Abstract
Acidic phospholipids are minor membrane lipids but critically important for signaling events. The main acidic phospholipids are phosphatidylinositol phosphates (PIPs also known as phosphoinositides), phosphatidylserine (PS) and phosphatidic acid (PA). Acidic phospholipids are precursors of second messengers of key signaling cascades or are second messengers themselves. They regulate the localization and activation of many proteins, and are involved in virtually all membrane trafficking events. As such, it is crucial to understand the subcellular localization and dynamics of each of these lipids within the cell. Over the years, several techniques have emerged in either fixed or live cells to analyze the subcellular localization and dynamics of acidic phospholipids. In this chapter, we review one of them: the use of genetically encoded biosensors that are based on the expression of specific lipid binding domains (LBDs) fused to fluorescent proteins. We discuss how to design such sensors, including the criteria for selecting the lipid binding domains of interest and to validate them. We also emphasize the care that must be taken during data analysis as well as the main limitations and advantages of this approach.
Keywords: Biosensor, phosphatidylinositol phosphate, phosphatidic acid, phosphatidylserine, genetically encoded probes, lipid binding domain, live imaging, PtdIns, lipid signaling, Phospholipase
Introduction
Anionic phospholipids have a negatively charged head group, which gives them specific properties, notably in terms of protein-lipid interactions. The main acidic phospholipids are phosphatidylserine (PS), phosphatidic acid (PA) and phosphatidylinositol (PI and PIPs). In erythrocytes, the PS/PA/PI proportions (by weight) are approximately 8.5%, 1.5% and 1.0%, respectively, but these may vary according to species or cell types [1].
Phosphatidylinositolphosphates (PIPs) are minor phospholipids, accounting less than one percent of total membrane lipids, yet they are of disproportionate importance for many membrane-associated signaling events: i) PIPs can be precursors of various second messengers (e.g. Inositol-3-Phosphate, Diacylglycerol), ii) they can activate many ion channels and enzymes, iii) they are involved in membrane trafficking and, iv) they can recruit proteins to the plasma membrane or intracellular compartments through several structured interaction domains (e.g. Pleckstrin Homology domain (PH), Phox homology domain (PX), Fab1/YOTB/Vac1/EEA1 domain (FYVE)) [1-4]. PIPs can be phosphorylated at different positions of the inositol head group, which can generate up to seven different PIP species that include three phosphatidylinositol monophosphates [PI3P, PI4P and PI5P], three phosphatidylinositol biphosphate [PI(3,4)P2, PI(3,5)P2 and PI(4,5)P2] and one phosphatidylinositol triphosphate [PI(3,4,5)P3]. PIP kinases and phosphatases modify the phosphorylation state of the inositol head group, and phospholipases hydrolyze PIPs to release the soluble head group into the cytosol [1,4]. The combined action of these enzymes produces the PIP signature of a cell, where certain membrane compartments are enriched or depleted of specific PIPs, contributing to their functional identity [1,3,4].
Phosphatidylserine (PS) is an important constituent of eukaryotic membranes and the most abundant acidic phospholipid (up to 10% of biological membrane) [1,5-7]. PS is involved in many signaling pathways, as it can recruit and/or activate proteins, notably through their stereospecific PS-binding domain and by regulating membrane surface charges [1,5,6,8]. One particularity of PS is its role as a lipid landmark in both extracellular and intracellular membranes leaflets. For instance, extracellular PS (exposed on the outer leaflet of the plasma membrane) serves as an “eat me” signal for the clearance of apoptotic cells [7,9]. Intracellular PS regulates a number of signaling pathways involving kinases, small GTPases and fusogenic proteins [5,8].
Phosphatidic acid (PA) is a precursor for the biosynthesis of many lipids [10,11]. Indeed, various enzymes add different chemical group on PA, such as Choline, Ethanolamine, Serine or Inositol to produce phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS) and phosphatidylinositol (PI). PA is also the substrate of Phospholipase D, which produces diacylglycerol, a second messenger involved in many signaling pathways [12]. Furthermore, the biophysical characteristics of PA influence membrane properties such as membrane curvature or membrane fusion [1,13,14]. In addition, PA itself recruits various proteins to membranes and PA-protein interaction activates many enzymes. As such, PA can be considered a bona fide lipid second messenger.
Subcellular localization of anionic phospholipids at a glance
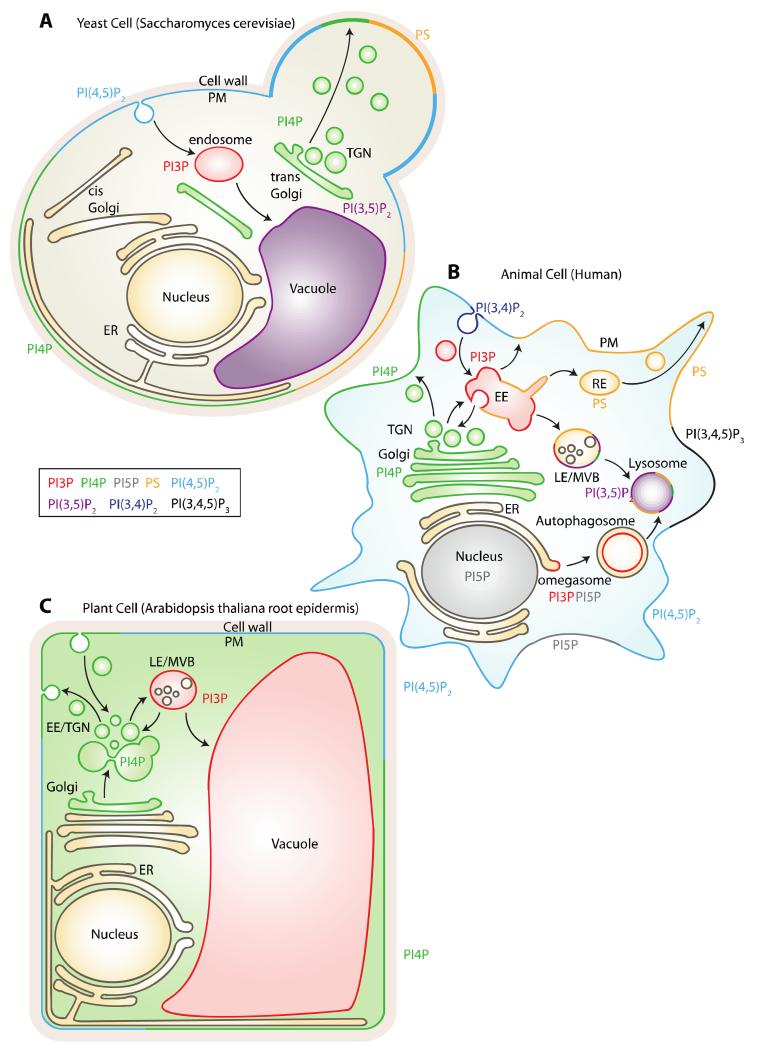
The localization of the various acidic phospholipid species has been an intense area of research [4,15,16]. Functional studies, together with biochemical and live-cell imaging, have built a relatively clear picture of the precise location of most acidic phospholipids in yeast (Figure 1A), cultured mammalian cell lines (Figure 1B), and plants (Figure 1C).
Figure 1. Summary of the subcellular localization of anionic phospholipids in yeast (A), animal (B) and plant (C) cells.
Note that the reported localization are not exhaustive and might vary depending on cell types or signaling activities. The cartoon representing the cell in panel B is adapted from Jean and Kiger 2012.
In animal cells, PI3P mainly resides in early endosomes, where it controls endosome maturation, cargo protein degradation/recycling and cell signaling notably through its interplay with Rab5 GTPases [3] (Figure 1B). During autophagy induction in animal (e.g. triggered by amino acid starvation), PI3P is transiently produced at the Endoplasmic Reticulum (ER) membrane by the PI3-kinase VPS34 [17] (Figure 1B). PI3P production in the ER supports the formation of the omegasome a specialized ER domain at the origin of the formation of the autophagophore (also known as the isolation membrane), that itself elongates to form the autophagosome (i.e., double membrane vesicles) [17,18].
In yeast and animals, PI4P is located in at least two different pools in the cell, one at the Golgi apparatus and the other one at the plasma membrane [19-21] (Figure 1A and B). Each pool of PI4P has separate and diverse functions. The main function of PI4P at the Golgi is to control membrane trafficking events, in particular, the sorting of proteins toward the plasma membrane or endosomes [3,22-24]. PI4P, together with other PIPs, recruits strong cationic proteins to the plasma membrane [25]. In yeast, the plasma membrane pool of PI4P controls ER-to-plasma membrane tethering sites that regulate cell signaling and ER morphology [26-28] (Figure 1A). Furthermore, plasma membrane-localized PI4P is a source of PI(4,5)P2 in animal cells [23,29]. A pool of PI4P has been recently described in late endosomes/lysosomes in animal cells but the function of PI4P in these compartments remains to be fully elucidated [20] (Figure 1B).
In mammals, the rare phosphoinositide, PI5P, accumulates in the nucleus and at the plasma membrane under certain stimuli, or during infection by certain pathogens such as the bacterium Shigella flexneri [30-34] (Figure 1B). Furthermore, it was recently showed that PI5P transiently accumulates at the ER during autophagy induction and can substitute PI3P at the omegasome [35] (Figure 1B).
In both animal and yeast, PI(3,5)P2 is thought to reside in late endosomes, where it regulates lysosome/vacuole biogenesis [36-38] (Figure 1A and B). In every eukaryotes, PI(4,5)P2 is localized at the plasma membrane where it has a large spectra of action such as anchoring signaling and membrane trafficking proteins [2,4,25,39-41] (Figure 1A-C). In addition, PI(4,5)P2 controls ion channel activation and is a substrate of Phospholipase C, which triggers synthesis of the second messengers inositol 1,4,5-trisphosphate and diacylglycerol [2,4,42]. PI(4,5)P2 is the source of PI(3,4,5)P3, which together with PI(3,4)P2, accumulate at the plasma membrane but only when specific signaling pathways are activated (e.g. growth factor signaling) [2,4](Figure 1B). PI(3,4)P2 also controls late-stage clathrin-coated pit formation, independent of PI(3,4,5)P3 [41,43].
PS is synthesized in the ER lumen and reaches the cytosolic leaflet through the action of P4-ATPases flipases [7,9]. Depending on the species, this translocation occurs either at the TGN and/or at the plasma membrane. This asymmetric PS distribution can be used as a signaling device by the regulated activation of scramblases, which rapidly exposes PS on the extracellular leaflet of the plasma membrane and plays important roles in blood clotting and apoptosis [7,16], as above-mentioned. On the cytosolic leaflet, PS mainly accumulates at the plasma membrane in yeast (Figure 1A), while it is present both at the plasma membrane and throughout the endosomal system in animal cells (Figure 1B) [5,8,44].
Like PS, PA is synthetized in the ER in all eukaryotic cells [10,11]. PA can also be synthesized de novo in other organelles such as for example mitochondria or chloroplasts [10]. However, the main pool of PA that is facing the cytosol is likely localized at the plasma membrane. This pool is locally produced by Phospholipase D and Diacylglycerol kinases [12].
Detection of acidic phospholipids by Lipid Binding Domains
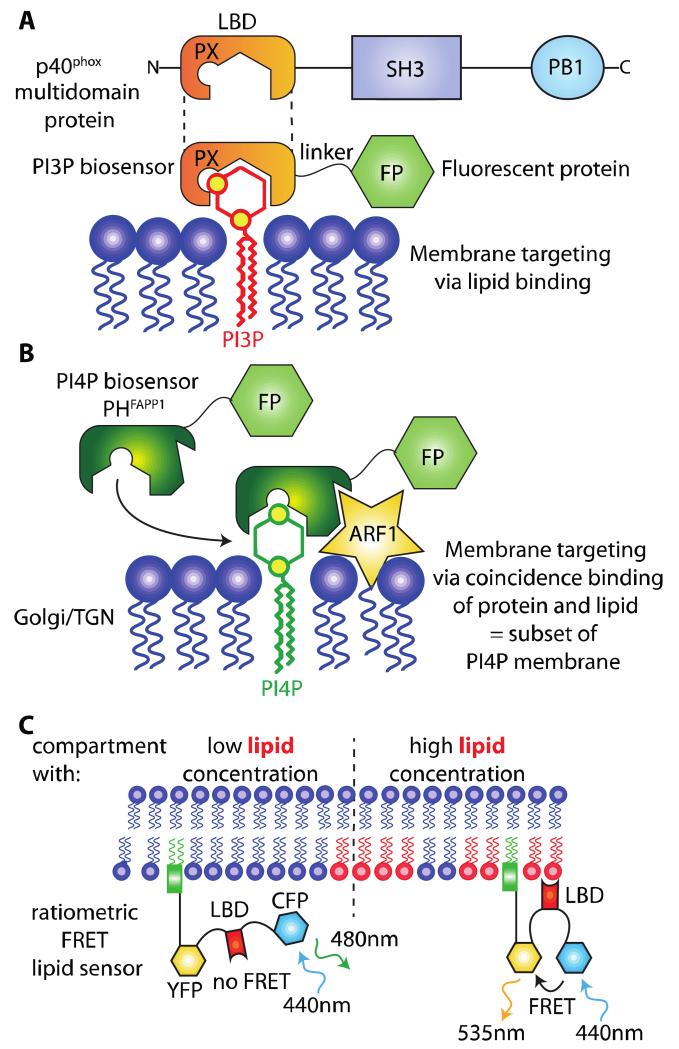
Anionic lipids such as phosphoinositides are markers of organelle identity. Moreover, because they act as second messengers, their quantity varies rapidly (i.e. within minutes) upon stimulation of various signaling pathways. It is therefore key to be able to track the amount of these lipids in real time and at subcellular resolution. However, the investigation of lipid subcellular localization has proven to be difficult for various reasons. First, it is obviously not possible to label lipids by direct tagging with fluorescent proteins (FPs). Second, common methods of cell or tissue fixation do not fix lipids and are therefore not compatible with the study of lipid subcellular localization. Yet, many techniques have been used over the years to uncover the subcellular localization of acidic phospholipids and their respective dynamics upon various stimulations. These techniques were used either in fixed cells, such as for example immuno-labeling with anti-PIP antibodies [19] or live cells, such as for example direct labeling of lipid molecules or the use of genetically encoded biosensors [45]. The later method has been extensively used to indirectly reveal the localization and dynamics of PIPs in intact living cells and, currently, is probably the most widespread technique used to localize acidic phospholipid species [4,40,45]. Importantly, this method is directly amenable to live imaging techniques. Genetically encoded biosensors consist of lipid-binding domains (LBDs) that interact specifically with known lipid species in vitro (Figure 2A and B). These domains localize in the compartments of the cell that accumulate the targeted PIPs and can be easily traced when fused with a fluorescent protein (Figure 2A and B). LBDs are globular domains that mostly bind to acidic phospholipids such as PIPs and PS [1,46]. Broadly, they fall into two categories: non-specific LBDs and stereospecific LBDs. Non-specific LBDs recognize general membrane properties, such as curvature, lipid packing defects or charges [1,14]. Examples of non-specific LBDs include the BAR domain that recognizes membranes with a specific curvature or the KA1 domain that binds highly electronegative membrane [1,47]. Stereospecific LBDs bind particular acidic lipids with sometime exquisite specificity. PH, PX, FYVE and some C2 domains belong to this category [1,46]. To date most LBDs that have been used to report on lipid localization are stereospecific LBDs, yet in recent years non-specific LBDs have also been exploited to probe some basic properties of the cytosolic leaflet of membrane compartments. For example, the KA1 domain has been used as a reporter of membrane surface charges in human cells [25].
Figure 2. General principle of genetically encoded lipid biosensors.
A) A lipid-binding domain (LBD) from a multidomain protein (p40phox in this example) is fused with a fluorescent protein (FP). This protein fusion acts as a biosensor for PI3P. B) Some LBDs require binding to both a lipid and another molecules (i.e., Ca2+, proteins). This coincidence binding specifies the localization of the corresponding biosensor to a subset of the lipid-enriched membrane, which also contains the target protein. In this example, the PH domain of FAPP1 binds PI4P and ARF1, hereby restricting its localization to the Golgi/TGN. C) Ratiometric FRET sensors are targeted to membranes independently of lipid binding (e.g., via a lipid anchor or a transmembrane segment) and report on the presence of the lipid based on the conformational changes induced in the sensor when the LBD binds its lipid (which increases or decreases the proximity between the two FPs and therefore their FRET ratio).
Design of genetically encoded acidic phospholipid probes
Construct strategy
To visualize a certain lipid species, the strategy is to fuse the LBD of interest with a fluorescent protein (FP) (Figure 2A). Most LBDs can be fused either to their N-terminal or C-terminal end without affecting their binding properties since they are derived from multi-domain proteins. To maximize the chances to obtain a stable and functional fusion protein, we usually place the LBD where it would be in its original protein context and separates it from the fluorescent protein by a short flexible linker (e.g. SAGGSAGG or GAGARS linkers). For example the PX domain of the p40phox protein is localized at its N-terminus. We therefore replaced the C-terminal part of this protein with fluorescent proteins, giving PXp40-FP constructs (Figure 2A). A fluorescent protein is usually sufficient to report each lipid, however methods based on Förster Resonance Energy Transfert (FRET) have also been used [48-51] (Figure 2C).
Most genetically encoded lipid sensors are soluble proteins and therefore are designed to report only the lipid species that are facing the cytosol. However, addition of a signal peptide to the probe has been generated to secrete the LBD and to follow the accumulation of its cognate lipid along the secretory pathway, such as for example its presence in the ER lumen [9]. However, because of the resolution limits of conventional light microscope, this approach requires Transmission Electron Microscopy (TEM) to distinguish between membrane-bound LBDs and soluble LBDs in the organelle’s lumen.
Choosing the appropriate LBD, consideration on LBD specificity
The most critical aspect in the design a genetically encoded sensor for a given lipid is to take into account binding specificity and affinity of the LBDs. If one wants to report the localization of a given lipid, the ideal probe should be highly specific for this lipid. However, very few, if any, LBDs are completely specific for only one lipid. Most of the time, their affinity is greater for a lipid than for the others, yet this is enough to confer a specificity of recognition in vivo. Nonetheless, this should be verified, if possible by several in vitro lipid-binding assays. Such assays include qualitative methods (e.g. lipid-protein overlay assays) and more quantitative techniques such as liposome-binding assays, surface plasmon resonance or isothermal titration calorimetry. Finally, the structure of the LBD-lipid complex (e.g. by x-ray crystallography or NMR spectroscopy) might help to rationalize how the domain specifically recognizes a particular phospholipid headgroup [1].
Moreover, it is common that LBDs require the coincidence detection of a given lipid together with another molecule to promote membrane binding. The most widespread examples are LBDs that bind their target lipid in a calcium-dependent manner (e.g., most C2 domain binds their lipids, mostly PS, only in the presence of Ca2+) [1]. Some LBDs also require the coincidence binding of another protein [1] (Figure 2B). For example, the PH domain of FAPP1 (and to a lesser extend the PH domain of OSBP) interacts with PI4P preferentially in the presence of the small GTPase ARF1 [21] (Figure 2B). This requirement for coincidence binding can lead to confounding results that are sometime difficult to evaluate. For example, the PH domain of FAPP1 is capable of binding PI4P alone, but in vivo membrane binding is enhanced by the presence of ARF1 [21]. Because ARF1 mainly localizes at the Golgi and TGN, two compartments that are enriched in PI4P, the PH domain of FAPP1 (and OSBP) preferentially localizes to these two compartments, although PI4P is also present at the plasma membrane [21] (Figure 1). This particular result led to the long-lasting belief that PI4P is mainly localized at the Golgi and TGN. Therefore, the PH domains of FAPP1 and OSBP are not optimum to report PI4P in all membranes. However, because PI4P association is required for membrane binding of these LBDs, they are suitable PI4P reporters in the Golgi and TGN and have been successfully used to this aim [52] (Figure 2B). When available, the use of probes that do not require coincidence binding with other molecules should be favored. Alternatively, if such LBD has not been characterized yet, the use of LBD requiring coincidence binding should not be discarded entirely, but the results should be interpreted accordingly.
Choosing the appropriate LBD, consideration on LBD affinity
The second parameter that one should take into account is the relative binding affinity of the LBD for its target lipid. This is also an important parameter, since difference in relative affinity might result in different subcellular localization of the probe. The first obvious caveat is when the binding affinity is too weak, which leads to mostly or exclusively soluble localization of the probe (their localization by default, in the absence of binding, being soluble in the cytosol). For example, a single PI3P-binding FYVE domain is soluble when express in mammalian cells and only a tandem dimer construct (2xFYVE domain) is localized to early endosomes, where PI3P accumulates [53]. This leads to the second caveat, which is when binding affinities are too high and high-affinity LBDs might outcompete the lipid binding of endogenous proteins, leading to toxicity upon expression of the probe. However, because any given cell expresses hundreds of proteins harboring LBDs at the same time, it is unlikely that transgenic expression of LBDs will outcompete all the other lipid-binding proteins. It is however common that expression of acidic phospholipid probes affects some signaling pathways. It is therefore advisable to test the toxicity due to the expression of the probe and to favor cells or transgenic organisms with relatively weak expression of the probe (for example by using promoters that confer mild expression).
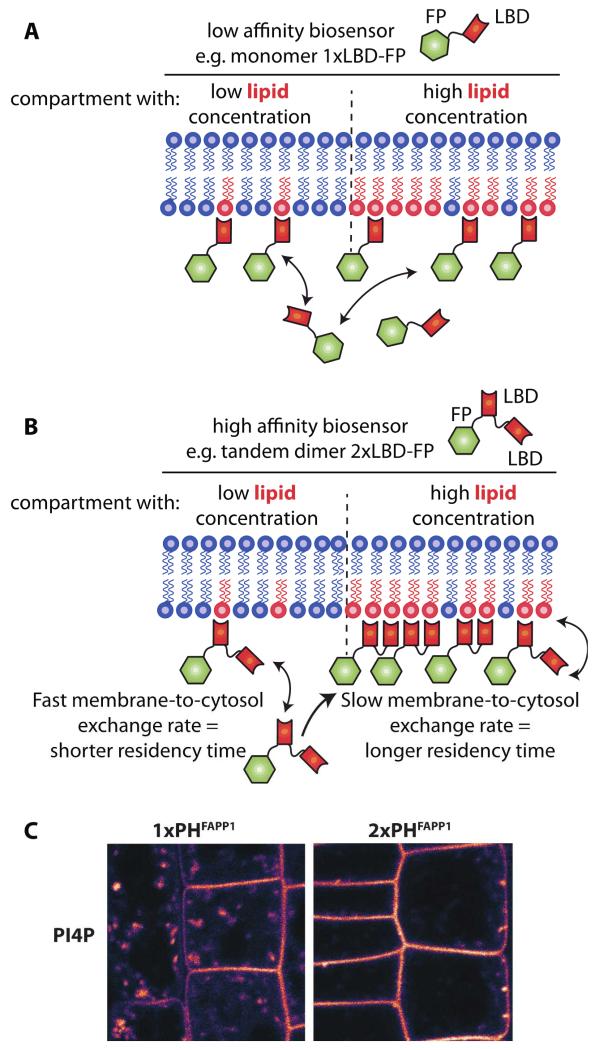
One should choose LBDs that have affinity ranging in between the two extreme scenarios discussed above. Because there is no way to predict in silico how a LBD will behave in vivo in a particular system, it is preferable to use, when available, several probes to report on the same lipid species. Because of slight changes in either binding affinity or specificity, we often observed that several reporters for the same lipid might harbor different, although overlapping, localization [40]. For example in Arabidopsis root, a 2xFYVE PI3P reporter is localized to late endosomes (where PI3P accumulates in plants, Figure 1C), while the PX domain of the p40phox protein, also a well characterized PI3P binding domain, localizes to both late endosomes and tonoplast (the membrane of the plant cell vacuole) [40] (Figure 1C). Although, it is not entirely understood how these differences in localization might be explained, these results are useful for several reasons. First, both probes localize to late endosomes, providing confirmation that PI3P is likely to accumulate in this compartment in plants. Second, because the PX domain also localizes to the tonoplast, this raised the possibility that PI3P might localized to this compartment. Although this conclusion should be taken with care, since it was confirmed with only one of the two LBD, it provided us with a new testable hypothesis. One way to explain the dissimilar localization of the FYVE and PX domains is to consider their difference in relative binding affinity. In fact, high affinity LBDs are expected to localize more specifically to the membrane compartment that accumulates the most its cognate lipid, while lower affinity LBDs are more likely to have a broader localization domain (Figure 3). Low affinity sensors are less efficient in discriminating between two membranes with two different concentrations of their targeted lipid species and as a result they might be targeted to both of these membranes (Figure 3A). By contrast, high affinity sensors will have increased dwell time at the membrane that is the most enriched in the targeted lipid and they will accumulate preferentially in this compartment (Figure 3B). In other words, high affinity sensors work like a “Velcro fastener”: they will grab more strongly to a surface with more spikes (in this case the spikes being an acidic lipid) (Figure 3B). Therefore, it is possible that the high affinity 2xFYVE probe mainly localizes to late endosomes because this could be the cell compartment where PI3P accumulates the most, while the PX-based probe localizes also to the tonoplast because this compartment might also have PI3P but to a lesser extent than late endosomes. This is further exemplified when comparing the localization of single versus tandem dimer LBDs. For example in Arabidopsis, we found that the high affinity PI4P sensor 2xPHFAPP1 was more strongly localized to the plasma membrane and less to endomembrane compartments than the low affinity sensor 1xPHFAPP1 [40] (Figure 3C). When kept in mind, these variations in localization can actually be exploited to address the relative concentration of a given lipid in several membranes. For example, the results presented Figure 3C suggest that the concentration of PI4P is greater at the plasma membrane than in intracellular compartments in plants [40].
Figure 3. LBD affinities influence the subcellular localization of the sensors.
When several pools of the same lipid exist within the cell, low or high affinity sensors will behave differently with respect to these pools. A) A low affinity sensor (e.g., 1xLBD) will localize to both membranes with slightly more sensor molecules at the compartment with the highest lipid concentration, while (B) a high affinity sensor (e.g., 2xLBD) will localize preferentially to the compartment with the highest lipid concentration. C) Example of low (1xPHFAPP1) and high (2xPHFAPP1) affinity sensor localization in Arabidopsis root cell (image from Simon et al., 2014 Plant Journal).
Validation of acidic phospholipid sensors
As mentioned in the previous section, it is important to test the in vitro binding specificity of a particular LBD. However, this apparent in vitro specificity does not necessarily reflect its localization in vivo or the localization of its cognate lipid in cells. In fact, a comprehensive study on all yeast PH domain suggest that in vitro binding specificity is not a good indicator of the localization of this domain in vivo and does not always predict whether the LBD will be a useful lipid probe or not [54]. Expression of each LBD has to be tested in vivo and if possible validated. A first screen will rapidly discard domains that do not properly accumulate, do not localizes to any membrane compartment or induce strong phenotypes [40]. It is then important to check whether the localization of the probe is in fact dependent on the presence of its cognate lipid. Among other approaches, this could be achieved by pharmacological or genetic inhibition of the lipid biosynthetic enzymes (e.g. phosphatidylinositol kinases, phosphatidylinositol phosphatases, phospholipases…). For example, a loss-of-function mutation in mss4, the yeast PI4P 5-kinase, leads to a soluble localization of a 2xPHPLC probe that normally highlights PI(4,5)P2 at the yeast plasma membrane [21]. An elegant approach is also the targeted recruitment of lipid kinases or phosphatases to a specific compartment using small molecules or light, because these approaches mediate rapid lipid modifications that are spatially restricted [20,25,39,42,55-59]. The localization of an ideal lipid reporter should be dependent on its cognate lipid in both loss- and gain-of-function experiment but not dependent on the production/loss of unrelated lipids. In other word, the probe should leave its endogenous membrane compartment upon loss of its cognate lipid at that membrane. Conversely, it should be recruited to a new membrane compartment upon production of its cognate lipid in this organelle. To date, very few probes have been tested extensively with such gain- and loss-of-function experiments. Besides, they are rarely so versatile, probably because of their requirement on coincidence binding to other molecules (see above the section on the design of genetically encoded acidic phospholipid probes). However, the recent characterization of the P4M PI4P reporter is a must read as an example on how to validate an acidic phospholipid sensor in vivo [20].
In order to validate the localization of a lipid sensor and therefore the cellular localization of a particular lipid, it is important to accumulate several lines of evidence to confirm this localization, such as for example the use of alternate techniques (immunolocalization, direct lipid labeling, …), the similar localization of independent LBDs known to bind the same lipid and/or the colocalization of the probe with endogenous lipid binding proteins.
Well-characterized acidic phospholipid sensors
Several LBDs have been used over the years in different systems and have been shown to behave robustly. In this section we will briefly describe these well characterized genetically encoded lipid sensors and, if applicable, point out their respective advantages and limitations. It is nonetheless important to consider the controls described above when using one of these reporters in a new biological context (e.g., new species, new cell type).
Phosphoinositide sensors
PI3P
The most widely used probe for PI3P are derived from the PX domain of the p40phox protein and the tandem dimer of the FYVE domains (2xFYVE) from the HRS or EEA1 proteins [1,46,53,60,61]. These domains have been extensively used over the years and are well-accepted PI3P reporters. In animal cells, they mainly report the localization of PI3P in early endosomes [53], but plasma membrane localization has been observed in certain conditions (e.g. insulin treatment [62,63]). However, they do not highlight the pool of PI3P at the ER upon autophagy induction.
PI4P
As discussed above (see “choosing the appropriate LBD, consideration on LBD specificity” section), the PH domain of FAPP1 and OSBP report on the localization of PI4P at the Golgi/TGN but not in other membrane compartments due to their requirement for ARF1 binding [21]. The PH domain of the yeast OSBP-like protein OSH2 is not dependent on ARF1 binding [64]. It is localized both at the Golgi and plasma membrane in yeast but it is localized mainly at the plasma membrane and only weakly at the Golgi in mammalian cells [20,64]. Therefore, PHOSH2 seems to be a better reporter of plasma membrane PI4P than PHFAPP1 or PHOSBP. The exact reasons for the plasma membrane preference of PHOSH2 are unknown, but might be due to residual PI(4,5)P2 binding [20,64]. The newly described PI4P reporter, called P4M, seems to be able to report both Golgi and plasma membrane PI4P localization in animal cells and it detects as well a previously uncharacterized pool of PI4P in late endosomes [20]. This reporter seems to be superior to the PH domains of FAPP1, OSBP and OSH2 since it is very specific to PI4P and does not require coincidence binding with other proteins. However, because it has been described fairly recently, it is not yet clear whether this probe will behave similarly in a broad range of cellular contexts.
PI5P
Few PI5P-binding domains have been characterized, including the PH domains of Dok-1 and Dok-2 [34,32] and the PHD domain of ING2. A triple repeat of this domain (3xPHDING2) has been used as a sensor of PI5P localization. It mainly localizes to the nucleus in animal cells [30,32]. However, immunolocalization and mass spectrometry methods suggest that PI5P localizes in membrane compartments such as the plasma membrane or endosomes [32,65]. 3xPHDING2 was recently found to accumulate in omegasomes during autophagy induction by glucose starvation [35]. However, 3xPHDING2 has not extensively been used over the years, perhaps because its expression inhibits PI5P-dependent processes [32]. Therefore, this reporter should be used with caution.
PI(4,5)P2
The PH domain of PLCdelta1 (hereafter referred to as PLC) was one of the first LBD to be used as a lipid biosensor [4,45,66]. It has an exquisite selectivity for PI(4,5)P2 and has been robustly expressed in many different cellular systems including yeast, mammalian and plant cells [4,21,32,40,66]. It allowed for example to monitor the reversible PI(4,5)P2 hydrolysis triggered upon PLC activation; i.e. relocalization of membrane-bound PHPLC into the cytosol upon PLC activation by agonists [66]. The C-terminal domain of the TUBBY protein has also been used as a PI(4,5)P2 reporter [40,67-69], however this protein domain binds PI(3,4)P2 in vitro in addition to PI(4,5)P2 [69]. Both reporters are localized exclusively to the plasma membrane, while PI(4,5)P2 has been found in Golgi and ER membrane. This point out to a possible limitation of these probes or simply to the fact that the concentration of PI(4,5)P2 in these compartments is not sufficient to trigger membrane binding at these sites. It is also possible that the physico-chemical properties of these compartments (such as their packing or curvature) are not compatible with binding of these domains. Finally, we cannot exclude that both LBD actually rely on coincidence binding of PI(4,5)P2 and a plasma membrane-resident protein. However, the fact that both reporters behave similarly in many different cellular contexts and species argues against this hypothesis. Altogether, PHPLCd1 and TUBBY-C are robust reporters of PI(4,5)P2 dynamics at the plasma membrane but might not reflect the possible pool of this lipid in other membrane compartments.
PI(3,5)P2
The ENTH domains of the yeast proteins Ent3p and Ent5p as well as the PROPPIN domains of Svp1p protein binds to PI(3,5)P2 in vitro [36,37,70]. These proteins localize to the membrane of the yeast vacuole suggesting that PI(3,5)P2 accumulates in this compartment [36,37,70], but expression of the isolated ENTH or PROPPIN domains does not give consistent results when express in heterologous systems such as animal cells or plants (personal communication). Recently, the cytoplasmic phosphoinositide-interacting domain (ML1N) of the transient receptor potential Mucolipin 1 (TRPML1) has been described to bind PI(3,5)P2 in vitro in the nanomolecular range [38]. A 2xML1N construct was used successfully to report on the localization of PI(3,5)P2 in late endosomes and lysosomes in animal cells [38]. Yet, this new tool remains to be tested in additional cellular contexts.
PI(3,4)P2
Some PX and PH domains are binding PI(3,4)P2 in vitro (e.g. the PX domain of p47 and the PH domains of TAPP1 and TAPP2) [60,71]. Mainly, PHTAPP1 has been used as a read-out of PI(3,4)P2 in vivo and revealed that this lipid mainly accumulates at the plasma membrane [43,72].
PI(3,4,5)P3
The PH domain of AKT recognizes both PI(3,4)P2 and PI(3,4,5)P3 and has been extensively used as a read out of type I PI3-kinase activity [4,45]. Several PH domains have also been described to recognize specifically PI(3,4,5)P3 but not PI(3,4)P2, such as the PH domains from BTK, GRP1, ARNO or cytohesin1 [1,4,45,46]. PI(3,4,5)P3 does not accumulate at the plasma membrane in the absence of specific stimulus but is synthetized upon stimulation by growth factor or insulin. For example, PHBTK has been used to detect PI(3,4,5)P3 generation at the plasma membrane upon stimulation of fibroblasts by EGF or PDGF [73].
PS
PS-binding C2 domains have been characterized early on, but in many cases, lipid binding occurs only in the presence of calcium [1]. This restricted the use of these domains to study PS localization in vivo. Nonetheless, the recombinant purified C2 domain of Annexin A5 has been used to detect the presence of PS on the plasma membrane outer leaflet, but this assay requires the presence of exogenous calcium and is not compatible with live imaging of intracellular events [8]. However, the C2 domain of Lactadherin Synthase 1 (LactC2) was shown to bind specifically PS in the absence of calcium and turned out to be an excellent PS reporter in many systems, including yeast and animal cells [8,9,15,74,75]. The PH domain of EVECTIN2, a protein localized to the recycling endosomes and involved in membrane traffic, was also shown to specifically bind PS in vitro and to report PS localization in vivo in human cells [44].
PA
To date, only PA-binding linear motifs but no PA-binding domains have been found and characterized [1]. These short stretches of sequences do not seem to have a particular globular structure and are often rich in basic amino acids. As such, these PA-binding motifs are relatively poorly stereospecific and are able to bind, although with various affinities, other acidic phospholipids [1,13]. Biosensors using these PA-binding motifs rather than LBDs have been used, such as the PA-binding sequence of the yeast SNARE protein, spo20p, or the yeast protein kinase, Raf1 [76]. Because of the questionable specificity of these motifs for PA, results obtained with these probes should be cautiously interpreted. Their use has nonetheless been instrumental to address some aspects of PA localization and dynamics [76-78].
Special care and caveat of the approach
We have highlighted some of the limitations and important controls that must be carried out while analyzing results deduced from genetically encoded lipid biosensors throughout this chapter. However, there are additional potential pitfalls of this approach that should also be considered. We have already covered potential problems due to toxicity. This toxicity might arise, in part, because of competition between endogenous protein and transgenically expressed LBDs for binding the same lipid. This situation is likely to occur when the transgene is overexpressed by strong constitutive promoters and we advocate for the use of mild promoters and/or for the selection of cells or organisms that express weak-to-intermediate level of the reporters. Another strategy is to use inducible expression systems and to study the localization of the lipid sensor at the onset of expression following transgene induction. Furthermore, overexpression of LBDs might induce feedback regulation on the synthesis of the lipid, leading to over-accumulation of this lipid. Systems for mild expression, or better, inducible expression, will reduce these potential feedbacks. It is likely that this lipid over-accumulation is involved in some of the toxicity, which can be observed upon LBD overexpression, possibly by displacing endogenous proteins to new pool of lipids. In addition, it is important to keep in mind that in some cases, phosphoinositide binding LBDs are able to recognize both the membrane bound lipid and its soluble inositol phosphate counterpart, which could influence membrane association. Lastly, it is unlikely that all phosphoinositides are freely available for LBDs binding. Rather, some lipid species might be synthesized locally and readily engage interactions with endogenous lipid binding proteins as they are being synthesized. For example PI(4,5)P2 is a very important lipid involved in clathrin mediated endocytosis (CME) and several proteins involved in this process are known to binds to this lipid, yet a PHPLC reporter does not localize to clathrin coated pits (CCP) [79]. It is fully conceivable that PI(4,5)P2 in CCPs are bound by the CME machinery and therefore not labeled by the PHPLC probe.
Altogether, it is important to keep in mind that the absence of labeling by a lipid reporter is by no mean a proof of the absence of this lipid. However, the detection of a certain lipid pool by a LBD reporter, if controlled adequately (see section “validation of acidic phospholipid sensors”) is a useful tool, directly amenable to live imaging and dynamic studies.
Acknowledgement
We thank Mathilde Simon, Marie-Cécile Caillaud and Marlene Dreux for commenting the manuscript. Y.J. has received funding from the European Research Council - ERC Grant Agreement n. [3363360-APPL] and from the Marie Curie Action – CIG Grant Agreement n. [PCIG-GA-2011-303601] under the European Union's Seventh Framework Programme (FP/2007-2013).
References
- 1.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature reviews Molecular cell biology. 2008;9(2):99–111. doi: 10.1038/nrm2328. doi:10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 2.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438(7068):605–611. doi: 10.1038/nature04398. doi:10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 3.Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nature reviews Molecular cell biology. 2012;13(7):463–470. doi: 10.1038/nrm3379. doi:10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- 4.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological reviews. 2013;93(3):1019–1137. doi: 10.1152/physrev.00028.2012. doi:10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kay JG, Grinstein S. Phosphatidylserine-mediated cellular signaling. Advances in experimental medicine and biology. 2013;991:177–193. doi: 10.1007/978-94-007-6331-9_10. doi:10.1007/978-94-007-6331-9_10. [DOI] [PubMed] [Google Scholar]
- 6.Kay JG, Koivusalo M, Ma X, Wohland T, Grinstein S. Phosphatidylserine dynamics in cellular membranes. Molecular biology of the cell. 2012;23(11):2198–2212. doi: 10.1091/mbc.E11-11-0936. doi:10.1091/mbc.E11-11-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hankins HM, Baldridge RD, Xu P, Graham TR. Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic. 2015;16(1):35–47. doi: 10.1111/tra.12233. doi:10.1111/tra.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319(5860):210–213. doi: 10.1126/science.1152066. doi:10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 9.Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. The Journal of cell biology. 2011;194(2):257–275. doi: 10.1083/jcb.201012028. doi:10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nature reviews Molecular cell biology. 2008;9(2):112–124. doi: 10.1038/nrm2330. doi:10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Athenstaedt K, Daum G. Phosphatidic acid, a key intermediate in lipid metabolism. European journal of biochemistry / FEBS. 1999;266(1):1–16. doi: 10.1046/j.1432-1327.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- 12.Cazzolli R, Shemon AN, Fang MQ, Hughes WE. Phospholipid signalling through phospholipase D and phosphatidic acid. IUBMB life. 2006;58(8):457–461. doi: 10.1080/15216540600871142. doi:10.1080/15216540600871142. [DOI] [PubMed] [Google Scholar]
- 13.Horchani H, de Saint-Jean M, Barelli H, Antonny B. Interaction of the Spo20 membrane-sensor motif with phosphatidic acid and other anionic lipids, and influence of the membrane environment. PloS one. 2014;9(11):e113484. doi: 10.1371/journal.pone.0113484. doi:10.1371/journal.pone.0113484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Developmental cell. 2012;23(5):886–895. doi: 10.1016/j.devcel.2012.10.009. doi:10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Kay JG, Grinstein S. Sensing phosphatidylserine in cellular membranes. Sensors (Basel) 2011;11(2):1744–1755. doi: 10.3390/s110201744. doi:10.3390/s110201744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annual review of biophysics. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. doi:10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 17.Dall'Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Current biology : CB. 2013;23(1):R33–45. doi: 10.1016/j.cub.2012.10.041. doi:10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of cell biology. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. doi:10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) The Biochemical journal. 2009;422(1):23–35. doi: 10.1042/BJ20090428. doi:10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond GR, Machner MP, Balla T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. The Journal of cell biology. 2014;205(1):113–126. doi: 10.1083/jcb.201312072. doi:10.1083/jcb.201312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Current biology : CB. 2002;12(9):695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 22.Cruz-Garcia D, Ortega-Bellido M, Scarpa M, Villeneuve J, Jovic M, Porzner M, Balla T, Seufferlein T, Malhotra V. Recruitment of arfaptins to the trans-Golgi network by PI(4)P and their involvement in cargo export. The EMBO journal. 2013;32(12):1717–1729. doi: 10.1038/emboj.2013.116. doi:10.1038/emboj.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szentpetery Z, Varnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(18):8225–8230. doi: 10.1073/pnas.1000157107. doi:10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daboussi L, Costaguta G, Payne GS. Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nature cell biology. 2012;14(3):239–248. doi: 10.1038/ncb2427. doi:10.1038/ncb2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337(6095):727–730. doi: 10.1126/science.1222483. doi:10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stefan CJ, Manford AG, Emr SD. ER-PM connections: sites of information transfer and inter-organelle communication. Current opinion in cell biology. 2013;25(4):434–442. doi: 10.1016/j.ceb.2013.02.020. doi:10.1016/j.ceb.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Developmental cell. 2012;23(6):1129–1140. doi: 10.1016/j.devcel.2012.11.004. doi:10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144(3):389–401. doi: 10.1016/j.cell.2010.12.034. doi:10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 29.Ling Y, Stefan CJ, Macgurn JA, Audhya A, Emr SD. The dual PH domain protein Opy1 functions as a sensor and modulator of PtdIns(4,5)P(2) synthesis. The EMBO journal. 2012;31(13):2882–2894. doi: 10.1038/emboj.2012.127. doi:10.1038/emboj.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114(1):99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 31.Viaud J, Lagarrigue F, Ramel D, Allart S, Chicanne G, Ceccato L, Courilleau D, Xuereb JM, Pertz O, Payrastre B, Gaits-Iacovoni F. Phosphatidylinositol 5-phosphate regulates invasion through binding and activation of Tiam1. Nature communications. 2014;5:4080. doi: 10.1038/ncomms5080. doi:10.1038/ncomms5080. [DOI] [PubMed] [Google Scholar]
- 32.Viaud J, Boal F, Tronchere H, Gaits-Iacovoni F, Payrastre B. Phosphatidylinositol 5-phosphate: a nuclear stress lipid and a tuner of membranes and cytoskeleton dynamics. BioEssays : news and reviews in molecular, cellular and developmental biology. 2014;36(3):260–272. doi: 10.1002/bies.201300132. doi:10.1002/bies.201300132. [DOI] [PubMed] [Google Scholar]
- 33.Ramel D, Lagarrigue F, Pons V, Mounier J, Dupuis-Coronas S, Chicanne G, Sansonetti PJ, Gaits-Iacovoni F, Tronchere H, Payrastre B. Shigella flexneri infection generates the lipid PI5P to alter endocytosis and prevent termination of EGFR signaling. Science signaling. 2011;4(191):ra61. doi: 10.1126/scisignal.2001619. doi:10.1126/scisignal.2001619. [DOI] [PubMed] [Google Scholar]
- 34.Guittard G, Gerard A, Dupuis-Coronas S, Tronchere H, Mortier E, Favre C, Olive D, Zimmermann P, Payrastre B, Nunes JA. Cutting edge: Dok-1 and Dok-2 adaptor molecules are regulated by phosphatidylinositol 5-phosphate production in T cells. J Immunol. 2009;182(7):3974–3978. doi: 10.4049/jimmunol.0804172. doi:10.4049/jimmunol.0804172. [DOI] [PubMed] [Google Scholar]
- 35.Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC. PI(5)P Regulates Autophagosome Biogenesis. Molecular cell. 2015;57(2):219–234. doi: 10.1016/j.molcel.2014.12.007. doi:10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eugster A, Pecheur EI, Michel F, Winsor B, Letourneur F, Friant S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Molecular biology of the cell. 2004;15(7):3031–3041. doi: 10.1091/mbc.E03-11-0793. doi:10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friant S, Pecheur EI, Eugster A, Michel F, Lefkir Y, Nourrisson D, Letourneur F. Ent3p Is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Developmental cell. 2003;5(3):499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RG, Weisman LS, Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(52):21165–21170. doi: 10.1073/pnas.1311864110. doi:10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoncu R, Perera RM, Sebastian R, Nakatsu F, Chen H, Balla T, Ayala G, Toomre D, De Camilli PV. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(10):3793–3798. doi: 10.1073/pnas.0611733104. doi:10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon ML, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, Dreux M, Munnik T, Jaillais Y. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. The Plant journal : for cell and molecular biology. 2014;77(2):322–337. doi: 10.1111/tpj.12358. doi:10.1111/tpj.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmid SL, Mettlen M. Cell biology: Lipid switches and traffic control. Nature. 2013;499(7457):161–162. doi: 10.1038/nature12408. doi:10.1038/nature12408. [DOI] [PubMed] [Google Scholar]
- 42.Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314(5804):1454–1457. doi: 10.1126/science.1131163. doi:10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Posor Y, Eichhorn-Gruenig M, Puchkov D, Schoneberg J, Ullrich A, Lampe A, Muller R, Zarbakhsh S, Gulluni F, Hirsch E, Krauss M, Schultz C, Schmoranzer J, Noe F, Haucke V. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499(7457):233–237. doi: 10.1038/nature12360. doi:10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- 44.Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, Wakatsuki S, Misaki R, Koike M, Uchiyama Y, Iemura S, Natsume T, Kuwahara R, Nakagawa T, Nishikawa K, Mukai K, Miyoshi E, Taniguchi N, Sheff D, Lencer WI, Taguchi T, Arai H. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(38):15846–15851. doi: 10.1073/pnas.1109101108. doi:10.1073/pnas.1109101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochimica et biophysica acta. 2006;1761(8):957–967. doi: 10.1016/j.bbalip.2006.03.019. doi:10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 46.Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nature chemical biology. 2010;6(7):507–513. doi: 10.1038/nchembio.390. doi:10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143(6):966–977. doi: 10.1016/j.cell.2010.11.028. doi:10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishioka T, Frohman MA, Matsuda M, Kiyokawa E. Heterogeneity of phosphatidic acid levels and distribution at the plasma membrane in living cells as visualized by a Foster resonance energy transfer (FRET) biosensor. The Journal of biological chemistry. 2010;285(46):35979–35987. doi: 10.1074/jbc.M110.153007. doi:10.1074/jbc.M110.153007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cicchetti G, Biernacki M, Farquharson J, Allen PG. A ratiometric expressible FRET sensor for phosphoinositides displays a signal change in highly dynamic membrane structures in fibroblasts. Biochemistry. 2004;43(7):1939–1949. doi: 10.1021/bi035480w. doi:10.1021/bi035480w. [DOI] [PubMed] [Google Scholar]
- 50.van der Wal J, Habets R, Varnai P, Balla T, Jalink K. Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer. The Journal of biological chemistry. 2001;276(18):15337–15344. doi: 10.1074/jbc.M007194200. doi:10.1074/jbc.M007194200. [DOI] [PubMed] [Google Scholar]
- 51.Sato M, Ueda Y, Takagi T, Umezawa Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nature cell biology. 2003;5(11):1016–1022. doi: 10.1038/ncb1054. doi:10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- 52.Niu Y, Zhang C, Sun Z, Hong Z, Li K, Sun D, Yang Y, Tian C, Gong W, Liu JJ. PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nature cell biology. 2013;15(4):417–429. doi: 10.1038/ncb2710. doi:10.1038/ncb2710. [DOI] [PubMed] [Google Scholar]
- 53.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. The EMBO journal. 2000;19(17):4577–4588. doi: 10.1093/emboj/19.17.4577. doi:10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Molecular cell. 2004;13(5):677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 55.Toth DJ, Toth JT, Gulyas G, Balla A, Balla T, Hunyady L, Varnai P. Acute depletion of plasma membrane phosphatidylinositol 4,5-bisphosphate impairs specific steps in endocytosis of the G-protein-coupled receptor. Journal of cell science. 2012;125(Pt 9):2185–2197. doi: 10.1242/jcs.097279. doi:10.1242/jcs.097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314(5804):1458–1461. doi: 10.1126/science.1134389. doi:10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153(7):1494–1509. doi: 10.1016/j.cell.2013.05.026. doi:10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(35):E2316–2323. doi: 10.1073/pnas.1211305109. doi:10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammond GR. Membrane biology: Making light work of lipids. Current biology : CB. 2012;22(20):R869–871. doi: 10.1016/j.cub.2012.09.005. doi:10.1016/j.cub.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nature cell biology. 2001;3(7):675–678. doi: 10.1038/35083070. doi:10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 61.Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PR, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nature cell biology. 2001;3(7):679–682. doi: 10.1038/35083076. doi:10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 62.Safi A, Vandromme M, Caussanel S, Valdacci L, Baas D, Vidal M, Brun G, Schaeffer L, Goillot E. Role for the pleckstrin homology domain-containing protein CKIP-1 in phosphatidylinositol 3-kinase-regulated muscle differentiation. Molecular and cellular biology. 2004;24(3):1245–1255. doi: 10.1128/MCB.24.3.1245-1255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maffucci T, Brancaccio A, Piccolo E, Stein RC, Falasca M. Insulin induces phosphatidylinositol-3-phosphate formation through TC10 activation. The EMBO journal. 2003;22(16):4178–4189. doi: 10.1093/emboj/cdg402. doi:10.1093/emboj/cdg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roy A, Levine TP. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. The Journal of biological chemistry. 2004;279(43):44683–44689. doi: 10.1074/jbc.M401583200. doi:10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- 65.Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. The Biochemical journal. 2010;428(3):375–384. doi: 10.1042/BJ20100129. doi:10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. The Journal of cell biology. 1998;143(2):501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ben El Kadhi K, Roubinet C, Solinet S, Emery G, Carreno S. The inositol 5-phosphatase dOCRL controls PI(4,5)P2 homeostasis and is necessary for cytokinesis. Current biology : CB. 2011;21(12):1074–1079. doi: 10.1016/j.cub.2011.05.030. doi:10.1016/j.cub.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 68.Szentpetery Z, Balla A, Kim YJ, Lemmon MA, Balla T. Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC cell biology. 2009;10:67. doi: 10.1186/1471-2121-10-67. doi:10.1186/1471-2121-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292(5524):2041–2050. doi: 10.1126/science.1061233. doi:10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 70.Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, Parker PJ, Lemmon MA. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. The EMBO journal. 2004;23(9):1922–1933. doi: 10.1038/sj.emboj.7600203. doi:10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. The Biochemical journal. 2000;351(Pt 1):19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marshall AJ, Krahn AK, Ma K, Duronio V, Hou S. TAPP1 and TAPP2 are targets of phosphatidylinositol 3-kinase signaling in B cells: sustained plasma membrane recruitment triggered by the B-cell antigen receptor. Molecular and cellular biology. 2002;22(15):5479–5491. doi: 10.1128/MCB.22.15.5479-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. The Journal of biological chemistry. 1999;274(16):10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- 74.Fairn GD, Hermansson M, Somerharju P, Grinstein S. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nature cell biology. 2011;13(12):1424–1430. doi: 10.1038/ncb2351. doi:10.1038/ncb2351. [DOI] [PubMed] [Google Scholar]
- 75.Yeung T, Heit B, Dubuisson JF, Fairn GD, Chiu B, Inman R, Kapus A, Swanson M, Grinstein S. Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. The Journal of cell biology. 2009;185(5):917–928. doi: 10.1083/jcb.200903020. doi:10.1083/jcb.200903020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kassas N, Tryoen-Toth P, Corrotte M, Thahouly T, Bader MF, Grant NJ, Vitale N. Genetically encoded probes for phosphatidic acid. Methods in cell biology. 2012;108:445–459. doi: 10.1016/B978-0-12-386487-1.00020-1. doi:10.1016/B978-0-12-386487-1.00020-1. [DOI] [PubMed] [Google Scholar]
- 77.Zhang F, Wang Z, Lu M, Yonekubo Y, Liang X, Zhang Y, Wu P, Zhou Y, Grinstein S, Hancock JF, Du G. Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Molecular and cellular biology. 2014;34(1):84–95. doi: 10.1128/MCB.00987-13. doi:10.1128/MCB.00987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bohdanowicz M, Schlam D, Hermansson M, Rizzuti D, Fairn GD, Ueyama T, Somerharju P, Du G, Grinstein S. Phosphatidic acid is required for the constitutive ruffling and macropinocytosis of phagocytes. Molecular biology of the cell. 2013;24(11):1700–1712. doi: 10.1091/mbc.E12-11-0789. doi:10.1091/mbc.E12-11-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Antonescu CN, Aguet F, Danuser G, Schmid SL. Phosphatidylinositol-(4,5)-bisphosphate regulates clathrin-coated pit initiation, stabilization, and size. Molecular biology of the cell. 2011;22(14):2588–2600. doi: 10.1091/mbc.E11-04-0362. doi:10.1091/mbc.E11-04-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]