Abstract
Sphingolipid metabolites are involved in the regulation of autophagy, a degradative recycling process that is required to prevent neuronal degeneration. Drosophila blue cheese mutants neurodegenerate due to perturbations in autophagic flux, and consequent accumulation of ubiquitinated aggregates. Here, we demonstrate that blue cheese mutant brains exhibit an elevation in total ceramide levels; surprisingly, however, degeneration is ameliorated when the pool of available ceramides is further increased, and exacerbated when ceramide levels are decreased by altering sphingolipid catabolism or blocking de novo synthesis. Exogenous ceramide is seen to accumulate in autophagosomes, which are fewer in number and show less efficient clearance in blue cheese mutant neurons. Sphingolipid metabolism is also shifted away from salvage toward de novo pathways, while pro-growth Akt and MAP pathways are down-regulated, and ER stress is increased. All these defects are reversed under genetic rescue conditions that increase ceramide generation from salvage pathways. This constellation of effects suggests a possible mechanism whereby the observed deficit in a potentially ceramide-releasing autophagic pathway impedes survival signaling and exacerbates neuronal death.
Defective sphingolipid metabolism, in particular misregulation of ceramide production and a failure to catabolize complex sphingolipids, leads to neurodegeneration, often accompanied by increased ceramide levels1,2. While excessive ceramide production can trigger apoptotic mitogen-activated protein kinase (MAPK)-c-Jun Kinase (JNK) activation3,4, ceramide metabolites are also positive regulators of autophagy, critical for maintaining neuronal health by ridding the cell of pathogenic aggregated proteins and aged mitochondria5,6. In non-pathological situations, autophagy provides a source of energy under nutrient stress7,8,9,10,11. For this reason, it has been proposed that ceramides act as a ‘rheostat’ that can lead to cellular catastrophe if pushed out of balance12.
Relatively little is known about causal relationships of ceramide misregulation to neurodegeneration in disease models, but genetic manipulation of sphingolipid metabolism has been shown to influence retinal and muscle degeneration in flies13,14,15. Drosophila blue cheese (bchs) mutants are neurodegenerative, and Alfy, the human homologue of bchs, acts as a scaffolding protein that ferries the aggresome via p62/sequestosome (fly Ref(2)p) with Atg5, Atg12, Atg16L, and Atg8/LC3 toward autophagosomal degradation16. Ubiquitin and p62 accumulate progressively in bchs mutants17,18, suggesting that some aspect of autophago/lysosomal degradation is faulty. Larval motor neurons also show increased lysosomal size and altered lysosomal transport in bchs mutants19,20. Bchs contains a conserved BEACH domain, involved in vesicle trafficking21. Although no specific function has been assigned to the BEACH domain, the same domain is required in FAN (Factor Activating Neutral-Sphingomyelinase) for its activity upon interaction with TNFα receptor22 and production of the signaling molecule ceramide. Based on this information, and because of the precedent of ceramide imbalance in several neurodegenerative situations, we hypothesized that the degeneration in bchs animals may be related to ceramides, which might then be associated with its known autophagic defects.
Here, we show by lipidomic analysis that ceramide levels in bchs mutant brains are unexpectedly increased. Furthermore, we demonstrate that ceramides are centrally involved in the bchs degenerative phenotype, by genetically manipulating ceramide levels, and quantitatively measuring the effect on bchs degeneration, autophagic clearance, and Akt, MAPK, and stress signaling. Surprisingly, although bchs brains have increased endogenous ceramides, and accumulate exogenous ceramide in autophagosomes, degeneration is rescued by genetic backgrounds that increase ceramide availability; conversely, it is exacerbated by catabolism of ceramide and by drugs that lower de novo ceramide production.
In this study, genetic modulation of sphingolipid catabolic pathways that increase ceramide load from salvage (recycling) pathways, not only rescued degeneration, but also improved autophagic clearance, and restored sphingolipid-regulated Akt, MAPK, and stress signaling that were dysregulated in bchs. This suggests that ceramides that are recycled through an autophagic pathway act positively on Akt- and MAPK survival pathways, while suppressing ER-stress in the degenerative background, and thereby prevent neuronal demise.
Results
Neurodegeneration in bchs mutants is modulated by genetically or pharmacologically altering ceramide availability
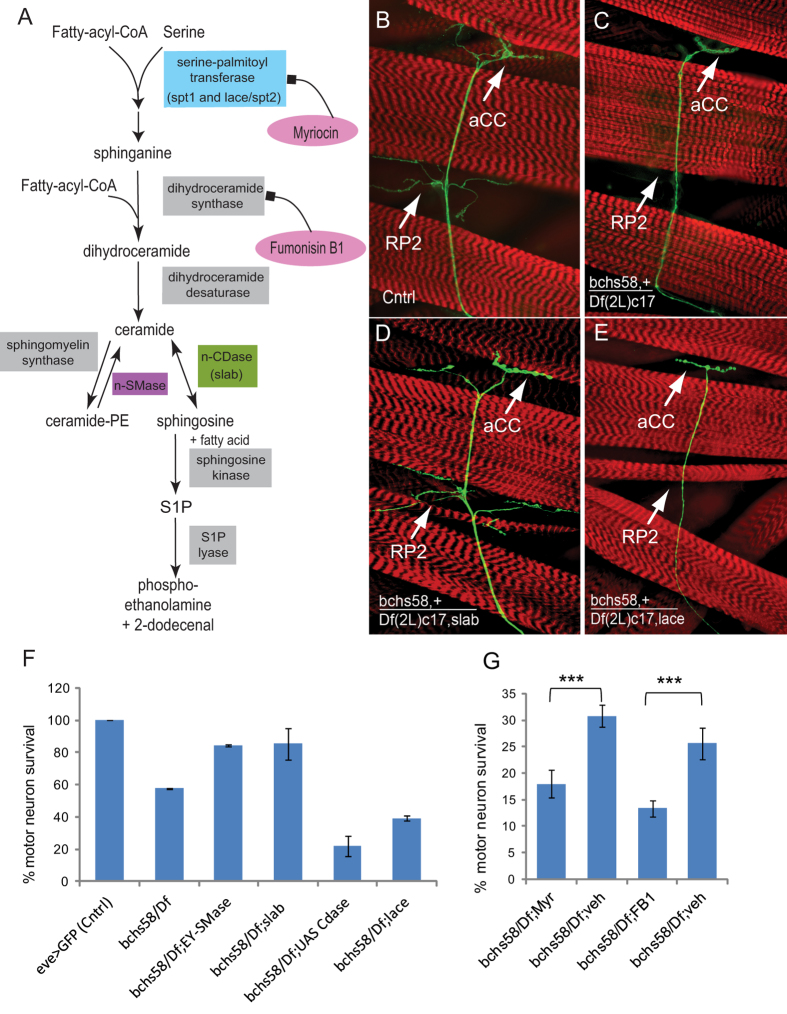
The potential link of the BEACH domain to sphingomyelin catabolism, perturbations in which can lead to neuronal death23,24,25, led us to ask whether bchs degeneration could arise from the mis-regulation of ceramides. If this were the case, we reasoned that bchs might be sensitive to genetic changes in sphingolipid biosynthesis or catabolism (Fig. 1A), which in the fly would involve hydrolysis of ceramide (CerPE), the functional equivalent of sphingomyelin. To test this, we used even-skipped Gal4 driving GFP (eve > GFP; Fig. 1B), to label two identified neurons, RP2 and aCC, which in bchs null mutants, e.g. bchs58/Df(2L)cl7 (denoted bchs58/Df) are lost with a penetrance of ~50%20, Fig. 1C,F). Using this assay, different genetic interactors and pharmacological agents could be tested for their effects on motor neuron loss (Fig. 1B–G).
Figure 1. Genetic and pharmacological manipulation of sphingolipid metabolism influences bchs neurodegeneration.
(A) The sphingolipid metabolic pathway showing enzymes that were targeted in colored boxes and their given Drosophila gene names. Pharmacological agents are indicated in pink ovals. (B–E) Larval fillets showing motorneuron aCC and RP2 termini expressing mCD8GFP (green) and muscles stained with rhodamine-Phalloidin (red). Loss of motor neurons is evident in bchs vs. control (C) vs. (B). (D,E) show rescue and exacerbation, respectively, by heterozygosity for slab2 (D) and lacek05305 (E). (F) Quantification of motor neuron loss in bchs58/Df(2L)c17 and in combination with modifiers. Bars indicate percent survival of RP2 motor neuron in >200 scored hemisegments. For (F,G) bars represent standard error of the mean. (G) Effect on motorneuron survival after feeding wtih 12.4 mM Myriocin (Myr) in vehicle (veh) 50 mM NaOH, or 1 mM Fumonisin (FB1) in water. ***P < 0.001, by Chi-square test.
The graph in Fig. 1F summarizes changes in the penetrance of the bchs motor neuron phenotype when components of the sphingolipid pathway are manipulated. Reduction of ceramide biosynthesis in the bchs null background was first achieved through recombination of lacek05305 (serine palmitoyl transferase 2; spt2; CG4162) with the deficiency for bchs, Df(2L)cl7. Secondly, UAS-nCeramidase (CDase; slab; CG1471)26, was recombined onto Df(2L)cl7, and driven simultaneously with eve > GFP13,27,28. Both these conditions exacerbated motor neuron loss (Fig. 1E,F).
Next, an increase in ceramide levels (evidence for which is shown in Fig. 2) in the presence of bchs was achieved by introducing heterozygosity for the CDase mutation slab2 or by driving EY00448 (inserted near the neutral-sphingomyelinase (nSMase) homologue CG12034) in the neurons being scored. Both these backgrounds resulted in rescue of the bchs phenotype (Fig. 1D,F), complementing the above findings. None of the modifiers affected neuronal survival significantly by themselves: neuronal survival in slab2/+ was 98.6%; lacek05305/+ 100%; driving CDase and nSMase (EY00448) resulted in 99.3% and 100% motor neuron survival respectively.
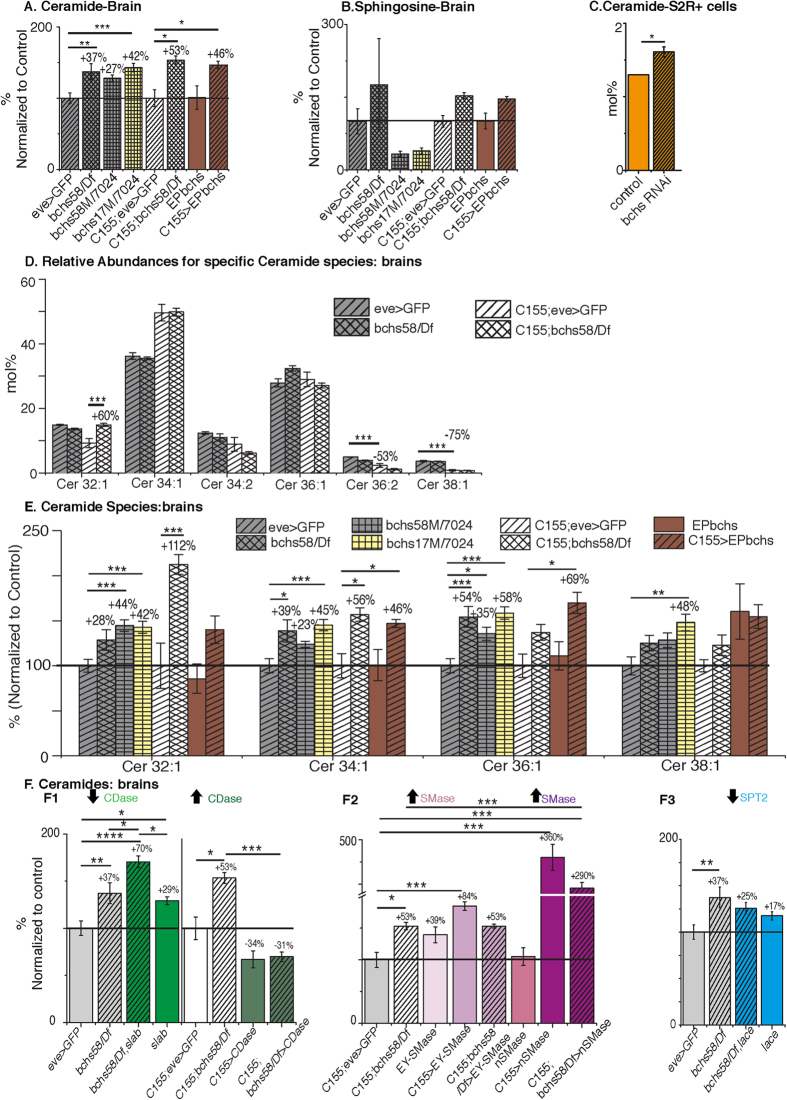
Figure 2. Increased ceramide levels with bchs reduction.
(A) Total ceramide levels are significantly increased in bchs58/Df, bchs17M/7024, C155; bchs58/Df larval brain, and by overexpression in CNS via C155 > EPbchs (EP2299). (B) Abundance of C14-sphingosine in larval brain as determined by LC-MS3 analysis. (C) Ceramide levels in S2R + cells treated with or without (control) bchs RNAi, (D) Relative distribution profiles for ceramide species in larval brain profiles are similar for bchs58/Df (double hatched bars) in the two genetic backgrounds, with or without C155. Cer 32:1 is changed in bchs in the C155; eve > GFP background. (E) Major ceramide species in brains compared across the allelic combinations show significant increases in one or more of the major species. (A,B,E) Bars represent mean ± SEM of percent change for lipids (quantified as picomoles/brain) with respect to the genetic controls, eve-Gal4 driving UASmCD8GFP (eve > GFP; grey hatched bars) alone or in combination with C155-Gal4 (C155; eve > GFP; white hatched bars). The control level of 100% is indicated by a horizontal line. (C-D) Bars represent percent molar ratios for total ceramides (normalized to phospholipids) measured from S2R + cells (C) and for ceramide species measured from larval brains (D,F). Modifiers of bchs degeneration change ceramide levels. F1. CDase heterozygote slab2/+ increases total ceramides and rescues the degeneration whereas CDase overexpression lowers ceramide levels. F2. nSMase overexpression markedly increases ceramide. F3. lacek05305/spt2 heterozygotes do not affect ceramide levels. Bars represent mean ± SEM of percent change with respect to genetic control of total ceramide (quantified as picomoles/brain) for manipulations of CDase (slab) (1), nSMase (2) and Spt2 (lace) (3). Color and hatching schemes represent different bchs allelic combinations or bchs combined with a given genetic background. Numbers represent percent change relative to the genetic control, set to 100% (indicated by horizontal line). Statistical significance between 2 genotypes is indicated by *p < 0.05, **p < 0.005 and ***p < 0.0005 as determined by ANOVA followed by post-hoc Tukey analyses. Lipidome-wide changes are shown in supplementary sections S1 and S2. Changes in specific ceramide species are summarized in supplementary Figures S3–S5.
From these genetic interactions, it can be inferred that the neurodegenerative phenotype in bchs mutants is highly sensitive to mutations in sphingolipid metabolism that alter ceramide availability (quantitated for the different genetic backgrounds in Fig. 2F). We went on to test whether bchs mutants responded in a similar way to pharmacological interference with the sphingolipid biosynthetic pathway (outlined in Fig. 1A) using myriocin, an inhibitor of29 and FumonisinB1 (FB1) a fungal inhibitor of dihydroceramide synthase30. In agreement with genetic exacerbation by the spt2(lacek05305)/+ background, which should also reduce de novo production of sphingolipids, we found that myriocin exacerbated bchs degeneration by 36% and FB1 exacerbated it by 48% (Fig. 1G). Both pharmacological treatments indicate that deficits in de novo synthesis of ceramide worsen the neurodegeneration.
Lipidomics analyses of bchs mutants reveal specific changes in ceramides
Since our genetic interaction experiments suggested sensitivity of bchs to changes in sphingolipid metabolism, a LC-MSn based lipidomic analysis was performed on brains of bchs mutants and interactor genotypes. Lipids were identified based on accurate mass measurements and retention time tags and/or formation of characteristic fragment ions of the sphingoid base30,31,32. Identified lipid classes are listed in Supplementary Table S1. The abundances of lipid species and lipid classes closely overlap with recently reported lipidomic profiles of third-instar larval brain33. No significant changes in the major abundant lipids phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylethanolamine ether (PE-O) and ceramide phosphorylethanolamine (CerPE) were detected across all genotypes (Supplementary Table S2). However, in line with results of our genetic interactions, ceramides, and the long-chain base sphingosine are significantly affected in bchs mutants and/or interactors (Fig. 2; Supplementary Table S2).
Unexpectedly in light of its sensitivity to genetic and pharmacological perturbation of ceramides, bchs reduction led to higher overall ceramides in larval brains and S2R + cell lines (Fig. 2A,C). The two stronger allelic combinations bchs58/Df and bchs17/7024 showed an increase of 37% and 42% respectively compared to their genetic background, eve > GFP (Fig. 2A). bchs58/Df and EP2299 also caused increases in ceramides of 53% and 46% in a C155 background (Fig. 2A), without significantly changing sphingosine (Fig. 2B).
Distribution profiles for ceramide species in larval brains were consistent with previous reports for the Drosophila lipidome33, Cer 34:1 and Cer 36:1 constituting ~70% of total ceramide (Fig. 2D), and the sphingadienes Cer 34:2, Cer 36:2 being one fifth as abundant. It should be noted that the two genetic (control) backgrounds exhibited significant differences for the very low abundant ceramide species- Cer 36:2 and Cer 38:1 (Fig. 2D; white and grey single hatched bars). The main contribution to increased total ceramide levels in bchs mutant brains originated from Cer 32:1, 34:1 and 36:1 (Fig. 2E), but significant changes in all species were seen in at least one bchs allelic combination. Increases seen in the sphingadiene ceramides were not significant (Suppl Fig. S3).
Neurodegeneration in bchs is sensitive to changes in metabolic availability of ceramides
Since the genetic interaction studies indicated that bchs neurodegeneration may be modulated by ceramide levels, we wanted to find out whether these genetic backgrounds indeed influenced sphingolipid levels in the expected ways.
First, we assessed the effect of the CDase mutant heterozygote with bchs58/Df, which showed a rescue of neurodegeneration (Fig. 1D,F). For bchs58/Df, slab2/+ an even higher, nearly additive increase of 70% in total ceramide was observed as compared to bchs58/Df (37%) and slab2/+ (29%) alone (Fig. 2-F1, green). Unlike this rescue, overexpression of CDase in bchs (C155; bchs58/Df > CDase) exacerbated the degenerative phenotype, and exhibited a reduction in total ceramide levels by 84% (53% + 31%) compared to C155; bchs58/Df alone, similar to the levels seen in CDase overexpression by itself (−34% vs. −31%; Fig. 2-F1, dark green). The genetic formalism that an additive contribution of individual mutants to a phenotype is evidence of parallel mechanisms, suggests that the additive increases in ceramides that we see when both bchs and one copy of CDase are removed point to parallel (rather than linear) roles of these two genes in a sphingolipid regulatory pathway.
Driving EY-nSMase, which rescued neurodegeneration, increased ceramides as expected, (by 84%) but in the presence of bchs this was decreased to the same level ( + 53%) as bchs alone (Fig. 2-F2, lavender). Again this is consistent with the genetic formalism of an epistatic relationship, wherein bchs may be required for full nSMase activity. UAS-nSMase directly driven by C155 gave a much stronger increase in ceramides, of 360% (Fig. 2-F2, purple). This line induced loss of visible GFP in aCC and RP2 neurons by itself, and was therefore not used for phenotypic interaction studies with bchs.
Finally, lacek05305 (spt2) heterozygosity, which exacerbated degeneration in bchs, reduced bchs’ ceramide increase to insignificance (Fig. 2-F3). This epistasis of lace over bchs suggests that bchs’ increase in ceramides may be partly due to an upregulation of de novo ceramide production via spt. However, the increases in ceramides in all three genotypes are also likely to be due in part to an upregulation of the salvage pathway from sphingosine34. Major ceramide species reflect the same trends that were seen for total ceramides (Supplementary Fig. S4). Ceramide sphingadienes were only significantly changed in the slab2/+ backgrounds with or without bchs (Supplementary Fig. S5).
We considered that a long chain base-phosphate (LCB-P), (referred to as sphingosine-1-phosphate, or S1P), given its known role as a signaling molecule, in particular with respect to MAPK signaling and autophagy (reviewed in5), could be a determinant of neuronal survival in bchs mutant neurons. However, the amounts of tissue required to perform the analysis of S1P were prohibitive for larval brain. Therefore, levels of S1Ps as well as sphingosines were measured in 4 day adult male fly heads from bchs alone and in combination with slab2/+ and CDase-overexpressing backgrounds. (Note that here in accordance with the Drosophila literature, we refer to these species as sphingosine (sph) and S1P, although they contain C14 and C16 bases rather than the mammalian C18 base). No change was seen in S1Ps in bchs mutant heads (Supplementary Fig. S6A). Moreover, the analysis shows no obvious correlation between levels of S1Ps or sphingosines with either the genetic rescue or exacerbation effect on neurodegeneration (Supplementary Fig. S6A, B). Therefore, we conclude that sphingosines and S1Ps do not strongly influence bchs degeneration.
In summary, the ceramide measurements across all genotypes indicate that manipulations that increase the available ceramide pool act as rescuers of bchs degeneration, whereas a reduction of de novo synthesis or increased ceramide catabolism exacerbates degeneration. Overall, the metabolic snapshot as assessed by lipidomics supports the findings of the genetic interaction study.
bchs neurons sequester exogenous ceramide in autophagosomes
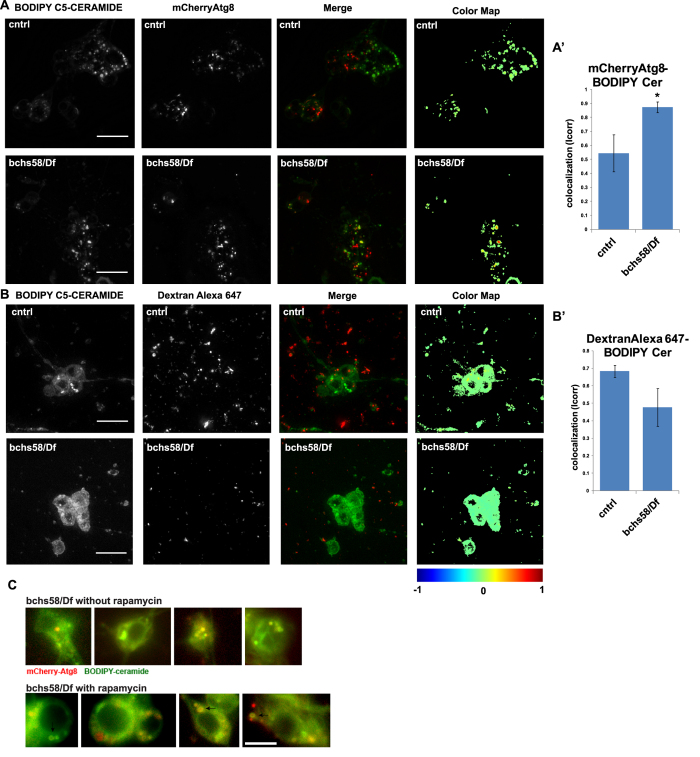
The fact that bchs brains accumulated excess ceramides, but were nonetheless rescued by increasing the pool of available ceramides genetically (and conversely were exacerbated by lowering ceramides pharmacologically or catabolically), led us to hypothesize that ceramides, although present in neurons, were perhaps being sequestered in a degradative compartment, unable to be released for signaling or other metabolic activities. Thus, we prepared primary neurons from bchs and wild type controls and allowed them to take up exogenous C5-ceramide labeled with fluorescent BODIPY-FL, such that the fate of ceramide could be traced in the cells. This was done in the background of compartmental markers mCherry-Atg8 (labeling autophagosomes35 or 10 kDa Dextran–Alexa647, which had been pulse-chased overnight to label late endo/lysosomal compartments31,36. While wild type neurons displayed relatively little colocalization of BODIPY-ceramide with mCherry-Atg8-labelled autophagosomes, bchs neurons had significantly more BODIPY-ceramide in these autophagic compartments (Fig. 3A,A’). The amounts of BODIPY-ceramide trafficking to degradative compartments labeled by Dextran-Alexa647, however, was not changed significantly between bchs and wild type (Fig. 3B,B’). This leads us to believe that ceramide, while present overall in higher amounts in neurons, is indeed sequestered and made unavailable in an autophagic recycling pathway that does not function properly (also see figs. 4,534,37).
Figure 3. BODIPY-C5 Ceramide in bchs primary neurons accumulates aberrantly in mCherry-Atg8-expressing autophagosomal compartments.
Live primary neurons incubated with BODIPY C5-ceramide and imaged after chase in label-free medium, in a bchs58/Df; UAS-mCherry-Atg8 expressing background (A) and primary neurons from bchs58/Df brains treated with dextran-Alexa647 (B) to label endolysosomes. Error bars represent mean ± SEM of correlation coefficient denoting extent of co-localization between ceramide and (A) Atg8, or (B) Dextran, from four and three experiments, respectively. Scale bar = 10 μm. Co-localization analysis was carried out using the Fiji co-localization color map plug-in (https://sites.google.com/site/colocalizationcolormap/). (C) mCherry-Atg8 compartments in bchs primary neurons incubated with rapamycin (representative cells in bottom row) appear to clear BODIPY-ceramide more effectively, resulting in hollow compartments rather than the solid BODIPY-ceramide spots seen in untreated neurons (top row).
BODIPY-ceramide labelled primary bchs mutant neurons were treated with rapamycin in order to determine whether the accumulated lipid would indeed be cleared from autophagosomes more effectively if autophagy were boosted in this way. Figure 3C shows “donut-shaped” mCherry-Atg8-labelled compartments which appear to be cleared of BODIPY-ceramide after rapamycin treatment, whereas the compartments appear solid without the treatment.
Sphingolipid-modulating genetic backgrounds rescue defects in autophagic clearance in bchs
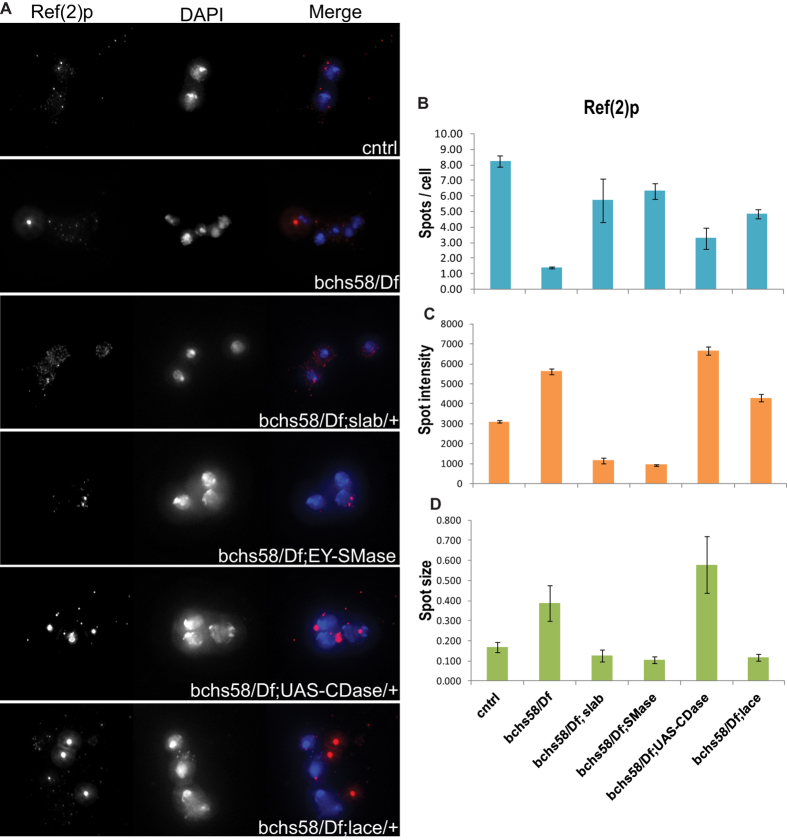
Ceramides have a known role in positively regulating autophagy through activation of stress-associated MAP kinase pathways11, and Bchs has been characterized as a pro-autophagic adaptor protein that is required for clearance of aggregated proteins16,38. We therefore wondered whether the excess ceramides detected in bchs brains were associated with an effect on autophagic initiation or clearance. In order to investigate this proposed interaction, we monitored expression of Atg8 and Ref(2)p (the Drosophila p62/sequestosome homolog)18 in primary neurons prepared from larval brains of bchs, or bchs combined with the exacerbating and rescuing sphingolipid-modulating genetic combinations tested in the previous experiments.
Atg8 and p62or/Ref(2)p punctae were analysed using multiple parameters. Analysis of Ref(2)p showed that bchs mutants had fewer punctae than wild type controls, but of larger size and higher intensity (Fig. 4A,B–D). Strikingly, these effects were rescued in the backgrounds lacking CDase (slab2/+) and overexpressing nSMase (Fig. 4A,B–D) the same manipulations that rescued neurodegeneration in larvae. In contrast, the exacerbating background UAS-CDase (Fig. 4A,B–D) made the bchs changes in intensity and size even more extreme. Combination with lacek05305/+ (Fig. 4A,B–D) reversed the bchs changes partially toward wild type.
Figure 4. bchs and its modifiers affect an autophagic flux marker in primary neurons.
Primary neuron cultures of bchs58/Df and bchs58/Df larval brain in sphingolipid modifying genetic backgrounds were immunostained with Ref(2) p antibody (A, left panel) and the images were quantified for number of punctae (spots) per cell (B), spot intensity (C) and spot size (D). DAPI was used for nuclear staining. Error bars represent ± SEM.
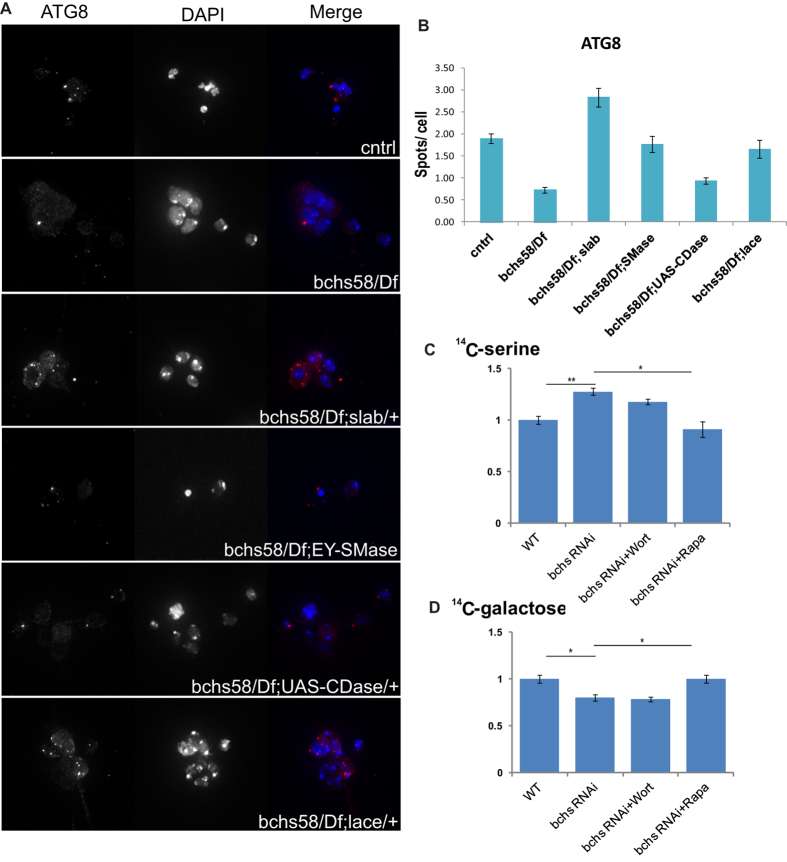
Atg8 punctae are shown in Fig. 5A,B quantified as spot number/cell in bchs and sphingolipid-altering backgrounds, since other parameters (intensity, size) did not vary significantly. A significant decrease in number of Atg8 punctae in bchs primary neurons was rescued back to wild type levels in the same rescuing combinations as were seen for Ref(2)p (slab2/+ and nSMase overexpression), but remained unchanged in the exacerbating background overexpressing UAS-CDase. Once again, the lacek05305/+ background rescued Atg8 spot number, similar to its effects on Ref(2)p. These results indicate that autophagic flux is perturbed in bchs animals, and that this can be strongly ameliorated specifically by enhanced ceramide salvage via an increase in nSMase or reduction of CDase (slab). Alternatively, reduction of de novo via lace appears to have similar rescuing effects on autophagic clearance.
Figure 5. bchs mutants alter autophagic induction and imbalance in recycling vs. de novo sources of ceramide.
Primary neuron cultures of bchs58/Df and bchs58/Df in sphingolipid modifying genetic backgrounds were immunostained with Atg8 antibody (A, left panel) and spot number per cell was quantified similarly to Ref(2)p in Fig. 4 (B). DAPI was used for nuclear staining. (C,D) TLC quantification of total sphingolipids extracted from S2R + cells treated with bchs dsRNA and grown in medium containing either 14C-serine (C) or 14C-galactose (D) for 24 hrs. For quantification, total lipid was normalized to total protein levels and 14C signal was calculated by densitometry of the autoradiograph.
We confirmed that the observed effects on spot number in Atg8, and spot number, size, and intensity in Ref(2)p were pertinent indicators of autophagic flux by treating primary wild type and bchs58/Df neurons with rapamycin (Supplementary Fig. S7A–C). Notably, rapamycin improved flux in wild type, but not in bchs, consistent with our findings that rapamycin does not rescue degeneration of strong bchs loss-of-function mutations (submitted elsewhere).
Additionally, no significant effects were seen on Atg8 and Ref(2)p in the CDase, nSMase, and lace interacting backgrounds by themselves (Supplementary Fig.7D), suggesting that elevated or reduced ceramide levels in these backgrounds are insufficient to induce changes in autophagic turnover on their own, even though they alleviate the defects in bchs.
Disturbed autophagic flux in bchs alters the balance between de novo and recycling sources of sphingolipids
Because of the loss of autophagic flux demonstrated in Figs 4 and 5, and the modulation of these phenotypes by genetic changes in salvage (recycling) vs. de novo sphingolipid production, we next tested whether the relative amounts of sphingolipids arising from these two pathways might be perturbed in bchs, using a method established in mammalian cells39. In order to monitor this, S2R + cells were treated with bchs RNAi, and de novo ceramides were labeled with radioactive 14C-serine. A parallel experiment was carried out using 14C-galactose, which may be incorporated into glycosphingolipids generated both from de novo and from recycled ceramides (i.e. the entire pool of sphingolipids); although it should be noted that glycosphingolipids in mammalian cells are predominantly from recycling sources40. Thus, comparing the change in both of these markers should give an indication of any relative change in de novo vs. total sphingolipids. Bchs knockdown resulted in a significant increase in 14C-serine incorporation, compared to a significant decrease in 14C-galactose incorporation (Fig. 5C,D), suggesting that there is indeed an increase in de novo sphingolipid production vs. that derived from recycling. To test whether this is related to autophagy, we treated the bchs RNAi cells with the autophagic inhibitor Wortmannin, or with rapamycin to increase autophagic turnover. Indeed, these cases showed that the de novo/salvage ratio could be restored to normal, dependent on autophagy (Fig. 5C,D).
CDase loss and nSMase overexpression “super-rescue” down-regulated Akt and MAPK pathways in bchs brains
To better understand how perturbed ceramide levels in bchs mutants might impinge upon survival pathways in neurons, we quantified several key downstream effectors of ceramide signaling in bchs larval CNS and adult heads. In light of the role of autophagy in bchs neurodegeneration16 we tested whether neuronal death may be due to alterations in the mitogen-activated protein kinase kinase MKK4, a sphingolipid-regulated pathway that can induce both autophagy and apoptosis11,41. Since ceramide also activates PP2A, and thereby negatively regulates Akt, the major convergence point of insulin and TOR growth pathways, we looked for changes in this pathway as well42.
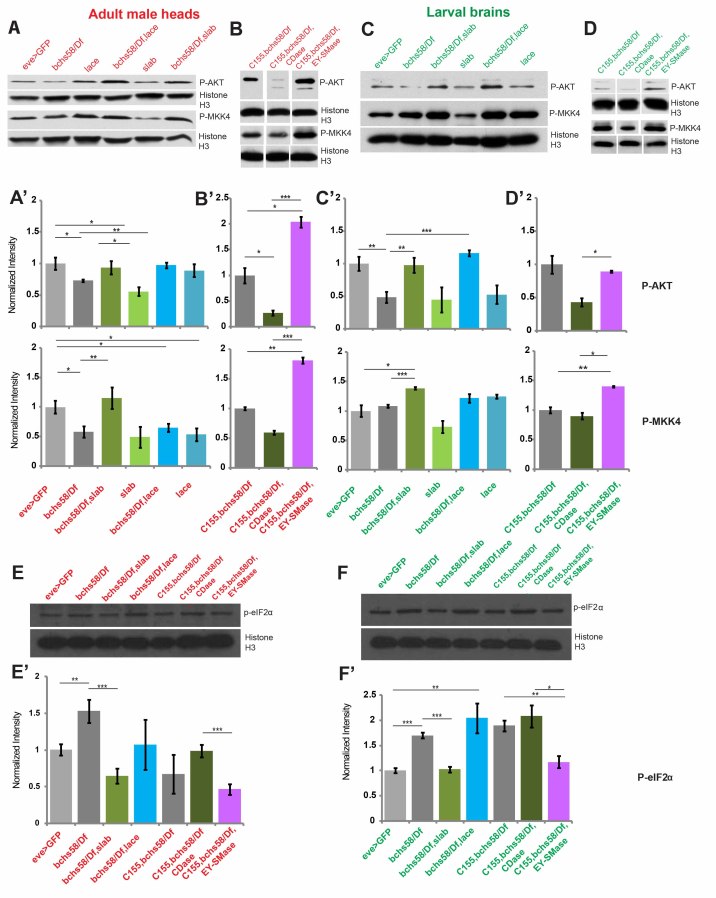
In accordance with increased ceramide levels, bchs adult and larval brains (Fig. 6A,A’) have reduced p-Akt levels compared to Akt (total Akt is unchanged across genotypes; Supplementary Fig. S8). This does not seem to be mediated by a drop in PP2A levels (data not shown), although we were not able to measure the more relevant target p-PP2A due to the unavailability of the antibody in Drosophila.
Figure 6. Genetic increases in sphingolipid salvage pathways rescue perturbations seen in ceramide effector pathways in bchs.
Western blots and quantification of adult heads (A,A’,B,B’) and 3rd instar larval brains (C,C’,D,D’) from bchs58/Df and bchs58/Df in sphingolipid-modifying genetic backgrounds, probed against p-Akt, p-MKK4, and the ER-stress marker p-eIF2α (E,E’,F,F’). For quantification, signals were normalized to histone H3 loading control and analyzed in Image J. Significance was calculated by Student’s-t test (*p < 0.05, **p < 0.01 and ***p < 0.001). Error bars represent ± SEM.
p-MKK4 was surprisingly also reduced in bchs adult heads (Fig. 6A,A’), even though increased ceramide would be expected to enhance MLK3 downstream signaling43, of which MKK4 is a component. Antibody recognizing Drosophila non-phosphorylated MKK4 was unavailable. Interestingly, reduced p-Akt is also observed in slab2 heterozygotes, which have elevated ceramides similarly to bchs (Fig. 2), while lacek05305 heterozygosity reduces p-MKK4 (Fig. 6A,A’). This observation that genetic manipulations of ceramide levels on their own can affect these signaling pathways, in addition to modulating bchs’ effects, suggests that it is indeed the changes in ceramides that cause the signaling changes.
Most strikingly, p-Akt and p-MKK4 are restored to levels at or even beyond those found in controls (“super-rescue”) when the rescuing backgrounds slab2/+ or overexpressing nSMase are combined with bchs (Fig. 6A,A’, Supplementary Fig. S8). In contrast, exacerbation by CDase overexpression further reduced p-Akt from its already low levels in bchs (Fig. 6B,B’). Thus, these sphingolipid-altering genetic backgrounds’ effects on signaling mirrored their effects on degeneration. The apparently rescuing effect of lacek05305/+ on p-Akt, however, contrasts with its exacerbation of degeneration.
In order to discover whether ER stress might contribute to (or be a consequence of) the bchs phenotype, we also measured p-eIf2α in bchs adult and larval CNS39. We found p-eIf2α to be consistently elevated in bchs, and again rescued by slab2/+ and nSMase overexpression. CDase overexpression and lacek05305/+ by contrast did not modify the bchs changes (Fig. 6E,F).
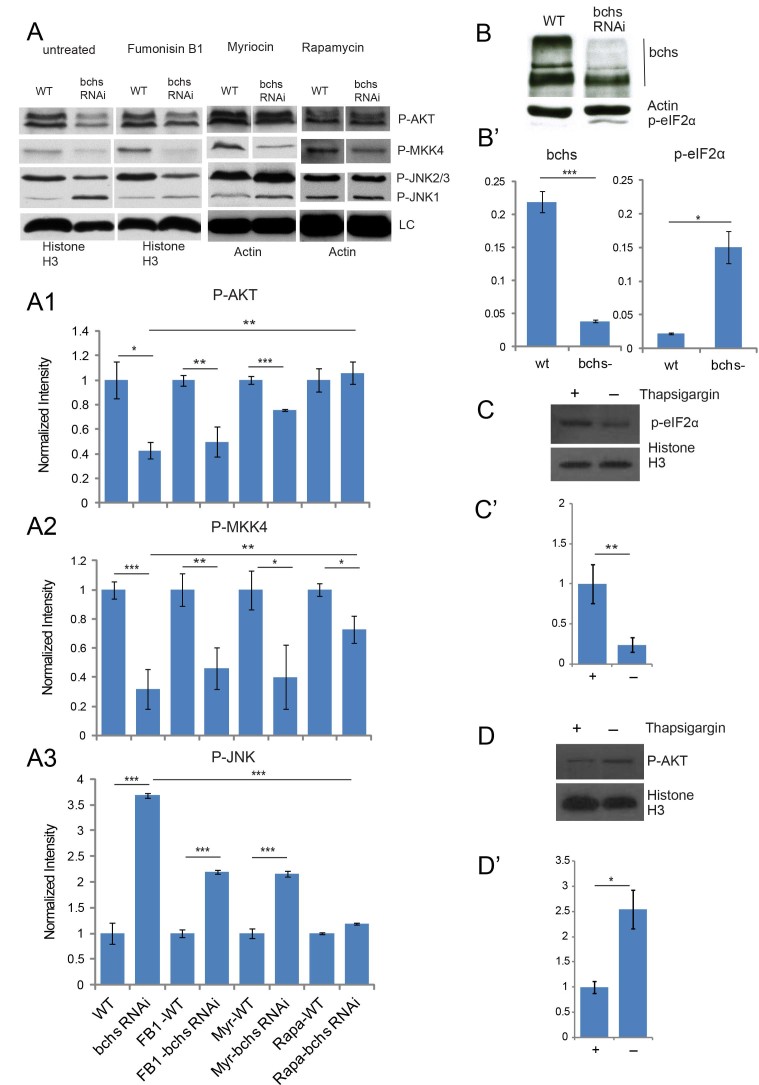
Bchs knockdown in Drosophila S2 cell line reduces activated Akt and MKK4, but increases stress signals
Although each case in the above Western studies was compared to a control strain of a similar genetic background (i.e. only single chromosomes were exchanged in pairwise comparisons), some genetic variability is difficult to avoid. In order to surmount this intrinsic variability between strains, which we note does affect the quantitative outcome of signaling pathways, we carried out Western analysis of the same markers, Akt, MKK4, and EIf2α, on Drosophila S2R + cells where the Bchs product had been knocked down with dsRNA (Fig. 7A,B). Here, we found similar overall trends in the reduction of survival-associated p-Akt and p-MKK4 (Fig. 7A1), whereas ER stress-associated p-eIF2α was clearly increased in cells (Fig. 7B). As a control, we used thapsigargin treatment of the cells to induce ER stress. As expected, this increased p-eIF2α (Fig. 7C,C’), and lowered p-Akt (Fig. 7D,D’) suggesting that the lower p-Akt seen in bchs could also be due to ER stress. Total Akt levels were unchanged across genotypes and treatments (Supplementary Fig. S8); total eIF2α antibody was unavailable.
Figure 7. Pharmacological intervention in de novo synthesis or autophagy rescues perturbations seen in ceramide effector pathways upon bchs knockdown in S2R + cells.
Western analysis of S2R + cells after bchs knockdown via dsRNA and treatment with Fumonisin B1, myriocin, or rapamycin (A) for p-Akt (A1), p-MKK4 (A2), p-JNK (A3) and p-eIF2α (B,B’). Knockdown of Bchs product by dsRNA was confirmed in B, B’, and control thapsigargin treatment of S2R + cells and its effect on p-eIF2α and p-Akt are shown in (C,D). Full blot images of adult heads, larval brains, S2R + cells, and total Akt can be found in suppl Fig. S8. Quantification of blots, shown in bar graphs, was done as in Fig. 6. Error bars represent ± SEM and significance was calculated by Student’s-t test (*p < 0.05, **p < 0.01 and ***p < 0.001).
We next examined whether pharmacological modulation of sphingolipid biosynthesis and/or autophagy using FB1 and myriocin (employed earlier) or rapamycin, also influenced these signaling readouts that were altered in bchs brains. There were no changes in these signaling pathways after FB1 and myriocin treatment alone, but these de novo synthesis inhibitors did significantly reverse the effects of bchs RNAi on p-Akt and p-JNK (Fig. 7A1)—but not MKK4 (Fig. 7A2)—suggesting that Akt and JNK misregulation in bchs are mediated at least in part by ceramides generated de novo, and that when synthesis is inhibited, these stress-related responses cannot be turned on; however, the stress response may in fact be protective, indicated by increased degeneration under these drugs, and in a lace heterozygous background (see Fig. 1).
Interestingly, rapamycin treatment, which increases autophagy44 also reversed all the signaling effects of bchs RNAi (Fig. 7A1–A3) (and rescued degeneration of a hypomorphic bchs mutation; manuscript submitted elsewhere), strongly suggesting that these effects come about as a result of defective autophagy, which has been reported to increase ER stress45.
Discussion
In this report, we demonstrate a decisive role for sphingolipid metabolism in autophagy-related brain degeneration in Drosophila. Even though total ceramide levels are elevated in bchs mutants, increasing them further through genetic manipulations rescues both neurodegeneration and survival signaling: neurons appear to survive better in spite (or because) of these higher ceramide levels. Conversely, decreasing ceramide content either by pharmacologically inhibiting synthesis, genetically lowering de novo synthesis, or increasing ceramide hydrolysis in a bchs deficient background, exacerbates neurodegeneration. This runs counter to the findings of many studies over the last decade documenting increases in ceramide production in degenerative conditions and aging brains32,33,46. Our finding that exogenous ceramide accumulates in autophagosomes, combined with the apparent increase in de novo sphingolipid production, provides an explanation for the observed ceramide increases, and suggests that compensatory mechanisms balance the loss of recycled sources of ceramides.
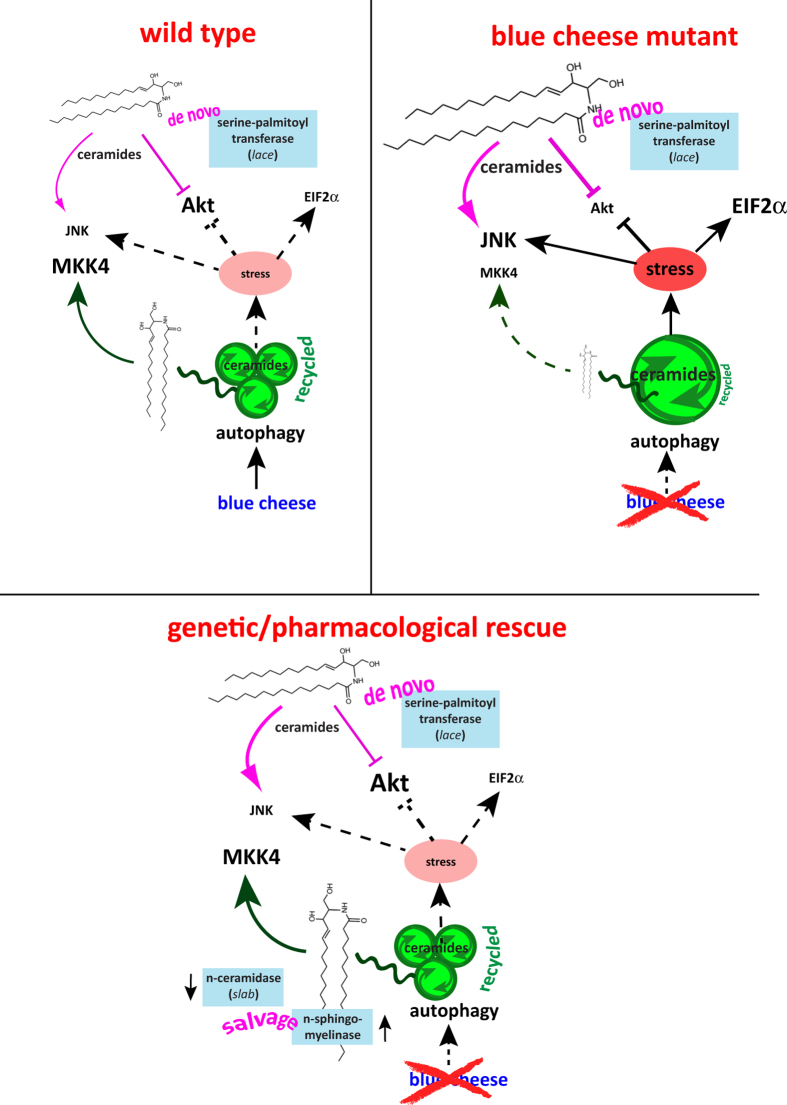
There does not appear to be a one-to-one correlation between total ceramide levels and severity of degeneration, perhaps because ceramides per se can influence a range of cellular outcomes, including growth and/or inflammatory signaling via ERK/MAPK-activation at one end of the spectrum47 and JNK-mediated induction of autophagy or apoptosis at the other5,11. An attractive possible correlate to sphingolipid-related degeneration, suggested by our findings, is that the potentially recyclable pool of ceramide is sequestered in dysfunctional autophagosomes, where it has limited access to signaling cascades mediated by the MAP kinases. Since it appears to be de novo ceramide that abnormally suppresses Akt and partially activates JNK in bchs, whereas autophagy induction or increases in salvaged ceramide restore all signaling conditions back to normal, we hypothesize that the autophagic defect is the driver of an imbalance in both ceramide sources and overall levels. This triggers stress signals such as JNK and eIf2α, and lowers Akt signaling, which impair neuronal survival. This idea is summarized in the model in Fig. 8.
Figure 8. Model summarizing key sphingolipid metabolic pathways contributing to de novo (pink) and salvaged ceramide pools (green), whose normal balance (‘wild type’ situation) may be perturbed by bchs’ blockage of normal autophagic clearance (‘blue cheese mutant’), which increases stress and impinges on survival pathways involving MKK4, JNK, Akt, and EIF2α.
Rescue of these signaling pathways, and of neuronal death (‘genetic/pharmacological rescue’), can be achieved by genetic increases in salvage pathways, or by increases in autophagic clearance. Suppression of de novo synthesis via spt (lace) reduction or treatment with pharmacological agents, only partially rescues signaling pathways (JNK and Akt) but does not rescue neuronal death.
Exacerbation of degeneration by suppression of de novo synthesis, which we found both genetically and pharmacologically, is consistent with the idea that the brain may compensate for inaccessible ceramide pools by increasing de novo production. If this is the case, rescue should be seen in backgrounds where less ceramide is broken down to sphingosine, and more is recycled from CerPE by the nSMase homolog, exactly as we observe. Mutants in sphingosine lyase in the mouse show the converse: de novo sphingolipid synthesis is down-regulated to compensate for an excess of salvaged ceramides40. That this also promotes degeneration implies that the balance of ceramides in the brain from de novo synthesis vs. salvage is important for neuronal survival.
Elevated (stored) ceramides may be inaccessible to some signaling pathways in bchs brains, since we see a consistent decrease in p-MKK4, even though this MAPK component is thought to be activated by ceramide48. p-Akt was also lower, however, as one would expect when ceramides are increased. Notably, both p-MKK4 and p-Akt are restored to normal levels, when the rescuing slab2 (CDase-) or nSMase backgrounds are introduced. This is presumably associated with the improved autophagic clearance also observed in these backgrounds when combined with bchs. Since ceramide is known to trigger JNK and reduce p-Akt by PP2A activation3,49, increased ceramide levels can cause neurons to die apoptotically. Alternatively, JNK can also phosphorylate Bcl2, leading to release of the beclin/Bcl2 complex, and autophagic initiation11. Stress-associated increases in p-JNK signaling may upregulate autophagy, even though this autophagy may be ineffectual, as seen in bchs. All of these effects are consistent with phenotypes we observe in bchs, including the defective clearance of Ref(2)p/p62-compartments, ceramide storage, increased p-eIF2α and p-JNK stress signaling, and decreased p-Akt. We postulate that when these stress pathways in bchs are blocked, e.g. by suppression of de novo ceramide synthesis via spt2 mutation or pharmacological inhibition, this leads to an inadequate response to these stresses while Akt-driven growth signals persist, and these conflicting signals exacerbate neurodegeneration.
The strong rescue of bchs neuronal survival and Akt/MAPK signaling by CDase loss may seem paradoxical when both of these genotypes have similar phenotypes (in terms of ceramide increase as well as signaling defects); such genetic interactions, however, are typical for genes that mutually oppose each other, suggesting that Bchs activity may suppress CDase. Consistent with this possibility, creating the opposite genetic situation via CDase overexpression, instead exacerbates degeneration, loss of p-Akt, and ER stress in bchs. Bchs’ known autophagic function has been limited to the clearance of aggregated proteins, but since lipids also recycle by autophagy50, and BODIPY-ceramide accumulates in autophagosomes in bchs neurons, it is conceivable that Bchs has a role in controlling ceramide turnover via CDase.
We note that the excess ceramide generated by overexpression of nSMase in our experiments was damped by loss of bchs, indicating that Bchs may be conducive to nSMase generation of ceramide, while it appears to act genetically in opposition to CDase. Thus, by regulating nSMase and/or CDase activity in turn, Bchs may alter the availability of a ceramide pool, and at the same time may encourage autophagic release of recycled sources of ceramide that carries out particular signaling functions, thereby influencing the survival of neurons.
Materials and Methods
Fly strains
Animals were raised in standard conditions on Bloomington semi-defined medium (http://flystocks.bio.indiana.edu/Fly_Work/media-recipes/media-recipes.htm). Loss of motor neurons was assayed using the RRa-Gal4(eve-gal4), mCD8GFP line27 in mutant alleles as described in20,51. bchs alleles 58M and 17M were generated from precise excision of the EP2299 element in bchs52 and bchs17 reported in51. Other fly stocks were: lacek05305, UAS-Ceramidase (CG1471)13 and slab2 (CG1471)13,28, and C155-gal4. nSMase (CG12034) was manipulated using the insertion allele EY00448 (Bloomington Drosophila Stock Center) and a UAS-nSMase line. A detailed description of genotypes used in the lipidomic study is found in Supplementary item S9.
Generation of UAS-nSMase
Full-length nSMase (CG12034) was amplified from cDNA (DGRC LD24865) using the 5′ primer GGCTCCGCGGCCGCCATGTTGCTGCTGGAGCTGAAC NotI site upstream of ATG and the 3′ primer GGCTCCGGCGCGCCCGTAGAAGTACTCGTACTTCTG AscI site. The product was cloned into Gateway pENTR using the introduced NotI and AscI sites and subcloned into pTW (DGRC #1129) using LR Clonase II (Life Technologies). Transgenic flies were made by BestGene, Inc (California, USA).
Larval Immunostaining and drug treatments
Larval fillets were immunostained as previously described20. Image stacks were acquired on a Deltavision fluorescence microscope (Applied Precision) at 20× (0.75 NA), and projected. Newly hatched larvae were raised in 500 μl of medium containing 10 μl of 12.4 mM Myriocin in 50 mM NaOH or 1 mM Fumonisin B1 in water.
Western Blotting
Three adult heads or larval brains per lane were crushed in 25 mM Tris pH7.5; TritonX100 0.5%; EDTA 5 mM; NaCl 250 mM with protease inhibitor cocktail and phosSTOP (Roche). Anti-p-SAPK/JNK (Thr183/Tyr185) (CST#9251,1:1000), p-SEK1/MKK4 (Ser257/Thr261) (CST#9156,1:1000), p-Akt (Ser473) (CST#4060,1:2000), Akt (CST#9272,1:1000), β-actin (CST#4967 1:1000), p-eIF2α (Ser 51) and Histone H3 (CST#4499,1:2000) and HRP-coupled goat anti-rabbit (Jackson ImmunoResearch,1:20,000) signals were detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, Illinois), scanned on an HP Scan jet G4050 and quantified with ImageJ (http://rsbweb.nih.gov/ij), by normalizing first to the loading control, then to the day average of total protein over all genotypes per experiment. Student’s t-test was performed to determine significance.
Larval brain primary neurons and immunostaining
Four to six 3rd instar larvae were washed thoroughly with 90% ethanol, then de-ionized water; brains were dissected in basal medium M3 + BYPE (without antibiotics/antimycotics, FBS or insulin) freed of imaginal discs, and briefly spun down in 1.5 ml Eppendorf tubes. Medium was replaced with 500 μl of Rinaldi’s saline, then incubated in 200 μl of 0.5 mg/ml collagenase Type I (Sigma) in Rinaldi’s saline for 1 h. 150 μl complete culture medium (M3 + BYPE, 10% FBS [HyClone], 20 μg/ml insulin [from bovine pancreas, Sigma] and antibiotics/antimycotics [Invitrogen]) was added to the brains and triturated ~50 times to dissociate the tissue. Cell suspension was loaded on the centre of the coated coverslip in a 25 mm culture dish, and incubated overnight in a humid chamber. Additional 900 μl of complete medium was then added. Primary neurons were fixed in 4% paraformaldehyde in PBS for 5 minutes at room temperature, washed twice in PBS and blocked in 5% normal goat serum, 0.05% Triton X-100 in PBS for 1 h. Primary antibodies rabbit anti-Atg8 (a generous gift of Katja Kohler) 1:500 and rabbit anti-Ref(2)p, (kindly donated by Ioannis Nezis) 1:1000 in normal goat serum block solution were incubated at 4 °C overnight. Atg8 and Ref(2)p were detected with Cy5 (1:500) and Alexa 488 (1:1500) conjugated rabbit secondary antibodies, respectively. DAPI (1:4000 in PBS) was added for 3 min, and washed in PBS, before mounting on glass slides in 90% glycerol/0.05% propyl gallate.
Cell labelling with BODIPY C5-Ceramide
5 mg BODIPY C5-ceramide (B-22650; Invitrogen) complexed with BSA in 150 μl of water (0.5 mM stock solution) was stored at −20 °C. 5 μM staining solution was prepared in HBSS/HEPES buffer. Primary neurons were incubated in 10,000 MW Dextran-Alexa647 2 h, washed, and chased overnight with fresh medium. Thereafter cells were rinsed in HBSS/HEPES, incubated with 5 μM BODIPY-ceramide 30 min at 4 °C, rinsed several times with cold medium, incubated in fresh medium at room temperature for up to 15 h, and imaged. Colocalisation analysis was carried out using the Fiji colocalisation color map plug-in (https://sites.google.com/site/colocalizationcolormap/).
Drosophila S2 cells RNAi, drug treatment
Drosophila S2R + were exposed to dsRNA vs. bchs after amplifying bchs genomic DNA by PCR using primers (forward) TTAATACGACTCACTATAGGGAGA TGGCGCAAGAACGCAGCTG and (reverse) TTAATACGACTCACTATAGGGAGACTTTTCATGTGTATCCGCTCTG. DNA was purified using QIAquick PCR purification kit and transcribed into dsRNA using T7 Megascript (Invitrogen #AM1334). ~10 μg of RNA was added to each well of a six-well plate and incubated for 3–4 days; knock-down was verified by Western blotting. 15 μM myriocin, 10 μM FB1 or 10 μM rapamycin were added to the RNAi-treated and control cells for 6, 2, or 2 h respectively. For BODIPY-ceramide labelling experiments, cells were incubated in rapamycin continually in the 6 h preceding and during imaging.
Lipidomic Analyses
Larval brain lipidomics
Analyses were carried out on all manipulations used to score motor neuron survival (genotypes listed in Supplementary item S9). Third-instar larval brains were pooled into groups of ten, washed in 20% methanol, freeze-dried, homogenized and extracted with Methyl-tert-butyl ether53. Homogenates were spiked with internal standard (Supplementary item, S10) before extraction. LC-MSn analysis was performed on a 1200 micro-LC-system (Agilent Technologies, Waldbronn, Germany) coupled to a hybrid LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) using the TriVersa Nanomate as ion source (Advion BioSciences Ltd, Ithaca NY, US). Lipids were identified based on accurate mass measurements and retention time tags and/or the formation of the characteristic fragment ions of the sphingoid base54,55,56. MS data interpretation was performed with Xcalibur (Thermo Fisher Scientific, Bremen, Germany). Lipid quantities for PE, PC, PE-ethers, CerPE, Cer and Sph were determined by extracting peak areas of high resolution LC-MS1 traces with a mass accuracy of ±5 ppm. For Ceramides, Cer 34:1, Cer 36:1, Cer 34:2, Cer 36:2 and C14-sphingosine, MS3 was applied to generate specific fragments. Lipids were quantified in picomoles of lipid/brain. Abundance of individual ceramide species is quantified as mol% (picomoles of individual ceramide/picomoles of total ceramide)*100. For a better representation of the MS results related genotypes were normalized to a suitable genetic control. ANOVA and Post-ANOVA pair wise comparisons using Tukey’s test and graphs were computed with OriginPro8.1 (OriginLab Corporation, USA).
S2 cell ceramide analysis
Ceramide quantification from S2 cells was analyzed by direct infusion. Samples were infused via nanoflow ESI source (Advion Triversa) into a quadrupole-orbitrap hybrid mass spectrometer (QExactive, Thermo Fisher Scientific). The nanospray source was operated by the Advion Chipsoft software and the spray settings were 0.8 psi and 1.2 kV in both positive and negative mode. Ion transfer tube temperature was set to 200 °C and S-Lens level was set to 50. Full MS spectra were acquired with 140000 resolution (at m/z 400) with an AGC setting of 1 × 106 and a maximum ion injection time of 50 ms. MS/MS-spectra were acquired with a resolution of 70000 (at m/z 400) with AGC setting of 1 × 105 and a maximum ion injection time of 1000 ms. For fragmentation, the precursor isolation window was set to 1 Th centered on each half integer m/z values using a targeted MS/MS (t-MS2) method in conjunction with an inclusion list covering the entire mass range between 420–900 in positive mode, so that every mass unit was subject to fragmentation. Normalized collision energy level was set to 20 in positive and 23 in negative mode.
Ceramide and phospholipid species were identified using LipidXplorer by matching the m/z values of the precursor and fragment masses to expected formulas. Ceramides were identified in negative mode by the presence of the a3 and b5 ions as described in57 and in positive mode by the long chain base fragments as described in58. Phospholipids were identified by the presence of the expected precursor mass and the corresponding fatty acids as described in52. The mass error tolerance for the data import was set to 5 ppm and the intensity threshold to 10-times the noise level reported by the Xcalibur software. Quantification of positively identified lipid species was done via normalization to the respective internal standard Cer 18:1/12:0 (20 pmol), PC-OO 36:0 (40 pmol) and PE-OO (50 pmol) which were added upon sample extraction56,59.
The analysis of LCB and LCB-P in adult head is described in Supplementary item S11.
Image Analysis
Images of primary neuron cultures were acquired as Z-stacks, with a step of 200 nm in between slices, using the same exposure, light intensity and filter settings for every condition. Collected stacks were then deconvolved in SoftWoRx 4.1.0 (Applied Precision Inc., WA, USA), and converted into 16-bit TIFF stacks in FIJI (http://fiji.sc) using LOCI BioFormats plug-in60. Each stack was then Z-projected using the maximum intensity method. Images were inspected under DAPI channel and only single cells, or small aggregates (<5 cells) with clearly separated cell nuclei were used for further spot counting. Because of varying spot intensity relative to cell body, and variable background signal, spots were counted manually, after setting the same display range of values. The cell number N used for calculations varied between conditions (Naverage ~100), but was not less than Nmin = 50. The average spot number per cell and the standard error of the mean (SEM) were calculated. To measure spot characteristics, projected images were masked using the Maximum Entropy threshold method61. Brightness and size distribution and SEM of selected spots were measured using the Analyze Particles FIJI command.
Radio-labeling of sphingolipids
Cellular sphingolipids were metabolically labeled according to a published protocol39 with [14C]-serine (2 μCi/ml) or D1-[14C]-galactose (2 μCi/ml). After 24 h, Drosophila S2 cells were harvested and extracted by incubating with 3 ml chloroform/methanol/water (10:5:1, v/v) 24 h at 48 °C. Phospholipids were degraded by alkaline hydrolysis with 100 mM methanolic NaOH, 2 h at 37 °C. Extracts were desalted by reverse-phase chromatography on an RP18 column and neutral lipids were separated from anionic lipids on a DEAE-Sephadex A-25 column, applied to silica gel plates (Merck, Darmstadt, Germany) and run in chloroform/methanol/0.22% CaCl2 (60:35:8, v/v) to separate glycolipids or in chloroform/methanol/acetic acid (190:9:1) to separate other sphingolipids. Radioactively labeled sphingolipids were visualized by autoradiography and quantified using ImageJ.
Preparation of Figures
All figures were assembled in Adobe Illustrator CS5.1 (Adobe Systems) by importing microscopy images (see image analyses) and graphs imported from OriginPro8.1 (OriginLab Corporation, USA) and Microsoft Excel 2011.
Additional Information
How to cite this article: Hebbar, S. et al. Ceramides And Stress Signalling Intersect With Autophagic Defects In Neurodegenerative Drosophila blue cheese (bchs) Mutants. Sci. Rep. 5, 15926; doi: 10.1038/srep15926 (2015).
Supplementary Material
Acknowledgments
This work in DS laboratory was supported by Wellcome Trust/DBT India Alliance; and the NCBS-Merck & Co International Investigator Award (DS). Work in the RK laboratory was supported through funding from A-STAR/BMRC grant #09/1/22/19/610, and the Ministry of Education, Singapore.
Footnotes
Author Contributions S.H., I.S., I.A., A.M., K.O., C.P., F.T., P.N., X.H.F., D.S. and R.K. carried out experiments and/or analyzed data. S.H. wrote original drafts of the manuscript, which were subsequently modified and edited by I.S., K.O., D.S. and R.K. M.W., A.S., D.S. and R.K. guided the study.
References
- Haughey N. J., Bandaru V. V. R., Bae M. & Mattson M. P. Roles for dysfunctional sphingolipid metabolism in Alzheimer’s disease neuropathogenesis. Biochimica et Biophysica Acta (BBA)—Molecular and Cell Biology of Lipids 1801, 878–886 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke M. M. et al. Plasma Ceramide and Glucosylceramide Metabolism Is Altered in Sporadic Parkinson’s Disease and Associated with Cognitive Impairment: A Pilot Study. PLoS ONE 8, e73094, 10.1371/journal.pone.0073094 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S. & Kolesnick R. Stress signals for apoptosis: ceramide and c-Jun kinase. Oncogene 17, 3277–3285, 10.1038/sj.onc.1202570 (1998). [DOI] [PubMed] [Google Scholar]
- Pettus B. J., Chalfant C. E. & Hannun Y. A. Ceramide in apoptosis: an overview and current perspectives. Biochim Biophys Acta 1585, 114–125, S1388198102003311 (2002). [DOI] [PubMed] [Google Scholar]
- Young M. M., Kester M. & Wang H.-G. Sphingolipids: regulators of crosstalk between apoptosis and autophagy. Journal of Lipid Research 54, 5–19, 10.1194/jlr.R031278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B. et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90, 1383–1435, 90/4/138310.1152/physrev.00030.2009 (2010). [DOI] [PubMed] [Google Scholar]
- Rabinowitz J. D. & White E. Autophagy and Metabolism. Science 330, 1344–1348, 10.1126/science.1193497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G. et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 281, 8518–8527, M506182200 10.1074/jbc.M506182200 (2006). [DOI] [PubMed] [Google Scholar]
- Signorelli P. et al. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer letters 282, 238–243 (2009). [DOI] [PubMed] [Google Scholar]
- Fyrst H. et al. Natural sphingadienes inhibit Akt-dependent signaling and prevent intestinal tumorigenesis. Cancer Res 69, 9457–9464, 0008-5472.CAN-09-234110.1158/0008-5472.CAN-09-2341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S. et al. Role of JNK1-dependent Bcl-2 Phosphorylation in Ceramide-induced Macroautophagy. Journal of Biological Chemistry 284, 2719–2728, 10.1074/jbc.M805920200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M. et al. Regulation of autophagy and its associated cell death by sphingolipid rheostat: reciprocal role of ceramide and sphingosine-1-phosphate in the mTOR pathway. Journal of Biological Chemistry, 10.1074/jbc.M112.416552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya U. et al. Modulating sphingolipid biosynthetic pathway rescues photoreceptor degeneration. Science 299, 1740–1743 (2003). [DOI] [PubMed] [Google Scholar]
- Yonamine I. et al. Sphingosine kinases and their metabolites modulate endolysosomal trafficking in photoreceptors. J Cell Biol 192, 557–567, jcb.20100409810.1083/jcb.201004098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Tran D.-H. et al. Molecular Mechanism of Sphingosine-1-Phosphate Action in Duchenne Muscular Dystrophy. Disease Models & Mechanisms, 10.1242/dmm.013631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M. et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 38, 265–279, S1097-2765(10)00284-4 10.1016/j.molcel.2010.04.007 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley K. D. et al. Blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci 23, 1254–1264 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T. H. et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344, 11226 (2010). [DOI] [PubMed] [Google Scholar]
- Kumarasamy S. K., Wang Y., Viswanathan V. & Kraut R. S. Multivariate profiling of neurodegeneration-associated changes in a subcellular compartment of neurons via image processing. BioData Min 1, 10, 1756-0381-1-10 10.1186/1756-0381-1-10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. & Kraut R. The Drosophila BEACH family protein, blue cheese, links lysosomal axon transport with motor neuron degeneration. J Neurosci 29, 951–963, 29/4/951 10.1523/JNEUROSCI.2582-08.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A. The role of BEACH proteins in Dictyostelium. Traffic 4, 6–12 (2003). [DOI] [PubMed] [Google Scholar]
- Perrotta C. & Clementi E. Biological Roles of Acid and Neutral Sphingomyelinases and Their Regulation by Nitric Oxide. Physiology 25, 64–71, 10.1152/physiol.00048.2009 (2010). [DOI] [PubMed] [Google Scholar]
- Grimm M. O. W., Grimm H. S. & Hartmann T. Amyloid beta as a regulator of lipid homeostasis. Trends in Molecular Medicine 13, 337–344 (2007). [DOI] [PubMed] [Google Scholar]
- Jana A., Hogan E. L. & Pahan K. Ceramide and neurodegeneration: susceptibility of neurons and oligodendrocytes to cell damage and death. J Neurol Sci 278, 5–15, S0022-510X(08)00601-1 10.1016/j.jns.2008.12.010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth B. M., Gustafson S. J. & Kuhn T. B. Neutral sphingomyelinase activation precedes NADPH oxidase-dependent damage in neurons exposed to the proinflammatory cytokine tumor necrosis factor-α. Journal of Neuroscience Research 90, 229–242, 10.1002/jnr.22748 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C. et al. CDase Is a pan-ceramidase in Drosophila. Mol. Biol. Cell, mbc.E10-05-0453, 10.1091/mbc.E10-05-0453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Emi-Sarker Y., Yusibova G., Goto T. & Jaynes J. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126(11), 2527–2538 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbough J. et al. Ceramidase regulates synaptic vesicle exocytosis and trafficking. J Neurosci 24, 7789–7803, 10.1523/JNEUROSCI.1146-04.200424/36/7789 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn S. C. et al. Regulation of de novo sphingolipid biosynthesis and the toxic consequences of its disruption. Biochem Soc Trans 29, 831–835 (2001). [DOI] [PubMed] [Google Scholar]
- Merrill A. H. Jr., van Echten G., Wang E. & Sandhoff K. Fumonisin B1 inhibits sphingosine (sphinganine) N-acyltransferase and de novo sphingolipid biosynthesis in cultured neurons in situ. J Biol Chem 268, 27299–27306 (1993). [PubMed] [Google Scholar]
- Hortsch R. et al. Glycolipid trafficking in Drosophila undergoes pathway switching in response to aberrant cholesterol levels. Mol Biol Cell 21, 778–790, E09-01-0005 10.1091/mbc.E09-01-0005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Echten-Deckert G. & Walter J. Sphingolipids: Critical players in Alzheimer’s disease. Prog Lipid Res 51, 378–393, S0163-7827(12)00039-210.1016/j.plipres.2012.07.001 (2012). [DOI] [PubMed] [Google Scholar]
- Grimm M. O. et al. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat Cell Biol 7, 1118–1123 (2005). [DOI] [PubMed] [Google Scholar]
- Kitatani K., Idkowiak-Baldys J. & Hannun Y. A. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal 20, 1010–1018, S0898-6568(07)00371-3 10.1016/j.cellsig.2007.12.006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O. & Neufeld T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7, 167–178, 10.1016/j.devcel.2004.07.009S153458070400245X (2004). [DOI] [PubMed] [Google Scholar]
- Sriram V., Krishnan K. S. & Mayor S. Deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J Cell Biol 161, 593–607 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A. et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184 (2008). [DOI] [PubMed] [Google Scholar]
- Simonsen A. et al. Genetic Modifiers of the Drosophila Blue Cheese Gene Link Defects in Lysosomal Transport With Decreased Life Span and Altered Ubiquitinated-Protein Profiles. Genetics 176, 1283–1297 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A. et al. Endoplasmic Reticulum Stress Response Mediated by the PERK-eIF2alpha-ATF4 Pathway Is Involved in Osteoblast Differentiation Induced by BMP2. Journal of Biological Chemistry 286, 4809–4818, 10.1074/jbc.M110.152900 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen-Euteneuer N., Lutjohann D., Park H., Merrill A. H. & van Echten-Deckert G. Sphingosine 1-Phosphate (S1P) Lyase Deficiency Increases Sphingolipid Formation via Recycling at the Expense of de Novo Biosynthesis in Neurons. Journal of Biological Chemistry 287, 9128–9136, 10.1074/jbc.M111.302380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Wang M. C. & Bohmann D. JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech Dev 126, 624–637, S0925-4773(09)01157-5 10.1016/j.mod.2009.06.1082 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowsky R. T., Kamibayashi C., Mumby M. C. & Hannun Y. A. Ceramide activates heterotrimeric protein phosphatase 2A. J Biol Chem 268, 15523–15530 (1993). [PubMed] [Google Scholar]
- Sathyanarayana P. et al. Activation of the Drosophila MLK by Ceramide Reveals TNF-α and Ceramide as Agonists of Mammalian MLK3. Molecular Cell 10, 1527–1533 (2002). [DOI] [PubMed] [Google Scholar]
- Berger Z. et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 15, 433–442 (2006). [DOI] [PubMed] [Google Scholar]
- Yin J., Gu L., Wang Y., Fan N., Ma Y. & Peng Y. Rapamycin Improves Palmitate-Induced ER Stress/NFκB Pathways Associated with Stimulating Autophagy in Adipocytes. Mediators of Inflammation 2015, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler R. G. et al. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. PNAS 101, 2070–2075 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke N. & Hannun Y. A. Bioactive sphingolipids: metabolism and function. J Lipid Res 50 Suppl, S91–96, R800080-JLR200 10.1194/jlr.R800080-JLR200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K. et al. TAK1 Mediates the Ceramide Signaling to Stress-activated Protein Kinase/c-Jun N-terminal Kinase. Journal of Biological Chemistry 272, 8141–8144, 10.1074/jbc.272.13.8141 (1997). [DOI] [PubMed] [Google Scholar]
- Ruvolo P. P., Deng X., Ito T., Carr B. K. & May W. S. Ceramide Induces Bcl2 Dephosphorylation via a Mechanism Involving Mitochondrial PP2A. Journal of Biological Chemistry 274, 20296–20300, 10.1074/jbc.274.29.20296 (1999). [DOI] [PubMed] [Google Scholar]
- Singh R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodosh R., Augsburger A., Schwarz T. L. & Garrity P. A. Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development 133, 4655–4665 (2006). [DOI] [PubMed] [Google Scholar]
- Schuhmann K. et al. Bottom-up shotgun lipidomics by higher energy collisional dissociation on LTQ Orbitrap mass spectrometers. Anal Chem 83, 5480–5487, 10.1021/ac102505f (2011). [DOI] [PubMed] [Google Scholar]
- Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A. & Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49, 1137–1146, D700041-JLR200 10.1194/jlr.D700041-JLR200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M. et al. Survival strategies of a sterol auxotroph. Development 137, 3675–3685, 137/21/3675 10.1242/dev.044560 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulman A. et al. High-resolution extracted ion chromatography, a new tool for metabolomics and lipidomics using a second-generation orbitrap mass spectrometer. Rapid Communications in Mass Spectrometry 23, 1411–1418, 10.1002/rcm.4015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwudke D. et al. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem 79, 4083–4093, 10.1021/ac062455y (2007). [DOI] [PubMed] [Google Scholar]
- Hsu F. F. & Turk J. Characterization of ceramides by low energy collisional-activated dissociation tandem mass spectrometry with negative-ion electrospray ionization. J Am Soc Mass Spectrom 13, 558–570, 10.1016/S1044-0305(02)00358-6 (2002). [DOI] [PubMed] [Google Scholar]
- Sullards M. C. et al. Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: “inside-out” sphingolipidomics. Methods Enzymol 432, 83–115, S0076-6879(07)32004-110.1016/S0076-6879(07)32004-1 (2007). [DOI] [PubMed] [Google Scholar]
- Schwudke D. et al. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem 78, 585–595, 10.1021/ac051605m (2006). [DOI] [PubMed] [Google Scholar]
- Linkert M. et al. Metadata matters: access to image data in the real world. The Journal of Cell Biology 189, 777–782, 10.1083/jcb.201004104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur J. N., Sahoo P. K. & Wong A. K. C. A new method for gray-level picture thresholding using the entropy of the histogram. Computer Vision Graphics and Image Processing 29, 273–285 (1985). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.