOXO3-RelA interaction on the activation of RelA. Using an NF-κB luciferase reporter system, where NF-κB RelA with luciferase is placed under an IL-6 promoter (a gift from Dr. Nancy Colburn, National Cancer Institute), we transduced the DC2.4 cell line to overexpress FOXO3 or an empty vector DNA as a control. Transfected cells were treated with rIL-6 (26) to activate NF-κB as demonstrated by an increase in luciferase activity. A significant decrease in luciferase was detected upon cotransfection with FOXO3 (Fig. 2D). The same decrease in NF-κB RelA activity was not detected upon cotransfection with the control DNA. These data confirm a previous report by Liu et al. (23) demonstrating FOXO3-suppressed NF-κB activation in T cells, where hyperactivation of NF-κB in FOXO3−/− mice resulted in autoimmune disease.
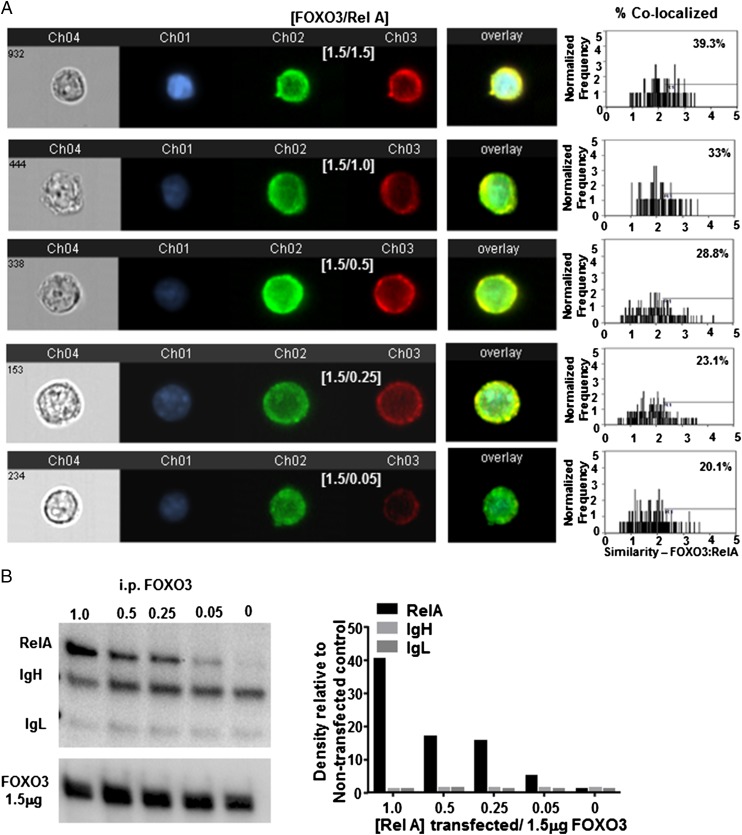
To more closely study the biochemical interactions of these proteins, we used HEK 293T cells, which, unlike DC cell lines, were easily and efficiently transfected with both FOXO3 and NF-κB RelA. First, we examined the expression level that was required for each protein for colocalization and/or binding to occur. HEK 293T were transfected with 1.5 μg FOXO3 plasmid DNA to induce overexpression and increasing concentrations from 0 to 1.5 μg RelA plasmid DNA. Transfected cells were imaged and quantified by Amnis Image Stream. At the highest concentrations of both proteins, ∼40% of the cells were measured to contain colocalized proteins. The frequency of cells with colocalization decreased as RelA concentrations decreased (Fig. 3A). Similarly, FOXO3 was immunoprecipitated from transfected cells and analyzed by Western blot. RelA coprecipitated with FOXO3 from cells transfected with 1.5 μg FOXO3 and 1 μg RelA; the quantity of protein coprecipitating decreased as concentrations of transfected RelA decreased (Fig. 3B). Tumors are known to induce increased levels of both of these proteins (reviewed in Refs. 27, 28). Therefore, these data support the hypothesis that at increased concentrations, which may occur in the TME, FOXO3, and NF-κB, RelA can form protein–protein complexes in the cytoplasm of TA-DCs.
FIGURE 3.
Colocalization diminishes when less protein is present. HEK 293T cells were transfected with 1.5 mg FOXO3 DNA and decreasing concentrations of RelA and analyzed (A) by Amnis Image Stream (original magnification ×40) for visual confirmation and (B) i.p. Western blot for quantitative analysis of cells containing colocalization. Results are representative of at least three individual experiments.
FOXO3 prevents NF-κB RelA activation by inhibiting nuclear translocation
To determine whether FOXO3−/− possessed increased RelA protein, we purified DCs from the spleen and tested them by Western blot for RelA expression. RelA protein was slightly, but not significantly, increased in FOXO3−/− mice compared with heterozygous control mice, suggesting that FOXO3 is not regulating the transcription or production of NF-κB RelA (data not shown). Therefore, we sought to further characterize the impact of FOXO3 on the activation of NF-κB RelA.
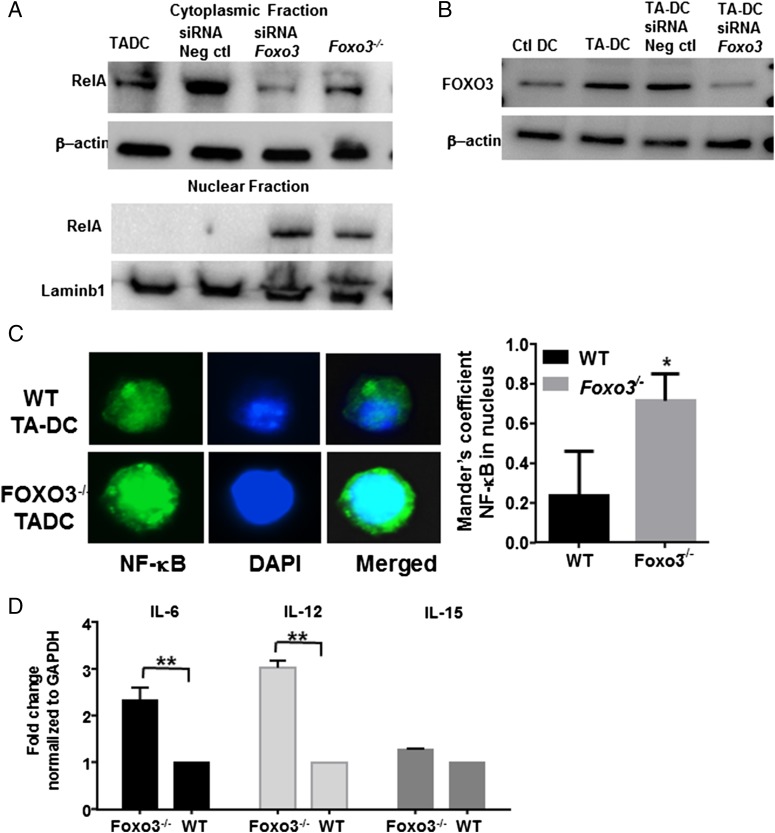
We tested whether the presence of FOXO3 influenced the nuclear localization of NF-κB RelA in TA-DCs. TA-DCs were purified from WT or FOXO3−/− B16 melanoma tumor-bearing mice, and protein lysates were further divided into cytoplasmic and nuclear fractions before Western blot analysis for RelA. To control for possible total gene deficiency abnormalities, we also treated WT TA-DCs with siRNA to silence FOXO3 expression before protein extraction. Analysis of protein fractions demonstrated that NF-κB RelA translocated to the nucleus in the absence of FOXO3 (Fig. 4A). Protein loading was controlled for by detection of β-actin for the cytoplasmic fraction and nuclear membrane protein Lamin-B1 for the nuclear fraction. Efficient knockdown of FOXO3 was confirmed by Western blotting; siRNA silencing did not completely abolish FOXO3 but did reduce expression to below levels expressed in Ctl DCs (Fig. 4B). To further confirm these results, we isolated DCs from tumors grown in WT or FOXO3−/− mice and stained for NF-κB RelA and DAPI. Significantly more NF-κB RelA was detected in the nucleus of FOXO3−/− TA-DCs compared with WT TA-DCs (Fig. 4C, Supplemental Fig. 4A). Given the increased nuclear translocation of RelA in the absence of FOXO3, we next tested the expression levels of RelA target genes in the presence or absence of FOXO3 as an indication of NF-κB RelA activation. DCs purified from FOXO3−/− mice had significantly increased mRNA expression of IL-6 and IL-12 and slightly increased IL-15 compared with DCs purified from FOXO3+/− control mice (Fig. 4D). Although the increase in these target genes was modest between 1.5- and 3-fold, it was consistent. These data suggest that an additional activation signal may be required to gain full activation potential. No significant changes in cell phenotype were observed. DCs from FOXO3−/− and WT tumors expressed similar levels of costimulatory molecules. FOXO3−/− TA-DCs did have a slight increase in MHC II expression (Supplemental Fig. 4B). Taken together, these data suggest that FOXO3 prevents NF-κB RelA activation by preventing RelA nuclear translocation, and silencing of FOXO3 allows for RelA nuclear expression and activation.
FIGURE 4.
FOXO3 inhibits NF-κB RelA nuclear localization. (A) Lysates from TA-DCs were fractionated into cytoplasmic or nuclear extracts and tested for RelA with or without FOXO3. (B) FOXO3 knockdown was confirmed by Western blot. (C) NF-κB RelA nuclear localization was determined by immunofluorescence at original magnification ×100. (D) NF-κB RelA target gene expression was tested in TA-DCs in the presence or absence of FOXO3 by RT-PCR. Results are displayed as one independent experiment and are representative of three trials. *p < 0.05, **p < 0.01.
NF-κB RelA promotes FOXO3 stability in the cytoplasm via a direct protein–protein interaction
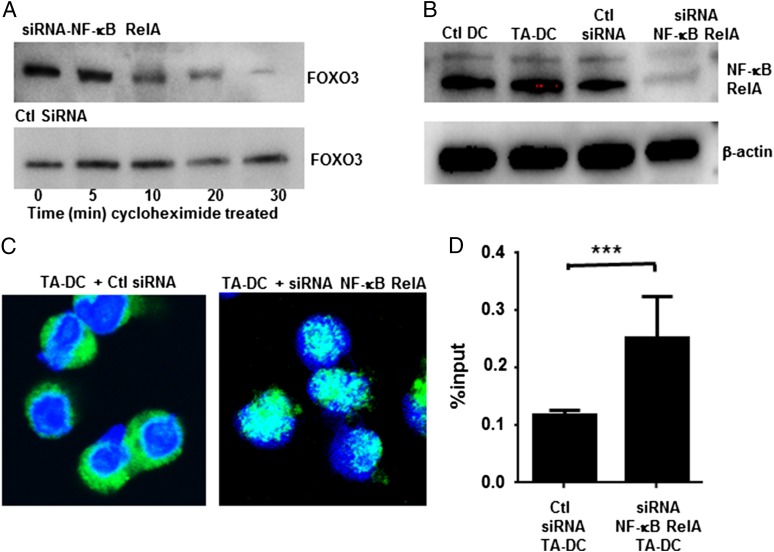
Given our initial findings that cytoplasmic FOXO3 was stable when DCs were treated with cycloheximide, we hypothesized that the formation of the FOXO3-RelA protein complex provided the necessary stability to prevent FOXO3 degradation, as typically happens to phosphorylated FOXO3 in the cytoplasm. To test this, we purified DCs from TRAMP tumors and transfected them with siRNA to silence NF-κB RelA or with scrambled siRNA controls. TA-DCs treated with control siRNA maintained consistent FOXO3 expression, whereas silencing NF-κB RelA led to the rapid degradation of FOXO3 upon cycloheximide treatment (Fig. 5A). NF-κB RelA knockdown was confirmed by Western blotting (Fig. 5B). In addition to undergoing degradation, immunofluorescence staining demonstrates that upon silencing NF-κB RelA, FOXO3 is also available to translocate to the nucleus (Fig. 5C). Similarly, using the ChIP assay, we found the quantity of FOXO3 binding to its known consensus sequence (androgen receptor promoter region) to be increased in the absence of NF-κB RelA, thus indicating an increase in functional FOXO3 in the nucleus (Fig. 5D). Taken together, these data demonstrate that FOXO3 and NF-κB RelA proteins interact in the cytoplasm in a way that both prevents nuclear translocation of either protein and provides protein stability for FOXO3.
FIGURE 5.
NF-κB RelA stabilizes cytoplasmic FOXO3 and prevents DNA binding. (A) TA-DCs were transfected with siRNA to silence NF-κB or with scrambled controls. FOXO3 stability was then tested by Western blot after cycloheximide treatment. (B) NF-κB RelA silencing was confirmed by Western blot. (C) DCs were stained with anti-FOXO3 FITC and DAPI to determine cellular localization of FOXO3 after transfection with siRNA to silence NF-κB RelA at original magnification ×40. (D) The amount of FOXO3 bound to its consensus sequences after NF-κB RelA silencing was quantified in TA-DCs by RT-PCR. Results represent two individual experiments with TA-DCs from five mice per group per experiment. ***p < 0.01.
FOXO3 and NF-κB RelA bind near the FOXO3 transactivation domain
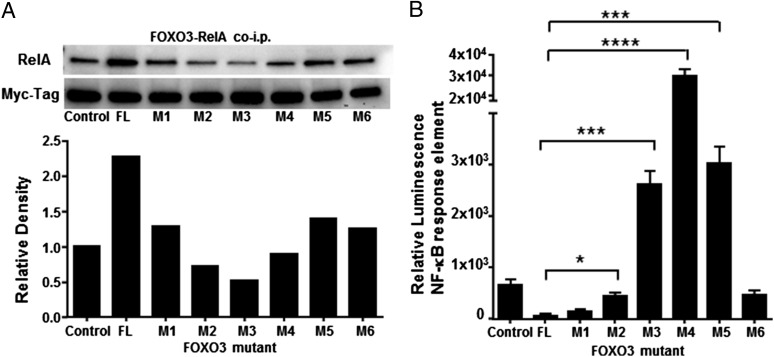
Our results demonstrate that there is a reciprocal repression of DNA binding and activation between FOXO3 and RelA. Therefore, it stands to reason that disruption of this protein–protein interaction may be an ideal target to enhance TA-DC function. NF-κB RelA activation is critical for proinflammation signaling, and as we demonstrated, removing NF-κB RelA from the protein complex allows for FOXO3 degradation. To attempt to identify the interaction site between these two proteins, we used plasmids containing either full-length FOXO3 or FOXO3 that contain mutations and/or deletions from the N terminus to the C terminus (kind gift of Dr. Mitsu Ikura, University of Toronto) (4) (Fig. 6). HEK 293 T cells were cotransfected with full-length FOXO3 or mutant FOXO3 along with NF-κB RelA. FOXO3 was then immunoprecipitated and proteins were tested for bound NF-κB RelA. Western blot analysis revealed decreased RelA coimmunoprecipitation with mutants 2, 3, and 4 (Fig. 6A). Notably, all mutants had reduced binding to NF-κB RelA than displayed by full-length FOXO3 (Fig. 6A). Further analysis using the NF-κB RelA reporter assay confirmed increased RelA activity in cells transfected with mutants 3, 4, and 5, with the highest level of activity detected in samples containing mutant 4 (Fig. 6B). Although the highest RelA response elements were detected in mutants 3, 4, and 5, it should be noted that all mutants except M1 had significantly higher luminescence than when the full-length FOXO3 was present (Fig. 6B). Mutant 4 had the highest activity of NF-κB RelA detected and was the only plasmid totally deficient in the FOXO3 transactivation domain, suggesting that FOXO3 and NF-κB RelA may interact at or near the transactivation domain of FOXO3. In this case, the location of the protein–protein interaction could disrupt space for DNA binding. These results are consistent with the fluorescence anisotropy findings, which demonstrate that binding of NF-κB RelA to FOXO3 inhibits the ability of FOXO3 to bind DNA.
FIGURE 6.
RelA binds FOXO3 near the transactivation domain. HEK 293T cells were cotransfected with full-length FOXO3 (FL) or a truncated mutant of FOXO3 and RelA. Lysates were tested for (A) coimmunoprecipitation and (B) NF-κB RelA reporter responses upon recombinant human IL-6 stimulation by luciferase detection. Results are displayed as one independent experiment and are representative of three trials. *p < 0.05, ***p < 0.01, ****p < 0.001.
Discussion
The function of FOXO3 as a transcription factor has been described to include critical roles in important cellular processes such as cellular metabolism, tumor suppression through apoptosis, and oxidative stress. This is the first report, to our knowledge, that FOXO3: 1) may have extranuclear functionality, and 2) interacts directly with NF-κB RelA. Although perhaps surprising, it is not uncommon for transcription factors to have roles in cellular processes in the cytosol. For example, another tumor suppressor gene, p53, was recently found to participate in controlling biological processes such as autophagy, metabolism, and oxidative stress from the cytoplasmic compartment (22, 29). Typically, cytoplasmic FOXO3 is phosphorylated, ubiquitinated, and degraded. Although cytoplasmic FOXO3 in TA-DCs was found to be phosphorylated, it was not degraded but was found instead to be colocalized and bound cytoplasmic NF-κB RelA. Our data further indicate that the interaction provided protein the stability for cytoplasmic FOXO3 and prevented both transcription factors from translocating to the nucleus or binding their respective DNA consensus sequences. A previous study also identified that NF-κB activation was impacted by FOXO3, where NF-κB was overactive in T cells in a model of autoimmunity due to the absence of FOXO3 (30). However, the mechanism of regulation seems to be different in the TME, or at least in TA-DCs. The previous report found that FOXO3 bound IκB to control NF-κB activation in T cells. In this study, we identified that FOXO3 could actually directly bind NF-κB RelA, but no other NF-κB family members. Further, IκB was not detected in the protein complex (Fig. 2A). FOXO3 that was phosphorylated at serine residues 318 and 293 also bound to NF-κB RelA, which suggests phosphorylation of FOXO3 via the Akt pathway does not preclude interaction in the cytoplasm (31, 32). Akt is known to promote cell survival by phosphorylating FOXO3 for nuclear exclusion (31). However, this is the first time, to our knowledge, it was found to remain stable in the cytoplasm and evade protein degradation. Importantly, we demonstrated that upon silencing FOXO3 expression, NF-κB was able to shuttle to the nucleus and turn on proinflammatory target genes (IL-6, IL-12, and IL-15). Similarly, upon silencing NF-κB, cytoplasmic FOXO3 was degraded. These data demonstrate a codependent relationship between these two proteins whereby FOXO3 binds NF-κB RelA, which provides cytoplasmic stability but also prevents proinflammatory signaling pathways from being activated.
The TME is a highly complex immune-suppressive network. The identification and description of specific regulatory protein complexes, such as the FOXO3–NF-κB RelA complex described in this report, provide both a mechanism of action for FOXO3 regulation of DC function and a potential target for the enhancement of immunotherapies against cancer. One avenue to target such interactions is the development of small-molecule inhibitors that can be targeted to specific cell populations by the use of a defined Ag tag, as seen with DEC-205 for targeting DCs (33). Small-molecule or kinase inhibitors have primarily been used to treat tumors directly such as MAPK, HIF-1a-p300, FAK-VEGFR3, and many others (34–36). The use of small molecules may be ideal to enhance current immune therapies in terms of both efficiency and duration of effect. For example, the Food and Drug Administration–approved Sipuleucel-T is an immunotherapy specifically using DCs. Recent studies indicate that Sipuleucel-T provides a 4.1-mo advantage in survival for patients with castration-resistant prostate cancer (37). Current research aims to improve this approach by boosting DC function, which may now be possible with the identification of the mechanism of FOXO3–NF-κB binding in DCs that infiltrate tumors. Disruption of this protein–protein complex in DCs may be a critical factor to boost DC support of CD8+ T cell effector and cytotoxic functions that would further enhance antitumor immunity.
Supplementary Material
Acknowledgments
We thank Dr. Michael Nishimura for research oversight and manuscript preparation. We also thank Drs. Jose Guevarra-Patino and Jill Suttles for critical review of the data and advice for manuscript preparation.
This work was supported by the National Cancer Institute, National Institutes of Health Grants R00CA151294 and GM06079 and a collaboration award from Trinity Health, Loyola University Chicago, and University of Notre Dame.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article contains supplemental material.
- BM-DC
- bone marrow–derived DC
- ChIP
- chromatin immunoprecipitation
- CT
- cycle threshold
- Ctl DC
- control DC population
- DC
- dendritic cell
- HKG
- housekeeping gene
- siRNA
- small interfering RNA
- TA-DC
- tumor-associated dendritic cell
- TME
- tumor microenvironment
- TRAMP
- transgenic adenocarcinoma of the mouse prostate
- WT
- wild type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Tenbaum S. P., Ordóñez-Morán P., Puig I., Chicote I., Arqués O., Landolfi S., Fernández Y., Herance J. R., Gispert J. D., Mendizabal L., et al. 2012. β-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat. Med. 18: 892–901. [DOI] [PubMed] [Google Scholar]
- 2.Zhou F., Wei H., Ding A., Qiu W., Feng L., Zhou Q., Liang J., Yue L. 2013. Different cellular localization of NF-κB p65 expression as an indicator of different prognoses of stage I-III gastric cancer patients. Clin. Transl. Sci. 6: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe M., Moon K. D., Vacchio M. S., Hathcock K. S., Hodes R. J. 2014. Downmodulation of tumor suppressor p53 by T cell receptor signaling is critical for antigen-specific CD4(+) T cell responses. Immunity 40: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F., Marshall C. B., Yamamoto K., Li G. Y., Plevin M. J., You H., Mak T. W., Ikura M. 2008. Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53. J. Mol. Biol. 384: 590–603. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y., Yu P., Li W., Ren G., Roberts A. I., Cao W., Zhang X., Su J., Chen X., Chen Q., et al. 2014. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene 33: 3830–3838. [DOI] [PubMed] [Google Scholar]
- 6.Liang X., Fu C., Cui W., Ober-Blöbaum J. L., Zahner S. P., Shrikant P. A., Clausen B. E., Flavell R. A., Mellman I., Jiang A. 2014. β-catenin mediates tumor-induced immunosuppression by inhibiting cross-priming of CD8⁺ T cells. J. Leukoc. Biol. 95: 179–190. [DOI] [PubMed] [Google Scholar]
- 7.Dejean A. S., Beisner D. R., Ch’en I. L., Kerdiles Y. M., Babour A., Arden K. C., Castrillon D. H., DePinho R. A., Hedrick S. M. 2009. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat. Immunol. 10: 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada Y., Harada Y., Elly C., Ying G., Paik J. H., DePinho R. A., Liu Y. C. 2010. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 207: 1381–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J. C., Espéli M., Anderson C. A., Linterman M. A., Pocock J. M., Williams N. J., Roberts R., Viatte S., Fu B., Peshu N., et al. UK IBD Genetics Consortium 2013. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 155: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerdiles Y. M., Stone E. L., Beisner D. R., McGargill M. A., Ch’en I. L., Stockmann C., Katayama C. D., Hedrick S. M. 2010. Foxo transcription factors control regulatory T cell development and function. Immunity 33: 890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang W., Beckett O., Ma Q., Paik J. H., DePinho R. A., Li M. O. 2010. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11: 618–627. [DOI] [PubMed] [Google Scholar]
- 12.Du, X., H. Shi, J. Li, Y. Dong, J. Liang, J. Ye, S. Kong, S. Zhang, T. Zhong, Z. Yuan, et al. 2014. Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. J. Immunol. 192: 1525–1535. [DOI] [PubMed]
- 13.Gingrich J. R., Barrios R. J., Morton R. A., Boyce B. F., DeMayo F. J., Finegold M. J., Angelopoulou R., Rosen J. M., Greenberg N. M. 1996. Metastatic prostate cancer in a transgenic mouse. Cancer Res. 56: 4096–4102. [PubMed] [Google Scholar]
- 14.Zhu Z., Singh V., Watkins S. K., Bronte V., Shoe J. L., Feigenbaum L., Hurwitz A. A. 2013. High-avidity T cells are preferentially tolerized in the tumor microenvironment. Cancer Res. 73: 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Watkins S. K., Zhu Z., Watkins K. E., Hurwitz A. A. 2012. Isolation of immune cells from primary tumors. J. Vis. Exp. 64: e3952 Available at: http://www.jove.com/video/3952/isolation-of-immune-cells-from-primary-tumors, [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Lakowicz J. R., Chowdhury M. H., Ray K., Zhang J., Fu Y., Badugu R., Sabanayagam C. R., Nowaczyk K., Szmacinski H., Aslan K., Geddes C. D. 2006. Plasmon-controlled fluorescence: a new detection technology. Proc. SPIE Int. Soc. Opt. Eng. 6099: 609909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mou Y., Tsai T. W., Chan J. C. 2007. Determination of the backbone torsion psi angle by tensor correlation of chemical shift anisotropy and heteronuclear dipole-dipole interaction. Solid State Nucl. Magn. Reson. 31: 72–81. [DOI] [PubMed] [Google Scholar]
- 18.Koorella C., Nair J. R., Murray M. E., Carlson L. M., Watkins S. K., Lee K. P. 2014. Novel regulation of CD80/CD86-induced phosphatidylinositol 3-kinase signaling by NOTCH1 protein in interleukin-6 and indoleamine 2,3-dioxygenase production by dendritic cells. J. Biol. Chem. 289: 7747–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calnan D. R., Brunet A. 2008. The FoxO code. Oncogene 27: 2276–2288. [DOI] [PubMed] [Google Scholar]
- 20.Yang L., Xie S., Jamaluddin M. S., Altuwaijri S., Ni J., Kim E., Chen Y. T., Hu Y. C., Wang L., Chuang K. H., et al. 2005. Induction of androgen receptor expression by phosphatidylinositol 3-kinase/Akt downstream substrate, FOXO3a, and their roles in apoptosis of LNCaP prostate cancer cells. J. Biol. Chem. 280: 33558–33565. [DOI] [PubMed] [Google Scholar]
- 21.Karube, K., S. Tsuzuki, N. Yoshida, K. Arita, F. Liu, E. Kondo, Y. H. Ko, K. Ohshima, S. Nakamura, T. Kinoshita, and M. Seto. 2012. Lineage-specific growth inhibition of NK cell lines by FOXO3 in association with Akt activation status. Exp. Hematol. 40: 1005–1015.e6. doi:10.1016/j.exphem.2012.08.005. [DOI] [PubMed]
- 22.Comel A., Sorrentino G., Capaci V., Del Sal G. 2014. The cytoplasmic side of p53’s oncosuppressive activities. FEBS Lett. 588: 2600–2609. [DOI] [PubMed] [Google Scholar]
- 23.Liu W., Gong Y., Li H., Jiang G., Zhan S., Liu H., Wu Y. 2012. Arsenic trioxide-induced growth arrest of breast cancer MCF-7 cells involving FOXO3a and IκB kinase β expression and localization. Cancer Biother. Radiopharm. 27: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakowicz J. R. 2006. Plasmonics in biology and plasmon-controlled fluorescence. Plasmonics 1: 5–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu J. L., Leemans A., Bai C. H., Lee C. H., Tsai Y. F., Chiu H. C., Chen W. H. 2008. Gender differences and age-related white matter changes of the human brain: a diffusion tensor imaging study. Neuroimage 39: 566–577. [DOI] [PubMed] [Google Scholar]
- 26.Kang M. I., Henrich C. J., Bokesch H. R., Gustafson K. R., McMahon J. B., Baker A. R., Young M. R., Colburn N. H. 2009. A selective small-molecule nuclear factor-kappaB inhibitor from a high-throughput cell-based assay for “activator protein-1 hits”. Mol. Cancer Ther. 8: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Neriah Y., Karin M. 2011. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12: 715–723. [DOI] [PubMed] [Google Scholar]
- 28.Kornblau S. M., Singh N., Qiu Y., Chen W., Zhang N., Coombes K. R. 2010. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin. Cancer Res. 16: 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chipuk J. E., Green D. R. 2004. Cytoplasmic p53: bax and forward. Cell Cycle 3: 429–431. [PubMed] [Google Scholar]
- 30.Fu C., Jiang A. 2013. β-catenin-mediated inhibition of cross-priming: a new mechanism for tumors to evade immunosurveillance. OncoImmunology 2: e26920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868. [DOI] [PubMed] [Google Scholar]
- 32.del Peso L., González V. M., Hernández R., Barr F. G., Núñez G. 1999. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene 18: 7328–7333. [DOI] [PubMed] [Google Scholar]
- 33.Ring S., Maas M., Nettelbeck D. M., Enk A. H., Mahnke K. 2013. Targeting of autoantigens to DEC205⁺ dendritic cells in vivo suppresses experimental allergic encephalomyelitis in mice. J. Immunol. 191: 2938–2947. [DOI] [PubMed] [Google Scholar]
- 34.Burslem G. M., Kyle H. F., Breeze A. L., Edwards T. A., Nelson A., Warriner S. L., Wilson A. J. 2014. Small-molecule proteomimetic inhibitors of the HIF-1α-p300 protein-protein interaction. ChemBioChem 15: 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gogate P. N., Ethirajan M., Kurenova E. V., Magis A. T., Pandey R. K., Cance W. G. 2014. Design, synthesis, and biological evaluation of novel FAK scaffold inhibitors targeting the FAK-VEGFR3 protein-protein interaction. Eur. J. Med. Chem. 80: 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachstetter A. D., Watterson D. M., Van Eldik L. J. 2014. Target engagement analysis and link to pharmacodynamic endpoint for a novel class of CNS-penetrant and efficacious p38α MAPK inhibitors. J. Neuroimmune Pharmacol. 9: 454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomella L. G., Petrylak D. P., Shayegan B. 2014. Current management of advanced and castration resistant prostate cancer. Can. J. Urol. 21(2Suppl. 1): 1–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.