Dear Editor,
Ferrets (Mustela putorius furo) have served as one of the most valuable animal models for recapitulating human disease since 19331. This is because they show many similarities to humans in terms of physiological features of brain function and reproductive biology as well as pathological characteristics of various diseases such as cancer, influenza infection and cystic fibrosis2,3,4. Despite these advantages, the current use of ferrets in research is significantly limited due to the lack of a method enabling precise genetic modification.
Here we report that by co-injecting Cas9 mRNA and sgRNAs into one-cell-stage embryos, founder ferrets with biallelic mutations in one of three genes (Dcx, Aspm and Disc1) can be generated with high efficiency up to 73.3%. Intriguingly, the founder animal carrying Dcx mutations showed a lissencephaly phenotype similar to human Doublecortin patients. Thus, we demonstrate for the first time that CRISPR/Cas9-mediated gene editing allows for efficient generation of gene knockout ferrets.
The CRISPR/CRISPR-associated (Cas) 9 system is a highly specific, efficient, and multiplexable approach for eukaryotic genome editing5,6,7. Due to the similarity of brain structure and development between ferret and human, we chose Dcx, Disc1 and Aspm, genes with well-known functions in brain development and gyrification, as the target genes8,9. Two sgRNAs were designed for each gene based on previously published studies5 and their target sites are shown in Supplementary information, Figure S1A. Since ferret cell lines are not yet available, we set out to test the efficiency of these sgRNAs with the Single Strand Annealing (SSA) luciferase reporter assay in 293T cells10. The targeting site for each sgRNA was sub-cloned into a reporter plasmid (Supplementary information, Figure S1B)10. We found that the Cas9/sgRNAs efficiently generated double-stranded breaks (DSBs) and homologous recombination repair was executed around their targeting sequences, indicated by the reconstitution of the luciferase activity (Supplementary information, Figure S1C). Based on these data, we chose sgRNAs with higher efficiency, Dcx-sgRNA1, Disc1-sgRNA2 and Aspm-sgRNA2, for the following embryo injection experiments.
Next, we tested whether ferrets carrying targeted genetic mutations could be generated by direct embryo injection of the CRISPR/Cas9 system. Cas9 mRNA and sgRNA targeting Dcx were injected into ferret one-cell-stage embryos. Mutations in human X-linked Dcx gene cause gross neocortical disorganization (lissencephaly or “smooth brain”) in males, whereas females heterozygous for Dcx mutations show a mosaic phenotype featuring the presence of a normal cortex and a second band of misplaced (heterotopic) neurons beneath the cortex (“double cortex syndrome”)9.
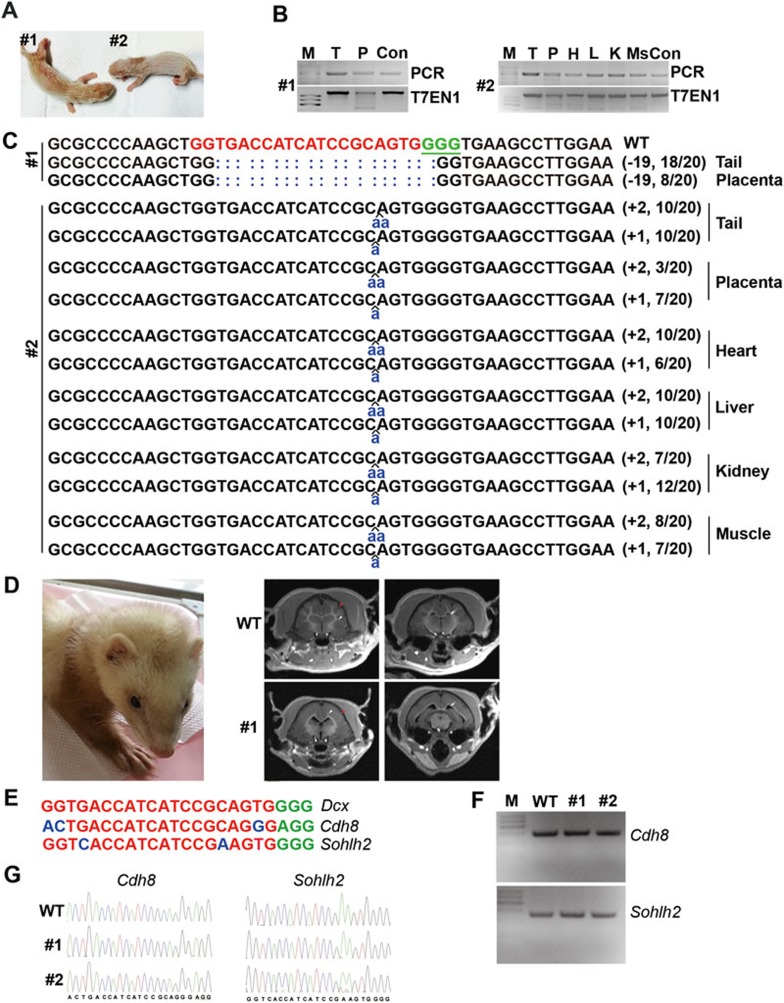
A total of 124 fertilized eggs at one-cell stage were collected (Supplementary information, Table S1A). Dcx-sgRNA1 (30 ng/μl) and Cas9 mRNA (20 ng/μl) were injected. 117 out of 124 injected zygotes were transferred into 10 surrogate females. Among the 10 surrogates, 5 had miscarriage 21 days after embryo transfer (Supplementary information, Table S1A). The first surrogate mother that went through the full-term pregnancy delivered one male (#1) and one female (#2) via caesarean section, but pup #2 died one day later (Figure 1A). Genomic DNAs from placenta and tail were isolated from both pups #1 and #2. Targeted genome modifications were screened by PCR amplification of regions surrounding the Dcx-sgRNA1 target sites, followed by the T7EN1 cleavage assay (Figure 1B) and sequencing analysis (Figure 1C). Different small insertions or deletions (indels) at the target site were detected by sequencing the PCR products (Figure 1C), demonstrating that the CRISPR/Cas9 system functions well in ferret embryos. The presence of gene modifications in pup #2 was further analyzed using genomic DNAs from heart, liver, kidney and muscle in addition to the tail and placenta, and the same mutant alleles were detected in these tissues, demonstrating that the target modifications have been ubiquitously and efficiently integrated into different tissues (Figure 1C). 13 more pups were delivered from the other 4 surrogate females later (Supplementary information, Figure S1D). In total, 11 out of 15 pups were detected to harbor Dcx modifications (Supplementary information, Figure S1D-S1E and Table S1A). Dcx is a brain-specific gene mutated in X-linked lissencephaly and the double cortex syndrome. In male patients, X-linked lissencephaly produces a smooth brain without normal folding of the brain surface due to lack of migration of immature neurons9. To assess whether a lissencephaly brain phenotype could be modeled in the Dcx-modified ferrets, we conducted magnetic resonance imaging (MRI) of ferret #1 (male) 14 weeks after its birth. MRI showed a smooth brain with thick cortex but no sulci, abnormal myelination of axonal fiber tracts and ventricular enlargement in this Dcx mutant ferret founder (Figure 1D), which resemble the typical phenotypes seen in male patients with Dcx mutations. This result demonstrates that the ferret can perfectly model the phenotypes of this human genetic disease, which was not achievable in rodent systems. Taken together, we have successfully achieved CRISPR/Cas9-mediated site-specific modifications in ferret genome by one-cell embryo microinjection.
Figure 1.
CRISPR/Cas9-mediated modification of Dcx in different tissues of ferrets. (A) Photograph of 1-day-old founder pups #1 and #2. (B) PCR products of the target region of Dcx in founders and the results of the T7EN1 cleavage assay. Genomic DNA from the indicated tissues was collected and the target region in the Dcx locus was amplified by PCR. The PCR products were then subjected to the T7EN1 cleavage assay. M, DNA maker; Con, control; T, tail; P, placenta; H, heart; L, liver; K, kidney; Ms, muscle. (C) Modified Dcx loci detected in founders #1 and #2. Genomic DNA from the indicated tissues was collected and the target region in Dcx was PCR-amplified. 20 TA clones of each PCR product were analyzed by DNA sequencing. The PAM sequence is underlined and highlighted in green; the targeting sequence in red; the mutations in blue, lower case; deletions (−), and insertions (+). N/N indicates positive clones out of total clones sequenced. (D) MRI reveals the lissencephalic brain of Dcx-modified founder #1. Arrow, ventricular; white arrowhead, white matter; red arrowhead, brain folds in wild-type (upper panel) but not in Dcx-mutated ferret (lower panel). (E) Two potential off-target sites of Dcx sgRNA1. The 12-bp seed sequence is labeled in red, mismatched sequence is labeled in blue and the NGG PAM sequence is labeled in green. Only two potential off-target loci (Cdh8 and Sohlh2) were identified in ferret genome for Dcx sgRNA1. (F) PCR products of Cdh8 and Sohlh2 fragments in ferrets #1 and #2. (G) Sequences of PCR products shown in F reveal no mutations in potential off-target loci Cdh8 and Sohlh2.
With the success of Dcx modification in ferret, we then sought to generate founder ferrets carrying mutations in Aspm and Disc1 genes. A total of 64 injected zygotes were transferred into 4 surrogate females for the Aspm targeting experiment, and 18 injected zygotes were transferred into 1 surrogate female for the Disc1 targeting experiment. In total, 12 pups for the Aspm experiment and 4 pups for the Disc1 experiment were delivered (Supplementary information, Table S1A). In each case, genomic DNA from the tail was collected and analyzed by PCR, T7EN1 cleavage assay and sequencing to examine the site-specific genome modifications. Characterization of the targeting sites by sequencing showed different indels with sizes ranging from −13 to +1 bp in ferret pups at these two target genes with an efficiency of 66.7% (8/12) for Aspm and 25% (1/4) for Disc1 (Supplementary information, Figure S1D-S1E and Table S1), suggesting that the CRISPR/Cas9 system functions efficiently in targeting different genes in ferret. To assess whether a microcephaly brain phenotype could be observed in ferrets with the Aspm gene modification, resembling patients with Aspm mutations, MRI was used to visualize the cortical structure. The ferret with Aspm mutation showed a smaller brain with gyrification defect compared to the wild type (Supplementary information, Figure S1F).
To test whether off-target mutations occurred in these genetically modified ferrets, we screened the ferret genome and identified two potential off-target loci, Cdh8 and Sohlh2, for Dcx-sgRNA1(Figure 1E); no potential off-target sites were identified for Aspm and Disc1 sgRNAs according to the predicting rule described previously7. The fragments around all the potential off-target loci were PCR amplified, and then subjected to sequencing assay. No mutation was detected at these two potential off-target loci, Cdh8 and Sohlh2 (Figure 1F-1G and Supplementary information, Figure S2A-S2B). These results demonstrate that the Cas9/sgRNA used in our experiment does not generate detectable mutations in potential off-target loci, suggesting that the CRISPR/Cas9 system could be a reliable genome modification tool for ferrets. However, we could not rule out the possibility of the existence of other off-target loci, which cannot be predicted based on the current rule.
Thus, our study demonstrates the high efficiency of the CRISPR/Cas9 system for gene editing in ferret and suggests the possibility of sophisticated genome engineering in many other species of which ES cells are not available. The establishment of gene-modified ferrets would facilitate the investigation of genes related to human diseases that are difficult to model in rodents and primates.
Acknowledgments
We thank William Walantus, Arnold Kriegstein for comments on the manuscript and members of the Wang and Gao laboratories for discussion. We thank Wensu Liu, Fangyu Shi, Jiayu Chen, Pingdong Hu for technical support. This work was supported by grants from the National Basic Research Program of China (2014CB964600, 2012CBA01300 and 2012CB966800), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01020309), the National Natural Science Foundation of China (NSFC; 31371100, 91332105, 31400937, 31400938 and 31400939), and the Thousand Young Talents Program of China.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Summary of embryo microinjection of Cas9 mRNA and sgRNAs
References
- 1Smith H, Sweet C. Rev Infect Dis 1988; 10:56–75. [DOI] [PubMed]
- 2McConnell SK, Ghosh A, Shatz CJ. Science 1989; 245:978–982. [DOI] [PubMed]
- 3Renegar KB. Lab Anim Sci 1992; 42:222–232. [PubMed]
- 4Maher JA, DeStefano J. Lab Anim (NY) 2004; 33:50–53. [DOI] [PubMed]
- 5Cong L, Ran FA, Cox D, et al. Science 2013; 339:819–823. [DOI] [PMC free article] [PubMed]
- 6Sander JD, Joung JK. Nat Biotechnol 2014; 32:347–355. [DOI] [PMC free article] [PubMed]
- 7Wang H, Yang H, Shivalila CS, et al. Cell 2013; 153:910–918. [DOI] [PMC free article] [PubMed]
- 8Bond J, Roberts E, Mochida GH, et al. Nat Genet 2002; 32:316–320. [DOI] [PubMed]
- 9Gleeson JG, Allen KM, Fox JW, et al. Cell 1998; 92:63–72. [DOI] [PubMed]
- 10Mashiko D, Fujihara Y, Satouh Y, et al. Sci Rep 2013; 3:3355. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of embryo microinjection of Cas9 mRNA and sgRNAs