Abstract
Understanding the genetic basis of natural phenotypic variation is of great importance, particularly since selection can act on this variation to cause evolution. We examined expression and allelic variation in candidate flowering time loci in Brassica rapa plants derived from a natural population and showing a broad range in the timing of first flowering. The loci of interest were orthologs of the Arabidopsis genes FLC and SOC1 (BrFLC and BrSOC1, respectively), which in Arabidopsis play a central role in the flowering time regulatory network, with FLC repressing and SOC1 promoting flowering. In B. rapa, there are four copies of FLC and three of SOC1. Plants were grown in controlled conditions in the lab. Comparisons were made between plants that flowered the earliest and latest, with the difference in average flowering time between these groups ∼30 days. As expected, we found that total expression of BrSOC1 paralogs was significantly greater in early than in late flowering plants. Paralog-specific primers showed that expression was greater in early flowering plants in the BrSOC1 paralogs Br004928, Br00393 and Br009324, although the difference was not significant in Br009324. Thus expression of at least 2 of the 3 BrSOC1 orthologs is consistent with their predicted role in flowering time in this natural population. Sequences of the promoter regions of the BrSOC1 orthologs were variable, but there was no association between allelic variation at these loci and flowering time variation. For the BrFLC orthologs, expression varied over time, but did not differ between the early and late flowering plants. The coding regions, promoter regions and introns of these genes were generally invariant. Thus the BrFLC orthologs do not appear to influence flowering time in this population. Overall, the results suggest that even for a trait like flowering time that is controlled by a very well described genetic regulatory network, understanding the underlying genetic basis of natural variation in such a quantitative trait is challenging.
Keywords: Candidate gene, Phenology, Gene expression, Climate change
Introduction
Genetic variation contributes to phenotypic variation and provides the raw material that natural selection acts upon to produce adaptive evolution. Despite a burgeoning amount of genetic and genomic information, we still know little about genetic variation in ecologically important traits in natural populations. One such trait in plant populations is the timing of first flowering. Flowering time is a key life-history trait that influences mating opportunities, reproductive fitness, gene flow and evolution (Elzinga et al., 2007; Franks, 2015; Primack, 1985). With changing climatic conditions, there have been widespread shifts to earlier flowering (Miller-Rushing & Primack, 2008; Parmesan & Yohe, 2003), with important implications for population and evolutionary dynamics. Plant populations can potentially respond to climate change through migration, plasticity or evolution, although their ability to do so may be limited (Franks, Weber & Aitken, 2014). To predict the ability of populations to evolve in response to climate change, it is particularly useful to understand the relationship between genetic variation and phenotypic variation in the traits of interest, since selection can act on this variation to produce evolutionary change (Hoffmann & Sgro, 2011). Although the genetic basis of phenotypic variation and evolutionary responses to climate change is rarely known, this is an emerging area of investigation, with the genetic basis of variation in flowering time particularly amenable to study (Franks & Hoffmann, 2012).
To investigate the genetic basis of phenotypic variation and evolutionary changes in flowering time, it is useful to work with a system where the phenotype is highly variable, and such an evolutionary shift has been documented. A rapid evolutionary shift to earlier flowering was shown to occur following a multi-year late season drought in California in two populations of the annual plant Brassica rapa L. (Franks, Sim & Weis, 2007). Within 7 generations during the drought, average flowering time in the Arboretum population, which is the focus of this study, shifted an average of 8.5 days earlier (Franks, Sim & Weis, 2007). Furthermore, there was a broad range in flowering time for selection to act upon within populations. For example, in the Arboretum population, grown in a greenhouse, the earliest flowering individuals initiated flowering 34 days after germination, while the latest flowering individual began flowering 112 days after germination. Flowering time was shown to be heritable, so variation in this trait has some genetic basis (Franks, Sim & Weis, 2007). Subsequent work showed that early flowering plants have lower water use efficiency and flower at a smaller size and earlier developmental stage (Franks, 2011). However, the genetic basis of this rapid evolutionary change in flowering time, as well as the genetic basis of flowering time variation within populations, remained unknown.
To investigate the genetic basis of flowering time variation and evolution in natural populations of B. rapa, we took advantage of the fact that there is a substantial amount of information known about genes, pathways, and processes involved in determining flowering time, mainly from work with the closely related plant Arabidopsis thaliana (for reviews, see e.g., Amasino & Michaels, 2010; Bastow & Dean, 2003; Michaels, 2009; Simpson & Dean, 2002). In Arabidopsis, flowering time is controlled by a complex integrated genetic regulatory network (Boss et al., 2004; Mouradov, Cremer & Coupland, 2002; Putterill, Laurie & Macknight, 2004) that promotes flowering at an appropriate time under suitable conditions, and suppresses flowering under environmental conditions that indicate inappropriate times to flower, such as too early or too late in the growing season. This effective regulation is the result of the integration of inputs from several internal and external signals through key genes that activate or suppress the flowering-promotion regulatory network (Boss et al., 2004). Thus, environmental factors and genes interact to influence flowering time, with environmental conditions serving as cues that signal appropriate times to flower, and also conditions such as stresses potentially inducing flowering (Riboni et al., 2014; Wada & Takeno, 2010; Ying, Chen & Cai, 2014). Differences in the activity of these key integrator genes could potentially underlie flowering time variation in natural populations. Two of these key central regulatory flowering time genes in Arabidopsis are FLC (FLOWERING LOCUS C; (Yan et al., 2010)) and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANTS 1; (Immink et al., 2012; Lee et al., 2004)), which are the focus of investigation in this study.
FLC is a MADS-box transcription factor that has been the subject of much research on flowering time regulation (Bastow et al., 2004; Lempe et al., 2005; Michaels & Amasino, 1999; Michaels & Amasino, 2001; Searle et al., 2006; Sheldon et al., 2000). In Arabidopsis, FLC suppresses flowering by repressing the expression of SOC1 and FT, which both promote flowering (Hepworth et al., 2002; Michaels & Amasino, 1999). When FLC is downregulated through the appropriate combination of signals, the key inducers of flowering are upregulated, and flowering is initiated. FLC is one of only a few flowering time genes that has been shown to vary in natural populations (Caicedo et al., 2004; Korves et al., 2007; Lempe et al., 2005; Scarcelli & Kover, 2009; Slotte et al., 2009; Stinchcombe et al., 2004). For example, previous studies in Arabidopsis have found latitudinal clines in frequencies of alleles of FLC and in flowering time (Caicedo et al., 2004; Gazzani et al., 2003; McKay, Richards & Mitchell-Olds , 2003), as well as a strong association between variation in FLC and variation in flowering time in a diverse panel of natural accessions (Lempe et al., 2005). Another study with Arabidopsis accessions found that expression in FLC was correlated with flowering time, although no genetic variation at FLC was detected in that study (Schläppi, 2001). These findings suggest that variation in FLC alleles may potentially influence flowering time in natural populations.
SOC1 is also a MADS-box gene that plays a central role in flowering time regulation (Immink et al., 2012). SOC1 promotes flowering (Liu et al., 2008; Moon et al., 2003) by activating the floral meristem identity genes (Immink et al., 2012). Recent research has characterized the mechanisms by which SOC1 interacts with other elements in the flowering time regulatory network in more detail, and has demonstrated that SOC1 is a key hub in the flowering time regulatory network (Immink et al., 2012).
Most of this previous work investigating the flowering time genetic regulatory network has focused on Arabidopsis, which is in the same family (Brassicaceae) as Brassica. Researchers working with Brassica have confirmed that many of the same genes and networks operate in both taxa (Kole et al., 2001; Lagercrantz et al., 1996; Lin et al., 2005; Osborn et al., 1997; Schranz et al., 2002; Schranz et al., 2007; Tadege et al., 2001). The genome of B. rapa has been sequenced and extensively annotated (Wang et al., 2011), facilitating work on flowering time genes in this species.
In contrast to Arabidopsis, which contains only one copy each of FLC and SOC1, the B. rapa genome possesses four copies of FLC and three of SOC1 (http://brassicadb.org/brad/). The four BrFLC genes (BrFLC1, BrFLC2, BrFLC3, BrFLC5) co-localize with flowering time QTL and have been shown to influence flowering time in an additive fashion in B. rapa (Kole et al., 2001; Li et al., 2009; Lou et al., 2007; Nishioka et al., 2005; Okazaki et al., 2007; Osborn et al., 1997; Schranz et al., 2002; Xiao et al., 2013; Zhao et al., 2010). In addition, studies have shown that allelic sequence variation, including splice site polymorphism, is correlated with transcript levels of BrFLC genes and with flowering time (Li et al., 2009; Yuan et al., 2009; Zhao et al., 2010). Overexpression of a B. rapa SOC1 ortholog (referred to as BrAGL20) in B. napus caused early flowering, suggesting that the function of this gene may be conserved (Hong et al., 2013). Quantitative gene expression analyses also indicate that at least two of the SOC1 orthologs may potentially play a role in flowering induction in B. rapa (Xiao et al., 2013).
In this study, we investigated the genetic basis of flowering time variation in plants derived from a natural population of Brassica rapa. Selection may have acted upon this underlying genetic variation to produce the evolutionary shifts to earlier flowering time observed previously (Franks, Sim & Weis, 2007). We focused on sequence and expression variation in orthologs of the key Arabidopsis flowering time regulatory genes FLC and SOC1, testing the hypothesis that such variation underlies the natural variation observed in flowering time. We investigated sequence variation in coding regions, introns, and upstream promoter regions in all paralogs of these genes, and quantified the expression of each paralog. We predicted that we would find lower BrFLC expression and greater BrSOC1 expression in early compared to late flowering plants. We looked for associations between allelic and expression variation at these genes and variation in flowering time, focusing on a set of the earliest and latest flowering individuals from the natural population grown under common conditions.
Materials & Methods
Sample collection and growing conditions
Seeds of Brassica rapa were collected in bulk from the Arboretum population in Irvine, California in the spring of 2008. The permit is #19699-21901 from the UC Reserve System (RAMAS) for collecting seeds of Brassica rapa at the San Joaquin Marsh Reserve, the University of California, Irvine. The Arboretum population is located on the grounds of the University of California Arboretum, adjacent to a wetland, and was previously shown to have a broad range in flowering time and to have evolved earlier flowering time in response to a natural drought (Franks, Sim & Weis, 2007). To determine the optimum tissue and developmental stage to sample for comparative gene expression analyses, we grew one set of plants (set 1) in controlled conditions to characterize changes in gene expression over time and among leaves. Once we had identified the appropriate stage and leaf for sampling, we grew two additional sets of plants (sets 2 and 3) in controlled conditions for the early–late flowering comparisons. Set one consisted of 16 plants; sets 2 and 3 consisted of 225 seeds selected haphazardly from the collection, at least 200 of which survived to first flowering. Because these seeds were haphazardly selected from the collection, they varied in flowering time. The seeds were planted in Sunshine mix #1 (Sungro Horticulture, Vancouver, Canada) in pots 6 cm × 6 cm × 9 cm deep, watered daily and fertilized once per week with 14-14-14 fertilizer. The plants were grown on light carts and given light 24 h per day, which allows flowering because Brassica rapa is a long-day plant (Salisbury, 1963). We recorded date of emergence (defined as the opening of the seed coat and emergence of the radicle) and date of first flowering (defined as the opening of the bud and visibility of both stigma and anthers) for all plants. We selected the earliest and latest flowering plants for all analyses of the association between flowering phenotype and genotype or gene expression level.
DNA and RNA extraction
We used set 1 plants for analysis of gene expression over time and among leaves, set 2 plants for comparative analysis of gene expression and sequence analysis of the coding regions of the genes, and set 3 plants for analysis of allelic variation in regulatory regions of our candidate genes. For set 1, we used a sterilized hole punch to collect leaf tissue from the first and second true leaf as soon as each leaf reached 2 cm in length, and every 4 days thereafter. Leaf discs were flash frozen in liquid nitrogen and stored at −80 ° C. The samples were ground in liquid nitrogen and RNA was extracted using the RNeasy Plant Mini kit (Qiagen, Venlo, Limburg) according to the manufacturer’s protocol. RNA was treated with DNAse (NEB, Ipswich, MA) to remove contaminant genomic DNA, and cDNA was synthesized from 1 µg of RNA using the Superscript II enzyme kit (Life Technologies, Norwalk, Connecticut, USA) with random hexamer primers.
For set 2, we collected ∼1 g of leaf tissue from the second true leaf of all plants 16 days from planting, before the plants had come into flower. Results from set 1 indicated that removal of this amount of leaf material did not alter flowering time (there was no difference in average flowering time in plants with tissue removed compared to control plants without tissue removed), and also that gene expression level at day 16 was a good predictor of expression at other times. The leaf tissue was immediately frozen in liquid nitrogen upon collection and then stored at −80 °C. After all plants had flowered, frozen samples from the 10 earliest and 10 latest flowering plants were selected. RNA was extracted and cDNA synthesized as above.
For set 3, which was used to evaluate regulatory sequence variability, leaf tissue was collected from all plants 16 days after planting and stored in silica gel at room temperature. After the plants had flowered, samples from the 20 earliest and 20 latest flowering plants were selected, and subsets (generally 10 each) of these were used for analyses. Samples were ground using a FastPrep (MP Biomedicals, Santa Anna, California, USA) and DNA was extracted using the DNeasy Plant Mini kit (Qiagen, Venlo, Limburg) according to the manufacturer’s instructions.
DNA amplification
CLC Main Workbench, v.6.8.2 (http://www.clcbio.com/products/clc-main-workbench) and Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) were used to design all primers described below (Table 1, Figs. S1 and S2).
Table 1. Primer information.
Primer coordinates and reference sequences are from th e BRAD database (http://brassicadb.org/brad/) except BrFLC5, which is from Genbank. Primers in exons were used for quantitative and semi-quantitative pcr, while primers in other regions were used for DNA sequencing. Locations of the primer attachment sites relative to the reference sequence are given in Figs. S1 and S2.
| Locus | Region | Name | Location | Sequence |
|---|---|---|---|---|
| BrSOC1 | Promoter | 2.p4928f | −1008,−990 | ATGAAGGGAAAAAGATGTG |
| Bra004928 | 2.p4928r | −308,−291 | CCGAAACAAAACAAACCA | |
| BrSOC1 | Promoter | 7.p9324f | −1100,−1082 | GGACATTTTCGACCATACT |
| Bra039324 | 7.p9324r | −275,−258 | ACCCAAAAACCAAACCAA | |
| BrSOC1 | Promoter | 16.pB0393f | −912,−894 | TTTGCTCTTCCTTTTTGCT |
| Bra000393 | 16.pB0393r | −184,−167 | TTCCTGGGGTTTGATTTT | |
| BrSOC1 | Promoter | 15.pA0393f | −580,−562 | CTCCTATATCTCTCTATCT |
| Bra000393 | 15.pA0393r | 217,234 | TTTCTCTCTTTCTCTCTC | |
| BrSOC1 | Exon6 | 67.c4928f | 467,487 | AGGAGAAAGCTCTAGCTGCAG |
| Bra004928 | UTR | 67.c4928r | 817,842 | ATTAGATTCTACAGAGGCAAGTATAC |
| BrSOC1 | Exon6 | 69.c9324f | 468,487 | GGAGAAAGCTCTAGCTGCAG |
| Bra039324 | UTR | 69.c9324r | 803.825 | AACATCTAGGTAGGCAACTGTAG |
| BrSOC1 | Exon6 | 71.c0393f | 495,514 | GAAACTCGCTGAAAAGTGGG |
| Bra000393 | UTR | 71.c0393r | 826,847 | AAGTGTATGAGAAATTGAGAAC |
| BrFLC2 | Promotor | FLC2f2 | −2083,−2064 | ACAGGTGGTATGAGTAATGA |
| Bra028599 | FLC2r2 | 232,250 | AAAGAGAAGAGGAACGGAA | |
| BrFLC3 | Promotor | FLC3f | −1179,−1158 | TTACTTACTGAGTTCAATTGGG |
| Bra006051 | GL1725r | −68,−49 | CGGTTCAAGTGGCCGGAGAT | |
| BrFLC5 | Promotor | FLC5f1 | −2762,−2742 | ACTGGCATCCGAACACCCATG |
| KBrH038M21 | FLC5r2 | −77,−57 | GTCGCCGGAGAGACTAAGCGT | |
| BrFLC1 | Exon1 | GL1132f | 33,52 | TGAGAACAAAAGTAGCCGAC |
| Bra009055 | Exon4 | GL1155r | 3291,3310 | GAACCCACACTTACATTATC |
| BrFLC2 | Exon1 | GL1132f | 33,52 | TGAGAACAAAAGTAGCCGAC |
| Bra028599 | Exon4 | GL1157r | 1791,1810 | GTCGACGCTTACATCAGAAT |
| BrFLC3 | Exon1 | GL1132f | 33,52 | TGAGAACAAAAGTAGCCGAC |
| Bra006051 | Exon4 | GL1156r | 1991,2009 | TGTCCACGCTTACACCACC |
| BrFLC5 | Exon1 | GL1132f | 33,52 | TGAGAACAAAAGTAGCCGAC |
| KBrH038M21 | Exon4 | GL1158r | 3304,3323 | ATCCACGCTTACATCATCAA |
| BrFLC1 | Exon4 | GL1036f | 3278,3297 | GGAATCAAATGTCGATAATG |
| Bra009055 | Exon7 | GL1125r | 4290,4310 | TTAAGCAGCGGGAGAGTYAC |
| BrFLC2 | Exon4 | GL1037f | 1780,1799 | TGTGGAATCAAATTCTGATG |
| Bra028599 | Exon7 | GL1125r | 3238,3256 | TTAAGCAGCGGGAGAGTYAC |
| BrFLC3 | Exon4 | GL1038f | 1978,1999 | GGAATCAAATGTCGGTGGTGTA |
| Bra006051 | Exon7 | GL1125r | 2922,2941 | TTAAGCAGCGGGAGAGTYAC |
| BrFLC5 | Exon4 | GL1039f | 3293,3312 | TGTGGAATCAATTGATGATG |
| KBrH038M21 | Exon7 | GL1125r | 4859,4878 | TTAAGCAGCGGGAGAGTYAC |
| BrFLC1 | Intron 1 | GL1319f | 203,224 | CTGGGGTTTTCCATTATTATTGT |
| Bra009055 | GL1319r | 2603,2626 | GTATGTTAGGATCAAAACTACCAG | |
| BrFLC2 | Intron 1 | GL1320f | 211,230 | TCCTTTATTTGCCCTTTTCG |
| Bra028599 | GL1321r | 1260,1288 | CAAAATAAGTTAAGATCAAAACAACTAGC | |
| BrFLC3 | Intron 1 | GL1322f | 214,236 | TTTATTAGCCTTTTAAGCTTCTG |
| Bra006051 | GL1323r | 1281,1308 | ACAATTAATGTTAAGAACAAAACTACTA | |
| BrFLC5 | Intron 1 | GL1325f | 216,236 | TGCCCTTTAAGCTTTCTTCTC |
| KBrH038M21 | GL1326r | 2584,2607 | GAGATCAAAAGTCAAAACTACTTG | |
| BrFLC1 | Exon4 | GL1099f | 3273,3298 | CTTGAGGAATCAAATGTCGATAATGT |
| Bra009055 | GL1100r | 3320,3341 | GTTCTCAAGGTGTTCCTCCAGC | |
| BrFLC2 | Exon3,Exon4 | GL1136f | 1685,1796 | AAGTAAGCTTGTGGAATCAAATTCTG |
| Bra028599 | Exon4,Exon5 | GL1137r | 1858,1953 | TCAACATTAGTTCTGTCTTCCTAGCTCTA |
| BrFLC3 | Exon4 | GL1101f | 1978,1999 | GGAATCAAATGTCGGTGGTGTA |
| Bra006051 | GL1102r | 2031,2052 | AGAGAGAGGGCATTTTCAAGGA | |
| BrFLC5 | Exon4 | GL1138f | 3287,3309 | CAAGCTTGTGGAATCAATTGATG |
| KBrH038M21 | GL1139r | 3338,3360 | GGGCAGTCTCAAGGTGATCTTCT |
Promoter regions and coding sequences were amplified and sequenced for all BrFLC and BrSOC1 paralogs. The first intron of the BrFLC loci was also sequenced, as evidence from Arabidopsis suggests it contains cis-regulatory elements (Sheldon et al., 2002). In many cases, not all of the 10 early and 10 late flowering individuals produced good quality sequence data. We attempted resequencing of individuals that initially did not produce good results, often several times. However, if no genetic variation was found in other individuals that did produce good results, and we were able to obtain good sequence from several early and late flowering individuals, we did not proceed beyond the earlier attempts at resequencing for individuals that did not produce good results. In addition, for some genomic regions, some paralogs proved difficult to amplify and sequence, therefore results are only presented for those loci for which clean sequence data was obtained for at least 5 early and 5 late flowering individuals, although in most cases our samples sizes were closer to 10 early and 10 late flowering individuals.
PCR reactions were performed as follows. For the promoter regions of the BrFLC paralogs, PCR reactions were performed on genomic DNA (gDNA). We used Taq 2x master mix (M0270; New England Biolabs, Ipswich, Massachusetts, USA) with a dNTP concentration of 200 µM, and final magnesium concentrations varying depending on the reaction. We used the following reaction conditions: an initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C, 30 s; annealing at variable temperatures, 30 s; elongation at 72 °C, variable times, and a final extension of 72 °C for 10 min.
Coding sequences and the first intron of all four BrFLC paralogs were amplified from cDNA and gDNA, respectively, using either (1) EconoTaq Plus Green 2X Master Mix (Lucigen, Middleton, Wisconsin, USA) in a reaction mix consisting of 7.5 µL EconoTaq, 4.8 µL water, 0.75 µL 10 mM primers, and 1.2 µL cNDA or gDNA, or (2) high activity Taq (Pluthero, 1993) in a reaction mix consisting of 9.4 µL water, 0.2 µL high activity Taq, 1.5 µL buffer, 0.6 µL MgCl2, 0.6 µL dNTPs (New England Biolabs, Ipswich, Massachusetts, USA), 0.75 µL 10 mM primers, and 1.2 µL cDNA or gDNA. PCR conditions were 94 °C for 5 min, 34–38 cycles of 94 °C for 30 s, annealing at appropriate temperature for 30 s, 64 °C or 72 °C for one minutes, and a final extension of 64 °C or 72 °C for 10 min. DNA was visualized on a 1% agarose gel stained with ethidium bromide. The coding sequences of all four BrFLC paralogs are similar, with a single variable region in the middle. This region was used to design paralog-specific reverse and forward primers. The reverse gene-specific primers were used with forward primers that annealed at the 5′ end of the coding sequence and were not paralog-specific; similarly, the forward paralog-specific primers were used with universal 3′ reverse primers to amplify the 3′ region of the genes. In this fashion all four paralogs were amplified in two sections with a gap in the middle where the primers annealed. All products of correct size were sequenced in both directions at the DNA Analysis Facility of Yale University (http://dna-analysis.research.yale.edu/). Sequences were analyzed, trimmed, and assembled in Sequencher (GeneCodes, Ann Arbor, Michigan, USA).
For amplifications of the BrSOC1 promoters, we used the following reaction conditions: an initial denaturation at 95 °C for 2 min, 32 cycles of denaturation at 95 °C, 30 s; annealing at variable temperatures, 30 s; elongation at 72 °C, variable times, and a final extension of 72 degrees for 10 min. We used 2–3 µM forward and reverse primers each. We used variable magnesium concentrations and Taq 2x master mix (M0270, New England Biolabs, Ipswich, Massachusetts, USA) to amplify a region of the BrSOC1 paralog coding sequences, and NEBNext High-Fidelity 2X PCR master mix (New England Biolabs, Ipswich, Massachusetts) to amplify a region of the BrSOC1 paralog promoters. DNA was visualized in 1% agarose gels pre-stained with GelRed dye (RGB-4103T; Phenix, Candler, North Carolina, USA).
We amplified the coding sequences of the BrSOC1 paralogs using the same reaction mixes and cycling parameters as for the BrFLC paralogs, with appropriate annealing temperatures.
DNA Sequencing and Alignment
Sanger sequencing was performed at Genewiz (http://www.genewiz.com), Cornell (http://www.biotech.cornell.edu) and Yale University (http://dna-analysis.research.yale.edu/). Promoters include sequence within 4 kb upstream of the gene transcription start site. Two regions of each of the BrFLC paralog promoters (within 4 kb upstream of the transcription start site), and one region of each BrSOC1 paralog promoter regions (within 4 kb upstream of the transcription start site, and including key regulatory elements such as the predicted BrFLC MADS box binding site) were sequenced. The contigs were assembled in CLC Main Workbench 7.6.2 (http://www.clcbio.com) and manually edited using overlapping (forward and reverse) sequence reads. Alignment of each paralog was performed using MUSCLE (Edgar, 2004) in CLC Main using BrFLC reference sequences Bra009055 (BrFLC2), Bra028599 (BrFLC3), Bra006051 (BrFLC5) and BrSOC1 reference sequences Bra000393, Bra039345, Bra004928 from the BRAD database (http://brassicadb.org/brad/). Exon 7 and the 3′ UTR were sequenced together to insure that a single paralog was amplified for qRT-PCR analysis and to confirm genome annotations. Alignments were visually inspected for proper codon alignment.
Quantitative expression analyses
Quantitative real-time PCR (qRT-PCR) was performed to quantify expression using set 1 hole punch material (to identify appropriate tissue and developmental stage for further analyses) and from the material collected from the 10 earliest and 10 latest flowering individuals of set 2 (to quantify expression of BrFLC and BrSOC1 paralogs in early- and late-flowering plants) on an ABI 7300 Real-Time PCR System (Life Technologies, Carlsbad, California, USA) using SYBR Green Master Mix (Life Technologies, Carlsbad, California, USA). Primers (Table 1) were designed using the ABI Primer Express program. The Brassica rapa serine/threonine-protein phosphatases PP2a catalytic subunit, which we determined to be expressed at a constant and appropriate level in our tissue samples (data not shown), was used as an endogenous control. Expression was quantified for all four BrFLC paralogs, the three BrSOC1 paralogs, and the control using three technical replicates for each sample and gene. All primers had comparable efficiencies. Reaction mixes consisted of 12.5 µL FastStart Universal SYBR Green Master Mix (Roche Diagnostics, Indianapolis, Indiana, USA), 2.4 µL forward and reverse primers (2.5 nmole), 15 ng cDNA template, and 3.5 µL sterile water. Reactions were run using the standard relative quantification cycling parameters: 95 °C for 20 s followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Relative expression was calculated using the ΔΔCT method using the 7300 System SDS Software provided with the 7300 Real-Time PCR System.
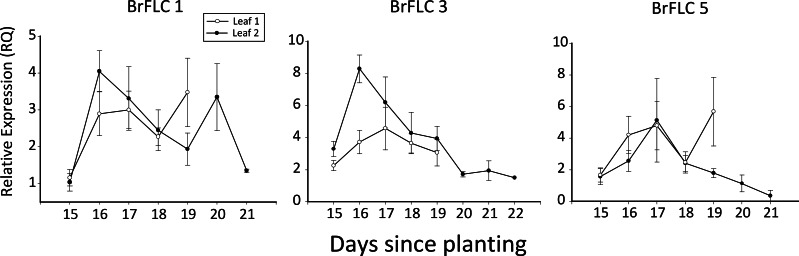
Approximately 3–5 hole punches were collected from leaf one and leaf two from the 16 plants of set 1. Expression of BrFLC3, which our preliminary analyses had shown to be strongly expressed, was quantified across all hole punches and leaves to determine a tissue and stage to sample. These results indicated that the second true leaf, collected 16 days after sampling, was appropriate for analysis of gene expression. The reason for this was that expression at this time was at or near peak, and was correlated with expression levels at other times (Fig. 3). For example, expression at day 16 and day 18 was highly correlated (r2 = 0.86, p = 0.0064).
Figure 3. BrFLC expression over time.
Shown are average relative expression (RQ) values from real-time quantitative PCR over time for the three BrFLC genes analyzed. Samples were taken from the first (white dots) and second (black dots) true leaves. The plants vary in flowering time. Bars represent 1 standard error.
Because BrSOC1 expression analysis was initially performed using general primers that amplified all BrSOC1 paralogs, a second set of expression analysis was performed with BrSOC1 paralog-specific primers (Table 1). We used semi-quantitative PCR to determine if there was a difference in expression between early and late flowering plants. For these assays, we performed PCR using cDNA for each of the BrSOC1 paralogs on the same set of early and late flowering plants as the qPCR assays, with one sample per plant and 10 replicates of early and 10 of late flowering plants. PCR reactions were run at 95 °C for 20 s followed by 36–38 cycles of 95 °C for 3 s and 60 °C for 30 s. Products were run on 1% agarose gels that included a ladder that served as a product size indicator as well as an intensity standard. The same amount of cDNA was used in each reaction and the same amount of product was loaded into each lane. Band intensity relative to the ladder was quantified from the gel image using the program GeneTools version 4.03 (Syngene, Frederick, Maryland). Quantified relative band intensity was used as our semi-quantitative measure of gene expression in these assays.
Statistical Analyses
Differences in gene expression between early and late flowering plants were analyzed with ANOVA. Differences in allele frequencies between early and late flowering plants were analyzed with Fisher exact tests and Wald two sample test of proportions.
Results
Flowering phenology
The Brassica rapa plants from the natural California population exhibited a broad range in flowering time when grown in the lab. We were able to sample plants that flowered in the early and late ends of the flowering time distribution and that were well above and below the mean flowering time. We examined flowering time in set 2 and set 3 plants.
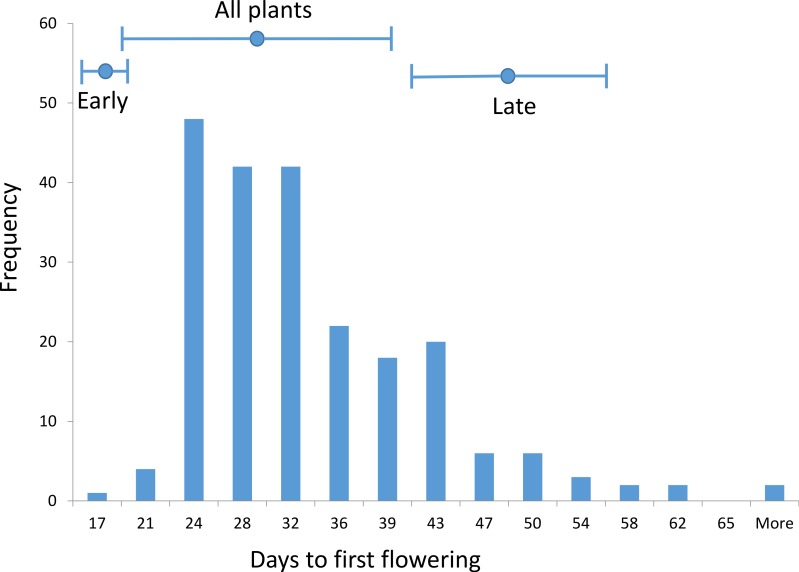
For set 2 plants, the average time to first flowering was 31.4 (±9.3) days (standard deviation in parentheses) (Fig. 1). The average time to first flowering was 20.7 (±1.2) days in the 20 earliest flowering plants and 51.9 (±7.4) days in the 20 latest flowering plants (Fig. 1). For set 3 plants, the average time to first flowering was 35.0 (±8.5) days. The average time to first flowering was 24.8 (±0.6) days in the 20 earliest flowering plants and 53.3 (±6.4) days in the 20 latest flowering plants.
Figure 1. Flowering time.
Shown is a histogram of flowering time for 203 Brassica rapa plants from the Arboretum population grown under common conditions on light carts in the lab. Above the histogram, the mean (dot) and standard deviation (bar) flowering time is shown for plants from the early flowering group, the late flowering group, and all plants. Plants from the early and late flowering groups were chosen for analyses.
Gene expression
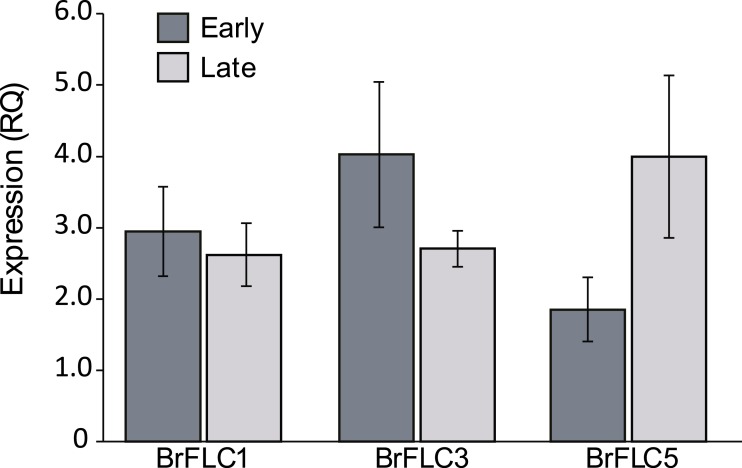
For the BrFLC genes, we designed primers specific to each paralog for quantitative reverse transcription PCR (qRT-PCR) analysis. We did not obtain sufficient sequence data for BrFLC2 for statistical evaluation. There was no difference in expression between early and late flowering plants (set 2) for BrFLC1 (F1,18 = 0.18, p = 0.68), BrFLC3 (F1,17 = 1.75, p = 0.20) or BrFLC5 (F1,7 = 2.53, p = 0.16). There was also no difference in expression between early and late flowering plants for the expression of the three BrFLC genes summed together (F1,18 = 0.40, p = 0.54). Trends showed greater expression in early than late flowering plants for BrFLC3, and greater expression in late than early plants in BrFLC5 (Fig. 2), but these were not statistically significant. There was variation in expression of the three BrFLC paralogs over time (set 1), with expression generally increasing at first and then showing an eventual decline, although there was variation in this pattern (Fig. 3).
Figure 2. Expression of BrFLC genes.
Shown are average relative expression (RQ) values from real-time quantitative PCR for the three BrFLC genes analyzed from individuals from the early (dark grey bars) and late (light grey bars) flowering groups. Bars represent 1 standard error.
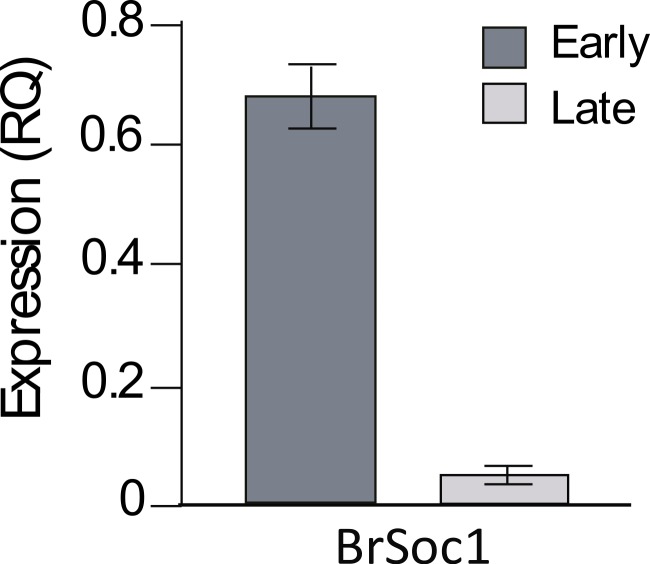
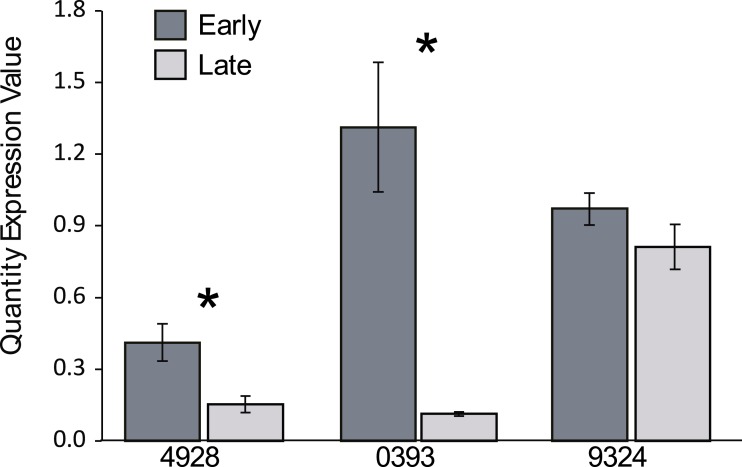
For the BrSOC1 genes, our initial primers amplified all paralogs together when used on cDNA prepared from set 2 plants. We found that combined BrSOC1 expression was significantly greater in early compared to late flowering plants (F1,18 = 49.2, p < 0.0001; Fig. 4), consistent with experiments in Arabidopsis showing that SOC1 promotes flowering (Immink et al., 2012). Paralog-specific primers were then designed and products quantified using semi-quantitative PCR. Expression was significantly greater in early compared to late flowering plants for Bra004928 (t = 3.03, d.f. = 18, p = 0.007) and Bra000393 (t = 4.44, d.f. = 18, p = 0.0003), but not for Bra039324 (t = 1.25, d.f. = 17, p = 0.230) (Fig. 5).
Figure 4. Total BrSOC1 expression.
Shown are average relative expression (RQ) values from real-time quantitative PCR for early (dark grey bars) and late (light grey bars) flowering plants using primers that amplified BrSOC1 generally and were not paralog-specific. Bars represent 1 standard error.
Figure 5. Paralog-specific BrSOC1 expression.
Shown are quantity expression values (QEV) derived from semi-quantitative pcr (see ‘Methods’ for details) for the three BrSOC1 paralogs (Br004928, Br000393, Br009324) for plants from the early (dark grey bars) and late (light grey bars) flowering groups. Bars represent 1 standard error. An * indicates that expression of early and late flowering individuals was significantly different for a given paralog at p < 0.05.
Allelic variation
Based on cDNA and gDNA sequencing of set 2 and set 3 plants, promoter regions, first introns, and coding sequences of all of four BrFLC genes showed no allelic variation. Sequences across the promoter and entire coding sequence appear to be fixed for all four BrFLC paralogs in B. rapa. Thus allelic variation at these loci does not explain variation in flowering time. Promoter regions of the BrSOC1 paralogs (set 3) did show allelic variation at several sites. In particular, Br009324 showed variation at 5 sites, with each of these sites a SNP with two alternate alleles. However, there was no statistically significant association between this allelic variation at any of the sites with flowering time variation (Table 2). No other regions sequenced showed variation, so these were not tested for associations with flowering time.
Table 2. Statistics table.
Statistical tests of association between allelic variation for each polymorphic nucleotide site in the promoter of locus Br009324, one of the BrSOC1 paralogs, and flowering time variation comparing the early to the late flowering plants. Numbers across the top refer to nucleotide sites relative to the start codon. Tests are Fisher exact tests and Wald two sample test for proportions. Shown are p-values for two-tailed tests.
| −952 | −920 | −904 | −852 | −639 | |
|---|---|---|---|---|---|
| Fisher | 0.1189 | 0.4667 | 0.1189 | 0.1189 | 0.1189 |
| Wald | 0.0880 | 0.2908 | 0.0880 | 0.0880 | 0.0880 |
Discussion
In this study, we were able to take advantage of variation in flowering time in plants from a natural population of Brassica rapa to explore the relationship between this phenological variation and allelic and expression variation at candidate flowering time loci. This genetic variation is important since selection can potentially act upon it to cause evolutionary changes in flowering time. We found a clear association between flowering time and expression in two BrSOC1 paralogs, but not with any of the BrFLC paralogs, and no relationship between flowering time and allelic variation at any of these loci in their coding or promoter regions.
Expression of two of the three BrSOC1 paralogs was greater in early than in late flowering plants. The trend for the third BrSOC1 paralog was in the same direction, but was not statistically significant. It thus appears that, as in Arabidopsis (Immink et al., 2012) and other species (Fu et al., 2014; Lei et al., 2013; Preston, Jorgensen & Jha, 2014), early upregulation of the BrSOC1 genes is indicative and predictive of early flowering in B. rapa. The cause of the differential expression in the BrSOC1 genes between early and late flowering plants remains unknown, because we were not able to detect any association between flowering time and allelic variation within the BrSOC1 promoter regions. We had hypothesized that variation in the promoter regions would influence flowering time, but this hypothesis was not supported. It is possible that some association could have been detected with a larger sample size, but a strong association would have been detected even with our modest sample. It is also possible that a region of the promoter that we did not sequence influences BrSOC1 expression. The absence of a relationship between flowering time variation and promoter variation suggests that there is an alternative explanation for the observed difference in expression levels between early- and late-flowering plants. Expression may be influenced by the products of activating and repressing upstream transcription factors. Although FLC is known to suppress expression of SOC1 in Arabidopsis (Hepworth et al., 2002), there was no relationship between expression of any of the BrFLC paralogs and flowering time in our study, suggesting that regulation of the BrSOC1 paralogs by the BrFLC paralogs is not a likely factor in the patterns that we observed. Other possibilities include orthologs of FT or FD, which upregulate SOC1 in Arabidopsis. However, our very preliminary investigations with BrFT paralogs did not uncover any genetic variation associated with flowering time variation, although we did find greater BrFT expression in early flowering than in late flowering plants. Additional possible explanations for the differences in expression are variation in potential enhancers that are not located within the 4 kb promoter region, or chromatin or DNA epigenetic modifications that would influence regulation but that are not detected with standard sequencing mechanisms.
Despite the fact that FLC is known to be a key regulator of flowering time in Arabidopsis, we found no association between flowering time and expression or allelic variation at any of the BrFLC orthologs in our population. It is worth noting that FLC operates though the autonomous and vernalization pathways, and the plants in our southern California population neither receive nor require vernalization to initiate flowering. If the vernalization pathway is not as important in populations that do not experience cold temperatures, then genes in this pathway might not play as large a role in influencing phenotypic variation in such populations. Such genes could influence variation in flowering time in temperate populations. They could also potentially be important in local adaptation, and may come under selection with changing environmental and climatic conditions.
Previous studies in Brassica species have detected flowering time QTLs, and some of these loci map to known flowering time genes (Axelsson, Shavorskaya & Lagercrantz, 2001; Lou et al., 2007). Other studies have shown associations between changes in expression of flowering time genes and flowering time phenotypes. For example, one recent study found that in Ambrosia artemisiifolia, expression of the orthologs of the genes AP1, FT and SOC1 changed during the course of flowering, and the genes CRY2 and SPY differed in expression between an early and a late flowering population (Li, Zhang & Liao, 2015). Other studies have shown that genetic variation in flowering time genes can influence the timing of flowering, but these genetic variants were generally major mutations that caused loss of function. For example, one study of Arabidopsis thaliana showed that variation between null and wild-type alleles of the gene FRI, along with interactions with FLC, resulted in a geographic cline in flowering time (Caicedo et al., 2004). Variation in these genes and their interactions was also found to influence flowering time in a broad survey of natural accessions of A. thaliana (Werner et al., 2005). We found associations between gene expression and flowering time, consistent with this previous work, but we did not find specific genetic variation that could be linked with flowering time variation.
Conclusions
Genetic regulatory networks are often highly integrated and complex, and can potentially greatly diverge from a simple additive model of genetic effects. The flowering time genetic regulatory network in Arabidopsis is well studied and contains over a hundred genes, regulatory elements and transcription factors that all work in concert to control the timing of flowering. How variation in such complex networks as this influences phenotypic variation in natural populations is unknown. The fact that a particular gene is part of this regulatory network does not necessarily mean that allelic or expression variation at that gene is responsible for variation in flowering time in natural populations. For example, BrFLC is known to play a central role in the flowering time regulatory network, but variation at this gene did not seem to influence variation in flowering time in the population examined in our study. Understanding how genetic variation influences phenotypic variation in natural populations is an emerging area of investigation, and is key to predicting how traits will evolve. This will be useful, for example, in predicting how traits such as flowering time will respond to selection by changing climatic conditions.
Supplemental Information
Relative locations of primers designed for sequencing promotor orthologs (A–C) and Exon 6 (D) of BrSoc1 flowering time genes in Brassica rapa. Solid blue arrow indicates the start codon. Where applicable, colors indicate matching sets of primers. For specific locations and primer sequence information, see Table 1.
Relative locations of primers designed for sequencing promotor regions (A) and coding regions (B) of BrFLC flowering time genes in Brassica rapa. Solid blue arrow indicates the start codon. Where applicable, colors indicate matching sets of primers. (Note for FLC2, the primer set shown in green covers Exon 3 (forward) and Exon 5 (reverse); for FLC1, FLC3 and FLC5 this set covers only Exon 4.) For specific locations, corresponding primer names and sequence information, see Table 1.
Acknowledgments
We thank the students who assisted with lab work, including Kim Barnum, Ricky Barnum, Rahitul Bhuiyan, Andrew Biello, India Brown, Nick Dovgala-Carr, Lolita Feld, Ellen George, Kerry Gunsalus, Jacqueline Gutkin, Mohammed Hosen, Eugene Kim, Ben Marsh, Kathryn Posocco, Maria Rodenberg, Diana Shao, Richard Stewart, Maren Toor, Jeanne Valente, Chris Yiachos, and Sandra Zajac. We thank two anonymous reviewers for comments.
Funding Statement
This research was supported by grant DEB-1142784 from the National Science Foundation to SJ Franks and funds from the Eppley Foundation for Scientific Research and The New York Botanical Garden to A Litt. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Steven J. Franks and Amy Litt conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Beatriz Perez-Sweeney performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables.
Maya Strahl, Anna Nowogrodzki and Kevin P. Jordan performed the experiments, analyzed the data.
Jennifer J. Weber performed the experiments, analyzed the data, prepared figures and/or tables, reviewed drafts of the paper.
Rebecca Lalchan performed the experiments.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
The permit is #19699-21901 from the UC Reserve System (RAMAS) for collecting seeds of Brassica rapa at the San Joaquin Marsh Reserve, the University of California, Irvine.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
The sequences can be found in the Supplemental Information.
References
- Amasino & Michaels (2010).Amasino RM, Michaels SD. The timing of flowering. Plant Physiology. 2010;154:516–520. doi: 10.1104/pp.110.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson, Shavorskaya & Lagercrantz (2001).Axelsson T, Shavorskaya O, Lagercrantz U. Multiple flowering time QTLs within several Brassica species could be the result of duplicated copies of one ancestral gene. Genome. 2001;44:856–864. doi: 10.1139/gen-44-5-856. [DOI] [PubMed] [Google Scholar]
- Bastow & Dean (2003).Bastow R, Dean C. Plant sciences—deciding when to flower. Science. 2003;302:1695–1697. doi: 10.1126/science.1092862. [DOI] [PubMed] [Google Scholar]
- Bastow et al. (2004).Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Boss et al. (2004).Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell. 2004;16:S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo et al. (2004).Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15670–15675. doi: 10.1073/pnas.0406232101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar (2004).Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga et al. (2007).Elzinga JA, Atlan A, Arjen B, Gigord L, Weis AE, Bernasconi G. Time after time: flowering phenology and biotic interactions. Trends in Ecology & Evolution. 2007;22:432–439. doi: 10.1016/j.tree.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Franks (2011).Franks SJ. Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytologist. 2011;190:249–257. doi: 10.1111/j.1469-8137.2010.03603.x. [DOI] [PubMed] [Google Scholar]
- Franks (2015).Franks SJ. The unique and multifaceted importance of the timing of flowering. American Journal of Botany. 2015;102:1401–1402. doi: 10.3732/ajb.1500234. [DOI] [PubMed] [Google Scholar]
- Franks & Hoffmann (2012).Franks SJ, Hoffmann AA. Genetics of climate change adaptation. Annual Review of Genetics. 2012;46:185–208. doi: 10.1146/annurev-genet-110711-155511. [DOI] [PubMed] [Google Scholar]
- Franks, Sim & Weis (2007).Franks SJ, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, Weber & Aitken (2014).Franks SJ, Weber JJ, Aitken SN. Evolutionary and plastic responses to climate change in terrestrial plant populations. Evolutionary Applications. 2014;7:123–139. doi: 10.1111/eva.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu et al. (2014).Fu J, Qi S, Yang L, Dai Y, Dai S. Characterization of Chrysanthemum ClSOC1-1 and ClSOC1-2, homologous genes of SOC1. Plant Molecular Biology Reporter. 2014;32:740–749. doi: 10.1007/s11105-013-0679-8. [DOI] [Google Scholar]
- Gazzani et al. (2003).Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiology. 2003;132:1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth et al. (2002).Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. Embo Journal. 2002;21:4327–4337. doi: 10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann & Sgro (2011).Hoffmann AA, Sgro CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Hong et al. (2013).Hong J, Kim S-Y, Kim K-S, Kwon S-J, Kim J, Kim J, Lee S, Lee Y-H. Overexpression of a Brassica rapa MADS-box gene, BrAGL20, induces early flowering time phenotypes in Brassica napus. Plant Biotechnology Reports. 2013;7:231–237. doi: 10.1007/s11816-012-0254-z. [DOI] [Google Scholar]
- Immink et al. (2012).Immink RG, Pose D, Ferrario S, Ott F, Kaufmann K, Valentim FL, De Folter S, Van der Wal F, Van Dijk AD, Schmid M, Angenent GC. Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiology. 2012;160:433–449. doi: 10.1104/pp.112.202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole et al. (2001).Kole C, Quijada P, Michaels SD, Amasino RM, Osborn TC. Evidence for homology of flowering-time genes VFR2 from Brassica rapa and FLC from Arabidopsis thaliana. Theoretical and Applied Genetics. 2001;102:425–430. doi: 10.1007/s001220051663. [DOI] [Google Scholar]
- Korves et al. (2007).Korves TM, Schmid KJ, Caicedo AL, Mays C, Stinchcombe JR, Purugganan MD, Schmitt J. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. American Naturalist. 2007;169:E141–E157. doi: 10.1086/513111. [DOI] [PubMed] [Google Scholar]
- Lagercrantz et al. (1996).Lagercrantz U, Putterill J, Coupland G, Lydiate D. Comparative mapping in Arabidopsis and Brassica, fine scale genome collinearity and congruence of genes controlling flowering time. Plant Journal. 1996;9:13–20. doi: 10.1046/j.1365-313X.1996.09010013.x. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2004).Lee SY, Kim J, Han JJ, Han MJ, An GH. Functional analyses of the flowering time gene OsMADS50, the putative suppressor of over expression of CO 1/agamous-like 20 (SOC1/AGL20) ortholog in rice. Plant Journal. 2004;38:754–764. doi: 10.1111/j.1365-313X.2004.02082.x. [DOI] [PubMed] [Google Scholar]
- Lei et al. (2013).Lei HJ, Yuan HZ, Liu Y, Guo XW, Liao X, Liu LL, Wang Q, Li TH. Identification and characterization of FaSOC1, a homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 from strawberry. Gene. 2013;531:158–167. doi: 10.1016/j.gene.2013.09.036. [DOI] [PubMed] [Google Scholar]
- Lempe et al. (2005).Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genetics. 2005;1:109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2009).Li F, Kitashiba H, Inaba K, Nishio T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Research. 2009;16:311–323. doi: 10.1093/dnares/dsp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Zhang & Liao (2015).Li X-M, Zhang D-Y, Liao W-J. The rhythmic expression of genes controlling flowering time in southern and northern populations of invasive Ambrosia artemisiifolia. Journal of Plant Ecology. 2015;8:207–212. doi: 10.1093/jpe/rtv008. [DOI] [Google Scholar]
- Lin et al. (2005).Lin SI, Wang JG, Poon SY, Su CL, Wang SS, Chiou TJ. Differential regulation of expression by vernalization FLOWERING LOCUS C in cabbage and Arabidopsis. Plant Physiology. 2005;137:1037–1048. doi: 10.1104/pp.104.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2008).Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development. 2008;135:1481–1491. doi: 10.1242/dev.020255. [DOI] [PubMed] [Google Scholar]
- Lou et al. (2007).Lou P, Zhao JJ, Kim JS, Shen SX, Del Carpio DP, Song XF, Jin MN, Vreugdenhil D, Wang XW, Koornneef M, Bonnema G. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. Journal of Experimental Botany. 2007;58:4005–4016. doi: 10.1093/jxb/erm255. [DOI] [PubMed] [Google Scholar]
- McKay, Richards & Mitchell-Olds (2003).McKay JK, Richards JH, Mitchell-Olds T. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Molecular Ecology. 2003;12:1137–1151. doi: 10.1046/j.1365-294X.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Michaels (2009).Michaels SD. Flowering time regulation produces much fruit. Current Opinion in Plant Biology. 2009;12:75–80. doi: 10.1016/j.pbi.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels & Amasino (1999).Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels & Amasino (2001).Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Rushing & Primack (2008).Miller-Rushing AJ, Primack RB. Global warming and flowering times in Thoreau’s Concord: a community perspective. Ecology. 2008;89:332–341. doi: 10.1890/07-0068.1. [DOI] [PubMed] [Google Scholar]
- Moon et al. (2003).Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant Journal. 2003;35:613–623. doi: 10.1046/j.1365-313X.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Mouradov, Cremer & Coupland (2002).Mouradov A, Cremer F, Coupland G. Control of flowering time: interacting pathways as a basis for diversity. Plant Cell. 2002;14:S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka et al. (2005).Nishioka M, Tamura K, Hayashi M, Fujimori Y, Ohkawa Y, Kuginuki Y, Harada K. Mapping of QTLs for bolting time in Brassica rapa (syn. campestris) under different environmental conditions. Breeding Science. 2005;55:127–133. doi: 10.1270/jsbbs.55.127. [DOI] [Google Scholar]
- Okazaki et al. (2007).Okazaki K, Sakamoto K, Kikuchi R, Saito A, Togashi E, Kuginuki Y, Matsumoto S, Hirai M. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theoretical and Applied Genetics. 2007;114:595–608. doi: 10.1007/s00122-006-0460-6. [DOI] [PubMed] [Google Scholar]
- Osborn et al. (1997).Osborn TC, Kole C, Parkin IAP, Sharpe AG, Kuiper M, Lydiate DJ, Trick M. Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana. Genetics. 1997;146:1123–1129. doi: 10.1093/genetics/146.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan & Yohe (2003).Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- Pluthero (1993).Pluthero FG. Rapid purification of high-activity Taq DNA polymerase. Nucleic Acids Research. 1993;21:4850–4851. doi: 10.1093/nar/21.20.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, Jorgensen & Jha (2014).Preston JC, Jorgensen SA, Jha SG. Functional characterization of duplicated SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1-like genes in Petunia. PLoS ONE. 2014;9:e1339. doi: 10.1371/journal.pone.0096108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primack (1985).Primack RB. Patterns of flowering phenology in communities, populations, individuals and single flowers. In: White J, editor. The population structure of vegetation. Dordrecht: Kluwer; 1985. pp. 571–593. [Google Scholar]
- Putterill, Laurie & Macknight (2004).Putterill J, Laurie R, Macknight R. It’s time to flower: the genetic control of flowering time. Bioessays. 2004;26:363–373. doi: 10.1002/bies.20021. [DOI] [PubMed] [Google Scholar]
- Riboni et al. (2014).Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L. Environmental stress and flowering time: the photoperiodic connection. Plant Signaling & Behavior. 2014;9:e1339. doi: 10.4161/psb.29036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury (1963).Salisbury FB. The flowering process. New York: Pergamon Press; 1963. [Google Scholar]
- Scarcelli & Kover (2009).Scarcelli N, Kover PX. Standing genetic variation in FRIGIDA mediates experimental evolution of flowering time in Arabidopsis. Molecular Ecology. 2009;18:2039–2049. doi: 10.1111/j.1365-294X.2009.04145.x. [DOI] [PubMed] [Google Scholar]
- Schläppi (2001).Schläppi M. RNA levels and activity of FLOWERING LOCUS C are modified in mixed genetic backgrounds of Arabidopsis thaliana. International Journal of Plant Sciences. 2001;162:527–537. doi: 10.1086/320141. [DOI] [Google Scholar]
- Schranz et al. (2002).Schranz ME, Quijada P, Sung SB, Lukens L, Amasino R, Osborn TC. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics. 2002;162:1457–1468. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz et al. (2007).Schranz ME, Song B-H, Windsor AJ, Mitchell-Olds T. Comparative genomics in the Brassicaceae: a family-wide perspective. Current Opinion in Plant Biology. 2007;10:168–175. doi: 10.1016/j.pbi.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Searle et al. (2006).Searle I, He YH, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Development. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon et al. (2002).Sheldon CC, Conn AB, Dennis ES, Peacock WJ. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. The Plant Cell. 2002;14:2527–2537. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon et al. (2000).Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC) Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3753–3758. doi: 10.1073/pnas.97.7.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson & Dean (2002).Simpson GG, Dean C. Flowering—Arabidopsis, the rosetta stone of flowering time? Science. 2002;296:285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- Slotte et al. (2009).Slotte T, Huang H-R, Holm K, Ceplitis A, Onge KS, Chen J, Lagercrantz U, Lascoux M. Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics. 2009;183:337–345. doi: 10.1534/genetics.109.103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe et al. (2004).Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4712–4717. doi: 10.1073/pnas.0306401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege et al. (2001).Tadege M, Sheldon CC, Helliwell CA, Stoutjesdijk P, Dennis ES, Peacock WJ. Control of flowering time by FLC orthologues in Brassica napus. Plant Journal. 2001;28:545–553. doi: 10.1046/j.1365-313X.2001.01182.x. [DOI] [PubMed] [Google Scholar]
- Wada & Takeno (2010).Wada KC, Takeno K. Stress-induced flowering. Plant Signaling & Behavior. 2010;5:944–947. doi: 10.4161/psb.5.8.11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2011).Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park B-S, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IAP, Batley J, Kim J-S, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon S-J, Choi S-R, Lee T-H, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Werner et al. (2005).Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics. 2005;170:1197–1207. doi: 10.1534/genetics.104.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao et al. (2013).Xiao D, Zhao JJ, Hou XL, Basnet RK, Carpio DP, Zhang NW, Bucher J, Lin K, Cheng F, Wang XW, Bonnema G. The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. Journal of Experimental Botany. 2013;64:4503–4516. doi: 10.1093/jxb/ert264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2010).Yan ZQ, Liang DW, Liu H, Zheng GC. FLC: a key regulator of flowering time in Arabidopsis. Russian Journal of Plant Physiology. 2010;57:166–174. doi: 10.1134/S1021443710020020. [DOI] [Google Scholar]
- Ying, Chen & Cai (2014).Ying L, Chen H, Cai W. BnNAC485 is involved in abiotic stress responses and flowering time in Brassica napus. Plant Physiology and Biochemistry. 2014;79:77–87. doi: 10.1016/j.plaphy.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Yuan et al. (2009).Yuan Y, Wu J, Sun R, Zhang X, Xu D, Bonnema G, Wang X. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. Journal of Experimental Botany. 2009;60:1299–1308. doi: 10.1093/jxb/erp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2010).Zhao JJ, Kulkarni V, Liu NN, Del Carpio DP, Bucher J, Bonnema G. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. Journal of Experimental Botany. 2010;61:1817–1825. doi: 10.1093/jxb/erq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative locations of primers designed for sequencing promotor orthologs (A–C) and Exon 6 (D) of BrSoc1 flowering time genes in Brassica rapa. Solid blue arrow indicates the start codon. Where applicable, colors indicate matching sets of primers. For specific locations and primer sequence information, see Table 1.
Relative locations of primers designed for sequencing promotor regions (A) and coding regions (B) of BrFLC flowering time genes in Brassica rapa. Solid blue arrow indicates the start codon. Where applicable, colors indicate matching sets of primers. (Note for FLC2, the primer set shown in green covers Exon 3 (forward) and Exon 5 (reverse); for FLC1, FLC3 and FLC5 this set covers only Exon 4.) For specific locations, corresponding primer names and sequence information, see Table 1.