Gene expression is regulated by complex coordinated processes, including chromatin remodeling, post-translational modifications of histones, and incorporation of non-allelic histone variants. The histone variant H2AZ constitutes only a few percents of the total H2A cellular pool (Ball et al., 1983; West and Bonner, 1980). However, H2AZ is essential in several multi-cellular organisms (Faast et al., 2001; Ren and Gorovsky, 2001; Ridgway et al., 2004; van Daal and Elgin, 1992) and required for normal proliferation in Schizosaccharomyces pombe and Saccharomyces cerevisiae (Carr et al., 1994; Santisteban et al., 2000). In mammalian cells, H2AZ is involved in embryonic stem (ES) cell biology (Faast et al., 2001; Creyghton et al., 2008). Recent reports define the importance of H2AZ and H2AZ-modifying enzymes in self-renewal of ES cells (Binda et al., 2013; Hu et al., 2013; Li et al., 2012). In addition, H2AZ is post-translationally modified by acetylation, SUMOylation, ubiquitination, and methylation of lysines (Binda et al., 2013). Herein, we summarize our perspective on the emergent role of H2AZ in the biology of ES cells, while bringing a particular emphasis on the functions of its post-translational modifications.
H2AZ acetylation
Histone acetylation and deacetylation constitute one of the most studied mechanisms of gene regulation. These reactions occur on the lysines at the amino terminal tail of histones and on the surface of the nucleosome core. In addition, the amino terminal tail of H2AZ is extensively modified by lysine acetylation (Fig. 1). More precisely, H2AZ is acetylated on lysines K4, K7, K11 (Bonenfant et al., 2007; Boyne et al., 2006; Dryhurst et al., 2009), and K13 (Bruce et al., 2005; Ishibashi et al., 2009; Valdés-Mora et al., 2012). In Saccharomyces cerevisiae, the histone acetyltransferases (HAT) ESA1 and GCN5 mediate the acetylation of Htz1 (yeast ortholog of H2AZ) (Keogh et al., 2006; Mehta et al., 2010; Millar et al., 2006) at lysine 14 (Millar et al., 2006). In mammalians, the ortholog of ESA1 is TIP60, a subunit of the NuA4 complex (Doyon and Côté, 2004).
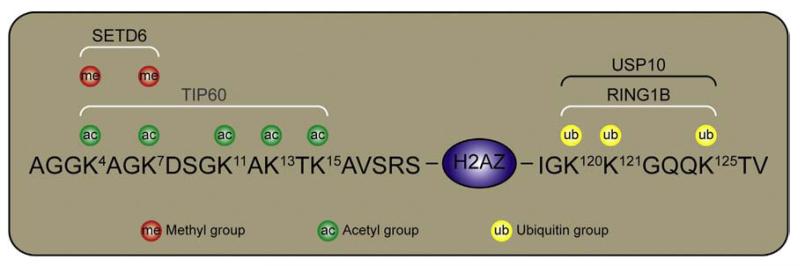
Figure 1.
Post-translational modifications of H2AZ lysines. H2AZ is methylated on lysines 4 and 7 by the methyltransferase SETD6 (Binda et al., 2013). H2AZ lysines 4, 7, 11, and 13 are acetylated, hypothetically by the acetyltransferase TIP60. Lysines 120, 121, and 125 of H2AZ are ubiquitinated by the E3 ubiquitin ligase RING1B (Sarcinella et al., 2007). H2AZub1 is deubiquitinated by the ubiquitin-specific peptidase USP10 (Draker et al., 2011).
TIP60 acetylates the canonical histones H4 and H2A, which is a prerequisite for the following incorporation of H2AZ into nucleosomes by the ATP-dependent SWR1/p400 histone exchange complex (Altaf et al., 2010). It is also known that silencing of TIP60 induces a reduction in acetylation of H2AZ (H2AZac) at the transcriptional start site and enhancer of CCND1 (Dalvai et al., 2013), suggesting that TIP60 may also be involved in the acetylation of H2AZ. Moreover, experimental evidence suggests that H2AZ is acetylated subsequently to its inclusion within nucleosomes (Keogh et al., 2006; Millar et al., 2006). Although TIP60 associates with H2AZ (Li et al., 2012), suggesting that TIP60 may acetylate H2AZ, more direct evidence, such as in vitro HAT assays using purified acetyltransferases, would be required to establish which enzymatic activities acetylate H2AZ.
Genome-wide surveys of H2AZ and H2AZac reveal that the acetylated form associates with transcriptionally active promoters in the LNCaP and PrEC human prostate cell lines (Valdés-Mora et al., 2012). Additionally, H2AZac is detected at euchromatin in the course of mouse pre-implantation development, but absent and undetectable upon embryonic genome activation (2-cell embryonic stage) (Bošković et al., 2012). This observation indicates that H2AZ is presumably involved in different gene expression programs at distinctive phases of embryonic development.
Interestingly, the dual bromodomain protein BRD2 associates with H2AZ-containing nucleosomes (Draker et al., 2012). Bromodomains are generally facilitating acetyl-lysine-dependent protein–protein interactions and the association of BRD2 with H2AZ-nucleosomes is dependent on the bromodomains and is enhanced by lysine acetylation (Draker et al., 2012). Competition assays with acetylated histone peptides suggest that H4K12ac is facilitating the association of BRD2 with H2AZ-nucleosomes (Draker et al., 2012). Moreover, H2AZ-nucleosomes contain more acetylated H3 and H4 than H2A-nucleosomes. Although H2AZ acetylation does not appear to be involved in the association with BRD2, the sequence of the amino terminal tail of the variant is somehow involved in the interaction. Precisely, BRD2 preferentially associates with H2AZ variant 1 (H2AFZ) over variant 2 (H2AFV), which slightly differ at residues 14 and 38. The threonine 14 in H2AFZ is an alanine in H2AFV and serine 38 in H2AFZ is a threonine in H2AFV. It would be interesting to investigate the acetylation of K13 in H2AFZ versus H2AFV and the association with BRD2.
H2AZ lysine methylation
The lysine methyltransferases SETD6 and SET7 efficiently methylate H2AZ in vitro (Binda et al., 2013). In particular, a detailed study clearly identified lysine 7 as the preferred site of methylation by SETD6 on H2AZ. This result was confirmed by mass spectrometric analysis and it also specified that H2AZ is monomethylated at lysine 7 (H2AZK7me1) (Binda et al., 2013). Interestingly, the amino terminal tail of H2AZ is monomethylated on both lysines 4 and 7 (H2AZK4me1K7me1) in vivo (Binda et al., 2013). Strikingly, upon induction of cellular differentiation using retinoic acid (RA), global levels of H2AZK4me1K7me1 increased by several folds in E14 mouse ES (mES) cells (Binda et al., 2013). However, the levels of SETD6 appear to lessen following the RA treatment (Binda et al., 2013), suggesting that perhaps SETD6 enzymatic activity is increased despite the lower amounts of SETD6 transcripts. SETD6 is ubiquitinated at the carboxy terminus within the substrate binding domain at lysine 441 (Kim et al., 2011). Therefore, ubiquitination of SETD6 could hypothetically regulate its methyltransferase activity or substrate specificity. Alternatively, SET7, or other KMTs, could also be responsible for the increased H2AZ methylation in differentiating E14 mES cells. Interestingly, the silencing of SETD6 in mES cells resulted in cellular differentiation, compromised self-renewal, and showed poor clonogenicity capacity (Binda et al., 2013). Since the methylation of H2AZK4 and H2AZK7 blocks the acetylation at those sites and the H2AZK4me1K7me1 and H2AZac marks are mutually exclusive (Binda et al., 2013), we propose that H2AZK4me1K7me1 is a transcriptional silencing mark. Indeed, upon RA treatment in E14 mES cells, H2AZK7me1 is removed along the silencing mark H3K27me3 from the promoters of Fgf5, FoxA2, and Hand2 (Binda et al., 2013), further suggesting that H2AZK7me1 is marking chromatin for silencing. Finally, although H2AZ and H2AZK7me1 occupy some promoters where H3K27me3 is present (Binda et al., 2013), the H2AZac and H3K27me3 marks do not overlap (Valdés-Mora et al., 2012), again supporting that H2AZK7me1 is involved in negative regulation of gene expression.
H2AZ ubiquitination
As essential components of chromatin, histones present many covalent modifications. Besides acetylation and methylation, histones can also be ubiquitinated. Interestingly, H2A was actually the first ubiquitinated protein identified (Goldknopf and Busch, 1977). Reminiscent of the canonical histone H2A, which is ubiquitinated on lysine 119 (H2AK119ub) (Cao et al., 2005; Wang et al., 2004), its variant, H2AZ, is ubiquitinated at lysine 121 (H2AZK121ub) (Draker et al., 2011; Sarcinella et al., 2007), as well as K120 and K125 (Sarcinella et al., 2007), but always occurs as a mono-ubiquitinated H2AZub1 form (Sarcinella et al., 2007) (Fig. 1). Interestingly, methyl-specific antibodies directed against H2AZK7me1 detect a slow-migrating form of H2AZ that, based on its molecular weight, may potentially be ubiquitinated (Binda et al., 2013), suggesting the coexistence of H2AZK7me1 and H2AZub1.
H2A is ubiquitinated by the polycomb repressive complex 1 (PRC1) subunit RING1B (Cao et al., 2005), an E3 ubiquitin ligase that is also reported to modify H2AZ at lysines 120, 121, and 125 (Sarcinella et al., 2007). The H2AZub1 mark is removed by the ubiquitin-specific peptidase USP10 (Draker et al., 2011). Cellular localisation experiments highlight that H2AZub1 is found at transcriptionally silent facultative heterochromatin on the inactive X chromosome (Sarcinella et al., 2007), suggesting that H2AZub1 is a silencing mark. Furthermore, the silencing of USP10 results in elevated levels of H2AZub1, while loss of H2AZub1 at KLK3 and PSA promoters correlates with transcriptional activation (Draker et al., 2011).
H2AZ SUMOylation
SUMO (Small Ubiquitin-like MOdifier) proteins are similar to ubiquitin and are covalently linked to lysines. In contrast to ubiquitin, SUMO does not mark proteins for proteosomal degradation. SUMOylation is a post-translational modification involved in diverse biological processes, including stress response, protein stability, cell cycle progression, apoptosis, nuclear-cytoplasm transport, and transcriptional control.
In Saccharomyces cerevisiae, the SUMOylation of Htz1 on lysines 126 and 133 plays an important role in DNA double-stranded-break by relocating the unrepaired chromosomal break to the nuclear periphery (Kalocsay et al., 2009). The very carboxy terminus of H2AZ is not well conserved with Htz1 and does not harbor the ψKxE SUMOylation motif found at Htz1K126. Interestingly, the SUMOylation of Htz1 does not happen to be conserved in mammals, whereas the ubiquitination of H2AZ appears to occur solely in mammals, suggesting that some post-translational modifications of H2AZ were lost while others were gained during evolution.
H2AZ histone exchange
H2AZ is incorporated within nucleosomes via ATP-dependent activities. The complexes mediating H2AZ-H2A exchanges are well characterized in Saccharomyces cerevisiae and include the Swr1-containing SWR complex (SWR-C). The mammalian orthologue of Swr1 is SRCAP. Both complexes share several evolutionarily conserved subunits. Purified SRCAP complex catalyses the ATP-dependent exchange of H2AZ/H2B and H2A/H2B dimers. This reaction leads to the loading of H2AZ into nucleosomes (Ruhl et al., 2006). The TIP60 complex contains another Swr1 orthologue, p400, which is found in the TIP60 complex. The Drosophila melanogaster Tip60 complex can exchange H2AZ and the complex seems to be conserved in mammals. Indeed, both the SRCAP and TIP60 complexes drive the exchange and deposition of H2AZ (Choi et al., 2009). However, the two complexes appear to operate through distinct mechanisms whereby H2A acetylation by TIP60 facilitates the exchange of H2AZ. Interestingly, the ATPases TIP48 and TIP49 on their own (in the absence of Swr1 orthologues SRCAP and p400) can mediate the exchange of H2AZ in vitro (Choi et al., 2009).
The histone acetyltransferase TIP60 modifies H2A, which enhances the removal of H2A and its replacement by H2AZ (Choi et al., 2009). The TIP48 and TIP49 subunits of the TIP60 complex are important for the integrity of the complex and TIP60 enzymatic activity. Furthermore, TIP48 is required for the estradiol-induced transcription of CCND1. In the absence of TIP48, the level of H2AZ at the transcriptional start site and enhancer is reduced. Consequently, the estrogen receptor ERα fails to bind DNA. Recent studies in yeasts also show the importance of histone acetylation in the exchange of H2AZ. Specifically, the acetylation of histone H3 at lysine 56 (H3K56ac) facilitates the removal of Htz1 from nucleosomes by SWR-C (Watanabe et al., 2013). Yeast strains harboring either a deletion of Htz1 (htz1Δ) or an acetyl-mimic of H3K56ac (H3K56Q) showed an overlapping of misregulated genes and loss of H2AZ at H2AZ-rich promoters (Watanabe et al., 2013).
Histone marks cross-talk
An additional level of complexity is conferred by the communication between different post-translational modifications and their interactions with other chromatin components.
For example, the SWR study conducted by Craig L. Peterson and colleagues clearly shows a cross-talk between H3K56ac and the eviction of H2AZ from nucleosomes (Watanabe et al., 2013). Their in vitro studies show that the exchange activity of SWR-C, but not the INO80 complex, was enhanced by H3K56ac (Watanabe et al., 2013). Furthermore, the genetic studies in yeast correlate the level of H3K56ac with Htz1 eviction (Watanabe et al., 2013).
The exchange of H2AZ is also regulated by the acetylation of the core histone H2A at lysine 5 (H2AK5ac). The pre acetylation of in vitro reconstituted nucleosomes using TIP60-containing H2AZ complex enhanced the incorporation of H2AZ by TIP48/TIP49 (Choi et al., 2009). In agreement with the role of TIP48/TIP49 in H2AZ exchange, the silencing of TIP49 results in decreased H2AZ levels at the p53 response element in the promoter of p21 (Choi et al., 2009).
Our laboratory observed that an H2AZK7me1-specific antibody recognized a slow migrating H2AZ species that could potentially be a ubiquitinated form (Binda et al., 2013). Immunoprecipitation of the FLAG-tagged H2AZK7R mutant followed by immuno-detection with an H2AZK7me1-specific antibody revealed that the slow-migrating form of H2AZ disappeared, suggesting cross-talk in cis between H2AZK7me1 and H2AZub (unpublished data). What still remains elusive is whether H2AZK7me1 or H2AZK4me1K7me1 blocks the interaction between H2AZ and RING1B or increases the affinity of USP10.
H2AZ embryonic stem cell biology
The histone variant H2AZ has emerged as a key regulator of chromatin signalling and plays an essential, but unknown role during mammalian development. Deletion of H2AZ in mice results in severe early developmental defects (Faast et al., 2001), suggesting a critical function for H2AZ in embryonic development, cellular differentiation, or cell fate decisions. Interestingly, H2AZ was recently found to facilitate the association of transcription factors with chromatin in mES cells (Hu et al., 2013) and be required for normal mES cell biology (Li et al., 2012). Complementarily, global lysine methylation levels of H2AZ on lysine 7 (H2AZK7me1) increased significantly in differentiating E14 mES cells (Binda et al., 2013). Moreover, the lysine methyltransferase SETD6 methylates H2AZ and is required for self-renewal of E14 mES cells (Binda et al., 2013). In addition, the silencing of H2AZ expression in CMTI-1 mES cells impaired self-renewal (Hu et al., 2013). However, the shRNA used for the depletion of H2AZ, which led to mES cell differentiation defects, also silenced the core histone H2A (Hu et al., 2013). Both histones are part of the same family and the results undeniably highlight the importance of the H2A family in the biology of mES cells. The Polycomb Repressive Complex 1 (PRC1) has ubiquitin ligase activity specific to H2A lysine 119 (Cao et al., 2005; Wang et al., 2004). Specifically, the E3 ubiquitin ligase activity of the PRC1 complex subunit RING1B on H2A is enhanced by the polycomb ring finger oncoprotein BMI1 (Cao et al., 2005). The catalysis of H2AK119ub by RING1B leads to transcriptional silencing of Hox genes (Cao et al., 2005; Wang et al., 2004). Furthermore, the silencing of the Jumonji demethylase UTX, which catalyses the removal of H3K27me3, led to increased H3K27me2/me3 at HOXA13 and HOXC4 loci in the human embryonic carcinoma NTERA2 cell line (Lee et al., 2007). Concomitantly with the H3K27me2/me3 increase, the recruitment of the PRC1 complex subunits BMI1 and RING1A was facilitated and H2AK119ub enhanced (Lee et al., 2007). Therefore, the loss of H2A expression in H2AZ-depleted mES cells could be a major contributor to self-renewal and pluripotency defects. Finally, previous attempts at silencing the expression of H2AZ in E14 mES cells failed to induce any phenotype, while several components of the TIP60/p400 histone exchange complex are required for the maintenance of mES cells identity (Fazzio et al., 2008). However, siRNA-mediated silencing of H2AZ impaired the differentiation of E14 mES cells towards endoderm hepatic progenitors (EHP) induced by activin A, Wnt3A, and DKK1 (Li et al., 2012). These inconsistencies in the requirement of H2AZ in mES cells biology may stem from the diverse cellular differentiation systems employed (none, RA, or EHP) and the various cell types used (CMTI-1, E14, or NTERA2). We hypothesize that these discrepancies quite possibly highlight distinct requirements for H2AZ during differentiation and in lineage decision.
Although H2AZK7me1 could be detected by ChIP at the transcriptional start site of a few differentiation markers (Fgf5, FoxA2, and Hand2) in E14 mES cells and the level of methylation decreased from those genes in response to retinoic acid-induced differentiation, we still do not know what H2AZK7me1 does. The decrease in H2AZK7me1 is accompanied by a loss of H3K27me3, suggesting that the two marks play a similar function. Do H2AZK7me1 and H2AZK4me1K7me1 facilitate an interaction between H2AZ and a histone mark reader? Do the methylation of H2AZ merely block the acetylation of the lysines?
H2AZ controls gene expression
Eukaryotic gene expression is highly conditioned by the local chromatin environment. The incorporation of histone variants, post-translational modifications of histones, and methylation of DNA determinate the accessibility of transcription factors to regulatory elements. In Saccharomyces cerevisiae, about 5% of the genes are misregulated in Htz1-null cells (Meneghini et al., 2003). In addition, Htz1 is incorporated at the chromatin of repressed promoters of inducible genes and removed upon activation of transcription (Santisteban et al., 2000). These early observations suggest an important role for H2AZ in the regulation of gene expression.
In AB1 mES cells, it was shown that the retinoic acid receptor RARγ is essential for the inclusion of H2AZ at RARγ-bound nucleosomes (Amat and Gudas, 2011). Interestingly, Hu et al. further demonstrate that RARα-bound genomic loci become H2AZ-enriched upon RA stimulation, and that H2AZ facilitates the association of the H3K4-specific methyltransferase MLL complex subunit RBBP5 and the PRC2 subunit SUZ12 with the chromatin (Hu et al., 2013). Specifically, RBBP5 was enriched at H2AZac- and H3K4me3-containing chromatin, while SUZ12 was mainly associated with H3K27me3 regions that harbor low H2AZac (Hu et al., 2013). These results are coherent with previous reports showing that H2AZ occupies PRC2-bound regions (Creyghton et al., 2008; Illingworth et al., 2012). Consistent with other reports showing that H2AZ is incorporated at chromatin bound by transcription factors such as ERα, MYC, and p53 (Gévry et al., 2007, 2009), H2AZ is suggested to facilitate the access of transcriptional machinery at the chromatin (Hu et al., 2013).
The expression of the cell cycle inhibitor p21 is regulated by an interplay between H2AZ, p53, and MYC at the chromatin level (Gévry et al., 2007). In p53+/+ cells, H2AZ is depleted from p21 upon DNA damage-induced transcriptional activation. Interestingly, the depletion of H2AZ by RNA interference does not induce the expression of p21 in p53−/− cells. Remarkably, inhibition of histone deacetylase activity with trichostatin A induces the expression of p21. This is accompanied by reduced level of H2AZ, but increased H2AZac level and recruitment of RNA polII at the promoter of p21 (Bellucci et al., 2013).
The increased H2AZ occupancy at TFF1 in response to estradiol-induced transcriptional activation observed by Gévry and colleagues (Gévry et al., 2009) was not observed at CCND1 (Dalvai et al., 2013). On the contrary, H2AZ level decreased from the transcriptional start site and downstream enhancer of CCND1 in response to estradiol and the recruitment of ERα (Dalvai et al., 2013). It thus appears that the mechanism of estradiol-induced transcriptional activation is distinctive from gene to gene and, although H2AZ is required for effective transcriptional activation, the histone variant plays different roles. Moreover, at some genes, H2AZ level increases (RARα- and ERα-regulated genes), whereas at other genes, H2AZ level decreases (p21 and CCND1) upon induction of gene expression. The recent studies on the regulation of p21 and CCND1 gene expression (Bellucci et al., 2013; Dalvai et al., 2013) suggest that H2AZac level, not H2AZ itself, seems to be a key component in gene activation.
Generally, histone modifications facilitate protein–protein interactions with effector proteins or readers. Bromodomains interact with acetyl-lysines, while chromodomains, plant homeodomains (PHD), and others associate with methyllysines. Interestingly, H2AZ-containing nucleosomes associate with several potential readers, including the chromodomain protein CHD3 (MI-2) (Fujimoto et al., 2012) and the bromodomain protein BRD2 (Draker et al., 2012).
H2AZ genome-wide
Genome-wide localisation experiments in protozoa, fungi, animals, and plants (Creyghton et al., 2008; Albert et al., 2007; Guillemette et al., 2005; Li et al., 2005; Petter et al., 2011; Raisner et al., 2005; Siegel et al., 2009; Whittle et al., 2008; Zilberman et al., 2008) demonstrate that H2AZ is highly enriched within the few nucleosome surrounding transcriptional start sites (TSS). In agreement with Valdés-Mora et al., who showed that H2AZac is solely found at the TSS of actively transcribed genes (Valdés-Mora et al., 2012), H2AZac is distributed at H3K27me3-depleted promoters (Hu et al., 2013). In addition, the level of H2AZac also correlates with levels of gene expression in v6.5 mES cells (Ku et al., 2012). Genome-wide ChIPseq surveys of H2AZ-containing nucleosomes in human and mouse ES cells show that H2AZ is enriched at promoters, enhancers, and intergenic regions marked by H3K4me3, as well as bivalent promoters marked by H3K27me3 and H3K4me3 (Ku et al., 2012), in agreement with previous studies (Creyghton et al., 2008; Illingworth et al., 2012). Interestingly, in v6.5 mES cells the H3K4me3/H3K27me3 bivalent domains contain the H2AZac/ub dual mark (Ku et al., 2012). Remarkably, H2AZub is portrayed as a silencing mark (Draker et al., 2011; Sarcinella et al., 2007), while H2AZac is associated with transcriptional activity (Bruce et al., 2005; Valdés-Mora et al., 2012), suggesting that bivalent domains are marked by antagonistic modifications on both H3 (H3K4me3 versus H3K27me3) and H2AZ (H2AZac versus H2AZub1).
H2AZ at enhancers
Enhancer regions underlay regulatory processes by which cells establish patterns of gene expression. In particular, the H2AZ and H3K4me1 marks are deposited at enhancer regions throughout the genome. The H2AZ mark has been used to identify tens of thousands of enhancers in hematopoietic stem and progenitor (HSP) cells and differentiated cell types. Remarkably, H2AZ was used to identify functional enhancers specific for distinct hematopoietic cell types. Specifically, in HSP cells, H2AZ was incorporated at specific enhancers with H3K4me1, but in differentiated B or T cells, the H2AZ and H3K4me1 marks were replaced by H3K27me3.
Genome-wide, H2AZ is found at H3K4me3-marked enhancers in both human and mouse ES cells (Ku et al., 2012). At the gene level, H2AZ is found at the downstream enhancer of CCND1 and is evicted upon estradiol-induced gene expression (Dalvai et al., 2013). Chromatin conformation capture (3C) studies at the CCDN1 locus reveal that the downstream enhancer interacts with the TSS and that these interactions are lost upon activation of transcription (Dalvai et al., 2013). In the absence of TIP48, the enhancer and TSS still interact, but upon estradiol induction of transcription, the two regions retain high level of interactions (Dalvai et al., 2013), suggesting that TIP48 is required for chromatin remodelling and disrupting the interactions between the enhancer and the TSS of CCND1 upon gene activation.
Concluding remark
The current emerging picture is that similarly to canonical histones, H2AZ biological functions are regulated by post-translational modifications. It is thus tempting to speculate that H2AZac, H2AZK7me1, H2AZK4me1K7me1, and H2AZK121ub mediate protein–protein interactions with as yet unidentified histone mark readers to convey chromatin signaling.
Acknowledgments
OB is supported by the Newcastle's Biomedical Fellowship Programme, which is in part funded through the Wellcome Trust's Institutional Strategic Support Fund.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, Côté J. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amat R, Gudas LJ. RARγ is required for correct deposition and removal of Suz12 and H2A.Z in embryonic stem cells. J. Cell. Physiol. 2011;226:293–298. doi: 10.1002/jcp.22420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball DJ, Slaughter CA, Hensley P, Garrard WT. Amino acid sequence of the N-terminal domain of calf thymus histone H2A.Z. FEBS Lett. 1983;154:166–170. doi: 10.1016/0014-5793(83)80896-5. [DOI] [PubMed] [Google Scholar]
- Bellucci L, Dalvai M, Kocanova S, Moutahir F, Bystricky K. Activation of p21 by HDAC inhibitors requires acetylation of H2A.Z. PLoS One. 2013;8:e54102. doi: 10.1371/journal.pone.0054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda O, Sevilla A, LeRoy G, Lemischka IR, Garcia BA, Richard S. SETD6 mono-methylates H2AZ on lysine 7 and is required for the maintenance of embryonic stem cell self-renewal. Epigenetics. 2013;8:177–183. doi: 10.4161/epi.23416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell. Proteomics. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- Bošković A, Bender A, Gall L, Ziegler-Birling C, Beaujean N, Torres-Padilla M-E. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics. 2012;7:747–757. doi: 10.4161/epi.20584. [DOI] [PubMed] [Google Scholar]
- Boyne MT, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Tsukada Y-I, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Carr AM, Dorrington SM, Hindley J, Phear GA, Aves SJ, Nurse P. Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol. Gen. Genet. 1994;245:628–635. doi: 10.1007/BF00282226. [DOI] [PubMed] [Google Scholar]
- Choi J, Heo K, An W. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 2009;37:5993–6007. doi: 10.1093/nar/gkp660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvai M, Bellucci L, Fleury L, Lavigne AC, Moutahir F, Bystricky K. H2A.Z-dependent crosstalk between enhancer and promoter regulates cyclin D1 expression. Oncogene. 2013;32:4243–4251. doi: 10.1038/onc.2012.442. [DOI] [PubMed] [Google Scholar]
- Dalvai M, Fleury L, Bellucci L, Kocanova S, Bystricky K. TIP48/Reptin and H2A.Z requirement for initiating chromatin remodeling in estrogen-activated transcription. PLoS Genet. 2013;9:e1003387. doi: 10.1371/journal.pgen.1003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Draker R, Sarcinella E, Cheung P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 2011;39:3529–3542. doi: 10.1093/nar/gkq1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8:e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryhurst D, Ishibashi T, Rose KL, Eirín-López JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, Hunt DF, Ausió J. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi: 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Seebart C, Guastafierro T, Prenni J, Caiafa P, Zlatanova J. Proteome analysis of protein partners to nucleosomes containing canonical H2A or the variant histones H2A.Z or H2A.X. Biol. Chem. 2012;393:47–61. doi: 10.1515/BC-2011-216. [DOI] [PubMed] [Google Scholar]
- Gévry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévry N, Hardy S, Jacques P-É, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf IL, Busch H. Isopeptide linkage between nonhis-tone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. U. S. A. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Botting CH, Grimes GR, Bickmore WA, Eskeland R. PRC1 and PRC2 are not required for targeting of H2A.Z to developmental genes in embryonic stem cells. PLoS One. 2012;7:e34848. doi: 10.1371/journal.pone.0034848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Dryhurst D, Rose KL, Shabanowitz J, Hunt DF, Ausió J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry. 2009;48:5007–5017. doi: 10.1021/bi900196c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Keogh M-C, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, Bernstein BE. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 2012;13:R85. doi: 10.1186/gb-2012-13-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan K-P, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta M, Braberg H, Wang S, Lozsa A, Shales M, Solache A, Krogan NJ, Keogh M-C. Individual lysine acetylations on the N terminus of Saccharomyces cerevisiae H2A.Z are highly but not differentially regulated. J. Biol. Chem. 2010;285:39855–39865. doi: 10.1074/jbc.M110.185967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter M, Lee CC, Byrne TJ, Boysen KE, Volz J, Ralph SA, Cowman AF, Brown GV, Duffy MF. Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter. PLoS Pathog. 2011;7:e1001292. doi: 10.1371/journal.ppat.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Ridgway P, Brown KD, Rangasamy D, Svensson U, Tremethick DJ. Unique residues on the H2A.Z containing nucleosome surface are important for Xenopus laevis development. J. Biol. Chem. 2004;279:43815–43820. doi: 10.1074/jbc.M408409200. [DOI] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Sarcinella E, Zuzarte PC, Lau PNI, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GAM. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 2012;22:307–321. doi: 10.1101/gr.118919.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A, Elgin SC. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell. 1992;3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013;340:195–199. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MHP, Bonner WM. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980;19:3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]