Abstract
Introduction
We examine interactive and intensification effects of type 2 diabetes (T2D) with APOE and an Alzheimer's disease genetic risk score (GRS) on neurocognitive speed performance and change in nondemented older adults.
Methods
In an accelerated longitudinal design, we used latent growth modeling to test moderators of level and change in a neurocognitive speed latent variable for 628 adults (baseline median age = 69.0) followed over 9 years. The GRS was compiled using the cumulative risk of APOE, CLU, CR1, and PICALM.
Results
First, T2D predicted slower speed performance at centering age (75). Second, no predictive effects were associated with APOE or GRS. Third, a significant interaction showed that high risk from both T2D and GRS was selectively associated with steeper longitudinal slowing than all comparison cross-domain risk groups.
Discussion
Higher AD-related genetic risk intensified deleterious effects of diabetes on neurocognitive slowing in nondemented aging beyond the independent influence of APOE.
Keywords: Alzheimer's disease genetic risk score, APOE, Type 2 diabetes, Neurocognitive speed, Victoria longitudinal study
1. Introduction
Risk factors associated with Alzheimer's disease (AD) can be identified several years before the onset of the disease (e.g., obesity [1]). Prominent clusters of risk factors for AD also influence patterns of nondemented cognitive aging. These include biological (genetic polymorphisms), medical (type 2 diabetes [T2D]), lifestyle, and environmental factors. T2D is a potentially modifiable risk factor that has been linked to increased risk of AD [2], [3] and to changes in the non-AD brain (e.g., exacerbated insulin dysregulation, disrupted Aβ clearance). These changes are associated with decrements in neurocognitive performance in cross-sectional and longitudinal studies [4], [5], [6], [7], [8], [9]. The effects of T2D on nondemented cognitive aging may be modified or intensified by other risk factors including genetic risk [10], [11], [12].
Although APOE is the gene most consistently linked to AD risk, genome-wide association studies have identified several additional genotypes associated with AD [13], [14], [15]. These include clusterin (CLU), complement component (3b/4b) receptor 1 (CR1), and phosphatidylinositol-binding clathrin assembly protein (PICALM). APOE has been associated with AD, mild cognitive impairment [16], [17], and nondemented cognitive decline [18], [19], [20]. Specifically, ɛ4 carriers are at higher, and ɛ2 carriers at lower, risk for cognitive deficits, including neurocognitive speed [18], [20], [21]. CLU, CR1, and PICALM all contribute to AD risk, but the effects on cognitive outcomes show mixed results. The CLU risk allele (i.e., C) has been linked to faster rates of memory decline in individuals who eventually convert to mild cognitive impairment or AD [22]. The CR1 risk allele (i.e., A) has been associated with faster decline in a five-domain global cognition measure [23]. For the PICALM risk allele (i.e., T), no significant associations with memory [24] or executive function [25] performance have been reported. This prompted us to test the minor allele (C) as a risk factor for cognitive decrements in nondemented aging. Independently, these genes present relatively low penetrance and consequently low effect sizes, but together they may account for substantial AD risk [13], [14], [26], [27], especially within the context of other risk factors [28], [29]. A joint or multilocus approach in the form of a genetic risk score (GRS) may be informative in representing combined AD genetic risk [26], [28], [30]. Although identification of multiple genetic risk factors via genome-wide association studies is very important, the indepth examination of selected individual risk variants, both separately and in combination, provides novel understanding of the pathways leading to cognitive decline and AD [28], [31]. Specifically for the present study, we examine APOE (rs429358, rs7412), CLU (rs11136000), CR1 (rs6656401), and PICALM (rs541458).
Neurocognitive speed is considered a basic cognitive ability influencing decline on multiple complex cognitive processes with aging [32], [33]. Speed may be an early indicator of normal or preclinical cognitive decline, possibly shaping the change profiles of more complex processes such as episodic memory or executive function. Speed has been reported to predict individual differences in global cognition and episodic memory [32], [33], mild cognitive impairment [34], [35], and risk of AD [36]. Important for the present study, T2D has been associated with typical speed deficits in older adults [7]. The present study uses a combinatorial candidate gene approach to identify an AD-related GRS. T2D status, APOE, and a GRS (i.e., APOE, CLU, CR1, PICALM) were analyzed independently and interactively (i.e., GRS × T2D status) using speed, modeled as a latent variable, as the outcome.
After first determining the best latent growth model representing the functional form of speed performance and change, we examined two research goals. Research goal 1 was to determine if T2D status, APOE, or GRS independently predicted latent speed level or 9-year longitudinal change. We hypothesized that both T2D status and GRS, but not APOE, would independently predict speed level and change. Research goal 2 was to determine if T2D status and APOE or GRS interactively predicted level of speed performance at age 75 (intercept) and 9-year slowing (slope). We hypothesized an intensification effect in that higher-risk GRS would magnify the negative associations of T2D with speed level and change above that of other risk combinations (including APOE independently). For validation, we checked the effects of T2D-associated factors as covariates and an alternate GRS (i.e., without APOE).
2. Methods
2.1. Participants
Participants were community-dwelling volunteer adults (initially aged 53–91 years) from the Victoria longitudinal study (VLS). The VLS is a longitudinal sequential study examining neurocognitive aging and impairment in relation to biomedical, genetic, health, lifestyle, and other aspects [37]. The VLS and all present data collection procedures are in full and certified compliance with prevailing human research ethics guidelines and boards. Informed written consent was provided by all participants. Using standard procedures (e.g., [38], [39]), we assembled longitudinal data consisting of three VLS samples, each with three available waves collected in the 2002–2012 period. The longitudinal period was 8.9 years, and the band of aging represented was about 40 years (53–95).
The eligible source sample consisted of 683 participants with genetic data (collected in 2009–2011). Several exclusionary criteria were then applied to this source sample as follows: (1) a diagnosis of AD or any other dementia, (2) a mini-mental status examination [40] score of less than 24, (3) a self-report of “severe” for potential comorbid conditions (e.g., epilepsy, head injury, depression), (4) a self-report of “severe” or “moderate” for potential comorbid diseases such as neurologic conditions (e.g., stroke, Parkinson's disease), and (5) insufficient cognitive data. From the remaining participants, we applied the standard and strict VLS multilevel diagnostic regimen for classifying T2D [7], [9]. Specifically, inclusion into the T2D group required the following conditions during any of the three data collection waves: (1) self-report of T2D diagnosis, (2) specified method of treatment (i.e., oral medication, insulin, diet and exercise, no control), (3) objective evidence of reported T2D medication, and (4) validation of T2D status (repeating the three previous steps) from the subsequent wave.
The final baseline sample for this study consisted of 628 nondemented adults; 422 were women and 206 were men (mean [M] age = 69.0 years, standard deviation [SD] = 7.57, range 53.2–91.0). See Table 1 for all background characteristics. The standard T2D diagnostic procedure resulted in 54 adults (8.6%) with T2D (at W1 M age = 70.0, SD = 7.57, range = 55.4–88.2 years; 29 women [53.7%]). Therefore, the W1 non-T2D group included 574 adults (M age = 68.9, SD = 7.57, range = 53.2–91.0 years; 393 women [68.5%]).
Table 1.
Participant characteristics categorized by time point
| Characteristics | W1 | W2 | W3 |
|---|---|---|---|
| n | 628 | 551 | 469 |
| Age (y) | 69.0 (7.57) | 73.5 (7.54) | 77.4 (7.19) |
| Range | 53–91 | 57–95 | 62–95 |
| Gender (% women) | 67.2 | 67.2 | 66.3 |
| Years between waves | — | 4.40 (0.50) | 4.50 (0.58) |
| Retention rate | 89% (W1–W2) | 85% (W2–W3) | |
| 75% (W1–W3) | |||
| Education (y) | 15.2 (3.01) | 15.3 (3.00) | 15.2 (3.14) |
| Health to perfect∗ | 1.76 (0.72) | 1.85 (0.74) | 1.89 (0.79) |
| Health to peers† | 1.58 (0.69) | 1.61 (0.67) | 1.68 (0.73) |
| Pulse pressure (mm Hg) | 51.7 (10.0) | 54.5 (12.4) | 56.9 (12.5) |
| Range | 31.1–95.2 | 26.2–120.9 | 29.0–102.6 |
| BMI (kg/m2) | 26.9 (4.49) | 26.7 (4.28) | 26.6 (4.34) |
| Range | 15.0-55.9 | 16.0-48.6 | 10.0-40.2 |
| Smoking status (%) | |||
| Present | 5.3 | 3.5 | 2.0 |
| Previous | 53.2 | 55.4 | 55.5 |
| Never | 41.4 | 41.2 | 42.5 |
| Alcohol use (%) | |||
| Presently | 87.1 | 90.5 | 88.1 |
| Previous | 5.7 | 5.1 | 9.5 |
| Never | 7.3 | 4.4 | 2.4 |
| T2D status (% T2D)‡ | 53 (8.5) | 45 (8.2) | 40 (8.5) |
| Characteristics | T2D | Control | T2D | Control | T2D | Control |
|---|---|---|---|---|---|---|
| Age | 70.0 (7.57) | 68.9 (7.57) | 74.2 (7.26) | 73.4 (7.57) | 78.2 (64.2) | 77.4 (7.23) |
| Range | 55–88 | 53–91 | 60–91 | 57–95 | 64–91 | 62–95 |
| Gender (% women) | 54.7 | 68.2 | 57.8 | 68.0 | 52.5 | 67.6 |
Abbreviations: W1, wave 1; W2, wave 2; W3, wave 3; BMI, body mass index; T2D, type 2 diabetes.
NOTE. Results presented as mean (standard deviation) unless otherwise stated. Smoking and drinking status are reported in percentages of participants who responded to the question (W1 n = 618; W2 n = 549; W3 n = 463).
Self-reported health relative to perfect.
Self-reported health relative to peers. Self-report measures are based on 1 “very good” to 5 “very poor.”
Reported at W1, W2, or W3.
2.2. Neurocognitive speed measures
Three multitrial computer-based reaction time measures were used to assess neurocognitive speed: (1) choice reaction time, (2) lexical decision, and (3) sentence verification (see Supplementary Material for description of tasks). All tasks were presented on a computer that controlled the presentation rate of the stimulus [35]. Correction procedures validated by the VLS were used to trim extreme outliers from raw latency scores for each of the reaction time measures (Supplementary Material).
2.3. DNA extraction, genotyping, and GRS development
Saliva was collected according to standard procedures from Oragene-DNA Genotek and stored at room temperature in the Oragene disks until DNA extraction. DNA was manually extracted from the saliva sample mix using the manufacturer's protocol, and genotyping was carried out using a PCR-RFLP strategy as described in the Supplementary Material. For all analyses including APOE, the genotype ε2/ε4 (n = 32) was removed. Genetic analyses included genotype categorization based on three degrees of risk. For distribution of risk by gene, see Table 2. The AD GRS was a simple count of allelic risk for APOE, CLU, CR1, and PICALM, according to a standard formula (i.e., no risk = 0 [no risk alleles], moderate risk = 1 [one risk allele], and most risk = 2 [two risk alleles; [41]]). The GRS was then grouped into low and high risk using a median split (median = 3.0; Table 2).
Table 2.
Allelic risk across genotypes and Alzheimer's disease-related genetic risk score
| Gene | No risk | Moderate risk | Full risk |
|---|---|---|---|
| APOE | ε2/ε2, ε2/ε3, ε3/ε3 (ɛ4−) | ε4/ε3 (ɛ4+) | ε4/ε4 (ɛ4+) |
| n | 449 | 134 | 13 |
| CLU | TT (C−) | TC (C+) | CC (C+) |
| n | 99 | 314 | 215 |
| CR1 | GG (A−) | GA (A+) | AA (A+) |
| n | 213 | 335 | 80 |
| PICALM | TT (C−) | TC (C+) | CC (C+) |
| n | 244 | 231 | 153 |
| Gene | Low risk | High risk |
|---|---|---|
| APOE, CLU, CR1, PICALM∗ | (0–3) | (4–8) |
| n | 385 | 211 |
| CLU, CR1, PICALM† | (0–3) | (4–6) |
| n | 452 | 176 |
Abbreviations: T2D, type 2 diabetes; GRS, genetic risk score.
Bivariate correlation between T2D and GRS = −0.011.
Bivariate correlation between T2D and GRS = 0.001.
2.4. Statistical analyses
2.4.1. Approach
Analyses pertaining to our research goals included confirmatory factor analysis and latent growth modeling. Statistical model fit for all analyses was determined using standard indexes (Supplementary Material). Using Mplus 7 [42], we identified a one-factor neurocognitive speed latent variable reflecting contributions from the three manifest indicators. We conducted invariance tests across three waves (for model goodness-of-fit indexes, see Supplementary Table 1). Using the best fitting speed model, we calculated factor scores and used these in all subsequent analyses. We analyzed the basic latent variable speed data to confirm expected sensitivity to differences in level and change. Age was a continuous variable centered at 75 years (the frequently used center point of the 40-year band of data [43]). Chronological age was used as the metric of change for these analyses. Higher scores indicated poorer performance. The results of the model showed that individuals (1) varied in speed performance at age 75 (b = 1.08, P < .001), (2) exhibited significant 9-year slowing (increased slope M = 0.021, P < .001), (3) showed variable patterns of decline (b = 0.006, P < .001), and (4) with better (lower) speed performance level at age 75 exhibited less 9-year slowing (r = 0.043, P < .001).
2.4.2. Analyses for research goal 1
2.4.2.1. Determine the independent effects of T2D status, APOE, and GRS
To determine the independent effect of each of our risk factors, T2D status, APOE, and GRS, we used the best unconditional growth model identified for speed and added each risk factor as an independent predictor of speed. The intercept (level of speed performance at age 75) and slope (9-year slowing) were regressed separately on these factors.
2.4.3. Analyses for research goal 2
2.4.3.1. Determine the interactive effects of T2D status, APOE, or GR
We computed a conditional growth model for speed with T2D status as a predictor using the APOE or GRS groupings (low-risk, high-risk). See Table 2 for distribution of the low- and high-risk groups.
2.4.4. Follow-up covariate analyses
For further clarification, we conducted two follow-up analyses. First, we tested covariates with possible associations to T2D (i.e., pulse pressure, body mass index, smoking, drinking, two measures of self-reported health, education, sex) in the model testing GRS × T2D interactions. Second, to further elucidate the role of APOE in predicting speed performance and change, we tested a model for which APOE was omitted from the GRS (i.e., CLU, CR1, PICALM).
3. Results
3.1. Research goal 1: Determine the independent effects of T2D status, APOE, and GRS
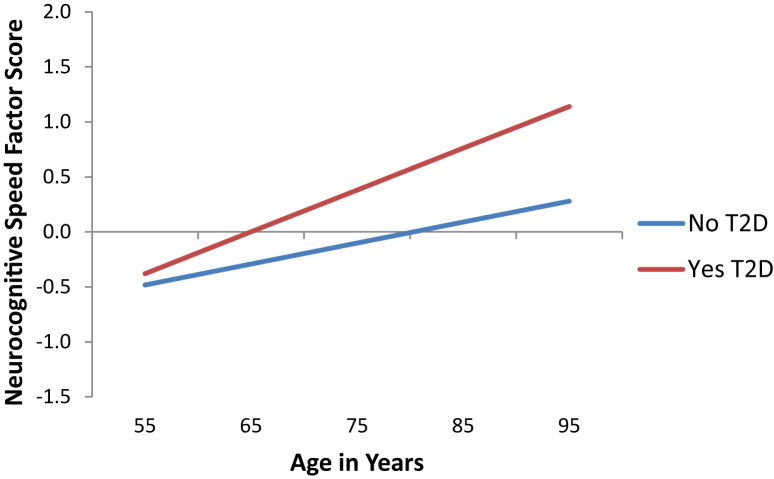
First, T2D status significantly predicted level of speed at age 75 (P = .005) but not 9-year change (P = .179; Fig. 1). Specifically, adults without T2D classification (M = −0.102) performed better on speed tasks at age 75 than adults classified with T2D (M = .380). Second, APOE showed no independent effect on speed level (P = .429) or 9-year change (P = .918). Third, no independent effect on speed level or 9-year change was observed for CLU (P = .304; P = .324), CR1 (P = .781; P = .962), or PICALM (P = .976; P = .149). Fourth, GRS did not confer an independent risk for speed performance at age 75 (P = .362) or 9-year decline (P = .898).
Fig. 1.
Predicted growth curve model of neurocognitive speed with T2D health status as predictor. Age used as a continuous variable centered at 75 years was the metric of change. Higher scores represent slower speed performance. Regression of intercept (level at age 75) b = 0.483 (SE = 0.174), P = .005. Regression of slope (9-year change) b = 0.019 (SE = 0.014), P = .179. Abbreviations: T2D, type 2 diabetes; SE, standard error.
3.2. Research goal 2: Determine the interactive effects of T2D status, APOE, or GRS
3.2.1. APOE
No significant interaction effect of T2D status on speed level and slowing was observed by APOE risk group (Supplementary Table 2).
3.2.2. GRS (APOE, CLU, CR1, and PICALM)
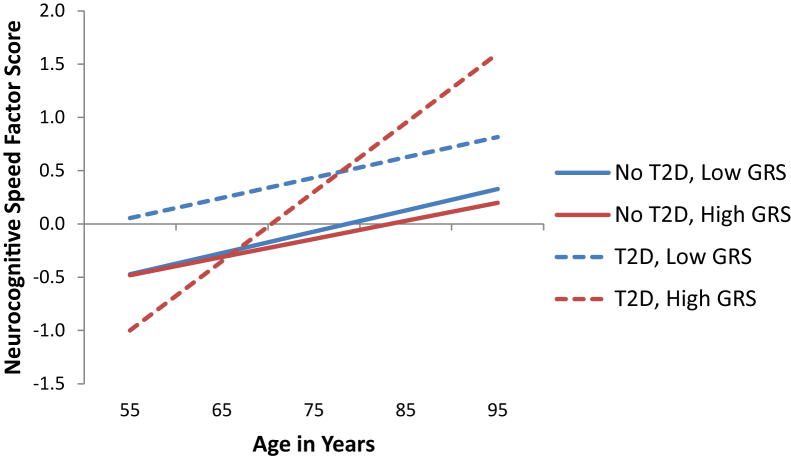
Significant interaction effects were observed for 9-year slowing (P = .029). The high-risk GRS group exhibited significant T2D health status differences in 9-year slowing (b = 0.048, P = .004; Fig. 2). Adults with high GRS risk and T2D (M = 0.065) exhibited more slowing than adults with high GRS risk and no T2D (M = .017). Adults in the low-risk GRS group exhibited no significant T2D status differences in slowing and were similar to adults in the high-risk GRS group with no T2D (i.e., low GRS risk, no T2D M = 0.019; low GRS risk, T2D M = 0.018).
Fig. 2.
Predicted growth curve model of neurocognitive speed using T2D status as predictor grouped by GRS (APOE, CLU, CR1, and PICALM) low and high risk. Higher scores represent slower speed performance. Low risk: Regression of intercept (level at age 75) b = 0.508 (SE = 0.270), P = .059. Regression of slope (9-year change) b = −0.001 (SE = 0.026), P = .979. High risk: Regression of intercept (level at age 75) b = 0.441 (SE = 0.252), P = .080. Regression of slope (9-year change) b = 0.048 (SE = 0.017), P = .005. Abbreviations: T2D, type 2 diabetes; GRS, genetic risk score; SE, standard error.
3.2.3. Follow-up analyses
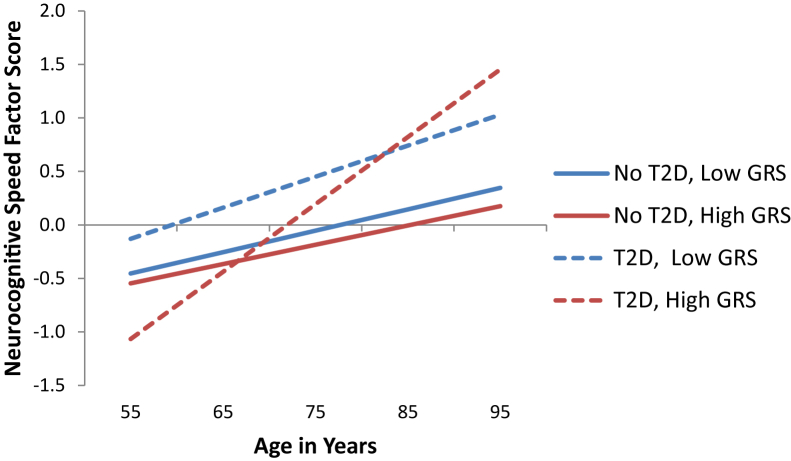
First, of the T2D-related covariates tested, only pulse pressure and education exhibited significant effects on speed (Table 3). Specifically, higher education was associated with better speed performance at age 75 (in both genetic risk groups), and poorer (higher) levels of pulse pressure were associated with more slowing in the low-risk genetic group. However, no change in the main results was observed: The GRS was still sensitive to T2D differences in the presence of these covariates. Second, although there were no independent effects observed for either GRS or APOE, the GRS (which included APOE) modified the T2D prediction of speeded performance. The follow-up analyses excluding APOE from the GRS revealed a similar basic pattern to that observed with the full GRS. Specifically, the high-risk genetic group with T2D exhibited the most 9-year slowing (b = 0.045, P = .023; Fig. 3). However, interaction analyses were not significant (P = .063). In addition, when pulse pressure was added as a covariate, the intensification interaction with the GRS (without APOE) was no longer significant, confirming the importance of the full GRS. We validated the simple count GRS by comparing it with an odds ratio GRS [28], [44]. We observed similar GRS × T2D effects on level of speed performance at age 75 and change for both versions of the GRS (Supplementary Table 3, Supplementary Figs. 1 and 2).
Table 3.
Genetic risk score × type 2 diabetes covariate models
| Type 2 diabetes plus covariates | Low genetic risk |
High genetic risk |
||
|---|---|---|---|---|
| Level β | Change β | Level β | Change β | |
| Genetic risk score all covariate model | ||||
| Type 2 diabetes | 0.435 | −0.003 | 0.239 | 0.039∗ |
| Gender | 0.060 | 0.006 | 0.184 | 0.004 |
| Pulse pressure | 0.001 | 0.002∗∗ | 0.005 | 0.001 |
| Body mass index | −0.011 | 0.000 | 0.004 | 0.000 |
| Smoking | −0.015 | 0.001 | −0.023 | −0.011 |
| Drinking | 0.089 | 0.002 | −0.017 | −0.002 |
| Education | −0.053∗∗ | −0.002 | −0.056∗ | −0.002 |
| Health relative to perfect | 0.194 | 0.000 | 0.182 | 0.028 |
| Health relative to peers | −0.070 | −0.012 | 0.078 | −0.027 |
| Genetic risk score pulse pressure, body mass index, smoking covariate model | ||||
| Type 2 diabetes | 0.535 | −0.006 | 0.397 | 0.040∗ |
| Pulse pressure | 0.002 | 0.002∗∗ | 0.010 | 0.002∗∗ |
| Body mass index | −0.010 | −0.001 | 0.002 | 0.000 |
| Smoking | 0.001 | 0.001 | −0.021 | −0.009 |
| Genetic risk score without APOE all covariate model | ||||
| Type 2 diabetes | 0.491 | 0.009 | 0.321 | 0.013 |
| Gender | 0.089 | 0.006 | 0.118 | 0.008 |
| Pulse pressure | 0.003 | 0.002 | 0.006 | 0.002∗∗ |
| Body mass index | −0.011 | 0.001 | −0.002 | −0.001 |
| Smoking | 0.028 | 0.002 | −0.021 | −0.006 |
| Drinking | 0.146 | 0.000 | −0.064 | 0.003 |
| Education | −0.054∗ | −0.002 | −0.061∗∗ | −0.002 |
| Health relative to perfect | 0.189 | 0.005 | 0.180 | 0.013 |
| Health relative to peers | −0.143 | −0.015 | −0.011 | −0.022 |
| Genetic risk score without APOE, pulse pressure, body mass index, smoking covariate model | ||||
| Type 2 diabetes | 0.575∗ | 0.007 | 0.402 | 0.012 |
| Pulse pressure | 0.004 | 0.002∗ | 0.009 | 0.002∗∗∗ |
| Body mass index | −0.009 | 0.001 | −0.002 | −0.002 |
| Smoking | 0.033 | 0.001 | −0.038 | −0.006 |
NOTE. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Fig. 3.
Predicted growth curve model of neurocognitive speed using T2D status as predictor grouped by GRS (CLU, CR1, and PICALM) low and high risk. Higher scores represent slower speed performance. Low risk: Regression of intercept (level at age 75) b = 0.505 (SE = 0.214), P = .019. Regression of slope (9-year change) b = 0.009 (SE = 0.020), P = .661. High risk: Regression of intercept (level at age 75) b = 0.379 (SE = 0.292), P = .194. Regression of slope (9-year change) b = 0.045 (SE = 0.007), P = .025. Abbreviations: T2D, type 2 diabetes; GRS, genetic risk score; SE, standard error.
4. Discussion
Our objective was to examine potential AD-related gene × health intensification of risk for longitudinal cognitive decline in nondemented older adults representing a 40-year band of aging. Specifically, we found that the interaction of an AD GRS with T2D status was associated with steeper longitudinal decline in neurocognitive speed. The key sequence of findings leading to the interpretation of risk intensification is as follows.
First, we confirmed the robustness of the neurocognitive speed latent growth model showing expected decremental change (slowing) over time. Second, we observed that adults with T2D performed slower (than adults without classified T2D) at the centering age of 75, but the groups did not differ in the rate of decline. These findings support research that suggests T2D may have a demarcated onset for cognitive dysfunction but not an ongoing acceleration of decline [5], [6]. Reduced insulin sensitivity and macrovascular disease associated with T2D may occur early in disease pathology, leading to cognitive deficits that predispose adults with diabetes for other downstream effects associated with dementia risk [5], [6], [45], [46], [47], [48]. Third, APOE was not independently associated with either speed performance (at the centering age) or decline. Some contrasting results have appeared regarding APOE-cognition associations in nondemented aging [20]. Unlike most studies, our results reflect statistical modeling and a latent speed variable, reflecting contributions of three manifest speed measures. Fourth, like APOE, the GRS had no independent effect on speed performance or change. The modest penetrance of especially three of the four genes comprising the GRS was not cumulatively sufficient to produce detectable effects on speed. Arguably, speed is not specific to AD [21], [28], [36] so the absence of an independent effect for both APOE and GRS (including APOE) is not surprising.
Fifth, our major result is that GRS intensified the deleterious effects of T2D on neurocognitive speed. Specifically, adults with high-risk GRS and T2D exhibited the steepest 9-year decline in neurocognitive speed. Notably, APOE alone did not exacerbate the effects of T2D on speed performance or change. Our findings suggest that the use of a cumulative AD-related GRS in combination with an AD-related health risk factor (T2D) can provide more information than single (candidate) genetic or health risk factors alone. We note that a GRS without APOE was also informative (suggesting the potential value of testing other genetic combinations) but was not as robust as the full GRS. The GRS is an efficient way of representing broader and deeper AD-related genetic risk, but its influence on cognitive (specifically, speed) change in nondemented adults is observed in synergistic combination with another AD-related risk factor [43], [49]. Overall, such an approach may also be valuable for predicting conversion to mild cognitive impairment and AD.
The pathways underlying the interactive effect of T2D × GRS on neurocognitive speed require detailed mechanistic studies. T2D effects may contribute risk upstream from the AD-pathology associated with AD genetic risk factors (e.g., increased Aβ, tau load) via vascular brain pathology in the form of increased cerebral infarcts or neuroinflammation [45], whole brain atrophy [46], white matter connectivity abnormalities [47], or acute hyperglycemic or insulin resistance effects for which genes associated with lipid control (i.e., APOE, CLU) may play a role [5]. Neurocognitive speed is known to decline systematically throughout adulthood and to be quite sensitive to subtle changes in brain health [32], [33], [35]. When well measured (e.g., at the latent variable level), it may prove to be an early behavioral indicator of accumulating biological changes associated with accelerated cognitive decline and preclinical impairment.
There are several limitations to note. T2D status was assessed using a strict and standard multistep process but did not include continuously distributed and relevant biomarkers (i.e., glycated hemoglobin). However, the VLS protocol for classifying T2D status is well developed and has been used successfully in previous studies [7], [8], [9]. In addition, although our T2D sample is consistent with national prevalence rates, the number of T2D participants in the sample limited the utilization of extreme genetic risk groupings. We therefore used dichotomized categorization of the GRS. Second, our sample was derived from a relatively healthy group of community-dwelling older adults with access to national health care. Although the sample may not be representative of all older adults, our findings represent a conservative estimation of the cumulative and interactive effects of genetic and health biomarkers for a growing segment of nondemented older adults. Third, the VLS data set does not include a full complement of AD-related genes. Although the AD-related GRS exhibited interesting results, future studies will include other genome-wide association study-identified AD-related genes examined individually and in synergistic combination.
There are also several strengths associated with this study. First, we used contemporary statistical approaches to analyze a set of research goals that systematically built the case for the final interaction analyses. Second, we examined the effect of continuously measured age in an accelerated longitudinal design that allowed us to determine the effects of T2D and the combined effects of four genes associated with AD risk across three data collection points spanning about 9 years. Third, our sample was relatively large (i.e., W1 n = 628) and well characterized. That this group comprised a band of about 40 years (55–91) is important to note.
In conclusion, consistent with a risk-intensification interpretation, adults with both high-risk genetic and T2D status risk declined differentially faster (than other risk combinations) in neurocognitive speed over three time points. Neither APOE alone nor GRS exhibited independent effects on speed performance and change. The observed association of T2D was limited to group difference in the level of performance and not 9-year slowing. Planned follow-up analyses clarified the role of APOE. In the absence of APOE, the three-gene GRS produced similar effects on speed, suggesting a cumulative genetic influence outside that contributed by APOE. Nevertheless, the three-gene GRS interaction effects were attenuated by vascular health, indicating that APOE played a risk-strengthening role in the context of the AD GRS. Predictions of cognitive change in nondemented aging, and perhaps transitions to cognitive impairment and AD, may benefit from consideration of specific multilocus genetic risk patterns. Moreover, the role of such risk scores may be substantial and detectable when they are examined in interaction with selected cross-domain biomarkers (i.e., health or lifestyle risk factors). Identification of specific subpopulations based on unique risk factors will be helpful for future clinical interventions.
Research in context.
-
1.
Systematic review: We reviewed the literature using online databases (e.g., Medline) and available sources. We consulted an emerging literature linking genetic and health risk factors for Alzheimer's disease (AD) and nondemented neurocognitive aging. We also used relevant citations from longitudinal design and analyses literature.
-
2.
Interpretation: Examining interactions among biomarker indices and health markers can substantially advance our understanding of trajectories and transitions leading to cognitive decline, impairment, and AD. An AD genetic risk score provided an empirically useful representation of genetic risk. Higher genetic risk intensified the deleterious effects of type 2 diabetes on neurocognitive slowing. The genetic risk score contributed predictive power beyond that of APOE alone.
-
3.
Future directions: Future research includes continued examination of independent and interactive predictions of AD risk from (1) genetic, (2) vascular health, and (3) lifestyle domains. We will use both candidate biomarkers and novel risk indices in longitudinal studies.
Acknowledgments
The Victoria longitudinal study is supported by a grant from the National Institutes of Health (National Institute on Aging) to R.A.D. (R01 AG008235), who is also supported by the Canada Research Chairs program. K.J.A. is supported by NHMRC Research Fellowship #1002560.
Authors' contributions: G.P.M., K.J.A., and R.A.D. contributed to the study concept. G.P.M. and R.A.D. did the design. G.P.M. did the analysis and drafting and revision of the article. G.P.M., K.J.A., and R.A.D. did the interpretation. S.A.W. did the analysis and interpretation consultation and revision of article for important intellectual content. D.V. did the genotyping and contribution of important genetic mechanism content. K.J.A. and R.A.D. did the critical revision of the article for important intellectual content. R.A.D. contributed to the data set assembly.
Footnotes
The authors have reported no conflicts of interest.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dadm.2015.08.002.
Supplementary data
References
- 1.Anstey K.J., Cherbuin N., Herath P.M. Development of a new method for assessing global risk of Alzheimer's disease for use in population health approaches to prevention. Prev Sci. 2013;14:411–421. doi: 10.1007/s11121-012-0313-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis Z., Wilson R.S., Bienias J.L., Evans D.A., Bennett D.A. Diabetes mellitus and risk of Alzheimer disease and decline in cognitive function. Arch Neurol. 2004;61:661–666. doi: 10.1001/archneur.61.5.661. [DOI] [PubMed] [Google Scholar]
- 3.Profenno L.A., Porsteinsson A.P., Faraone S.V. Meta-analysis of Alzheimer's disease risk with obesity, diabetes, and related disorders. Biol Psychiatry. 2010;67:505–512. doi: 10.1016/j.biopsych.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Awad N., Gagnon M., Messier C. The relationship between impaired glucose tolerance, type 2 diabetes, and cognitive function. J Clin Exp Neuropsychol. 2004;26:1044–1080. doi: 10.1080/13803390490514875. [DOI] [PubMed] [Google Scholar]
- 5.Biessels G.J., Strachan M.W., Visseren F.L., Kappelle L.J., Whitmer R.A. Dementia and cognitive decline in type 2 diabetes and prediabetic stages: Towards targeted interventions. Lancet Diabetes Endocrinol. 2014;2:246–255. doi: 10.1016/S2213-8587(13)70088-3. [DOI] [PubMed] [Google Scholar]
- 6.McCrimmon R.J., Ryan C.M., Frier B.M. Diabetes and cognitive dysfunction. Lancet. 2012;379:2291–2299. doi: 10.1016/S0140-6736(12)60360-2. [DOI] [PubMed] [Google Scholar]
- 7.McFall G.P., Geall B.P., Fischer A.L., Dolcos S., Dixon R.A. Testing covariates of type 2 diabetes-cognition associations in older adults: Moderating or mediating effects? Neuropsychology. 2010;24:547–562. doi: 10.1037/a0019246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McFall G.P., Wiebe S.A., Vergote D., Westaway D., Jhamandas J., Dixon R.A. IDE (rs6583817) polymorphism and type 2 diabetes differentially modify executive function in older adults. Neurobiol Aging. 2013;34:2208–2216. doi: 10.1016/j.neurobiolaging.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung S.E., Fischer A.L., Dixon R.A. Exploring effects of type 2 diabetes on cognitive functioning in older adults. Neuropsychology. 2009;23:1–9. doi: 10.1037/a0013849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaraman A., Pike C.J. Alzheimer's disease and type 2 diabetes: Multiple mechanisms contribute to interactions. Curr Diab Rep. 2014;14:476. doi: 10.1007/s11892-014-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindenberger U., Nagel I.E., Chicherio C., Li S.C., Heekeren H.R., Bäckman L. Age-related decline in brain resources modulates genetic effects on cognitive functioning. Front Neurosci. 2008;2:234. doi: 10.3389/neuro.01.039.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaik M.M., Gan S.H., Kamal M.A. Epigenomic approach in understanding Alzheimer's disease and type 2 diabetes mellitus. CNS Neurol Disord Drug Targets. 2014;13:283–289. doi: 10.2174/18715273113126660181. [DOI] [PubMed] [Google Scholar]
- 13.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 15.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brainerd C.J., Reyna V.F., Petersen R.C., Smith G.E., Taub E.S. Is the apolipoprotein e genotype a biomarker for mild cognitive impairment? Findings from a nationally representative study. Neuropsychology. 2011;25:679–689. doi: 10.1037/a0024483. [DOI] [PubMed] [Google Scholar]
- 17.Dixon R.A., DeCarlo C.A., MacDonald S.W., Vergote D., Jhamandas J., Westaway D. APOE and COMT polymorphisms are complementary biomarkers of status, stability, and transitions in normal aging and early mild cognitive impairment. Front Aging Neurosci. 2014;6:236. doi: 10.3389/fnagi.2014.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laukka E.J., Lovden M., Herlitz A., Karlsson S., Ferencz B., Pantzar A. Genetic effects on old-age cognitive functioning: a population-based study. Psychol Aging. 2013;28:262–274. doi: 10.1037/a0030829. [DOI] [PubMed] [Google Scholar]
- 19.Small B.J., Rosnick C.B., Fratiglioni L., Bäckman L. Apolipoprotein E and cognitive performance: A meta-analysis. Psychol Aging. 2004;19:592. doi: 10.1037/0882-7974.19.4.592. [DOI] [PubMed] [Google Scholar]
- 20.Wisdom N.M., Callahan J.L., Hawkins K.A. The effects of apolipoprotein E on non-impaired cognitive functioning: A meta-analysis. Neurobiol Aging. 2011;32:63–74. doi: 10.1016/j.neurobiolaging.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Ready R.E., Baran B., Chaudhry M., Schatz K., Gordon J., Spencer R.M. Apolipoprotein E-e4, processing speed, and white matter volume in a genetically enriched sample of midlife adults. Am J Alzheimers Dis Other Demen. 2011;26:463–468. doi: 10.1177/1533317511421921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thambisetty M., An Y., Nalls M., Sojkova J., Swaminathan S., Zhou Y. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol Psychiatry. 2013;73:422–428. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chibnik L.B., Shulman J.M., Leurgans S.E., Schneider J.A., Wilson R.S., Tran D. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gui H., Jiang C.Q., Cherny S.S., Sham P.C., Xu L., Liu B. Influence of Alzheimer's disease genes on cognitive decline: The Guangzhou Biobank Cohort Study. Neurobiol Aging. 2014;35 doi: 10.1016/j.neurobiolaging.2014.04.022. 2422.e3–2422.e8. [DOI] [PubMed] [Google Scholar]
- 25.Sweet R.A., Seltman H., Emanuel J.E., Lopez O.L., Becker J.T., Bis J.C. Effect of Alzheimer's disease risk genes on trajectories of cognitive function in the Cardiovascular Health Study. Am J Psychiatry. 2012;169:954–962. doi: 10.1176/appi.ajp.2012.11121815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barral S., Bird T., Goate A., Farlow M.R., Diaz-Arrastia R., Bennett D.A. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012;78:1464–1471. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertram L., Lill C.M., Tanzi R.E. The genetics of Alzheimer disease: Back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Adams H.H.H., de Bruijn R.F.A.G., Hofman A., Uitterlinden A.G., van Duijn C.M., Vernooij M.W. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia. Alzheimers Dement. 2015 doi: 10.1016/j.jalz.2014.12.008. http://dx.doi.org/10.1016/j.jalz.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Josefsson M., Luna X., Pudas S., Nilsson L.G., Nyberg L. Genetic and lifestyle predictors of 15-year longitudinal change in episodic memory. J Am Geriatr Soc. 2012;60:2308–2312. doi: 10.1111/jgs.12000. [DOI] [PubMed] [Google Scholar]
- 30.RodriguezRodriguez E., SanchezJuan P., VazquezHiguera J.L., Mateo I., Pozueta A., Berciano J. Genetic risk score predicting accelerated progression from mild cognitive impairment to Alzheimer's disease. J Neural Transm. 2013;120:807–812. doi: 10.1007/s00702-012-0920-x. [DOI] [PubMed] [Google Scholar]
- 31.Carrasquillo M.M., Crook J.E., Pedraza O., Thomas C.S., Pankratz V.S., Allen M. Late-onset Alzheimer's risk variants in memory decline, incident mild cognitive impairment, and Alzheimer's disease. Neurobiol Aging. 2015;36:60–67. doi: 10.1016/j.neurobiolaging.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hertzog C., Dixon R.A., Hultsch D.F., MacDonald S.W. Latent change models of adult cognition: Are changes in processing speed and working memory associated with changes in episodic memory? Psychol Aging. 2003;18:755–769. doi: 10.1037/0882-7974.18.4.755. [DOI] [PubMed] [Google Scholar]
- 33.Salthouse T.A. The processing-speed theory of adult age differences in cognition. Psychol Rev. 1996;103:403–428. doi: 10.1037/0033-295x.103.3.403. [DOI] [PubMed] [Google Scholar]
- 34.Christensen H., Dear K.B., Anstey K.J., Parslow R.A., Sachdev P., Jorm A.F. Within-occasion intraindividual variability and preclinical diagnostic status: Is intraindividual variability an indicator of mild cognitive impairment? Neuropsychology. 2005;19:309–317. doi: 10.1037/0894-4105.19.3.309. [DOI] [PubMed] [Google Scholar]
- 35.Dixon R.A., Garrett D.D., Lentz T.L., MacDonald S.W.S., Strauss E., Hultsch D.F. Neurocognitive markers of cognitive impairment: Exploring the roles of speed and inconsistency. Neuropsychology. 2007;21:381–399. doi: 10.1037/0894-4105.21.3.381. [DOI] [PubMed] [Google Scholar]
- 36.Bäckman L., Jones S., Berger A., Laukka E.J., Small B.J. Cognitive impairment in preclinical Alzheimer's disease: A meta-analysis. Neuropsychology. 2005;19:520–531. doi: 10.1037/0894-4105.19.4.520. [DOI] [PubMed] [Google Scholar]
- 37.Dixon R.A., de Frias C.M. The Victoria longitudinal study: From characterizing cognitive aging to illustrating changes in memory compensation. Aging, Neuropsychology, and Cognition. 2004;11:346–376. [Google Scholar]
- 38.Dixon R.A., Small B.J., MacDonald S.W.S., McArdle J.J. Yes, memory declines with aging-but when, how, and why? In: Naveh-Benjamin M., Ohta N., editors. Memory and aging: Current issues and future directions. Psychology Press; New York, NY: 2012. pp. 325–347. [Google Scholar]
- 39.Small B.J., Dixon R.A., McArdle J.J. Tracking cognition-health changes from 55 to 95 years of age. J Gerontol B Psychol Sci Soc Sci. 2011;66:153–161. doi: 10.1093/geronb/gbq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Folstein M.F., Folstein S.E., McHugh P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 41.Che R., Motsinger-Reif A.A. Evaluation of genetic risk score models in the presence of interaction and linkage disequilibrium. Front Genet. 2013;4:138. doi: 10.3389/fgene.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muthén L.K., Muthén B.O. Muthén & Muthén; Los Angeles, CA: 1998. Mplus User's Guide. [Google Scholar]
- 43.McFall G.P., Wiebe S.A., Vergote D., Jhamandas J., Westaway D., Dixon R.A. IDE (rs6583817) polymorphism and pulse pressure are independently and interactively associated with level and change in executive function in older adults. Psychol Aging. 2014;29:418–430. doi: 10.1037/a0034656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 45.Bangen K.J., Gu Y., Gross A.L., Schneider B.C., Skinner J.C., Benitez A. Relationship between type 2 diabetes mellitus and cognitive change in a multiethnic elderly cohort. J Am Geriatr Soc. 2015;63:1075–1083. doi: 10.1111/jgs.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moran C., Phan T.G., Chen J., Blizzard L., Beare R., Venn A. Brain atrophy in type 2 diabetes: Regional distribution and influence on cognition. Diabetes Care. 2013;36:4036–4042. doi: 10.2337/dc13-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reijmer Y.D., Leemans A., Brundel M., Kappelle L.J., Biessels G.J., Utrecht Vascular Cognitive Impairment Study Group Disruption of the cerebral white matter network is related to slowing of information processing speed in patients with type 2 diabetes. Diabetes. 2013;62:2112–2115. doi: 10.2337/db12-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reitz C., Mayeux R. Alzheimer disease: Epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem Pharmacol. 2014;88:640–651. doi: 10.1016/j.bcp.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sapkota S., Vergote D., Westaway D., Jhamandas J., Dixon R.A. Synergistic associations of catechol-O-methyltransferase and brain-derived neurotrophic factor with executive function in aging are selective and modified by apolipoprotein E. Neurobiol Aging. 2015;36:249–256. doi: 10.1016/j.neurobiolaging.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.