Abstract
Background. MicroRNAs (miRNAs) are short noncoding RNAs integral for regulating gene expression at the posttranscriptional level. However, experimental methods often fall short in finding miRNAs expressed at low levels or in specific tissues. While several computational methods have been developed for predicting the localization of mature miRNAs within the precursor transcript, the prediction accuracy requires significant improvement. Methodology/Principal Findings. Here, we present MatPred, which predicts mature miRNA candidates within novel pre-miRNA transcripts. In addition to the relative locus of the mature miRNA within the pre-miRNA hairpin loop and minimum free energy, we innovatively integrated features that describe the nucleotide-specific RNA secondary structure characteristics. In total, 94 features were extracted from the mature miRNA loci and flanking regions. The model was trained based on a radial basis function kernel/support vector machine (RBF/SVM). Our method can predict precise locations of mature miRNAs, as affirmed by experimentally verified human pre-miRNAs or pre-miRNAs candidates, thus achieving a significant advantage over existing methods. Conclusions. MatPred is a highly effective method for identifying mature miRNAs within novel pre-miRNA transcripts. Our model significantly outperformed three other widely used existing methods. Such processing prediction methods may provide important insight into miRNA biogenesis.
1. Introduction
MicroRNAs (miRNAs) are short (generally 19–27 nucleotides), single-stranded noncoding RNAs that regulate gene expression at the posttranscriptional level [1]. miRNAs play important roles in a variety of biological processes, including organism development, tissue differentiation, and cell cycle [2, 3]. Dysregulation of miRNA processing is associated with many diseases, including cancer [4].
The processes of miRNA biogenesis in animals have been extensively studied. During this process, the long primary miRNA transcripts (pri-miRNAs) are cropped into double-stranded precursor miRNAs (pre-miRNAs) by a microprocessor complex, including Drosha and its cofactor DGCR8/Pasha [5]. Pre-miRNAs are usually ~70 nt and have a typical hairpin structure with an overhang of 2 nt on their 3′ ends [6]. The pre-miRNAs are then exported from the nucleus by carrier proteins such as exportin-5 [7] and most are cleaved into ~22 bp miRNA:miRNA∗ duplexes through the endonuclease Dicer [8, 9]. The miRNA:miRNA∗ duplex usually has two stable strands. While previous studies suggested one strand is the mature miRNA (the “guide strand”) that is incorporated into the RNA-induced silencing complex (RISC) for mRNA transcript target recognition [10–12], more recent studies showed that both mature miRNA and mature miRNA∗ species may exhibit inhibitory activity [13]. Consequently, the biogenesis of miRNAs is a very complex process, and knowledge of its various components/mechanisms would be very beneficial to the field of molecular physiology [14, 15].
Current experimental techniques, such as cloning and constructing short RNA libraries [16–18], can be used to identify miRNAs, but they often cannot find miRNAs that are expressed at low levels or in specific tissues. Moreover, not all miRNAs can be cloned. Recently, the emergence of next-generation sequencing (NGS) technologies, such as deep-sequencing, makes it possible to localize tissue-specific and development stage-specific miRNAs on a genome-wide scale. As NGS methods generate hundreds of millions of reads, separating miRNAs from other small RNA species or RNA degradation products is challenging [19, 20]. Consequently, computational approaches need to be developed to complement experimental techniques.
Several computational approaches have been reported for identifying mature miRNAs from their pre-miRNA transcripts. For instance, MiRmat [21] was designed to predict Drosha and Dicer cleavage sites in vertebrates using a Random Forest method. MiRRim2 [22] is designed for predicting human mature miRNA candidates using a conditional random fields (CRFs) method, based on evolutionary conserved features upstream of Drosha cleavage sites. MatureBayes [23] uses a Naive Bayes classifier to identify the location of human and mouse mature miRNAs based on information specific to individual nucleotide. Microprocessor uses a support vector machine (SVM) [24] classifier to identify Drosha cleavage sites based on 686 sequence- and structure-related features. MaturePred [25] finds mature miRNAs by integrating the structure-based free energy, and other features extracted from miRNA:miRNA∗ duplexes, based on a SVM model for plants. Similarly, another approach, miRdup [19], uses Random Forest algorithm integrated with Adaptive Boost (Adaboost) and can be used to detect the positions of miRNAs based on features from five lineages of cleavage sites on the miRNA:miRNA∗ duplexes. MiRPara [20] constructs two models to predict the location of mature miRNAs from genome-scale sequences for both plants and mammals. MirExplorer [26] is designed to predict pre-miRNAs and miRNAs with transition probability matrices and miRNA biogenesis vectors utilizing the Adaboost method for 16 species, while earlier methods, MIRcheck [27] and ProMir [5], are older tools for predicting mature miRNA candidates.
Although many methods are available for mature miRNA location prediction, they suffer from various limitations. For instance, many of these methods cannot be used to predict the miRtrons (miRNAs derived from the introns of protein-coding genes) [14], miRNAs without Drosha-mediated cleavage [21, 22, 24], or miRNAs that do not show strong evolutionary conservation [22]. In addition, since some approaches use a large number of features, it is difficult to achieve a possible biological explanation on which features contribute to the determination of the mature miRNA location [24]. Moreover, most of these prediction algorithms are based on early data resources [20, 26, 27], and several methods suffer from the inaccurate assumption that there is just one mature miRNA within a specific pre-miRNA transcript [5, 21, 24, 28]. Instead, only a few methods considered the double-stranded nature during miRNA maturation by integrating structure and sequence information. Thus, accurate prediction of mature miRNA locations remains a challenge [29].
In this study, we introduce a computational method, MatPred, which uses an RBF-SVM (radial basis function kernel/support vector machine) algorithm to predict the starting position of the mature miRNA in a pre-miRNA transcript. The parameters in the prediction model were trained using a set of experimentally validated mature miRNAs in the miRBase and further evaluated using a dataset that does not overlap with the training dataset. By systematically reviewing genomic features that can potentially affect the location of mature miRNAs, a more biologically relevant feature set was selected that includes the length of the mature and precursor miRNA transcript, the distances from the stem loop, and the minimum free energy of the RNA complex. More importantly, we innovatively design a series of features that represent the nucleotide and structure identities of each nucleotide for both miRNA:miRNA∗ complex and flanking regions. Comparison with existing tools suggests that MatPred achieved the highest prediction accuracy, according to the annotation documented in the latest version of miRBase (version 20).
2. Materials and Methods
2.1. Training and Test Datasets
Our training and test datasets were derived from 871 human pre-miRNAs documented in miRBase. These pre-miRNAs contained standard hairpin structures with single stem loops. In order to capture the features associated with experimentally validated starting positions of the mature miRNA molecules in the pre-miRNA sequences, we randomly selected 671 pre-miRNAs as our training dataset and further created two test sets, named Test set 1 and Test set 2, each containing 100 pre-miRNAs. In particular, Test set 2 is randomly selected from the newest dataset that only belongs to version 20. The list of training and test miRNA datasets can be found in Supplementary Table 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2015/546763. In the training set (671 pre-miRNAs), the starting position of the experimentally validated mature miRNA was defined as the positive dataset, while all the other putative positions were considered the negative dataset. Overall, our training set contained 671 and 11,677 positive and negative data points, respectively.
2.2. RNA Secondary Structure Prediction
The secondary structure of the pre-miRNA was predicted using RNAfold [31], with default parameters. Since our tool, MatPred, was designed to identify mature miRNAs from experimentally identified novel pre-miRNAs, we did not use the secondary structure feature included in miRBase, thus allowing generalization of our algorithm.
2.3. Mature miRNA Loci within the Pre-miRNA Structure
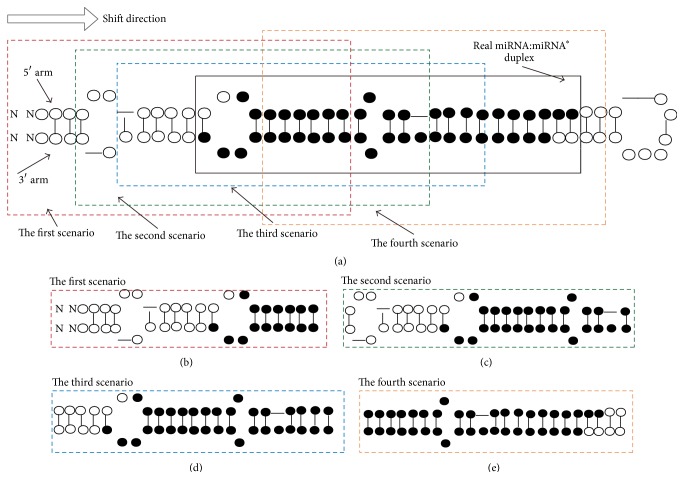
As shown in Figure 1, the goal of our algorithm was to identify the precise location of mature miRNAs (black circles) inside pre-miRNA structures. Without losing generalization, the mature miRNAs were assumed to be 22 nt in length. A sliding window scanning approach was taken by evaluating which nucleotide position was the most likely starting position of a box containing the mature miRNA:miRNA∗ duplex. Such a sliding box is defined as a duplex window.
Figure 1.
Definition of duplex window. (a) Four scenarios associated with duplex window selection. The solid black dot represents the nucleotides of the known mature miRNA. The hollow circles represent the nucleotides that are not part of the mature miRNA in the pre-miRNA transcript. All boxes represent duplex windows. The black box represents the known miRNA:miRNA∗ duplex. The four colored dashed boxes represent four scenarios while defining duplex window. (b)–(e) Detailed schematics corresponding to each scenario.
Based on the known theory of miRNA biogenesis, the starting position of the duplex window is design as follows: the start position of mature miRNA on the 5′ arm shifted 2 nucleotides to the left [20]. The ending position of this window was the ending position of the mature miRNA on its 5′ arm, as extracted from the predicted secondary structure of the pre-miRNA. In addition, the starting and ending positions could not be “-” (i.e., a missing nucleotide, which would create a bulge structure with no paired base).
While searching for the duplex window (i.e., the mature miRNA location), hypothetically, four different scenarios could emerge (as shown in the dashed boxes in Figure 1(a)).
Scenario 1 . —
The mature miRNA locates at the first nucleotide of the pre-miRNA. In this case, the duplex window will include 2 nucleotides before the starting position of 5′ arm in the pre-miRNA (Figure 1(b)).
Scenario 2 . —
When the ending position base of 5′ arm in the duplex window was “-”; it will be right-shift by 1 nucleotide excluding “-” (Figure 1(c)).
Scenario 3 . —
When the starting position character of the 5′ arm in the duplex window is “-”, the duplex window would be right-shifted by 1 nucleotide. The ending position would be dependent on the last nucleotide of the mature miRNA on its 5′ arm, which would be 22 nt from the preshifted position (Figure 1(d)).
Scenario 4 (standard scenario). —
When both starting and ending positions are nucleotides, the defined window will be specified as duplex window (Figure 1(e)).
2.4. Features Describing the Duplex Window
We designed 94 features to describe each duplex window, including position-specific structure features, distances to the stem loop, and minimum free energy characterizing the stability of the RNA molecule.
Duplex Window Region. Duplex window region is a window which contains the mature miRNA:miRNA∗ duplex. For each nucleotide in the duplex, we assigned one of 9 values to describe its sequence-structure characteristics, Ap, Cp, Gp, Up, A-, C-, G-, U-, and - -, where A, C, G, and U indicate nucleotide identity and p and “-” denote pairing or not. If the 5′- and 3′-arms of the nucleotide sequences in the duplex window perfectly match, this region would contain 48 nucleotides. However, in order to accommodate bulge structures (“-”), we devoted 60 features to characterize the position-specific structure. If the total nucleotide number in the duplex window was less than 60, the unused allocation was assigned to be 0 (data missing).
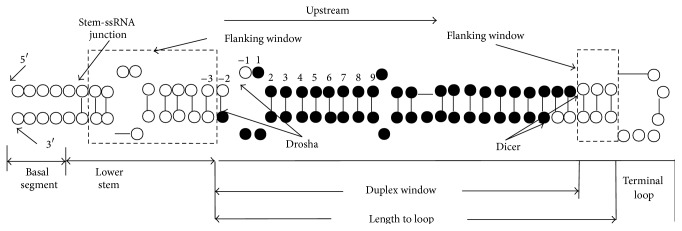
Lower Stem Loop (Figure 2). During miRNA biogenesis, the stem-ssRNA junction in the lower stem loop is often considered to determine the position of Drosha-processing sites, a key feature for localizing the mature miRNA in the pre-miRNA complex. Such junctions often locate ~11–13 nucleotides upstream of the duplex window [32]. In addition, the ~6–9 nucleotides flanking the mature miRNA usually include an internal loop and tend to have a specific sequence motif [33]. To capture such features, we extracted 18 sequence-structure characteristics of the upstream lower stem loop. Similarly, 6 additional features were assigned to the 3 pairs of nucleotides downstream the duplex window.
Figure 2.
Schematic of pre-miRNA compartments. According to Han et al. [32], we divided the pre-miRNA hairpin loop into four main regions: basal segment, lower stem, miRNA:miRNA∗ duplex, and the terminal loop. The black circles represent the miRNA:miRNA∗ duplex which we extracted using a flexible sliding window of 22 nt nucleotides. The dashed boxes represent flanking regions. The hollow circles represent the nucleotides of the immature miRNA in the pre-miRNA.
Minimum Free Energy. Minimum free energy is important for RNA stability. Under a certain temperature, RNA molecules reach thermodynamic equilibrium for having the minimum free energy and forming the most stable structure through adjusting the conformation. The duplex window usually has stable double-stranded structures [32, 34], and it is considered to have the minimum free energy. Usually, the most stable structure is observed on the duplex window plus 3 nt upstream of the Drosha-processing sites, while 6 nt upstream of that (9 nt upstream of the Drosha-processing site) is the most unstable region. In addition, for the mature miRNA in the 3′ arm, the 19-20th nucleotides have the highest free energy and thus the most instability. Meanwhile, within the miRNA of the 5′ arm, the 12th base pair is relatively unstable [32]. In order to capture these known and other unknown characteristics regarding minimum free energy, we designed 5 measurements for different regions relative to the duplex window, including MFE1 (duplex window), MFE2 (duplex window + 3 bp upstream of Drosha site), MFE3 (duplex window + 5 bp upstream of Drosha site), MFE4 (duplex window + 9 bp upstream of Drosha site), and MFE5 (duplex window + 3 bp downstream of end of duplex window). In addition, 4 other features were designed to measure the differences in minimum free energy for various regions, MFE6 = MFE2 − MFE1, MFE7 = MFE3 − MFE2, MFE8 = MFE4 − MFE3, and MFE9 = MFE5 − MFE1. In addition, we also used the distance from the beginning of the duplex window to the terminal loop as one feature. Overall, 94 features were used to evaluate one putative duplex window.
2.5. RBF-SVM
Due to its superior performance on prediction accuracy, we chose RBF-SVM (support vector machine with the radial basis function kernel as the decision function) to identify the mature miRNA location. The parameters in the model are determined using a standard support vector classification (SVC) algorithm.
Given the training data, positive data χ i ∈ S, i = 1,…, m, and negative data χ j ∈ S, j = 1,…, n, and for each vector of training data corresponding class label Z i ∈ {1, −1}, α i is the coefficient that must be determined. SVC solves the following optimization problem:
| (1) |
where χ i ∈ S, i = 1,…, m, and χ j ∈ S, j = 1,…, n, represent data points in the m positive and n negative training dataset, respectively, and α i is coefficient to be learnt (0 ≤ α i ≤ c).
k(χ, χ i) is the RBF kernel; this data classification method has been applied to several biological problems [35] and is an effective method to map data onto an infinite-dimensional feature space. The RBF kernel function is as follows:
| (2) |
The penalty parameter c and the RBF kernel parameter γ are implemented using the grid tool in the libSVM library [36].
This classifier offers the mature miRNAs candidates for the 5′ and 3′ arms. Every candidate is shown with two probabilities of belonging to any one category (positive or negative).
2.6. Prediction Outcome and Evaluation
For every nucleotide position in the pre-miRNA, a likelihood score will be calculated using the SVM model, based on the 94 features that are associated with the duplex window starting from its position. The accuracy of the model prediction is evaluated by comparing the distance between the predicted miRNA starting nucleotide from experimentally validated starting nucleotide on the 5′ arm; such distance is defined as position deviation (D(x)), where 0 means perfect prediction. A smaller position deviation indicates better prediction outcome.
3. Results and Discussions
3.1. Result
3.1.1. Model Performance
We tested the performance of MatPred using two separate datasets (each contains 100 pre-miRNAs) that are independent of training set. Specifically, Test set 2 is exclusively selected from pre-miRNAs that are in miRBase V20, but not in V19. This test is designed to examine whether MatPred is capable of detecting newly discovered miRNAs.
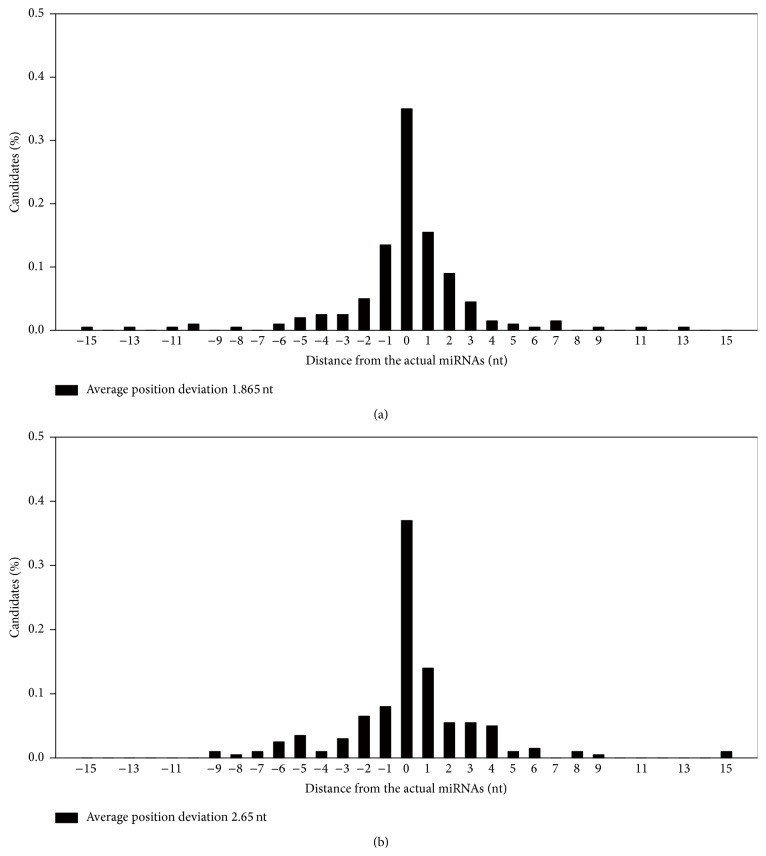
As shown in Figure 3 and Table 1, MatPred achieved fairly good prediction accuracy. In Test set 1, 35% of predicted mature miRNA loci matched perfectly with the experimentally validated starting nucleotides. In addition, predicted starting positions of 92% and 97% of miRNAs are within 5 nt and 10 nt of the experimentally validated positions, respectively. The analysis on Test set 2 dataset achieved similar prediction accuracy (Figure 3 and Table 1). This suggests that MatPred can be used to predict locations of newly discovered miRNAs. The overall position deviations, distance distribution (DS), for the two test datasets are 1.9 and 2.7 nucleotides, respectively. Figure 3 shows the average distance (AD) distributions over the Test 1 and test new datasets, illustrating the performance of MatPred for predicting the mature miRNA location.
Figure 3.
Distribution of the distances between predicted and known starting sites of mature miRNAs. (a) Test set 1 (randomly selected from nontraining dataset) and (b) Test set 2 dataset (selected from newly discovered miRNAs that are documented exclusively in miRBase V20).
Table 1.
Cumulative percentage of position deviation, DS, between predicted and known mature miRNA starting site.
| DS | ±0 | ±1 | ±2 | ±3 | ±4 | ±5 | ±6 | ±7 | ±8 | ±9 | ±10 | AD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Test 1 | 0.35 | 0.64 | 0.78 | 0.85 | 0.89 | 0.92 | 0.94 | 0.95 | 0.96 | 0.96 | 0.97 | 1.865 |
| Test 2 | 0.37 | 0.59 | 0.71 | 0.79 | 0.85 | 0.90 | 0.94 | 0.95 | 0.96 | 0.98 | 0.98 | 2.650 |
3.1.2. Comparisons with Other Methods
In order to compare the performance of our method with other published algorithms, we selected three methods that integrated a variety of sequence and structure features, MaturePred [25], MatureBayes [23], and miRdup [19]. These three algorithms are publicly available and actively maintained. In order to ensure the fair comparison with other methods, we used Test set 2, which contains only newly discovered miRNAs that are not documented in miRBase V20. This will guarantee that the entries in our test set do not overlap with the records in the training set of other algorithms and therefore avoid bias.
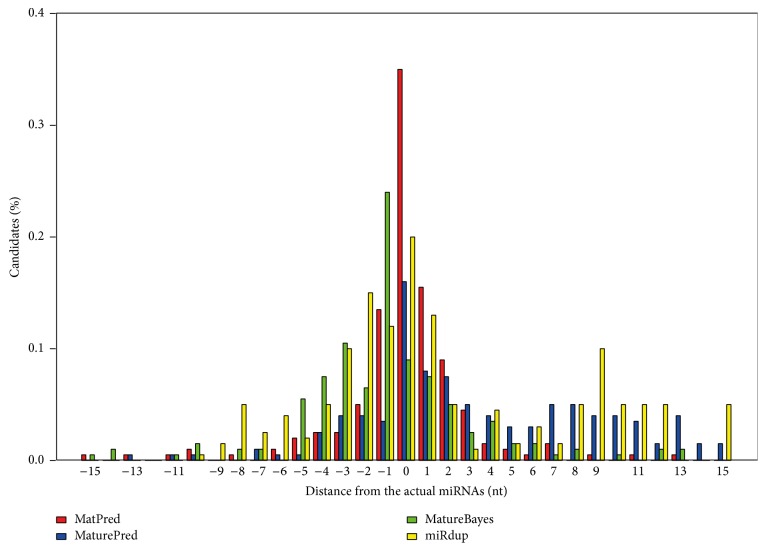
As shown in Figure 4 and Table 2, MatPred significantly outperformed the other three methods. The average position deviation, DS, for MatPred is 2.65 nt, as in comparison to 4.0 nt, 5.0 nt, and 3.0 nt for MaturePred, MatureBayes, and miRdup, respectively. In addition, our algorithm predicted 37% of the starting positions that coincided with those of the known miRNAs, significantly better than the 16%, 7%, and 19% predicted by MaturePred, MatureBayes, and miRdup, respectively.
Figure 4.
Distribution of distances between known mature miRNA starting sites and the ones predicted by MatPred, MaturePred, MatureBayes, and miRdup, based on newly discovered miRNAs (Test set 2).
Table 2.
Cumulative percentage of position deviation between known mature miRNA starting site and predicted ones based on four prediction algorithms.
| DS | ±0 | ±1 | ±2 | ±3 | ±4 | ±5 | ±6 | ±7 | ±8 | ±9 | ±10 | AD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MatPred | 0.37 | 0.59 | 0.71 | 0.80 | 0.86 | 0.90 | 0.94 | 0.95 | 0.97 | 0.98 | 0.98 | 2.650 |
| MaturePred | 0.16 | 0.29 | 0.36 | 0.42 | 0.49 | 0.58 | 0.64 | 0.74 | 0.82 | 0.85 | 0.88 | 4.005 |
| MatureBayes | 0.07 | 0.28 | 0.47 | 0.64 | 0.71 | 0.77 | 0.81 | 0.82 | 0.85 | 0.86 | 0.86 | 5.101 |
| miRdup | 0.19 | 0.42 | 0.59 | 0.65 | 0.74 | 0.82 | 0.86 | 0.91 | 0.94 | 0.96 | 0.98 | 3.004 |
Comparison between Test set 1 and other algorithms showed similar trend: MatPred outperformed all the other tools for prediction accuracy. Briefly, MatPred predicted that 35% of the starting positions coincided with those of known miRNAs, which is significantly better than the 16%, 9%, and 20% as predicted by MaturePred, MatureBayes, and miRdup, respectively. In addition, the average position deviation between predicted and known miRNA starting site is 1.865 nucleotides, while the respective values for MaturePred, MatureBayes, and miRdup were 5.603 nt, 4.22 nt, and 4.045 nt.
In order to compare the performance of our method for precise identification of the position of mature miRNAs with other published algorithms, we selected hsa-miR-6855, hsa-miR-6764, hsa-miR-7114, and hsa-miR-6894 which are the newly discovered pre-miRNA in miRBase V20 and hsa-miR-504 which is known pre-miRNA and experimentally verified having one miRNA in miRBase V19 as the test data. For the novel pre-miRNAs, the identification start positions of hsa-miR-6855-5p, hsa-miR-6764-5p, hsa-miR-7114-5p, and hsa-miR6894-5p using our method were supported by present reports [37, 38]. MatureBayes, MaturePred, and miRdup identify the start position of hsa-miR-6855-5p with −1 nt, −1 nt, and 1 nt deviation, hsa-miR-6764-5p with 2 nt, 1 nt, and 1 nt deviation, hsa-miR-7114-5p with 3 nt, 2 nt, and 2 nt deviation, and hsa-miR6894-5p with 2 nt, −1 nt, and −1 nt deviation. For the known pre-miRNAs, our method discovers the new miRNA and identifies the start position of hsa-miR-504-3p which is proved by the present reports [37, 39]. And MatureBayes, MaturePred, and miRdup predicted its position with −3 nt, −1 nt, and 3 nt deviation.
3.2. Discussions
Although the secondary structure properties of pre-miRNA transcripts have been long exploited in miRNA biology, they have only been effectively used in some mature miRNA prediction methods [19, 23, 30]. In this study, we report a new prediction algorithm, MatPred, for predicting the location of mature miRNAs within the novel pre-miRNA transcripts, based on RBF-SVM. The major purpose of our algorithm is to identify mature miRNA loci from precursor miRNA sequences, not to distinguish pre-miRNAs from pseudo-pre-miRNAs which include sequence hairpin structures derived from the 3′UTR regions. Therefore, the noncanonical mature miRNA loci will serve as a base for false prediction.
Based on the secondary structure prediction for the pre-miRNA transcripts, MatPred generates a feature list that integrates the RNA secondary structure properties of each nucleotide within and flanking the mature miRNA region. In addition, several other features that are highly relevant to miRNA biogenesis were selected, including length of mature and pre-miRNA, distances from stem loop, and minimum free energy. Such features not only described the base-pairing structures of miRNA:miRNA∗ complex but also captured RNA stability characteristics that associate with different regions on the pre-miRNA transcripts; for instance, a bulge at lower stem region (5′-flanking region of the duplex window in Figure 2) may help to create higher free energy for Drosha cleavage.
As evaluated by two independent datasets, our model showed significant improvement on the prediction accuracy compared to several existing methods. Remarkably, ~35% predictions accurately reported the known locations of mature miRNAs, and over 90% of the predicted positions were within 5 bp of known sites.
It should be noted that our prediction is based on static pre-miRNA sequences presented in the miRBase database, which is derived from the reference sequence assembly. This model does not consider the genetic variations in the human population, which may affect the accuracy of the prediction. In addition, it has been widely reported that mature miRNAs have significant end-heterogeneity. This may further underestimate the accuracy evaluation on our method.
4. Conclusion
In conclusion, our method suggests that the biologically relevant features extracted from extending duplex windows, combined with RBF-SVM, generate an effective classifier for mature miRNA location identification. Such modeling could provide key insight into miRNA processing and biogenesis.
Supplementary Material
Supplementary Table 1: proposes the training and test datasets.
Acknowledgments
The authors would like to acknowledge the financial support of the National Key Scientific Instrument and Equipment Development Projects of China (2012YQ04014001 and 2012YQ04014010), the National Natural Science Foundation of China (61471139), the Fundamental Research Funds for the Central Universities (HEUCFT1102, HEUCFT1302, and HEUCFX41303), the Science and Technology Research Program of Heilongjiang Educational Committee (12541898), the Applied Technology Research and Development Project of Heilongjiang Province (no. GC13A308), and the Natural Science Fund of Heilongjiang Province (F201331).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Fahlgren N., Jogdeo S., Kasschau K. D., et al. Microrna gene evolution in Arabidopsis lyrata and Arabidopsis thaliana . Plant Cell. 2010;22(4):1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts T. C. The microRNA biology of the mammalian nucleus. Molecular Therapy—Nucleic Acids. 2014;3, article e188 doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang Q., Wang Y., Hao Y., et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Research. 2009;37(supplement 1):D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero-Cordoba S. L., Salido-Guadarrama I., Rodriguez-Dorantes M., Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biology and Therapy. 2014;15(11):1444–1455. doi: 10.4161/15384047.2014.955442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nam J.-W., Shin K.-R., Han J., Lee Y., Kim V. N., Zhang B.-T. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Research. 2005;33(11):3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi R., Qin Y., Macara I. G., Cullen B. R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tijsterman M., Plasterk R. H. A. Dicers at RISC; the mechanism of RNAi. Cell. 2004;117(1):1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y. S., Nakahara K., Pham J. W., et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117(1):69–81. doi: 10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 9.Brennecke J., Stark A., Russell R. B., Cohen S. M. Principles of microRNA-target recognition. PLoS Biology. 2005;3(3, article e85) doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Research. 2006;34(supplement 2):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stark A., Brennecke J., Russell R. B., Cohen S. M. Identification of Drosophila microRNA targets. PLoS Biology. 2003;1(3, article E60) doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamura K., Phillips M. D., Tyler D. M., Duan H., Chou Y. T., Lai E. C. The regulatory activity of microRNA∗ species has substantial influence on microRNA and 3′ UTR evolution. Nature Structural & Molecular Biology. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finnegan E. F., Pasquinelli A. E. MicroRNA biogenesis: regulating the regulators. Critical Reviews in Biochemistry and Molecular Biology. 2013;48(1):51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha M., Kim V. N. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 16.Friedländer M. R., Chen W., Adamidi C., et al. Discovering microRNAs from deep sequencing data using miRDeep. Nature Biotechnology. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 17.Li S.-C., Chan W.-C., Ho M.-R., et al. Discovery and characterization of medaka miRNA genes by next generation sequencing platform. BMC Genomics. 2010;11(supplement 4):p. S8. doi: 10.1186/1471-2164-11-s4-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo J. K., Kim C.-H., Jung H. Y., Lee D. R., Kim J. K. Discovery and characterization of miRNA during cellular senescence in bone marrow-derived human mesenchymal stem cells. Experimental Gerontology. 2014;58:139–145. doi: 10.1016/j.exger.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Leclercq M., Diallo A. B., Blanchette M. Computational prediction of the localization of microRNAs within their pre-miRNA. Nucleic Acids Research. 2013;41(15):7200–7211. doi: 10.1093/nar/gkt466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., Wei B., Liu H., Li T., Rayner S. MiRPara: a SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinformatics. 2011;12, article 107 doi: 10.1186/1471-2105-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C., Li L. Y.-X., Zhang G., et al. MiRmat: mature microRNA sequence prediction. PLoS ONE. 2012;7(12) doi: 10.1371/journal.pone.0051673.e51673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terai G., Okida H., Asai K., Mituyama T. Prediction of conserved precursors of miRNAs and their mature forms by integrating position-specific structural features. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0044314.e44314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gkirtzou K., Tsamardinos I., Tsakalides P., Poirazi P. MatureBayes: a probabilistic algorithm for identifying the mature miRNA within novel precursors. PLoS ONE. 2010;5(8) doi: 10.1371/journal.pone.0011843.e11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helvik S. A., Snøve O., Jr., Sætrom P. Reliable prediction of Drosha processing sites improves microRNA gene prediction. Bioinformatics. 2007;23(2):142–149. doi: 10.1093/bioinformatics/btl570. [DOI] [PubMed] [Google Scholar]
- 25.Xuan P., Guo M., Huang Y., Li W., Huang Y. MaturePred: efficient identification of microRNAs within novel plant Pre-miRNAs. PLoS ONE. 2011;6(11) doi: 10.1371/journal.pone.0027422.e27422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan D.-G., Liao J.-Y., Qu Z.-H., Zhang Y., Qu L.-H. mirExplorer: detecting microRNAs from genome and next generation sequencing data using the AdaBoost method with transition probability matrix and combined features. RNA Biology. 2011;8(5):922–934. doi: 10.4161/rna.8.5.16026. [DOI] [PubMed] [Google Scholar]
- 27.Jones-Rhoades M. W., Bartel D. P. Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Molecular Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 28.Sætrom P., Snøve O., Jr., Nedland M., et al. Conserved microRNA characteristics in mammals. Oligonucleotides. 2006;16(2):115–144. doi: 10.1089/oli.2006.16.115. [DOI] [PubMed] [Google Scholar]
- 29.Hertel J., Langenberger D., Stadler P. F. Computational prediction of microRNA genes. Methods in Molecular Biology. 2014;1097:437–456. doi: 10.1007/978-1-62703-709-9_20. [DOI] [PubMed] [Google Scholar]
- 30.Liu X., He S., Skogerbø G., Gong F., Chen R. Integrated sequence-structure motifs suffice to identify microRNA precursors. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0032797.e32797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auyeung V. C., Ulitsky I., McGeary S. E., Bartel D. P. Beyond secondary structure: primary-sequence determinants license Pri-miRNA hairpins for processing. Cell. 2013;152(4):844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J., Lee Y., Yeom K.-H., et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 33.Saçar M. D., Hamzeiy H., Allmer J. Can MiRBase provide positive data for machine learning for the detection of MiRNA hairpins? Journal of Integrative Bioinformatics. 2013;10(2):p. 215. doi: 10.2390/biecoll-jib-2013-215. [DOI] [PubMed] [Google Scholar]
- 34.Krol J., Sobczak K., Wilczynska U., et al. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. The Journal of Biological Chemistry. 2004;279(40):42230–42239. doi: 10.1074/jbc.m404931200. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z. R. Biological applications of support vector machines. Briefings in Bioinformatics. 2004;5(4):328–338. doi: 10.1093/bib/5.4.328. [DOI] [PubMed] [Google Scholar]
- 36.Chen P.-H., Fan R.-E., Lin C.-J. A study on SMO-type decomposition methods for support vector machines. IEEE Transactions on Neural Networks. 2006;17(4):893–908. doi: 10.1109/tnn.2006.875973. [DOI] [PubMed] [Google Scholar]
- 37.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research. 2011;39(1):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ladewig E., Okamura K., Flynt A. S., Westholm J. O., Lai E. C. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Research. 2012;22(9):1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meunier J., Lemoine F., Soumillon M., et al. Birth and expression evolution of mammalian microRNA genes. Genome Research. 2013;23(1):34–45. doi: 10.1101/gr.140269.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: proposes the training and test datasets.