Abstract
Asymmetric cell division (ACD) is a simple and evolutionary conserved process whereby a mother divides to generate two daughter cells with distinct developmental potentials. This process can generate cell fate diversity during development. Fate asymmetry may result from the unequal segregation of molecules and/or organelles between the two daughter cells. Here, I will review how fate asymmetry is regulated in the sensory bristle lineage in Drosophila and focus on the molecular mechanisms underlying ACD of the sensory organ precursor cells (SOPs). WIREs Dev Biol 2015, 4:299–309. doi: 10.1002/wdev.175
For further resources related to this article, please visit theWIREs website.
Conflict of interest: The author has declared no conflicts of interest for this article.
INTRODUCTION
Asymmetric cell division (ACD) can be viewed as a four-step process. In a first step, the mother cell acquires and/or re-orients a polarity axis. Second, cell fate determinants, i.e. molecules and/or organelles that are unequally inherited and that have the ability to influence the fate of the daughter cells, are distributed in a polar manner in the mitotic cell. Third, the mitotic spindle lines up along the cell polarity axis so that fate determinants become asymmetrically segregated at cytokinesis. In a fourth step, these fate determinants regulate a binary fate choice to implement fate asymmetry (Figure 1). Many excellent reviews have discussed the process of ACD and the molecular mechanisms underlying these steps in model systems, particularly the early Caenorhabditis elegans embryo and the Drosophila neuroblasts.1–6 Here, I will review how an epithelial cell, known as the sensory organ precursor (SOP) cell, divides asymmetrically within the plane of the single-layered epithelium of the Drosophila pupal thorax to produce two distinct daughter cells.7,8 This simple experimental model has been used to study ACD using genetics and live imaging. Its analysis has provided key insights into the ACD process. For instance, the unequal segregation of Numb, the first cell fate determinant to be discovered, was observed in SOPs.9 Also, planar cell polarity (PCP) mediated by Frizzled signaling was first shown to orient cell division in this context.10 Importantly, SOPs remain integrated within the notum epithelium following their specification and divide asymmetrically in the presence of cell–cell junctions within the plane of the epithelium. Thus, SOPs differ from both worm embryos that have no junctions and fly neuroblasts for which junction disassembly accompanies neuroblast delamination and precedes ACD in fly embryos. Therefore, SOPs provide an interesting model to study how cell polarity is regulated in the context of ACD within epithelia.
Figure 1.

Key steps in asymmetric cell division. In a first step, a polarity axis is set up in the mother cell (M), as indicated by the polarized distribution of polarity proteins (red) at the cell cortex prior to mitosis. In a second step, the polarization of cell M leads to the polar distribution of cell fate determinants (blue). In a third step, cortical cues localized along the polarity axis capture astral microtubules and orient the mitotic spindle (green; chromatin in gray) along the polarity axis such that the cell fate determinants are unequally inherited at cytokinesis. In a fourth step, cell fate determinants regulate the binary A versus B cell fate decision.
The fly body is covered with sensory organs. On the dorsal thorax of the adult fly, mechano-sensory organs are found at regular space intervals (Figure 2(a)). Each of these sensory organs comprises only four different cells (Figure 2(b) and (c)). These four cells are produced via a stereotyped lineage from a single SOP (Figure 2(d)).7,8,11 SOPs are specified a few hours after the onset of metamorphosis within a single-layered epithelium called the notum. SOPs are selected from proneural clusters via Notch-mediated lateral inhibition.12 The notum comprises only two types of cells, i.e. epidermal and sensory organ cells. While SOPs and epidermal cells divide within the plane of the epithelium, SOPs divide asymmetrically in an oriented manner along the fly body axis to generate an anterior pIIb cell (precursor of the sensory organ internal cells) and a posterior pIIa cell (precursor of the sensory organ external cells) whereas epidermal cells divide symmetrically in a randomly oriented manner to produce two epidermal cells (Figure 2(e)–(f″)).10 The a–p orientation of the SOP division is regulated by the planar polarity of the tissue.10 In the absence of PCP, SOPs divide with a random orientation within the plane of the epithelium. Nevertheless, SOPs divide asymmetrically and produce differentiated sense organs. Thus, PCP regulates the orientation of SOP polarity but is not essential for SOP asymmetry at mitosis. Additionally, PCP specifically regulates SOP orientation and does not input into other dividing cells of this planar polarized tissue. These observations raised a number of questions: how is the pIIa/pIIb binary fate decision regulated? How is asymmetry established in dividing SOPs? How is the anterior–posterior (a–p) orientation of the SOP division regulated? I will review below what is currently known and will highlight remaining issues. I will not, however, discuss the asymmetry of the subsequent divisions in the bristle lineage (see Refs 7, 13–14). I will also not review here how PCP is established and regulated and will refer the reader to recent reviews.15–17
Figure 2.
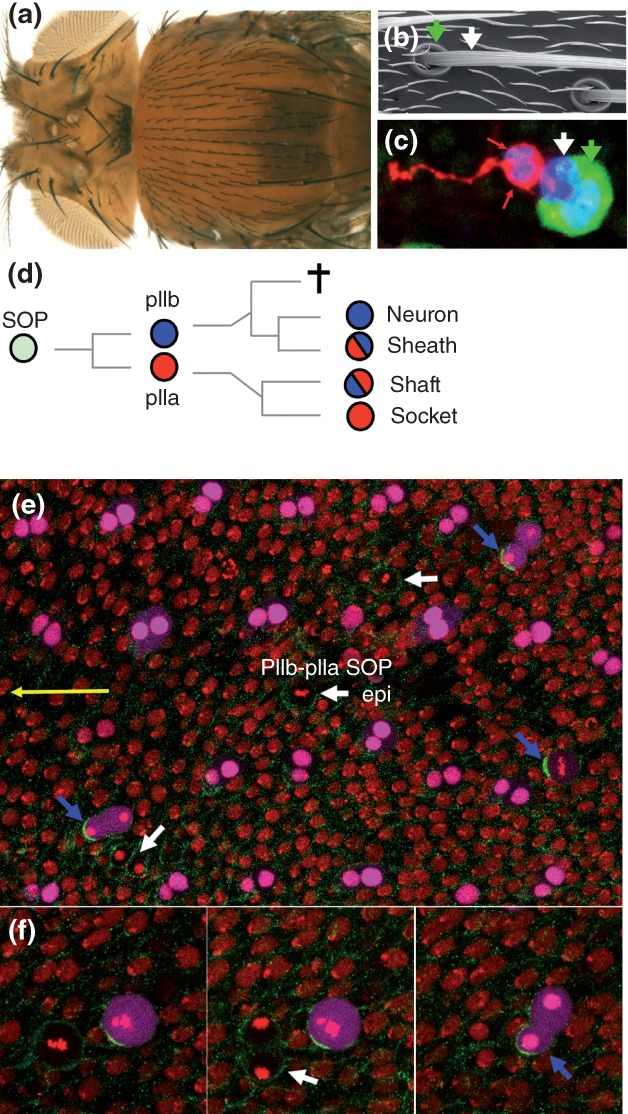
The bristle lineage. (a) Dorsal view of an adult fly, showing the dorsal thorax, or notum, covered with sensory bristles. (b) Electron micrograph showing a bristle shaft (white arrow) and its socket (green arrow) at the base of the shaft. Small hairs mark epidermal cells. (c) Each bristle sensory organ is composed of four cells (sensory organ nuclear marker, blue; socket cell marker, green; internal cell marker, red): the neuron and sheath cells are shown with red arrows, the shaft cell with a white arrow and the socket cell with a green arrow. (d) The sensory bristle lineage: the SOP divides asymmetrically to generate two distinct secondary precursor cells, pIIa (red, Notch ON) and pIIb (blue, Notch OFF). The pIIb cell generates the two internal cells, i.e. the neuron (Notch OFF again) and its associated sheath cell (Notch ON), together with a cell fated to die by apoptosis whereas the pIIa cell produces the two external cells, i.e. the shaft (Notch OFF) and socket (Notch ON again) cells. (e–f″) snapshot views of living pupa (yellow arrow points toward the head in (e) showing all notum cells at 17 h APF. A time series is shown in higher magnification views (f–f″). Histone2A-RFP (red) marks all epithelial cells. Sensory cells, i.e. SOPs and their pIIb-pIIa progeny cells, express a nuclear eqFP670 (magenta). SOPs divide along the a–p axis so that pIIb is anterior and pIIa posterior. NumbGFP (green) localizes asymmetrically in dividing SOPs and is inherited by the anterior cell (blue arrows) whereas it is symmetric in dividing epidermal cells (epi; white arrows). Note that sensory organs form a regular pattern of rows. In this and all other figures, anterior is to the left.
NOTCH REGULATES THE BINARY pIIa/pIIb DECISION
The pIIa/pIIb binary fate decision is regulated by Notch. Loss of Notch activity leads to a pIIa-to-pIIb transformation18,19 and, conversely, expression of activated Notch results in a pIIb-to-pIIa transformation. Thus, high Notch activity directs SOP daughters toward a pIIa fate whereas inhibition of Notch is required for adoption of the pIIb fate (Figure 3(a)). Activation of Notch in pIIa was recently monitored via the detection of endogenous intracellular Notch, i.e. activated Notch, in the nucleus of pIIa. Moreover, live imaging of a functional GFP-tagged Notch receptor showed that nuclear Notch is present in pIIa, but not pIIb, soon after division, i.e. during cytokinesis20 (Figure 3(b)–(c‴)). Activation of Notch in pIIa requires the redundant activities of Delta (Dl) and Serrate (Ser).21 While it is generally viewed that Dl (and Ser) signal from the surface of pIIb to activate Notch in pIIa, it cannot be excluded that Dl (and Ser) from epidermal cells also contribute to Notch activation in pIIa. Also, where Notch receptors are activated is not clear. Are Dl and Ser present and active at apical junctions and/or microvilli of pIIb22 to activate apical Notch in pIIa? Are Dl and Ser active along the basal-lateral membrane of pIIb? Detailed analysis of Notch, Dl and Ser localization may provide insights into this question. Finally, the key target genes of Notch for the pIIa/pIIb decision remain to be characterized. While some HLH genes of the Enhancer of split Complex [E(spl)-C] are specifically expressed in pIIa (unpublished observation), deletion of the entire complex did not prevent the specification of pIIa cells.23 Thus, additional Notch target genes likely act redundantly with E(spl)-HLH genes to specify the pIIa fate.
Figure 3.
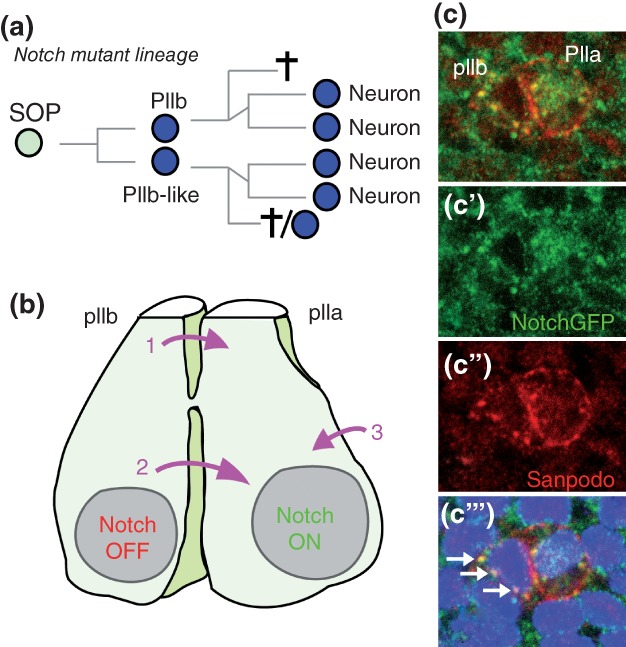
Fate asymmetry is regulated by Notch. (a) The lineage of Notch mutant SOPs: defective fate asymmetry results in the specification of two pIIb-like cells that generate neuron-like cells. (b) Notch is ‘ON’ in pIIa and ‘OFF’ in pIIb. Notch receptors in pIIa are activated by ligands present at the surface of the pIIb cell (pink arrows 1 and 2) and/or of neighboring epidermal cells (pink arrow 3). Whether activation of Notch receptors occur apically (1) and/or along the basal cytokinetic furrow (2) remains to be determined. (c) The signaling activity of Notch can be monitored using a functional NotchGFP receptor that allowed the detection of nuclear activated Notch (anti-GFP, green) in pIIa.20 Sanpodo (red) is a SOP-specific protein that localizes mostly at the plasma membrane of pIIa whereas it accumulates in endosomes in pIIb. Note that Notch and Sanpodo co-accumulate into apical endosomes in pIIb (white arrows in C‴’).28
NUMB INHIBITS THE RECYCLING OF NOTCH IN pIIb
How is directional Notch signaling established at cytokinesis? A key insight came from the genetic and molecular analysis of Numb. In numb mutants, the anterior SOP daughter cell adopts a pIIa fate, resulting in a pIIb-to-pIIa transformation9 (Figure 4(a)–(b’)). This transformation depends on the presence of Notch, indicating that Numb antagonizes Notch.24 Importantly, Numb localizes asymmetrically in dividing SOPs: it localizes at the basal-anterior cortex of SOPs and is specifically inherited by the pIIb cell (Figure 4(c)–(c″″)),9 indicating that Numb acts in pIIb to inhibit Notch. These findings identified Numb as a cell fate determinant.
Figure 4.

Regulation of cell fate by Numb. (a) The lineage of numb mutant SOPs: defective fate asymmetry results in the specification of two pIIa-like cells that generate socket-like cells. (b and b′) In the absence of numb (in a clone of mutant cells positively marked by the expression of a nuclear GFP in green), sensory cells adopt a socket-like fate: multiple sockets but no shaft are seen at the surface of the fly (arrow). (c–C″) Snapshots of a live imaging movie showing the unequal segregation of Numb in dividing SOPs. The Numb crescent (NumbGFP, green) forms at the anterior cortex of dividing SOPs (marked by Histone2B-RFP, red, used here as a SOP-specific mitotic marker).28 The mitotic spindle (followed here using Asterless-RFP, red, to mark the centrosomes) rotates to line up along the polarity axis (red arrow in c).73 At metaphase (c″), the two centrosomes are nicely aligned along the polarity axis such that NumbGFP is specifcally segregated into the anterior pIIb cell (left) at anaphase (c‴). At cytokinesis, the NumbGFP crescent disassembles (c″″) and Numb relocalizes at apical endosomes in pIIb.28
Numb is a conserved multifunctional protein regulating a wide range of processes, from intracellular trafficking to proteasome-mediated degradation.25 It is a membrane-associated phospho-protein that localizes at endosomes and along the baso-lateral membrane in epithelial cells.26–28 In mammals, flies and worms, Numb interacts with the adaptor protein complexes AP-1 and AP-2 as well as other endocytic factors to regulate the endocytosis, endosomal sorting and/or recycling of various cargoes.29–34 In the context of SOP ACD, Numb appears to regulate the endosomal sorting of Notch-Sanpodo (Spdo) complexes. Spdo is a SOP-specific regulator of Notch endocytosis that directly interacts with Numb,20,28,35–39 suggesting that Notch may interact with Numb via Spdo. Notch and Spdo co-localize with Numb at apical sorting endosomes in pIIb, but not pIIa28 (Figure 3(c‴)). Since Numb is dispensable for the internalization of Notch and Spdo but is required to inhibit the recycling of Spdo, Numb was proposed to inhibit the recycling of Notch-Spdo complexes back to the cell surface of pIIb (Figure 5)28,33 and to regulate the sorting of Notch and Spdo toward late endosomes.40 This ‘recycling inhibition’ model is consistent with earlier data showing that Numb can regulate the post-endosomal sorting of Notch1 in mammals.41 However, if Spdo was required for the inhibitory activity of Numb, the loss of spdo activity should result in a numb-like mutant phenotype. Actually, the loss of spdo activity leads to the opposite phenotype, i.e. a pIIa-to-pIIb transformation. This is because Spdo is also required for Notch activity in pIIa where, in the absence of Numb, Spdo facilitates the γ-secretase cleavage of Notch.42 Nevertheless, a mutational analysis of Spdo showed that the Numb-binding motif of Spdo was required together with an endocytic sorting signal for the inhibition of Notch in pIIb, indicating that Spdo acts together with Numb to inhibit Notch in pIIb.42 In summary, a current model proposes that Numb inhibits the recycling of Notch in pIIb, hence creating an asymmetry of Notch at the surface of the anterior (low Notch) and posterior (high Notch) daughter cells, leading to the binary pIIa/pIIb fate choice (Figure 5).
Figure 5.

Regulation of Notch trafficking by Numb in pIIb. Numb (red dots) is specifically segregated into the anterior pIIb cell (left) where it inhibits Notch. Numb relocalizes from the basal cortex of pIIb to apical endosomes where it colocalizes with Notch (blue bars) and sanpodo (green dots).28 Two models for Numb mode of action have recently been discussed. In a first ‘internalization model’, Numb acts in pIIb to positively regulate, i.e. speed-up, the internalization of Notch-Sanpodo oligomers (large green arrow). In a second ‘recycling inhibition model’, Notch-sanpodo oligomers are internalized in a Numb-independent manner and Numb acts to inhibit the recycling of Notch and Sanpodo back to the cell surface (red stop sign). Recent experimental evidence favors the ‘recycling inhibition model’ (see text).
Several questions, however, remain. For instance, how does Numb act at the molecular level to inhibit the recycling of Notch and Spdo and promote their late endosomal sorting? What is the role of the Numb-AP-2 interaction which is essential for Notch inhibition in pIIb?34 Is this Numb-AP-2 interaction important to relocalize Numb from the basal cortex to apical endosomes, as suggested by the live imaging of Numb in α-adaptin mutant cells?28 Also, to what extent does the asymmetry of Numb contribute to the directionality of Notch signaling? In other words, could symmetric (yet fully active) Numb support a binary pIIa/pIIb fate decision due to redundant mechanisms? Future studies will certainly shed new light on how Numb inhibits Notch at the molecular level.
NEURALIZED PROMOTES DELTA ENDOCYTOSIS AND SIGNALING IN pIIb
Indeed, additional regulatory mechanisms have been proposed to contribute to fate asymmetry. The E3 ubiquitin ligase Neuralized (Neur) localizes at the anterior pole of mitotic SOPs, is unequally inherited by the pIIb cell at cytokinesis and contributes to high Notch activity in pIIa.43 In pIIb, Neur positively regulates the ubiquitination and endocytosis of Dl, a direct molecular target of Neur.43–47 It is generally viewed that endocytosis of ubiquitinated Dl promotes the trans-activation of Notch.48,49 Indeed, the activation of Notch involves a conformation change of the Notch extracellular negative regulatory region (NRR). In its closed auto-inhibited conformation, the NRR prevents access to the metalloprotease cleavage site known as the S2 site.50,51 Since ligand-dependent S2 cleavage depends on the unmasking of this cleavage site, it is thought that this conformation change can be induced by mechanical forces associated with the endocytosis of Dl bound to Notch. Thus, Neur acts in pIIb to activate Notch in pIIa (Figure 6). Interestingly, the SOP-specific expression of Neur and its unequal segregation into pIIb suggest that pIIb is the main source of ligands activating Notch in pIIa (see Figure 3(b)). The contribution of Neur asymmetry to fate asymmetry may not, however, be essential since expression of the E3 ubiquitin ligase Mindbomb1 can largely suppress the neur mutant pIIa-to-pIIb transformation.52 Nevertheless, the unequal segregation of Neur was also observed in asymmetrically dividing neuroblasts in Drosophila,53 indicating that this regulatory mechanism may be more generally used to establish fate asymmetry. Finally, Neur also acts in a Notch-independent manner to stabilize the apical domain in epithelial cells.54 This novel function of Neur raises two questions. First, does Neur regulate apical–basal polarity in SOPs and, second, is the Neur-dependent signaling by Dl functionally coupled to a change in apical–basal polarity in SOPs. Addressing these issues may shed new light on how signaling and cell polarity are coupled in the context of ACD.
Figure 6.

Regulation of Delta endocytosis and signaling by Neuralized in pIIb. Notch (green, at the surface of pIIa) is activated by Delta (red, at the surface of pIIb) in a Neur-dependent manner. Activation of Notch requires ligand binding (1). The E3 ubiquitin ligase Neur acts in pIIb to add ubiquitin (black) to the intracellular tail of Dl (2). This modified form of Dl is then recognized and targeted for internalization by the endocytic machinery. The internalization of Dl bound to Notch (3) provides a pulling force that alters the conformation of the NRR region of Notch. This conformation change renders the S2 cleavage site accessible to the metalloprotease Kuzbanian (orange). This ligand-dependent cleavage generates a membrane-tethered form of Notch that is recognized and cleaved by the γ-secretase complex (4). This releases the activated form of Notch that acts in the nucleus to regulate gene expression in pIIa.
DO OTHER MECHANISMS CONTRIBUTE TO FATE ASYMMETRY?
Additional mechanisms may act in parallel to Numb and Neur to reinforce the directionality of Notch signaling. First, the subcellular distribution of recycling endosomes marked by the GTPase Rab11 differs between pIIa and pIIb, suggesting that the rapid recycling of Dl in pIIb may contribute to high Dl activity in pIIb.55 Consistent with this possibility, the Rab11 interactor Sec15 promotes the exocytosis of Dl and is essential for Notch activation in pIIa.56 However, whether Dl is the relevant target of Sec15 in pIIb, whether Rab11 regulates the activity of Dl in pIIb and whether the recycling of Dl is necessary for its signaling activity57 remain to be clarified. Thus, further investigation may be needed to test the relevance of this model.
Second, an endosomal pool of Dl and Notch has been proposed to traffic into pIIa at cytokinesis to contribute to Notch activity in pIIa.58 (Figure 4(b)). This model is in part based on a live imaging analysis of internalized Notch and Dl that were detected using an antibody uptake assay. These experiments suggested that internalized Notch and Dl localized into Sara-positive endosomes that traffic into pIIa at cytokinesis. While Notch localizing into these Sara-positive endosomes is predicted to traffic into pIIa, live imaging of a functional version of Notch showed that the endosomal pool of Notch is equally distributed at cytokinesis.40 Also, disrupting the asymmetry of Sara segregation at cytokinesis appeared to have no effect on the pIIa/pIIb decision and only correlated with low penetrant socket/shaft and neuron/sheath decision defects upon loss of a Notch regulator.59 Thus, further studies are needed to test the role of the directional trafficking of Sara-positive endosomes for Notch activity in pIIa.
Third, additional cell fate regulators, including Brat60 or Miranda,61 may also contribute to the pIIa/pIIb decision. Likewise, the unequal segregation of the cortical proteins Vang, Prickle, Gαi, Pins, and Discs-large into pIIb may regulate various cellular activities in pIIb.62–64 Conversely, cortical proteins localizing at the posterior pole of dividing SOPs and that may be enriched into pIIa, such as Frizzled, atypical protein kinase C (aPKC), Bazooka (Baz, the fly homolog of Par3) and Par6 (see below), may also affect various processes.62,63,65 Also, whether centrosome asymmetry and inheritance occur in SOPs and contribute to fate asymmetry remains to be studied.66 Thus, intracellular organization, cytoskeleton dynamics and/or vesicular trafficking are expected to differ between pIIa and pIIb possibly independently of the activation/inhibition status of Notch.67
NUMB ASYMMETRY IN SPACE AND TIME
The unequal segregation of Numb and Neur relies on their polar localization at mitosis. The mechanism whereby Numb localizes at the anterior cortex is relatively well understood and involves an inhibitory phosphorylation mechanism.65 Numb is a membrane-associated protein and phosphorylation of Numb inhibits its localization at the cell cortex.27 One of the kinases phosphorylating Numb is aPKC, a conserved kinase that plays an essential function in cell polarity. aPKC contains an auto-inhibitory domain with a pseudo-substrate motif blocking the active site. This auto-inhibition can be released via the interaction of aPKC with Par6.68 The activity of Par6-aPKC is increased at mitosis by the phosphorylation of Par6 by the mitotic kinase AuroraA (AurA). It is also restricted in space within the cell by several Par6-aPKC interactors, including Lethal(2) giant larvae (Lgl), Baz, and/or Cdc42.65,69–71 In SOPs, prior to mitosis, Par6-aPKC interacts with and is inhibited by Lgl.65 Upon mitosis, phosphorylation of Par6 by AurA promotes the dissociation of Lgl from Par6-aPKC and favors the formation of Baz-Par6-aPKC complexes65 that localizes at the posterior-apical cortex.63 Baz recruits Numb to this posterior complex, promotes its phosphorylation by aPKC and thereby excludes Numb from the posterior cortex.65 As a result, the localization of Numb is restricted to the anterior cortex (Figure 7).
Figure 7.
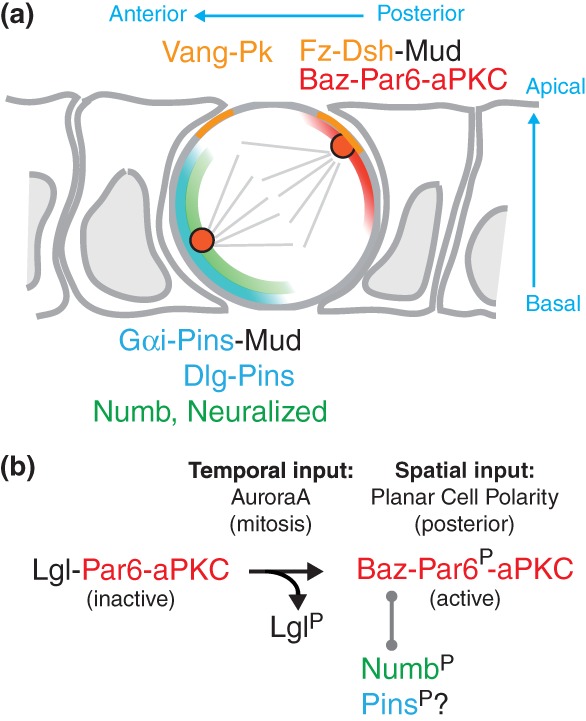
Numb anterior localization and orientation of the spindle by PCP and asymmetric aPKC activity. (a) Diagram of a dividing SOP at metaphase. The posterior PCP complex includes Fz and Dsh (light orange). The Baz-Par6-aPKC complex (red) also localizes apically at the posterior cortex. Numb (green) and Pins (blue) co-localize at the anterior basal cortex. Dsh, at the posterior-apical cortex, and Pins, recruits Mud. Mud interacts with dynein and pulls on astral microtubules to line up the mitotic spindle along the anterior–posterior axis, with a slight anterior basal tilt. (b) At interphase, Lgl inhibits the active Par6-aPKC complex. Phosphorylation of Par6 by AurA leads to the aPKC-dependent release of Lgl at mitosis and to the formation of the active Baz-Par6-aPKC complex. This complex localizes at the posterior cortex in a PCP-dependent manner. This complex interacts with Numb, a target of aPKC. Phosphorylated Numb is excluded from the posterior cortex. A similar mechanism may account for the exclusion of Pins from the posterior cortex at mitosis. Thus, PCP provides a spatial input and mitosis provides a temporal input for the asymmetric localization of the Baz-Par6-aPKC complex that regulates, together with PCP, both the asymmetric localization of Numb and the orientation of the mitotic spindle.
The cortical exclusion of Numb by a phosphorylation-dependent mechanism regulated by aPKC provides an elegant control mechanism for the polar distribution of Numb at mitosis. Indeed, since aPKC localizes at the posterior cortex of dividing SOPs, it regulates where Numb localizes at mitosis. Also, since the interaction of Baz with Par6-aPKC is regulated by AurA, it controls when Numb accumulates at the anterior cortex. Thus, the phosphorylation of Numb by aPKC integrates both temporal and spatial cues provided by the AurA-dependent formation of the Baz-Par6-aPKC complex at the posterior pole of SOPs.65 Whether a similar mechanism regulates the anterior accumulation of Neur remains to be tested. This mechanism may, however, be general since the localization of the cell fate determinant Miranda is regulated by this mechanism.61
Since Baz is also a substrate of aPKC,72 Baz might inhibit the phosphorylation of Numb by blocking access to the active site. However, since phosphorylated Baz still interacts with Par6-aPKC,72 it is conceivable that phosphorylated Baz interacts with the active Par6-aPKC complex at the posterior cortex of dividing SOPs, i.e. no longer blocks the active site of aPKC. Whether aPKC phosphorylates Baz in SOPs, whether phosphorylated Baz forms an active complex with Par6-aPKC at the posterior cortex and whether this regulation is important for the recruitment and phosphorylation of Numb remain to be studied.
COUPLING MITOTIC SPINDLE ORIENTATION WITH FATE DETERMINANT ASYMMETRY
The SOP divides within the plane of the epithelium and along the body axis. The a–p orientation of the mitotic spindle depends on PCP.10 Live imaging showed that the mitotic spindle rotates to line up along the a–p axis, thereby ensuring the proper segregation of Numb and Neur into pIIb.73 At prophase, the two centrosomes appear to be randomly positioned relative to the a–p axis. At prometaphase, the centrosome located closest to the anterior cortex appears to be pulled toward this cortical domain. The resulting spindle rotation aligns the mitotic spindle along the a–p polarity axis.73 The existence of a centrosome-attracting activity located at the anterior cortex was further inferred from the live imaging of SOPs with an anterior domain of increased size: when the two centrosomes appear to interact with this domain via astral microtubules, they are pulled toward this cortical domain at anaphase74 (Figure 7). At the molecular level, a Pins-Mud-Dynein complex localizes at the anterior cortex and provides a pulling force on astral microtubules.75,76 This complex is, at least in part, anchored at the anterior plasma membrane via the interaction of Pins with Gαi.64 Since Pins is a phosphorylation target of aPKC in mammals,77 aPKC-mediated phosphorylation of Pins might lead to its exclusion from the posterior cortex of SOPs. Consistent with this hypothesis, aPKC is required to exclude Pins from the apical cortex of dividing epidermal cells and to promote the planar orientation of these divisions in Drosophila.78 Pins is also a phosphorylation target of the mitotic kinase AurA79 and phospho-Pins might interact with the PDZ-containing protein Disc-large at the anterior cortex of SOPs.73,79 Thus, a phosphorylation-based mechanism could provide spatial and temporal cues for the asymmetric localization of Pins (Figure 7).
However, the complete loss of pins activity had no significant effect on the orientation of the mitotic spindle in SOPs.73 Thus, other mechanisms must act in a manner redundant with the anterior Pins-Mud complex to orient the mitotic spindle in SOPs. Consistent with this, analysis of PCP mutant clones suggested that anterior and posterior PCP complexes provide redundant inputs within the SOP to orient the mitotic spindle.74 Indeed, Fz and Dishevelled (Dsh) were shown to act at the posterior cortex to orient the mitotic spindle by directly recruiting Mud76,80 (Figure 7). Additionally, a Rho- and Formin-mediated actin polymerization mechanism appears to act downstream of Fz-Dsh to cooperate with the Mud-dynein complex in the capture and stabilization of astral microtubules at the posterior cortex of dividing SOPs.80 Thus, Pins at the anterior cortex and Dsh at the posterior cortex recruit the Mud-dynein complex, thereby accounting for the redundant inputs orienting the spindle along the a–p axis.76 These combined inputs also account for the slight anterior-basal/posterior-apical tilt of the spindle that results from the basal localization of Pins and apical localization of Dsh76 (Figure 7).
PLANAR POLARIZATION OF THE aPKC ACTIVITY AT THE APICAL CORTEX
The data reviewed above highlight the importance of the posterior localization of Par6-aPKC for asymmetric SOP division. A key and largely unanswered question remains: when and how does the Baz-Par6-aPKC complex localize at the posterior cortex? In particular, whether planar polarization of active aPKC is established at mitosis or prior to mitosis remains to be determined. Obviously, PCP must be involved in this process. Since in PCP mutants, however, Baz-Par6-aPKC was found to be largely asymmetric albeit randomly oriented relative to the body axis,62,63 it is thought that a PCP-independent mechanism breaks the planar symmetry of SOPs. The nature of this symmetry-breaking cue is unknown. Since Pins acts redundantly with Fz to localize Baz asymmetrically63 and since Baz acts redundantly with Dsh to localize Pins asymmetrically,62 a mutual antagonism might underlie this symmetry-breaking activity to create two opposite Pins- and Baz-containing complexes. The molecular basis of this antagonism and the mechanisms ensuring that this antagonism operates within the plane of the epithelium (rather than along the apical–basal axis) and at mitosis (and not before) are not known. Since Pins is a target of aPKC in mammals,77 one model is that phosphorylation of Pins by aPKC leads to its cortical exclusion. Whether this antagonism is sufficient to establish asymmetry at mitosis in the absence of PCP remains to be determined. Also, whether mitotic kinases, such as AurA that phosphorylates Par6 and thereby promotes the activity of aPKC upon mitosis, might regulate symmetry breaking by modulating this mutual antagonism remains to be investigated.
In summary, PCP provides a spatial bias to localize Pins and Baz-Par6-aPKC at the anterior and posterior SOP cortex, respectively, but PCP is not essential to establish asymmetry at mitosis. How PCP provides this bias is unclear but molecular interactions between core PCP proteins and components of the Baz-Par6-aPKC complex may provide entry points into this question.81,82
CONCLUSION
Studying a simple and stereotyped lineage in model organism has brought significant insights into how fate asymmetry is regulated in the context of a dividing progenitor cell. Obviously, the simplicity and stereotyped nature of this lineage greatly facilitated genetic and live imaging analysis of SOP ACD. Interestingly, SOPs maintain epithelial characteristics as they divide asymmetrically within the plane of the epithelium. Keeping junctions may be important for the SOP to interpret tissue polarity cues that operate at the level of junctions. Thus, keeping junctions may be important to orient SOP asymmetry relative to the a–p axis and to produce differentiated sensory cells in a patterned manner at the body surface. However, junctions have been proposed to inhibit asymmetric division in the embryonic epithelium.83 Specifically, AJs were proposed to antagonize the activity of Baz to regulate spindle orientation along the apical–basal axis of epithelial cells. Whether junction remodeling is required for the planar polarization of SOPs and, more generally, for ACD in epithelia, is an open question.
REFERENCES
- Cowan CR, Hyman AA. Acto-myosin reorganization and PAR polarity in C. elegans. Development. 2007;134:1035–1043. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- Motegi F, Seydoux G. The PAR network: redundancy and robustness in a symmetry-breaking system. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130010. doi: 10.1098/rstb.2013.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Inaba M, Yamashita YM. Asymmetric stem cell division: precision for robustness. Cell Stem Cell. 2012;11:461–469. doi: 10.1016/j.stem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocheteau P, Le Roux I. Asymmetric cell divisions and asymmetric cell fates. Annu Rev Cell Dev Biol. 2009;25:671–699. doi: 10.1146/annurev.cellbio.24.110707.175415. [DOI] [PubMed] [Google Scholar]
- Gho M, Bellaiche Y, Schweisguth F. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development. 1999;126:3573–3584. doi: 10.1242/dev.126.16.3573. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Gho M, Schweisguth F. Frizzled signalling controls orientation of asymmetric sense organ precursor cell divisions in Drosophila. Nature. 1998;393:178–181. doi: 10.1038/30265. [DOI] [PubMed] [Google Scholar]
- Fichelson P, Gho M. The glial cell undergoes apoptosis in the microchaete lineage of Drosophila. Development. 2003;130:123–133. doi: 10.1242/dev.00198. [DOI] [PubMed] [Google Scholar]
- Simpson P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development. 1990;109:509–519. doi: 10.1242/dev.109.3.509. [DOI] [PubMed] [Google Scholar]
- Orgogozo V, Schweisguth F, Bellaiche Y. Lineage, cell polarity and inscuteable function in the peripheral nervous system of the Drosophila embryo. Development. 2001;128:631–643. doi: 10.1242/dev.128.5.631. [DOI] [PubMed] [Google Scholar]
- Roegiers F, Younger-Shepherd S, Jan LY, Jan YN. Two types of asymmetric divisions in the Drosophila sensory organ precursor cell lineage. Nat Cell Biol. 2001;3:58–67. doi: 10.1038/35050568. [DOI] [PubMed] [Google Scholar]
- Abley K, De Reuille PB, Strutt D, Bangham A, Prusinkiewicz P, Maree AF, Grieneisen VA, Coen E. An intracellular partitioning-based framework for tissue cell polarity in plants and animals. Development. 2013;140:2061–2074. doi: 10.1242/dev.062984. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Strutt D. Principles of planar polarity in animal development. Development. 2011;138:1877–1892. doi: 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert JR, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–138. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol. 1990;142:13–30. doi: 10.1016/0012-1606(90)90147-b. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Couturier L, Vodovar N, Schweisguth F. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat Cell Biol. 2012;14:131–139. doi: 10.1038/ncb2419. [DOI] [PubMed] [Google Scholar]
- Zeng C, Younger-Shepherd S, Jan LY, Jan YN. Delta and Serrate are redundant Notch ligands required for asymmetric cell divisions within the Drosophila sensory organ lineage. Genes Dev. 1998;12:1086–1091. doi: 10.1101/gad.12.8.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Tien AC, Haueter CM, Schulze KL, Bellen HJ. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Exp Cell Res. 2010;316:900–906. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Smith CA, Dho SE, Donaldson J, Tepass U, McGlade CJ. The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol Biol Cell. 2004;15:3698–3708. doi: 10.1091/mbc.E04-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier L, Mazouni K, Schweisguth F. Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Curr Biol. 2013;23:588–593. doi: 10.1016/j.cub.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP. Numb is an endocytic protein. J Cell Biol. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Conradt B, Ruaud AF, Chen CC, Hatzold J, Bessereau JL, Grant BD, Tuck S. Caenorhabditis elegans num-1 negatively regulates endocytic recycling. Genetics. 2008;179:375–387. doi: 10.1534/genetics.108.087247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger JR, Taylor P, Gajadhar AS, Guha A, Moran MF, McGlade CJ. Identification and selected reaction monitoring (SRM) quantification of endocytosis factors associated with Numb. Mol Cell Proteomics. 2013;12:499–514. doi: 10.1074/mcp.M112.020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton M, Benhra N, Le Borgne R. Numb inhibits the recycling of Sanpodo in Drosophila sensory organ precursor. Curr Biol. 2013;23:581–587. doi: 10.1016/j.cub.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich J. The endocytic protein α-adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev Cell. 2002;3:221. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- Babaoglan AB, O’Connor-Giles KM, Mistry H, Schickedanz A, Wilson BA, Skeath JB. Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development. 2009;136:4089–4098. doi: 10.1242/dev.040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Zitserman D, Serebriiskii I, Andrake M, Dunbrack R, Roegiers F. Numb independently antagonizes Sanpodo membrane targeting and Notch signaling in Drosophila sensory organ precursor cells. Mol Biol Cell. 2010;21:802–810. doi: 10.1091/mbc.E09-09-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A, Knoblich JA. Numb and α-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 2005;6:836–842. doi: 10.1038/sj.embor.7400500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor-Giles KM, Skeath JB. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev Cell. 2003;5:231–243. doi: 10.1016/s1534-5807(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Kandachar V, Roegiers F. Endocytosis and control of Notch signaling. Curr Opin Cell Biol. 2012;24:534–540. doi: 10.1016/j.ceb.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier L, Trylinski M, Mazouni K, Darnet L, Schweisguth F. A fluorescent tagging approach reveals late endosomal trafficking of Notch and Sanpodo. J Cell Biol. 2014;3:351–363. doi: 10.1083/jcb.201407071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Upadhyay A, Kandachar V, Zitserman D, Tong X, Roegiers F. Sanpodo controls sensory organ precursor fate by directing Notch trafficking and binding γ-secretase. J Cell Biol. 2013;201:439–448. doi: 10.1083/jcb.201209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Unequal segregation of neuralized biases notch activation during asymmetric cell division. Dev Cell. 2003;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Lai EC, Kintner C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev Cell. 2001;1:795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. Neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev Cell. 2001;1:807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- Daskalaki A, Shalaby NA, Kux K, Tsoumpekos G, Tsibidis GD, Muskavitch MA, Delidakis C. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. J Cell Biol. 2011;195:1017–1031. doi: 10.1083/jcb.201105166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloty-Kapella L, Shergill B, Kuon J, Botvinick E, Weinmaster G. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev Cell. 2012;22:1299–1312. doi: 10.1016/j.devcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaster G, Fischer JA. Notch ligand ubiquitylation: what is it good for? Dev Cell. 2011;21:134–144. doi: 10.1016/j.devcel.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Tiyanont K, Wales TE, Aste-Amezaga M, Aster JC, Engen JR, Blacklow SC. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure. 2011;19:546–554. doi: 10.1016/j.str.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- Bhat KM, Gaziova I, Katipalla S. Neuralized mediates asymmetric division of neural precursors by two distinct and sequential events: promoting asymmetric localization of Numb and enhancing activation of Notch-signaling. Dev Biol. 2011;351:186–198. doi: 10.1016/j.ydbio.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet S, Schweisguth F. Regulation of epithelial polarity by the E3 ubiquitin ligase neuralized and the bearded inhibitors in Drosophila. Nat Cell Biol. 2012;14:467–476. doi: 10.1038/ncb2481. [DOI] [PubMed] [Google Scholar]
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric rab11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Banks SM, Cho B, Eun SH, Lee JH, Windler SL, Xie X, Bilder D, Fischer JA. The functions of auxilin and Rab11 in Drosophila suggest that the fundamental role of ligand endocytosis in notch signaling cells is not recycling. PLoS One. 2011;6:e18259. doi: 10.1371/journal.pone.0018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumailleau F, Furthauer M, Knoblich JA, Gonzalez-Gaitan M. Directional delta and notch trafficking in Sara endosomes during asymmetric cell division. Nature. 2009;458:1051–1055. doi: 10.1038/nature07854. [DOI] [PubMed] [Google Scholar]
- Loubery S, Seum C, Moraleda A, Daeden A, Furthauer M, Gonzalez-Gaitan M. Uninflatable and notch control the targeting of Sara endosomes during asymmetric division. Curr Biol. 2014;24:2142–2148. doi: 10.1016/j.cub.2014.07.054. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Atwood SX, Prehoda KE. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaiche Y, Beaudoin-Massiani O, Stuttem I, Schweisguth F. The planar cell polarity protein Strabismus promotes Pins anterior localization during asymmetric division of sensory organ precursor cells in Drosophila. Development. 2004;131:469–478. doi: 10.1242/dev.00928. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O’Kane CJ, Bryant PJ, Schweisguth F. The partner of inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell. 2001;106:355–366. doi: 10.1016/s0092-8674(01)00444-5. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Shevchenko A, Knoblich JA. A protein complex containing inscuteable and the Gα-binding protein pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, Yamashita YM. Centrosome asymmetry and inheritance during animal development. Curr Opin Cell Biol. 2012;24:541–546. doi: 10.1016/j.ceb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauffred B, Llense F, Sommer B, Wang Z, Martin C, Bellaiche Y. Regulation of centrosome movements by numb and the collapsin response mediator protein during Drosophila sensory progenitor asymmetric division. Development. 2013;140:2657–2668. doi: 10.1242/dev.087338. [DOI] [PubMed] [Google Scholar]
- Graybill C, Wee B, Atwood SX, Prehoda KE. Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J Biol Chem. 2012;287:21003–21011. doi: 10.1074/jbc.M112.360495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Tepass U. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu Rev Cell Dev Biol. 2012;28:655–685. doi: 10.1146/annurev-cellbio-092910-154033. [DOI] [PubMed] [Google Scholar]
- Henrique D, Schweisguth F. Cell polarity: the ups and downs of the Par6/aPKC complex. Curr Opin Genet Dev. 2003;13:341–350. doi: 10.1016/s0959-437x(03)00077-7. [DOI] [PubMed] [Google Scholar]
- Morais-De-Sa E, Mirouse V, Johnston DS. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509–523. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol. 2001;3:50–57. doi: 10.1038/35050558. [DOI] [PubMed] [Google Scholar]
- Gomes JE, Corado M, Schweisguth F. Van Gogh and Frizzled act redundantly in the Drosophila sensory organ precursor cell to orient its asymmetric division. PLoS One. 2009;4:e4485. doi: 10.1371/journal.pone.0004485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Segalen M, Johnston CA, Martin CA, Dumortier JG, Prehoda KE, David NB, Doe CQ, Bellaiche Y. The Fz-Dsh planar cell polarity pathway induces oriented cell division via Mud/NuMA in Drosophila and zebrafish. Dev Cell. 2010;19:740–752. doi: 10.1016/j.devcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilgur LG, Prudencio P, Ferreira T, Pimenta-Marques AR, Martinho RG. Drosophila aPKC is required for mitotic spindle orientation during symmetric division of epithelial cells. Development. 2012;139:503–513. doi: 10.1242/dev.071027. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–1163. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Manning L, Lu MS, Golub O, Doe CQ, Prehoda KE. Formin-mediated actin polymerization cooperates with Mushroom body defect (Mud)-Dynein during Frizzled-Dishevelled spindle orientation. J Cell Sci. 2013;126:4436–4444. doi: 10.1242/jcs.129544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane A, Yogev S, Mlodzik M. The apical determinants aPKC and dPatj regulate Frizzled-dependent planar cell polarity in the Drosophila eye. Cell. 2005;121:621–631. doi: 10.1016/j.cell.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Wasserscheid I, Thomas U, Knust E. Isoform-specific interaction of Flamingo/Starry night with excess Bazooka affects planar cell polarity in the Drosophila wing. Dev Dyn. 2007;236:1064–1071. doi: 10.1002/dvdy.21089. [DOI] [PubMed] [Google Scholar]
- Lu B, Roegiers F, Jan LY, Jan YN. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]


