Abstract
Alternative splicing (AS) is an important mechanism used to generate greater transcriptomic and proteomic diversity from a finite genome. Nearly all human gene transcripts are alternatively spliced and can produce protein isoforms with divergent and even antagonistic properties that impact cell functions. Many AS events are tightly regulated in a cell-type or tissue-specific manner, and at different developmental stages. AS is regulated by RNA-binding proteins, including cell- or tissue-specific splicing factors. In the past few years, technological advances have defined genome-wide programs of AS regulated by increasing numbers of splicing factors. These splicing regulatory networks (SRNs) consist of transcripts that encode proteins that function in coordinated and related processes that impact the development and phenotypes of different cell types. As such, it is increasingly recognized that disruption of normal programs of splicing regulated by different splicing factors can lead to human diseases. We will summarize examples of diseases in which altered expression or function of splicing regulatory proteins has been implicated in human disease pathophysiology. As the role of AS continues to be unveiled in human disease and disease risk, it is hoped that further investigations into the functions of numerous splicing factors and their regulated targets will enable the development of novel therapies that are directed at specific AS events as well as the biological pathways they impact. WIREs RNA 2015, 6:311–326. doi: 10.1002/wrna.1276
For further resources related to this article, please visit the http://wires.wiley.com/remdoi.cgi?doi=10.1002/wrna.1276WIREs website.
Conflict of interest: The authors have declared no conflicts of interest for this article.
INTRODUCTION
The majority of human protein-coding genes contain multiple exons and introns. The precise excision of introns and ligation of exons from pre-messenger RNA (pre-mRNAs) to produce mature messenger RNAs (mRNAs) is fundamentally a crucial step in gene expression. The splicing machinery is directed by consensus sequences at or near the exon/intron boundaries; the 5′ splice site, the 3′ splice site, and the branchpoint sequence (BPS) that is located upstream of the 3′ splice site (Figure 1(a)). The 5′ splice site is a nine nucleotide sequence element in which the GU dinucleotide at the beginning of the intron is essential for splicing whereas matches are not required at all of the other positions. Similarly, the 3′ splice site consists of an invariant AG dinucleotide at the 3′ end of the intron and a polypyrimidine-enriched upstream sequence element known as the polypyrimidine tract (PPT). In general, 5′ or 3′ splice sites that have more matches to these consensus elements are more likely to be spliced and sequences with greater or lesser matches are thus often referred to as strong or weak splice sites. These elements are recognized by components of the major spliceosome, a complex and dynamic macromolecular machine that consists of the small nuclear ribonucleoproteins (snRNPs; U1, U2, U4, U5, and U6) and at least 150 additional proteins.1 Complementary base pairing interactions between U1 and the 5′ splice site, base pairing of U2 with the BPS, and binding of the splicing protein U2AF with the PPT and 3′ splice site are involved in recognition of the splice sites and early steps of spliceosome assembly. Following splice site recognition, there is step-wise assembly of RNA–protein complexes that undergo structural reorganization prior to and during splicing catalysis.2 The intron is removed in two catalytic transesterification reactions in which an adenosine residue of the BPS carries out a nucleophilic attack at the 5′ end of the intron to form a branched intron intermediate and then the upstream exon is ligated to the downstream exon with removal of the branched intron (Figure 1(a)). In addition to introns that are spliced by the major spliceosome (referred to as U2-type introns), a minor spliceosome is responsible for the removal of approximately 800 human introns (U12-type introns) that have a different set of core consensus sequences. The minor spliceosome shares use of the U5 snRNP, and also uses distinct U11, U12, U4atac, and U6atac snRNPs and has both overlapping and unique protein components.3
Figure 1.
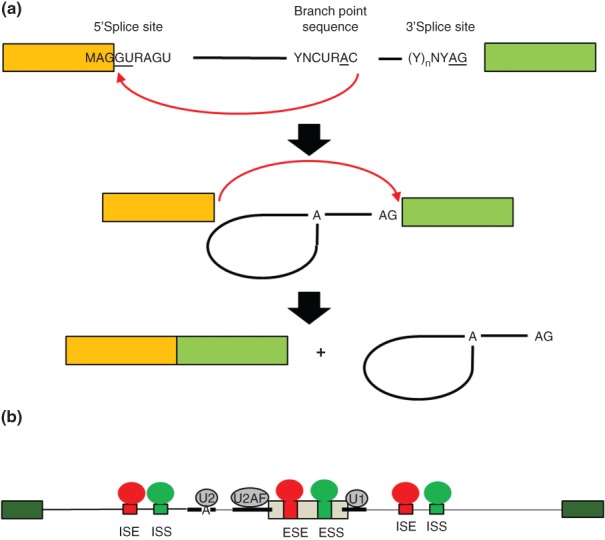
The biochemical steps of intron splicing and mechanisms of alternative splicing regulation. (a) Removal of an intron occurs in two catalytic steps that are directed by the core splice sites. In the first catalytic step of splicing, an adenosine residue in the branchpoint sequence (BPS) carries out nucleophilic attack at the 5′ end of the intron to form a branched 2′→5′ phosphodiester bond and lariat intermediate (middle). In the second catalytic step, the 3′ OH of the 5′ exon carries out nucleophilic attack at the 3′ splice site to ligate the exons in a 5′→3′ phosphodiester bond. The intron is released as a lariat product that is debranched and degraded. (b) Schematic of a model of combinatorial control of alternative splicing. An alternatively spliced cassette exon (gray) is flanked by two constitutively spliced exons (green). The positions of the core splice sites and BPS are indicated by thick lines. Intronic splicing enhancer (ISE) and exonic splicing enhancer (ESE) sequence elements are indicated in red together with splicing regulatory proteins (SRP) that bind them to promote exon splicing. Intronic splicing silencer (ISS) and exonic splicing silencer (ESS) are indicated in blue along with corresponding SRPs that promote exon skipping. The net combined activities of factors that promote splicing or skipping determine the level of exon splicing. These activities determine, at least in part, whether U1 binds to the 5′ splice site, U2AF to the PPT and 3′ splice site, and U2 to the BPS and subsequent steps of spliceosome assembly.
In addition to the core splicing elements, there are also diverse sequence elements located within exons as well as flanking introns that are involved in the recognition of the correct splice sites. These typically more degenerate splicing regulatory elements are referred to as exonic splicing enhancers or silencers (ESEs, ESSs) or intronic splicing enhancers or silencers (ISEs, ISSs). In addition to contributing to the splicing of constitutive exons, these auxiliary cis-elements also play central roles in the regulation of alternatively spliced exons. A general model has emerged wherein these auxiliary cis-elements are combinatorially bound by splicing regulatory factors and the splicing outcome is determined by their collective function to promote or inhibit splicing (Figure 1(b)).
AS is a critical mechanism that vastly expands proteomic diversity in humans. The importance of AS is highlighted by recent studies showing that nearly all human multi-exon genes produce multiple alternatively spliced mRNAs.4,5 The different types of AS are shown schematically in Figure 2. The most common type of AS consists of a single cassette exon that is either included or skipped in the spliced mRNA. Cassette exons can also be spliced or skipped in tandem or spliced in a mutually exclusive manner. Alternative 5′ or 3′ splice sites result in short and long forms of an exon. AS can also lead to alternative polyA sites through the use of alternative 3′ or 5′ splice sites. These types of events not only have the potential to alter the protein sequence, but by generating alternative 3′ untranslated regions (UTRs) they can also subject the mRNAs to differential regulation by microRNAs and RNA-binding proteins that control mRNA stability or translation. Alternatively, spliced transcripts produce protein isoforms with widely divergent functions including changes in subcellular localization, protein–protein interactions, and posttranslational modifications.6 Furthermore, many AS events are tightly regulated in a cell-type or tissue-specific manner, and at different developmental stages. AS is regulated by RNA-binding proteins, including cell- or tissue-specific splicing factors.7–9 An emerging concept in the field is that, similar to transcriptional regulators, tissue-specific splicing regulators coordinate programs of AS involving transcripts that encode proteins that function in biologically coherent pathways. In the past few years, technological advances such as high throughput sequencing and splicing sensitive microarrays have led to the elucidation of global programs of AS for an expanding number of splicing factors. In addition, methods that couple ultraviolet (UV) crosslinking with high throughput sequencing (e.g., HITS-CLIP, CLIP-Seq, PAR-CLIP, and iCLIP) allow for genome-wide determination of the binding sites for splicing factors and determination of direct targets of regulation.10–12 Armed with these technologies, many investigators are now refocused on understanding human diseases that result from alterations in genome-wide programs of AS. In this review, we describe several mechanisms by which disruptions in normal splicing can lead to disease, but with a primary emphasis on cases of diseases where mutations or alterations in the expression or function of AS regulators have been directly linked to or implicated in human disease. This review is not intended to be comprehensive, but focuses on several examples in which specific altered splicing regulators have been convincingly implicated in human diseases or pathophysiologic processes.
Figure 2.
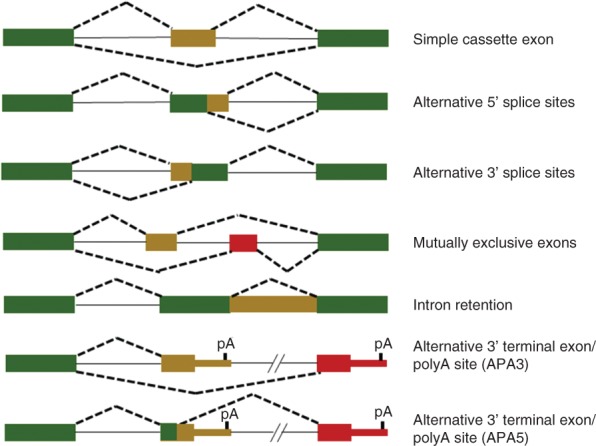
Schematic study of the different types of alternative splicing. Green boxes indicate constitutive exon sequences and red or brown boxes are alternatively spliced exons or regions. Solid lines indicate introns and dashed lines indicate alternative patterns. The hashmarks in the APA3 and APA5 events indicate that there can also be numerous additional cassette exons between the proximal and distal 3′ terminal exons.
SPLICING AND DISEASE
The most common means by which aberrant splicing is known to lead to disease is through mutations of the core splicing consensus sequences. Splice site mutations are among the most common mutations identified in studies of genetic diseases.13 For example, mutations that disrupt the invariant AG of the 3′ splice site or GT of the 5′ splice site will block use of the splice site and induce exon skipping, intron retention, or the use of otherwise cryptic splice sites. Mutations in other positions of the consensus that reduce splice site strength can similarly result in partial or complete inhibition of splice site use. Less commonly, mutations in the BPS, which is relatively degenerate in humans, can also impair splicing. In addition, mutations can result in the creation of new splice sites that lead to disease when they are used. Taken together, mutations that result in aberrant splicing have the potential to induce greater changes to the protein amino acid sequence that would be predicted to be more likely to induce disease than missense point mutations. However, altered splicing can also induce frame shifts leading to premature downstream stop codons. In most such cases, the resulting mRNA will be targeted for degradation through nonsense-mediated decay and thereby lead to effective loss of function. Because the number of genes and diseases associated with splice site mutations is legion, we do not describe specific examples here.
In addition to mutations in the splice site consensus sequence, there have also been numerous examples of genetic diseases in which aberrant splicing results from alterations in auxiliary cis-elements described above. The most commonly described examples, such as those in the neurofibromatosis (NF1) gene, occur in known or presumptive ESE elements and commonly lead to exon skipping. The types of exonic mutations that lead to exon skipping include missense and premature stop codons as well as synonymous nucleotide changes that would otherwise not be expected to disrupt gene function. As high throughput sequencing methodologies are increasingly being used to identify disease causing mutations or disease risk alleles, it remains a challenge to identify mutations that lead to aberrant RNA splicing using DNA analysis alone. Because of the degenerate nature of many splicing regulatory elements, it is often not possible to predict mutations that affect splicing and thus RNA-level analyzes will need to be more frequently incorporated into studies of genetic disease. Although there are now many examples of changes in splicing due to alterations in these regulatory elements, many studies of disease phenotypes that are attributed to mutations that change the protein sequence overlook the possibility that they may instead be primarily due to altered splicing. Mutations in splicing regulatory elements can also lead to disease by altering patterns of AS. For example, both exonic and intronic mutations in the MAPT gene encoding tau protein alter the ratio of mRNAs that contain alternatively spliced exon 10. These changes in AS predispose to aggregation of tau proteins in the inherited neurodegenerative disorder frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17). Several excellent previous reviews describe these and other examples of mutations in splicing regulatory elements that cause disease by inducing aberrant or altered regulated splicing in greater detail.13–15
Mutations in Core Spliceosome Components
As described above, all multi-exon genes require pre-mRNA splicing of the introns to assemble a mature mRNA which can be translated into protein. Thus, the splicing of all introns involves proper function of the snRNAs and associated snRNP proteins as well as other core spliceosomal proteins. Mutations in core components of the spliceosome that are associated with human diseases have recently been extensively reviewed13,16,17 and hence will not be described in detail here, but several illustrative examples will be briefly discussed. Retinitis pigmentosa, a photoreceptor neuronal degenerative syndrome, can be caused by mutations in PRPF3, PRPF8, PRPF31, or SNRNP200, which are all components of the core major spliceosome.18 Spinomuscular atrophy (SMA) is caused by inactivating mutations in the SMN1 gene. While SMN1 is not splicing factor, it is essential for small ribonuclear protein (snRNP) biogenesis.13 Mutations in the U4atac gene that inhibit proper formation of the minor spliceosome, cause Taybi-Linder syndrome/microcephalic osteodysplastic primordial dwarfism type 1 (TALS/MOPD1).19 Myelodysplastic syndrome (MDS), a hematopoetic stem cell disorder that increases the risk developing acute myeloid leukemia, has been associated with mutations in splicing factors including the core spliceosome components U2AF1 and the U2 snRNP component SF3B1.20 Mutations in these and other spliceosomal components are also observed in other myeloid diseases, including chronic lymphocytic leukemia (CLL). While the evidence for splicing defects in these diseases is sound, it bears mentioning that a largely unanswered question is how mutations in the core machinery, that are expected to impair splicing of most if not all introns, lead to very specific disease phenotypes that are often limited to specific organs or cell types.
Myotonic Dystrophy Types 1 and 2 (DM1 and DM2)
Among the best characterized examples of disease due to alterations in the function of AS factors are DM1 and DM2. While these diseases have been extensively covered in several recent reviews, they bear brief discussion because they constitute one of the best examples of genetic disease in which genome-wide changes in AS have been directly linked to alterations in the function of specific splicing factors.13,21,22 Myotonic dystrophy (DM) is a multisystemic disorder with muscular, cardiac, and neurologic defects that is caused at least in part by the deregulation of two AS factors, MBNL1 and CELF1 (also known as CUGBP1). These altered activities are not due to mutations in the MBNL1 or CELF1 genes themselves, but from a toxic RNA gain of function due to microsatellite expansions present in transcripts from other genomic loci. DM1 is caused by expansion of CTG repeats in the 3′ UTR of the dystrophia myotonica protein kinase (DMPK) gene, whereas DM2 is caused by expansion of CCTG repeats in the intron 1 of CCHC-type zinc finger, nucleic acid-binding protein (CNBP, also known as ZNF9). CUG repeat RNAs were shown to form nuclear aggregates and transgenic mice harboring CTG repeats inserted into the skeletal α-actin 3′ UTR (HSALR) form nuclear foci and hallmark symptoms of DM1 such as myotonia.23 These and other findings fueled the hypothesis that it was the RNA producing the disease by interfering with the function of one or more RNA-binding proteins. The identification of numerous changes in AS in DM1 ultimately led to the discovery that MBNL1 binds to the CUG repeats, which sequester it and inhibit its availability to regulate AS. Splicing factors of the MBNL family (MBNL1, MBNL2, and MBNL3) were shown to bind to CUG repeats and colocalize with nuclear aggregates. Further evidence supporting the role of MBNL proteins in DM1 was provided by Mbnl1 KO mice that exhibited myotonia and the demonstration that overexpression of Mbnl1 could reverse myotonia in the HSALR DM1 model.24,25 Furthermore, changes in splicing in muscle of Mbnl1 KO mice recapitulated changes in AS observed in patients with DM1. While Mbnl1 KO mice did not demonstrate neurologic deficits, a more recent study showed that the Mbnl2 KO mice display features of DM1 neurologic disease and show changes in splicing that are also observed in brains of human DM1 patients.26 These data suggest that sequestration and inhibition of both Mbnl1 and 2 functions contribute to DM1 pathogenesis albeit in a tissue-specific manner.
Another splicing factor involved in regulating AS in DM1 is CELF1.27 Interestingly, while a potential role for CELF1 in DM1 was based on its binding to CUG repeat elements, in contrast to sequestration and loss of function as determined for MBNL1, its expression and activity were paradoxically elevated. MBNL1 and CELF1 have antagonistic functions in the regulation of AS of overlapping target exons, where MBNL1 promotes adult splice forms and CELF1 promotes a fetal splicing program in muscle. The elevation of CELF1 activity in DM1 was shown to be due to aberrant PKC activation and CELF1 hyperphosphorylation as well as downregulation of miR-23a/b that normally repress CELF1 protein expression in adult cardiac tissue.28,29 Additional evidence for CELF1 in mediating splicing transitions in DM1 is that mice that overexpress it in skeletal muscle reproduce defects observed in DM1 tissue.30 The compound effect of MBNL1/2 sequestration and CELF1 activation is to promote an embryonic-like splicing pattern of a large set of AS targets in diseased tissues.31 The CCTG repeats that cause DM2 also sequester MBNL1 leading to similar yet less severe symptoms as in DM1; the role of CELF1 in DM2 is still emerging.13 While some of the pathophysiologic changes associated with DM1 can be attributed to specific splicing events (e.g., AS of the chloride channel CLCN1 causes myotonia and AS of the insulin receptor contributes to insulin resistance), the degree to which most components of genome-wide programs of AS disrupted in DM1 or DM2 contribute to disease remains to be determined.
A similar RNA gain of function phenomenon has also been invoked to link AS with disease phenotypes of the neurologic disease fragile X-associated tremor ataxia syndrome (FXTAS). Microsatellite expansions of >200 CGG repeats in the 5′ UTR of the FMR1 gene were initially associated with the more severe fragile X syndrome (FXS) that lead to methylation-dependent gene silencing. However, it was later shown that intermediate levels of the same expansions in FMR1 (5–200) lead to later onset FXTAS characterized by ataxia, tremor, Parkinsonism, and cerebral atrophy.32 In contrast to FXS, these repeats lead to elevated expression of FMR transcripts that form intranuclear inclusions.33 These observations suggested that FXTAS may arise from an RNA gain-of-function mechanism similar to that for DM. While a number of proteins were identified that are present in these intranuclear inclusion or that bind to CGG repeats, a potential role of alterations in AS was suggested based on a study that showed rapid association of the splicing factor Sam68 with inclusions formed in vitro, followed by Mbnl1 and HnRNPG.34 Splicing of two Sam68 target exons in ATP11B and SMN2 was shown to be altered in brain samples from FXTAS patients. These findings suggested that changes in AS due to Sam68 AS may underlie FXTAS pathophysiology. However, further evidence is needed to support this model, including a larger-scale analysis of AS in FXTAS brain and a more direct link to altered function of Sam68 or other potential AS factors that might be altered.
AMYOTROPHIC LATERAL SCLEROSIS (ALS)
ALS is an adult-onset neurodegenerative syndrome leading to paralysis and death within 1–5 years of diagnosis; while the majority of cases are sporadic, ∼10% of ALS is dominantly inherited.35,36 A seminal finding was that the RNA-binding protein TDP-43 (TARDBP) is a major component of cytoplasmic inclusions in motor neurons of ALS patients, as well as affected neurons in patients with frontotemporal lobar degeneration (FTLD).37 These inclusions are present in most ALS/FTLD patients and TDP-43 mutations are found in about 5% of all cases (familial and sporadic combined).38,39 TDP-43 is predominantly localized to the nucleus under normal conditions, where it is involved in RNA processing, including AS, and neurons with cytoplasmic aggregates showed depletion of nuclear TDP-43.37 This suggests that while TDP-43 may play a role in ALS pathogenesis owing to its involvement in ubiquitinylated cytoplasmic toxic protein aggregates, ALS disease manifestations may also be a consequence of a loss of its nuclear localization and function in RNA processing. Recent studies using CLIP-Seq, splicing sensitive microarrays, and RNA-Seq identified numerous TDP-43-regulated AS events, including direct targets, in the brain. Antisense oligo-mediated depletion of TDP-43 in mouse brain or in vitro knockdown of TDP-43 in a neuroblastoma cell line induced splicing switches of an array of transcripts, which included genes involved in neuronal function or neurological disease, such as Mef2D, Sortillin, tau, BIM, AP2, and parkin.16,40,41 Splicing switches involving several TDP-43 target exons were also observed in spinal cords from ALS patients relative to controls.42 Evidence linking these TDP-43-regulated AS programs to ALS arise from mouse models, where either transgenic expression of ALS-associated TDP-43 mutants (Q331K and M337V) or TDP-43 depletion in mice, which both induced changes in AS of TDP-43 target exons, led to neurodegeneration and ALS-like phenotypes in the absence of cytoplasmic inclusions.42,43 These data suggest that the loss of TDP-43 nuclear functions, including its role of AS, is a contributing factor in ALS.
Further evidence suggesting roles of AS in ALS came from the discovery of mutations in another RNA binding Protein (RBP) and splicing factor, FUS/TLS, that have been associated with ∼4% of familial ALS cases; similar to TDP-43, the role of FUS in ALS pathogenesis involves cytoplasmic inclusion bodies and a proposed loss of nuclear function.39,44,45 Transgenic mice expressing an ALS-associated FUS-mutant (R521C) displayed profound motor defects and altered splicing, including multiple retained introns.46 CLIP-seq for FUS- and ASO-mediated knockdown in mouse brain followed by splicing sensitive microarrays revealed mostly unique splicing changes relative to TDP-43 knockdown with some overlapping targets, such as Ndrg2, Tia1, and Kcnd3.47 As both TDP-43 and FUS mutations can lead to ALS, one could reason that the common target transcripts of the two factors should be at the forefront of future investigations aimed at further characterizing the molecular mechanisms involved in this disease.
A more recent discovery that also suggests a role of altered RNA processing, including splicing, has been the identification of GGGGCC expansions in the first intron of the C9ORF72 gene in ALS patients.48,49 These RNAs form nuclear foci and several RNA-binding proteins have been identified that colocalize with these foci and/or bind to GGGCC repeat RNA.22 These observations have suggested that a toxic RNA gain of function similar to that observed in DM1 and DM2 might lead to large-scale changes in RNA processing. Among RBPs associated with these foci is the well-described splicing factor hnRNP H suggesting that sequestration of hnRNP H or other splicing regulators may cause changes in splicing that account for ALS.50 However, further studies are needed to determine whether there are changes in splicing of targets of hnRNP H or other splicing factors in patient tissues with these expansions. Finally, it must be emphasized that while a loss of function of RNA-binding proteins, including those that function as AS regulators, is implicated in neurogenerative disorders associated with nuclear or cytoplasmic aggregates, the precise pathogenesis of these disorders remains unclear. It is certainly possible that a toxic gain of function of these aggregates plays a more direct role in these diseases and/or that both mechanisms together contribute to disease severity. In addition, repeat-associated non-ATG (RAN) translation of GGGGCC repeats has been proposed to yield peptides that form nuclear and cytoplasmic aggregates in the brains of patients with C9ORF72-associated ALS/FTLD.51,52 Further studies are clearly needed to resolve the roles of the different proposed disease mechanisms in these neurodegenerative diseases.
DILATED CARDIOMYOPATHY (DCM) AND RBM20
Dilated cardiomyopathy (DCM) is associated with substantial cardiac morbidity and mortality and has a strong genetic basis. In a remarkable convergence of genetic studies, mutations in the sarcomeric proteins Titin and the RBP RBM20 have been shown to cause familial DCM.53–56 While several mutations in the Titin gene itself have been linked to DCM, it was also known that Titin transcripts undergo complex AS that undergoes developmental transitions from fetal to adult heart.57 Furthermore, changes in the ratios of these isoforms have been observed in patients with congestive heart failure and DCM. 53 Intriguingly, a spontaneous rat mutant strain was identified in which there was a switch toward larger Titin splice isoforms (N2BA-G) that were more similar to fetal isoforms than the shorter N2BA and N2B isoforms observed in adult heart.58 Positional cloning identified a deletion of nearly the entire Rbm20 gene and it was shown that Rbm20 was a direct regulator of Titin AS.59 This study complemented work that identified mutations in RBM20 as a cause of human DCM and suggested that larger-scale disruptions in RBM20-regulated AS are likely to play a role in cardiac disease pathogenesis. These authors showed that a human DCM patient with an RBM20 mutation also showed a switch toward expression of a larger Titin isoform. The mechanism by which these larger Titin isoforms might contribute to cardiomyopathy was proposed to involve increased compliance or elasticity of the sarcomeres as the larger isoforms include addition exons in the ‘spring’ region of the protein that is one determinant of myocardial stiffness (Figure 3). Subsequent RNA-Seq analysis of cardiac tissue from WT and Rbm20−/− rats and from patients with DCM with or without RBM20 mutation identified 30 additional Rbm20-regulated AS events that were shared between rats and humans. Importantly, the genes comprising this splicing network were enriched for those encoding proteins with important cardiac functions as well as being enriched for terms linked to heart failure and cardiomyopathy. Among the genes that are a part of the RBM20 splicing network was Lim domain binding 3 (LDB3) that also has been shown to be mutated in patients with DCM. These observations illustrate and support the concept that components of splicing regulatory networks directed by specific splicing factors that are linked to physiologic processes and human diseases are themselves highly likely to be involved in those functions and disease pathophysiology. It will therefore be of interest to determine if mutations in other RBM20 targets also become linked to DCM or other cardiovascular diseases. The fact that RBM20 is predominantly expressed in striated muscle further suggests that it regulates coordinate programs of AS that fine tune gene expression programs that are important for the maintenance of cardiac and skeletal functions. Subsequent studies confirmed that many if not most of the RBM20-regulated AS events represent direct targets, and also that it functions primarily as a repressor of exon and intron splicing.60,61
Figure 3.
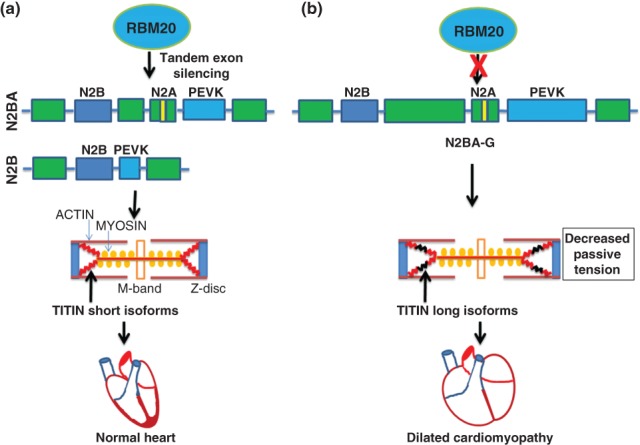
RBM20 regulates Titin alternative splicing. (a) RBM20 promotes the skipping of large cassettes of tandem exons in the Titin pre-mRNA via multiple binding sites. This results in the expression of shorter Titin isoforms, N2B and N2BA in normal heart, that are stiffer and thus require more force to stretch. (b) Loss of RBM20 function by deletion or from mutations in RBM20 that are associated with human dilated cardiomyopathy impede its ability to repress the tandem exons in the middle tandem Ig segment and the PEVK (rich in proline, glutamic acid, valine, and lysine residues) region. As a result larger Titin isoforms (N2BA-G) are produced that are more compliant and distensible. Green boxes indicate regions with tandem Ig-like repeats.
A recent study of RBM24, another RBP preferentially expressed in striated muscle, showed that mouse embryos in which Rbm24 was ablated died during embryogenesis with numerous cardiac malformations and defective sarcomere formation.62 It was shown that RBM24 also functions as a regulator of AS events in the heart. Numerous RBM24 splicing targets encode proteins that are important for cardiac development and sarcomerogenesis. Furthermore, several RBM24 targets, such as Abcc9 and Slc25a3 have also been shown to be mutated in human patients with cardiomyopathy. Hence, it will be of interest to determine if future studies identify mutations in RBM24 or other RBM24 target genes that may lead to cardiomyopathies or other cardiac or muscular diseases. Of note, there was only one shared alternatively spliced target between RBM20 and RBM24 suggesting that they coordinate separate but integrated programs of AS that are important for cardiac development and function.
AUTISM SPECTRUM DISORDER (ASD) AND RBFOX1
Several recent studies have drawn a link between genomic structural defects, SNPs, and alterations in the expression of RBFOX1 (also known as A2BP1) and autism spectrum disorder (ASD) and other neurological disorders. RBFOX1 is a paralog of RBFOX2 with more restricted expression in the nervous system, heart, and muscle. Together with neural-specific RBFOX3, all three paralogs have a single highly conserved RNA recognition motif (RRM) that mediates high affinity binding to (U)GCAUG sequence motifs and they have some overlapping AS targets in the nervous system.63,64 A set of RBFOX1/RBFOX2-predicted targets, including some that were validated, were shown to be enriched for genes with important neuromuscular functions suggesting that changes in splicing of some targets might impact neurological and muscular diseases.65 Mice with nervous system-specific knockout (KO) of Rbfox1 showed no gross anatomic or structural brain abnormalities, but were more prone to spontaneous seizures and mice with either homozygous or heterozygous KO had enhanced susceptibility to induced seizures that was attributed to altered synaptic function and increased excitability of neurons.63 The same study used splicing sensitive microarrays to define alterations in AS in Rbfox1 KO neurons and identified numerous targets that encode proteins with functions in synaptic transmission, including several that are directly linked to epilepsy. However, because of a compensatory upregulation of Rbfox2 in Rbfox1 null brains, many shared Rbfox1/Rbfox2 targets with roles in synaptic transmission and nervous system development may have eluded detection in this study. Another study used short-hairpin RNAs (shRNAs) to knockdown RBFOX1 in differentiated primary human neural progenitor cells and RNA-Seq to further define changes in splicing and total expression with RBFOX1 downregulation.66 Genes with changes in splicing were enriched for functions in neuronal development and synaptic functions. In addition, this study showed that genes downregulated at the total transcript level with RBFOX1 knockdown were also enriched for neuronal development and synapse function, although it was not clear whether these changes represented indirect changes in transcription or were due to altered mRNA stability. More direct links between RBFOX1 and human disease arose from studies that identified translocations that disrupted the RBFOX1 gene in several patients with epilepsy, mental retardation, and/or autism.67,68 Copy number variations (CNVs) in RBFOX1 have also been identified in ASD cohorts presumed to cause happloinsufficiency. However, the same CNVs were also observed in unaffected relatives suggesting that potential relations to disease include other genetic or environmental contributions.69,70 Structural disruptions in RBFOX1 have further been associated with epilepsy, developmental delay, and schizophrenia.69 Further evidence suggesting an association of RBFOX1 with ASD came from a study that used microarray to examine expression differences between post-mortem brain samples from a set of autistic and control brains.71 This study showed that RBFOX1 was a hub in a gene coexpression network that was underexpressed in autistic versus control brain cortex. Using a subset of autistic and control brains, RNA-Seq analysis showed large-scale differences in splicing between these brain samples, involving numerous examples of known RBFOX AS target genes, including many examples of genes with synaptic functions. A more recent study used HITS-CLIP to define genome-wide binding sites in mouse brain for RBFOX1, RBFOX2, and RBFOX3.64 Coupled with datasets of AS targets identified by KO or RNA interference, integrative modeling was used to define direct RBFOX target transcripts and regulatory networks. Substantial overlap was observed for conserved RBFOX target genes and genes with differences in AS in autistic versus control brains. Interestingly, it was observed that autistic brains also showed lower expression of RBFOX2 and RBFOX3, suggesting that more profound changes in splicing of overlapping RBFOX targets may result from decreases in the expression of all three paralogs. Many RBFOX targets were present in databases of candidate autism susceptibility genes, suggesting that other RBFOX targets may also include genes associated with autism and other neurologic disorders. While further evidence supporting direct links between RBFOX1 expression and AS with autism are needed, these studies suggest that global alterations in splicing in the brain that impact neural development and synaptic function may impact autism, epilepsy, as well as other diseases due to defective neurobiology.
Cancer
Alternative pre-mRNA splicing in cancer leads to the aberrant expression of transcripts that can contribute to tumor cell survival, proliferation, invasion, and metastasis. There is a wealth of literature describing specific examples of alterations in splicing that can promote cancer progression. For example, the AS of FAS exon 6 produces a dominant negative protein that inhibits FAS-mediated cell death and alternative 5′ splice sites of exon 2 in the BCL-X gene lead to two isoforms that oppositely regulate apoptosis.72,73 The multifaceted roles of aberrant or AS in cancer have been the subject of several comprehensive reviews.74–78 We will therefore limit our discussion in this section to a few well-characterized examples of splicing factors that regulate AS in cancer as well as those involved in the epithelial to mesenchymal transition (EMT). However, it bears mention that AS has been shown to impact all of the processes described as ‘hallmarks of cancer’ including cell proliferation, resistance to cell death, angiogenesis, immune system evasion, resistance to growth suppressors, and tissue invasion and metastasis.
SRSF1
In terms of splicing factors involved in cancer, SRSF1 is arguably the most extensively investigated. SRFS1 is amplified in different types of human tumors and its overexpression has been shown to induce transformation, indicating that it can function as a proto-oncogene.79,80 It regulates the AS of numerous transcripts that are relevant to cancer biology, such as BIN1, MNK2, S6K1, BIM, BCL2L11, and RON.76,77 SRSF1 induces inclusion of exon 12a in the BIN1 transcript, an inhibitor of the proto-oncogene cMyc, the resulting protein isoform lacks the ability to interact with and suppress cMYC transcriptional activity. cMYC can transcriptionally upregulate SRSF1 expression, which represents a potential feed forward mechanism of regulation. For S6K1, the isoform that was induced by SRSF1 overexpression was itself also shown to be oncogenic.78,79 SRSF1 also regulates splicing of RON, a receptor tyrosine kinase for the macrophage stimulating protein. Through binding to exon 12 SRSF1 promotes skipping of exon 11 of RON to produce a constitutively active kinase, known as delta-RON, which can promote in vitro cell motility and invasion.81 SRSF1 itself was also shown to enhance cell motility suggesting that it does so by inducing delta-RON expression.82 Taken together, these studies indicate that SRSF1 can promote several steps of cancer progression through coordinated changes in splicing. SRSF1 expression is also regulated itself at the posttranscriptional level by AS-and nonsense-mediated decay (NMD). The splicing of a 3′ UTR intron, which is retained in stable transcripts, turns the normally occurring stop codon into a premature termination codon (PTC) that induces NMD. ERK1/2-mediated phosphorylation of the splicing factor SAM68 induces inclusion of the intron and stabilization of SRSF1 mRNA.83
Lung Cancer [RBM10 and Quaking (QKI)]
A study using splicing sensitive microarrays identified increased inclusion of NUMB exon 9 as the most common change in AS in lung tumors compared with normal lung tissue.84 AS of NUMB exon 9 (also referred to as exon 12) produces two protein isoforms with divergent functions in regulating the Notch pathway. While isoforms that skip the exon lead to inhibition of Notch signaling and decreased cell proliferation, protein isoforms that include exon 9 are less stable and is associated with Notch activation and increased proliferation. Two recent studies have shed light on splicing factors that regulate splicing of NUMB exon 9 and are thereby implicated in lung cancer. Bechara et al. showed that RBM10 promotes skipping of NUMB exon 9 leading to reduced Notch activity and decreased cell proliferation.85 Together with observations of frequent RBM10 mutations in lung tumors, this study suggested that a switch in NUMB splicing due to decreased RBM10 function may contribute to cancer progression. Zong et al. similarly showed that QKI is downregulated in lung cancer and that it also regulates splicing of the same NUMB exon, providing another mechanism leading to increased cell proliferation via the same splicing switch86 (Figure 4). This group also showed that ectopic expression of QKI in the lung tumor cell line A549, markedly suppressed tumors in a xenograft assay.86 It will be interesting to test if the perturbation of the function or levels of endogenous RBM10 or QKI can lead to enhanced tumor growth in vivo. It is also worth noting that these studies used a combination of splicing sensitive microarrays, CLIP-Seq, and RNA-Seq to define global programs of AS regulated by each splicing regulator. It will thus also be of interest to identify other AS events associated with a loss of either protein that might also promote cancer development and progression.
Figure 4.
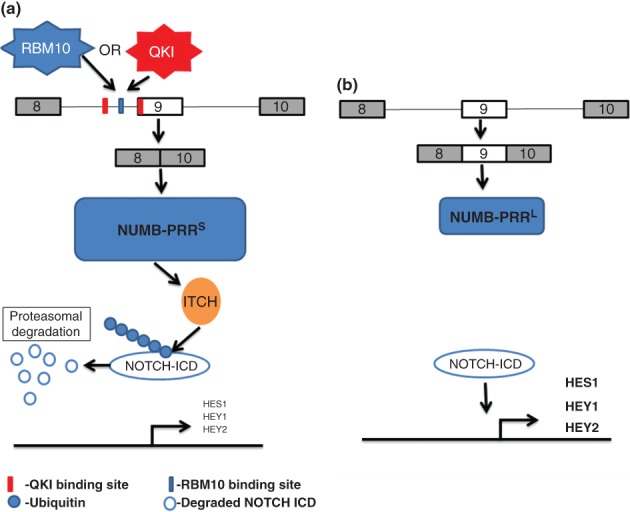
RBM10 and QKI regulate NUMB alternative splicing to suppress NOTCH signaling. (a) RBM10 and QKI bind to distinct cis-ISS regulatory elements in the intron upstream of and at the 5′ end of exon 9 leading to exon skipping. The resulting NUMB-PRRS protein isoform lacking the domain encoded by exon 9 is expressed at higher levels (due to an unknown mechanism) leading to suppression of NOTCH signaling through ITCH-mediated ubiquitination and degradation of the NOTCH intracellular domain (NICD). (b) In the absence of functional RBM10 or QKI, exon 9 is included leading to less expression of NUMB protein and enhanced transcriptional activation of NOTCH target genes (HES1, HEY1, and HEY2) and increased cell proliferation.
EPITHELIAL TO MESENCHYMAL TRANSITION (EMT)
Additional examples of AS factors and programs of AS that impact cancer pathophysiology have arisen from studies of the EMT. While both EMT and the reverse process of mesenchymal to epithelial transition (MET) are of fundamental importance in vertebrate development, these processes can also be hijacked by tumors to promote cancer progression, metastasis, and therapeutic resistance.87,88 A classic view has been that the acquisition of mesenchymal properties during EMT by tumors of epithelial origin allows some tumor cells to undergo tissue invasion and metastasis.87,89 On the other hand, recent data also implicates MET as a mechanism that enables colonization of cancer cells that escape the primary tumor at distant sites.90,91 Hence, it has been proposed that cells with general features of epithelial–mesenchymal plasticity (EMP) may be empowered to undergo reversible changes in cell phenotype to complete the steps of metastasis.92,93 More recently, EMT has also been proposed to contribute to the generation of CD44hi/CD24 low cancer stem cells (CSCs) or tumor initiating cells (TICs) that are also predictive of a poor response to conventional cancer therapies.94–96 Studies using human breast cancers, mouse breast cancer models, and breast cancer cell lines defined a gene signature that defines a ‘claudin-low’ cancer subtype that is associated with decreased survival and higher recurrence or resistance after chemotherapy in human patients.97,98 These studies further showed that this signature was associated with EMT markers as well as CD44hi/CD24 low cancer stem cell (CSC)/TIC characteristics. Taken together, studies showing the association of EMT with metastasis, formation of CSC/TIC cells, and drug resistance suggest an important need to better understand it at a fundamental molecular level.
The molecular and cellular mechanisms that account for epithelial–mesenchymal interconversions have been a focus of intense investigation. However, while transcriptional programs and signaling pathways associated with EMT have been well studied, until recently, the roles of AS in EMT were largely overlooked.87,88,99 Nonetheless, a splicing switch in FGFR2 during EMT was described nearly 20 years ago and hinted at a more global role of AS in EMT. AS of FGFR2 mutually exclusive exons known as exon IIIb and exon IIIc gives rise to receptor isoforms known as FGFR2-IIIb and FGFR2-IIIc that are expressed in epithelial and mesenchymal cells, respectively. These exons are located in a region encoding the extracellular domains that determine FGF ligand-binding specificities. As such, the exquisite cell type-specific expression of these receptor isoforms underlies paracrine epithelial–mesenchymal crosstalk that is crucial during development.100 Initial work from several groups identified a number of splicing factors involved in the regulation of FGFR2 splicing, yet most were expressed in both epithelial and mesenchymal cell types and thus the means by which cell type-specific expression was achieved was wanting. However, the discovery of the epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2), being necessary and sufficient for the expression of FGFR2-IIIb in epithelial cells, opened new avenues of investigation into the role AS plays in EMT.101 Subsequent studies using splicing sensitive microarrays and RNA-Seq defined genome-wide programs of AS regulated by the ESRPs.102–104 Importantly, the ESRPs are among the most highly downregulated transcripts in multiple models of EMT; as a consequence, ESRP target transcripts switch splicing from epithelial to mesenchymal splice variants.105 Studies that used RNA-Seq to directly define AS switches in one model system of EMT confirmed ESRP inactivation and showed substantial overlap in the EMT-associated splicing switches with ESRP target transcripts.106 Our lab and other groups subsequently showed overlap between ESRP-regulated AS events in EMT and targets of RBFOX2.102,106–109 For most shared targets, the ESRPs and RBFOX2 promoted the opposite pattern of splicing with RBFOX2 promoting mesenchymal splicing. However, in many cases, RBFOX2 promoted the expression of epithelial splice variants including key targets associated with EMT, such as FGFR2 and ENAH. Although some studies have identified modest increases in RBFOX2 expression in certain EMT systems, it should be noted that its expression is not highly mesenchymal specific. These observations suggest that the combinatorial regulation of AS in the EMT involves roles of cell type-specific factors as well more generally expressed factors. Additional roles of several other more ubiquitously expressed factors, such as SFRS1, MBNL1, PTB, Sam68, and hnRNPA2/B1 have been proposed (reviewed in Ref 110). AS of CD44 has also been shown to play a key role in EMT, where a switch toward the standard (CD44s) isoform was shown to be obligate for complete EMT in one model system.111 The ESRPs were shown to be required for the expression of epithelial CD44 splice variants in contrast to the mesenchymal CD44s variant that skips a set of tandem variable exons.101 A recent study showed that another ubiquitously expressed splicing factor hnRNP M promoted the expression of CD44s, but only when the ESRPs were downregulated.112 While there was limited overlap between hnRNP M-regulated splicing events and AS switches those previously shown to be involved in EMT, its regulation of this key event clearly indicates relevance to EMT. Interestingly, another epithelial cell type-specific RNA-binding protein, RBM47, was only recently shown to also be a splicing factor.113 It will thus be of interest to investigate the intersection of ESRP and RBM47 targets in promoting global programs of splicing in epithelial cells that are lost in EMT. A greater challenge will be to determine the functional consequences of EMT-associated splicing switches both at the level of individual genes as well as how they collectively contribute to changes in cell function and phenotype in EMT. The isoform-specific differences in function of several gene products that switch in EMT that impact these cell behaviors have been characterized, such as CD44, ENAH, p120-catenin, and Exo70 (EXOC7).105,110,114 Because the transcriptional and AS splicing programs in EMT have little overlap, it will be a high priority to identify the roles of both transcriptional and posttranscriptional events in EMT in order to identify key pathways that can be modulated by future therapies.
CONCLUSIONS
In just a few years technological explosions have led to massive new genome-scale information that has the potential to provide greater insights into genetic regulation that impacts embryonic development and human pathology. AS represents one important layer of gene expression whose impact on disease is now gaining increased recognition, yet, we still have only just begun to explore how global alterations of AS affect disease pathophysiology. Armed with high throughput sequencing technologies and improved splicing sensitive microarrays, it is now remarkably feasible to identify genome-wide splicing regulatory networks that differ between cell types as well as those that are directed by specific splicing factors. However, there remain many challenges in transforming this information into a more meaningful understanding of how AS affects pathways and processes that impact human disease. One major task is to define the functional consequences of AS and to dissect out the contributions of specific changes in splicing that impact disease processes. That is, it is important to investigate how different splice variants produce protein isoforms with differential activities and to further probe how the gain or loss of either isoform promotes disease. While detailed characterizations of isoform-specific functions have been carried out for a number of disease-relevant AS events, most have not. Furthermore, whereas several examples of specific splicing switches have been described that by themselves appear to promote processes leading to disease, it is more likely that the integrated changes in the functions of numerous targets of splicing factors contribute to human diseases that result from changes in function of AS regulators. It is also increasingly becoming apparent that most RNA-binding proteins are multifunctional and regulate posttranscriptional gene expression at several steps in the life cycle of an RNA transcript. Just as many RBPs traditionally considered to be, splicing regulators have now also been shown to regulate cytoplasmic functions, such as RNA localization, stability, and translation, the opposite is also frequently true. While the focus of this review has been on diseases associated with altered functions of AS regulators, it is almost certainly the case that some of the disease manifestations associated with this altered activity reflects changes at other posttranscriptional steps. It is also likely that the number of currently known human splicing factors is likely a fraction of the total proteins that function to regulate splicing. As we expand our collection of AS regulators and the programs of splicing that they control, it will be important to further investigate the combinatorial mechanisms by which they control AS in different cells and tissues and how deregulated splicing can lead to disease.
Acknowledgments
We thank Tom Bebee and Auinash Kalsotra for review of the manuscript. Work in the Carstens Lab is supported by NIH grants GM088809, HG006892, and AR066741.
REFERENCES
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, Reed R, Luhrmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;284:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. WIREs: RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. WIREs: RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, Konig J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA. New connections between splicing and human disease. Trends Genet. 2012;28:147–154. doi: 10.1016/j.tig.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P, Jr, Wu JY. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis. 2006;12:1259–1271. [PMC free article] [PubMed] [Google Scholar]
- Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos MG, Batra R, Charizanis K, Swanson MS. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol. 2011;3:a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A, Goodwin M, Swanson MS. RNA-protein interactions in unstable microsatellite diseases. Brain Res. 2014;1854:3–14. doi: 10.1016/j.brainres.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy [see comments] Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman R, Hagerman P. Advances in clinical and molecular understanding of the FMR1 premutation and fragile X-associated tremor/ataxia syndrome. Lancet Neurol. 2013;12:786–798. doi: 10.1016/S1474-4422(13)70125-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, Leehey M, Hagerman PJ. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Janssens J, Van Broeckhoven C. Pathological mechanisms underlying TDP-43 driven neurodegeneration in FTLD-ALS spectrum disorders. Hum Mol Genet. 2013;22:R77–R87. doi: 10.1093/hmg/ddt349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang H, Qiao T, Yang B, Aliaga L, Qiu L, Tan W, Salameh J, McKenna-Yasek DM, Smith T, et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2014;111:E1121–E1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci USA. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S, Barmada SJ, Finkbeiner S, Lui H, Carlton CE, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeWinter MM, Granzier HL. Titin is a major human disease gene. Circulation. 2013;127:938–944. doi: 10.1161/CIRCULATIONAHA.112.139717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Morales A, Gonzalez-Quintana J, Norton N, Siegfried JD, Hofmeyer M, Hershberger RE. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3:90–97. doi: 10.1111/j.1752-8062.2010.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells QS, Becker JR, Su YR, Mosley JD, Weeke P, D'Aoust L, Ausborn NL, Ramirez AH, Pfotenhauer JP, Naftilan AJ, et al. Whole exome sequencing identifies a causal RBM20 mutation in a large pedigree with familial dilated cardiomyopathy. Circ Cardiovasc Genet. 2013;6:317–326. doi: 10.1161/CIRCGENETICS.113.000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CM, Krzesinski PR, Campbell KS, Moss RL, Greaser ML. Titin isoform changes in rat myocardium during development. Mech Dev. 2004;121:1301–1312. doi: 10.1016/j.mod.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Greaser ML, Warren CM, Esbona K, Guo W, Duan Y, Parrish AM, Krzesinski PR, Norman HS, Dunning S, Fitzsimons DP, et al. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. J Mol Cell Cardiol. 2008;44:983–991. doi: 10.1016/j.yjmcc.2008.02.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Guo W, Dewey CN, Greaser ML. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013;41:2659–2672. doi: 10.1093/nar/gks1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatz H, Jens M, Liss M, Schafer S, Heinig M, Kirchner M, Adami E, Rintisch C, Dauksaite V, Radke MH, et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–3430. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hung LH, Licht T, Kostin S, Looso M, Khrameeva E, Bindereif A, Schneider A, Braun T. RBM24 is a major regulator of muscle-specific alternative splicing. Dev Cell. 2014;31:87–99. doi: 10.1016/j.devcel.2014.08.025. [DOI] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill BR, Lowe JK, Dybuncio CT, Fogel BL. Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. Int Rev Neurobiol. 2013;113:251–267. doi: 10.1016/B978-0-12-418700-9.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat F, Musthafa AA, Johnson D, Kramer AA, Shoffner D, Eliason M, Henry K, Spurlock B. Effect of a rapid response system for patients in shock on time to treatment and mortality during 5 years. Crit Care Med. 2007;35:2568–2575. doi: 10.1097/01.CCM.0000287593.54658.89. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- Liu S, Cheng C. Alternative RNA splicing and cancer. Wires: RNA. 2013;4:547–566. doi: 10.1002/wrna.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol. 2014;32C:30–36. doi: 10.1016/j.semcdb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collesi C, Santoro MM, Gaudino G, Comoglio PM. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol Cell Biol. 1996;16:5518–5526. doi: 10.1128/mcb.16.10.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol. 2010;191:87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misquitta-Ali CM, Cheng E, O'Hanlon D, Liu N, McGlade CJ, Tsao MS, Blencowe BJ. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol. 2011;31:138–150. doi: 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52:720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- van Denderen BJ, Thompson EW. Cancer: the to and fro of tumour spread. Nature. 2013;493:487–488. doi: 10.1038/493487a. [DOI] [PubMed] [Google Scholar]
- Ford HL, Thompson EW. Mammary gland studies as important contributors to the cause of epithelial mesenchymal plasticity in malignancy. J Mammary Gland Biol Neoplasia. 2010;15:113–115. doi: 10.1007/s10911-010-9182-0. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari N, Gheldof A, Tatari M, Christofori G. EMT as the ultimate survival mechanism of cancer cells. Semin Cancer Biol. 2012;22:194–207. doi: 10.1016/j.semcancer.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drasin DJ, Robin TP, Ford HL. Breast cancer epithelial-to-mesenchymal transition: examining the functional consequences of plasticity. Breast Cancer Res. 2011;13:226. doi: 10.1186/bcr3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT) Semin Cancer Biol. 2012;22:417–427. doi: 10.1016/j.semcancer.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Brosseau JP, Gadea G, Klinck R, Prinos P, Beaulieu JF, Lapointe E, Durand M, Thibault P, Tremblay K, et al. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol. 2013;33:396–405. doi: 10.1128/MCB.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, Hu Z, Simpson K, Pachter L, Durinck S, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res. 2010;8:961–974. doi: 10.1158/1541-7786.MCR-09-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2014;33:1082–1092. doi: 10.1038/onc.2013.50. [DOI] [PubMed] [Google Scholar]
- Biamonti G, Bonomi S, Gallo S, Ghigna C. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT) Cell Mol Life Sci. 2012;69:2515–2526. doi: 10.1007/s00018-012-0931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, et al. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191–1203. doi: 10.1101/gad.241968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, Darnell RB, Massague J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. elife. 2014;3:e02734. doi: 10.7554/eLife.02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Liu J, Liu S, Zeng J, Ding D, Carstens RP, Cong Y, Xu X, Guo W. Exo70 isoform switching upon epithelial-mesenchymal transition mediates cancer cell invasion. Dev Cell. 2013;27:560–573. doi: 10.1016/j.devcel.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


