Abstract
CD4+ regulatory T cells (Tregs) are essential for controlling immune responses and preventing autoimmunity. Their development requires regulation of gene expression by microRNAs (miRNAs). In order to understand miRNA function in Treg development, we searched for important miRNAs and their relevant target genes. Of the more abundantly expressed miRNAs in Tregs, only miR-15b/16, miR-24, and miR-29a impacted the in vitro induction of Tregs (iTregs) in overexpression and blocking experiments. miRNA mimics for these significantly enhanced the induction of iTregs in Dicer−/− CD4+ T cells. Furthermore, the overexpression of miR-15b/16 in conventional CD4+ T cells adoptively transferred into Rag2−/− mice increased the in vivo development of peripheral Tregs and diminished the severity of autoimmune colitis. In searching for targets of miR-15b/16, we observed that the mammalian target of rapamycin (mTOR) signaling pathway was enhanced in Dicer−/− CD4+ T cells, and its pharmacological inhibition restored induction of iTregs. Suppression of mTOR signaling is essential for induction of iTregs from naïve CD4+ T cells, and the mTORC2 component, Rictor contained a functional target site for miR-15b/16. Rictor was more abundantly expressed in Dicer−/− T cells as was mTOR, and their expression was downregulated by the overexpression of miR-15b/16. This led to a reduction in mTOR signaling as measured by phosphorylation of the downstream target, ribosomal protein S6. Finally, knockdown of Rictor by siRNAs enhanced Treg induction in Dicer−/− CD4+ T cells. Therefore, an important mechanism of miRNA regulation of Treg development is through regulation of the mTOR-signaling pathway.
INTRODUCTION
Regulation of the immune response and the prevention of autoimmunity require a class of helper T (Th) cells called Regulatory T cells (Tregs). Because of their essential function in maintaining peripheral tolerance, they play important roles in many immune-related disorders. Therefore, a significant interest exists in understanding their development and function in order to develop new therapeutic strategies for diseases involving the immune response (1).
The majority of circulating Tregs are produced in the thymus (tTregs) at the double positive stage. Current thought is that they enter the Treg lineage because of an intermediate affinity of their T cell receptor to self-antigens, which is above that normally required for positive but not sufficient for negative selection. This is hypothesized to induce a suppressor phenotype that protects the body against any subsequent immune response that might develop against self-antigens (1). Tregs can also develop in the periphery (pTregs) from naïve T CD4+ T cells when activated under the influence of specific factors (such as TGF-β and retinoic acid) produced by antigen-presenting and other cell types present at the site of antigen challenge. Both thymus and peripherally derived Tregs are important in preventing autoimmune disease (2, 3). Each is defined by the expression of the Treg specific transcription factor Foxp3, which plays an essential role in regulating the gene expression profile required for Treg development and function. Loss of Foxp3 results in the absence of Tregs and subsequent lethal autoimmunity (4).
MicroRNAs (miRNAs) are critical regulators of Treg development and function. These are small double-stranded RNAs of approximately 22 nucleotides in length that negatively regulate gene expression at a post-transcriptional stage (5). miRNAs are encoded in the genome within pol II transcripts. Their critical feature is a stem loop structure made up of the complementary strands of the miRNA, which is recognized and cleaved by protein complexes containing the RNases Drosha and Dicer to give the mature miRNA. This is incorporated into an effector complex called the RNA-induced silencing complex (RISC). Here, one strand is degraded, and the other can then target the complex to messages through imperfect base pairing, which typically occurs in the 3′-untranslated region (UTR) of the gene. This results in inhibition of translation and the subsequent destabilization of the message (6). The loss of miRNAs at the double positive stage of T cell development through a conditional knockout of Drosha or Dicer significantly inhibits the development of Tregs and ultimately leads to autoimmunity (7, 8). In addition, a deletion of either RNase specifically in Tregs disrupts their suppressor function and results in an acute autoimmune response that is as severe as the loss of Foxp3 (8-10). Therefore, miRNAs are important both in the steps leading up to commitment towards the Treg lineage and also downstream in mediating their suppressor function. Several studies have examined the roles of miRNAs in Treg function and have identified individual miRNAs that regulate key genes required for the function and stability of Tregs (11-17). However, it is still unknown how miRNAs regulate the expression of genes involved in the developmental steps leading to the Treg lineage.
We set about to examine miRNA function in Treg development by first determining functional miRNAs, then identifying critical genes they regulate. We found three miRNAs (miR-15b/16, miR-24, and miR-29a) that regulated the induction of Tregs from naïve CD4+ T cells with miR-15b/16 having the greatest effect in overexpression and blocking experiments. Important genes regulated by miR-15b/16 were Rictor and Mtor, which encode components of the mammalian target of rapamycin (mTOR) signaling pathway. Downregulation of the mTOR signaling pathway is important for the induction of Tregs (18-20). Therefore, miR-15b/16 plays an important role in regulating the mTOR signaling pathway and controlling the development of Tregs.
MATERIALS AND METHODS
Mouse strains, antibodies, and plasmids
C57BL/6 (Charles River, UK), Rag2−/− (kindly donated by Dr. Jian-Guo Chai, Imperial College London, UK), and CD4-Cre Dicerlox/lox (7) mice were used for the experiments and kept in a conventional specific pathogen-free facility. All animal work was performed according to the Animals (Scientific Procedures) Act 1996, UK under the animal Project License 70/6965.
Antibodies for flow cytometry experiments were all directed against mouse antigens. They were: CD4-FITC/PE/PerCP/APC (clone: GK1.5), CD8a-PerCP (clone: 53-6.7), CD25-PE (clone: PC61.5), Foxp3-APC (clone: FJK-16s), IFN-γ-FITC or -eFlour450 (clone: XMG1.2), IL-17-PE (all from eBioscience, UK), and IL-17a-PE (clone: TC11-18H10) (BD Biosciences). Western blot antibodies (all rabbit monoclonal antibodies) were: phospho-SMAD2 (S465/467), Total-SMAD2/3, PTEN (clone: D4.3) XP®, PDK-1, phospho-PDK-1 (S241), phospho-AKT (S473) (clone: D9E) XP®, phospho-AKT (T308) (clone: C31E5E), Total-AKT (pan) (clone: C67E7), mTOR (clone: 7C10), Rictor (clone: 53A2), phospho-FoxO1 (S256), Total-FoxO1a (clone: C29H4), phospho-FoxO3a (S253), total-FoxO3a (clone: 75D8), phospho-S6 Ribosomal Protein (Ser235/236) (clone: D57.2.2E)XP®, GAPDH (clone: D16H11) XP®, and β-actin (clone: 8H10D10) (all from Cell Signaling, UK).
miRNA expression vectors were derived by cloning the genomic region encoding specific miRNAs into the retroviral pMIG vector. Expression of miRNAs was confirmed by miR-qRT-PCR. miRNA decoy or sponge transcripts were designed as described (21, 22). Decoy sequences were cloned into the pSIF lentiviral vector for Pol III expression. Sponge sequences were cloned into pMIG. miRNA reporter constructs contained the 3′UTR of Rictor inserted downstream of luciferase in pGL3. Details of vectors are available upon request.
Isolation of naïve T cells and activation into Th subsets
Naïve CD4+ CD62Lhigh CD25− T cells were isolated from spleen and lymph nodes (inguinal, axillary, brachial, mediastinal, superficial cervical, mesenteric) of six-eight week-old female C57BL/6 mice using magnetic beads for CD4+ selection (Dynabeads® Untouched™ Mouse CD4 cells kit, Invitrogen) followed by CD25 and CD62L selection using biotinylated-anti-CD25 (7D4 clone; BD Biosciences, UK) or biotinylated-anti-CD62L (clone MEL-14; BD Biosciences) and streptavidin-Microbeads with MACS separation columns (Miltenyi Biotech, Germany).
Naïve CD4+ CD62Lhigh CD25− T cells were cultured in complete medium (R10: RPMI1640, pencillin/streptomycin, L-glutamine, HEPES and β-mercaptoethanol) (all from Sigma, UK) and 10% fetal bovine serum. They were activated in the presence of plate-bound anti-CD3/anti-CD28 antibodies (eBiosciences, UK) at a ratio of 1:2 (1ug/ml anti-CD3: 2ug/ml anti-CD28) for Th1, Th2 and iTregs and 1:10 (1ug/ml anti-CD3: 10ug/ml anti-CD28) for Th17. Activated T cells were differentiated into: Th1 using 20ng/ml recombinant-IL-12 (eBiosciences, UK) and anti-IL-4 (5ug/ml; BD Bioscience, UK); Th2 using 20ng/ml recombinant-IL-4 (BD Bioscience, UK) and anti-IFN-γ (5ug/ml; BD Bioscience, UK); Th17 using 2.5ng/ml recombinant-TGF-β, 50ng/ml recombinant-IL-6 (eBiosciences, UK), anti-IFN-γ (5ug/ml), anti-IL-4 (5ug/ml) and anti-IL-2 (5ug/ml; BD Bioscience, UK); and iTregs using 2.5ng/ml recombinant-TGF-β and 5ng/ml recombinant-IL-2 (eBiosciences, UK). Analysis was performed on cells three-four days of culture.
Retroviral transduction and transfection of miRNA mimics and siRNAs into T cells
Retroviruses were produced by calcium phosphate transfection of human embryonic kidney (HEK) 293T cells with the retroviral expression and the pcL-Eco helper virus vectors. Culture supernatants were harvested and used to spin-infect naïve CD4+ T cells that were activated overnight by plate bound anti-CD3/anti-CD28 antibodies. Cells were then differentiated into iTregs as described above. For comparing the effects of different miRNAs, an average Z score was calculated from individual experiments as described (23) where Z = (x – y)/SD. In this calculation, x = iTreg index, which is the percentage of Foxp3 positive cells induced in the control or with the expression of an individual miRNA, y = the mean of all the iTreg indices in a given experiment, and SD = the standard deviation of all these iTreg indices.
miRNA mimics or siRNAs were transfected into naïve CD4+ T cells using DharmaFECT3 miRNA mimic transfection reagent or siRNA Accell delivery medium, respectively, as recommended by manufacturer’s guidelines (Dharmacon, USA). For the miRNA mimic transfections, the negative control oligo, consisting of a random sequence, contained a Dy547 fluorescent tag for determining transfection efficiency, which was approximately 50% in each experiment.
Immunoassays
Immuno-staining for flow cytometric analysis was performed using the respective antibodies. For intracellular staining of cytokines, cells were treated with 1μg/ml phorbol 12-myristate 13-acetate (PMA), 1μg/ml ionomycin and 1μg/ml brefeldin A for two hours prior to staining. For Foxp3 staining, cells were fixed with Foxp3 fixation/permeabilization buffer (eBioscience, UK) for 30 minutes prior to staining, and if GFP was also detected, cells were fixed with 2% paraformaldehyde for five minutes prior to fixation/permeabilization. Data were acquired using a FACS Canto II (BD) and analyzed using FlowJo software (Tree Star).
Cytokine detection in culture supernatants from differentiated T cells was detected using the Flowcytomix™ multiple analysis detection kit for Th1/Th2 cytokines (eBioscience, UK).
Western blot analysis was performed from total cell extracts prepared from naïve CD4+ T cells that were either un-activated or activated for 30 minutes with plate bound anti-CD3/anti-CD28 (1:2) with or without the addition of 2.5ng/ml TGF-β. Proteins were detected using the indicated antibodies and a horseradish peroxidase-conjugated secondary antibody and then visualized using the enhanced chemiluminescence detection system. Quantitation was performed by densitometric analysis of exposed films.
miRNA and mRNA qRT-PCR
RNA was isolated from CD4+ T cells using miRNAeasy (for miRNAs) or mRNAeasy (for mRNAs) isolation kits (QIAGEN, Germany). For miRNA detection, cDNA synthesis and subsequent qPCR with locked nucleic acid primers for specific miRNAs was performed using miRCURY LNA™ universal reverse transcriptase microRNA cDNA synthesis and qPCR kit (EXIQON, Denmark). For mRNA detection, cDNA was prepared using the miRscript cDNA synthesis kit (QIAGEN, Germany). qPCR was performed using IQ™ BioRad SYBR green master mix (BioRad, USA) with the following primers for Rictor (Forward - 5′-ACCGGGCTTCTGACCATTAAA-3′ and Reverse - 5′-TTGTATGAACCGCCGACACT-3′). Measurements were performed using Chromo 4 BioRad PCR machines. The relative expression level of miRNAs was normalized to that of 5S rRNA and U6 snRNA, and of mRNAs to that of β-actin as described previously (24).
Adoptive T cell-transfer colitis model
C57BL/6 Rag2−/− mice were injected into the peritoneal cavity with 106 GFP FACS sorted naïve CD4+ CD62Lhigh CD25− T cells that were transduced with either control or miR-15b/16-expressing retroviruses. Weight was monitored every week, and mice were sacrificed after eight weeks (or when weight was reduced by 20%) for analysis. Histopathological review was performed as previously described (12, 25). The large intestine (from the ileocecocolic junction to anorectal junction) was removed and processed. Sections were stained with hematoxylin and eosin and were coded and assigned a score in a ‘blinded’ fashion. Severity of colitis was graded semi-quantitatively on an overall scale of 0-5, assessing the degree of epithelial hyperplasia and goblet cell depletion, leukocyte infiltration in the lamina propria, area of tissue affected, and the presence of markers of severe inflammation, such as crypt abscesses, submucosal inflammation, and ulcers. The total colonic score was calculated as the median of the individual scores of sections of proximal colon, mid-colon and distal colon.
3′UTR luciferase reporter assay
The 3′UTR of Rictor was cloned downstream of the firefly luciferase gene in the pGL-3 vector. HEK 293 T cells were transfected with this vector (or the mutated miR-15/16 target site vector), internal control pRL renilla luciferase vector, and the miRNA over-expression vector. After 48 hours, luciferase activity was measured using the Dual-Luciferase® reporter assay system (Promega, Madison, USA). Luciferase activity was measured with a Wallac 1420 VICTOR2™ multilabel plate reader (Perkin Elmer, UK).
Statistical analysis
Prism software (GraphPad) was used for statistical analyses employing the student’s t-test for significance. Figures were made in Excel and GraphPad prism software. P values equal to or less than 0.05 were considered significant.
RESULTS
Identification of important miRNAs
To analyze miRNA function in Treg development, a model system was needed in which individual miRNAs could be manipulated and tested. Therefore, we utilized an in vitro model of Treg induction (iTreg) involving the activation of naïve CD4+ T cells in the presence of TGF-β plus IL-2 and measuring the differentiation into the Treg lineage by the expression of Foxp3. This model represents the development of pTregs, which along with tTregs are critical in suppressing the immune response and preventing autoimmune disease (2, 3). Furthermore, the induction of iTregs is significantly reduced in Dicer−/− CD4+ T cells just like the in vivo development of Tregs in the mutant mice (7). For this model we hypothesized that important miRNAs would be both abundant and more highly expressed in iTregs when compared to conventionally activated CD4+ T cells or those polarized to other Thsubsets. miRNA profiling experiments previously done by us (7) had identified multiple miRNAs that are more highly expressed in ex vivo-derived Tregs compared to activated conventional CD4+ T cells. These miRNAs were felt to be the most likely candidates to have function for the induction of iTregs so their relative level was determined by qPCR in iTregs and compared to other Th subsets. These included resting naïve CD4+ T cells and those activated under non-polarizing or polarizing conditions towards Th1, Th2, or Th17 subsets. miRNAs that were more abundantly expressed in ex vivo Tregs (miR-15b, 16, 21, 24, 29a, 92b, 142 5′, 142 3′ 146a, 150, and 223) were also found to be most abundant in iTregs compared to T cells cultured in other conditions; whereas those that were not more abundantly expressed in ex vivo Tregs (miR-23a, 30c, 99b, 125a, 191, and 326) were also not more abundantly expressed in iTregs (Fig 1A). Therefore, we focused on the function of the miRNAs most abundantly expressed in iTregs.
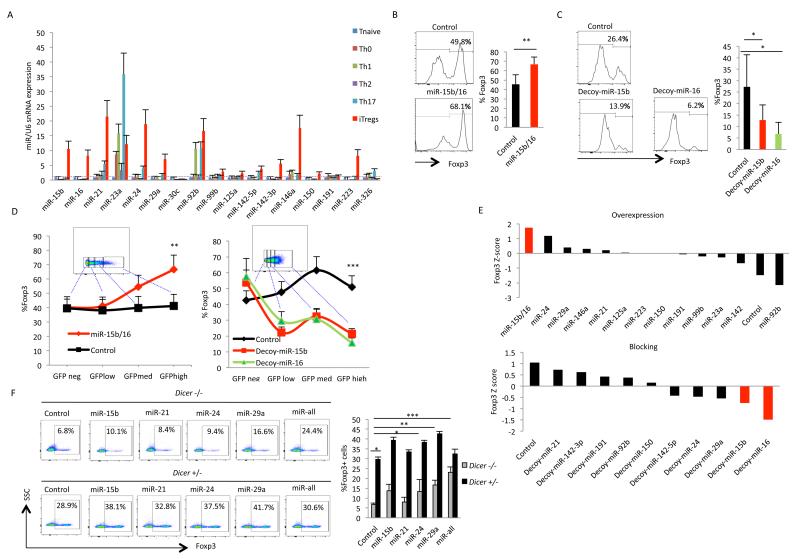
Figure 1. miR-15b/16, miR-24, and miR-29 are important for the in vitro induction of Tregs.
(A) Relative expression levels of miRNAs (as determined by qPCR) in naïve Th cells (CD4+ CD62L+ CD25− CD44−) from lymph nodes and spleen of C57BL/6 mice that were either un-activated or activated under the indicated polarizing conditions. (B) The effect of overexpression of miR-15b/16 on the induction of iTregs. Naïve CD4+ T cells were transduced with GFP-expressing retroviruses that either did not express any miRNA (control) or expressed miR-15b/16. iTreg induction was measured in GFP+ cells by the expression of Foxp3. On the left is a representative experiment, and on the right is the mean and standard deviation of six independent experiments, which demonstrated a significant difference between control and miR-15b/16 overexpressing cells (** p=0.005). (C) The effect of blocking of miR-15b/16 on iTreg induction. Similar to above, naïve CD4+ cells were transduced with control or miR-15b or miR-16 decoy-expressing lentiviruses, and iTreg induction was measured by the expression of Foxp3. On the left is a representative experiment, and on the right are the mean and standard deviation values from six independent experiments showing a significant difference control and decoy expressing cells (miR-15b, * p=0.045; and miR-16, * p=0.02). (D) The effect of the level of expression of miR-15b/16 or its decoys on iTreg induction. The level of expression of the miRNAs or their decoys was deduced from GFP expression. iTreg induction (measured by Foxp3 expression) was determined in populations of cells within gates of no, low, medium, or high GFP expression as indicated in the insets, and statistically significant differences were found when comparing no versus high GFP expression (overexpression, ** p=0.005; decoy expression, *** p=0.001). (E) Comparison of the effects of individual miRNAs in overexpression and blocking experiments using a Z-score analysis as described in the Materials and Methods. Only miR15b/16, miR-24, and miR-29a were found to impact iTreg induction. Data are derived from six individual experiments for each miRNA. (F) Restoration of iTreg induction in Dicer−/− CD4+ T cells by miR-15b/16, miR-24, and miR-29a. miRNA mimics for the indicated miRNAs were transfected into naïve Dicer+/− or Dicer−/− CD4+ T cells, and the effect on iTreg induction was determined by Foxp3 expression (miR-all indicates a combination of miR-15b, miR-24, and miR-29). On the left is a representative experiment, and on the right is the mean and standard deviation values from three independent experiments, which demonstrated a significant difference in Dicer−/− CD4+ T cells between the positive acting miRNAs (but not the negative acting miR-21) and the negative control miRNA mimic (miR-15b, * p=0.02; miR-24, * p=0.05; miR-29, ** p=0.002; all three miRNAs together, *** p=0.0005).
The function of individual miRNAs was examined by measuring the effect of their overexpression or blocking in iTreg induction. Overexpression was achieved through the use of retroviruses that expressed genomic sequences encoding the miRNAs. Blocking was achieved through the use of lentiviruses or retroviruses that expressed miRNA decoy or “sponge” target sequences, respectively. These viruses were transduced into CD4+ T cells, and iTregs were induced by the addition of TGF-β plus IL-2. Since the viruses also encoded GFP to track their expression, GFP-positive cells were analyzed for the induction of iTregs by the expression of Foxp3. The miRNAs that had the greatest effect in these experiments were miR-15b and miR-16, which are encoded within the same primary transcript and are closely related such that they target the same sequences in mRNAs. Overexpression of miR-15b/16 significantly increased the induction of iTregs as compared to cells transduced with a control retrovirus lacking any miRNA sequences (Fig 1B). Likewise, expression of decoys for either miR-15b or miR-16 inhibited the induction of iTregs (Fig 1C). These effects were dependent on the level of expression of the miRNAs or decoys, as deduced from GFP expression. Cell populations within gates of increasing GFP expression were more affected in iTreg induction (Fig 1D).
Comparing the effects of all the miRNAs tested using a Z score analysis (as described in the Materials and Methods) revealed that the only other miRNAs to have a significant positive effect on the induction of iTregs were miR-24 and miR-29a (Fig. 1E). All these positive-acting miRNAs could also partially reverse the defect in iTreg induction in Dicer−/− CD4+ T cells. Transfection of miRNA mimics for miR-15b, miR-24, or miR-29a enhanced induction of iTregs in Dicer−/− CD4+ T cells whereas miR-21 (which did not have an effect in overexpression and blocking experiments) failed to do so. In addition, transfection of a combination of miR-15b, miR-16, and miR-29a into Dicer−/− CD4+ T cells brought the level of iTreg induction nearly back to that seen in Dicer+/− CD4+ T cells (Fig. 1F). Therefore, these miRNAs appear to be most important in regulating the induction of iTregs. It is interesting to note that in contrast to the importance of miR-15b/16 in the overexpression and blocking studies, miR-29a was most effective in the complementation studies. However, the biological significance of this observation remained unclear. The difference could have been due to the different experimental systems, or it could have been due to differences in expression of the miRNAs between transfection and retroviral transduction.
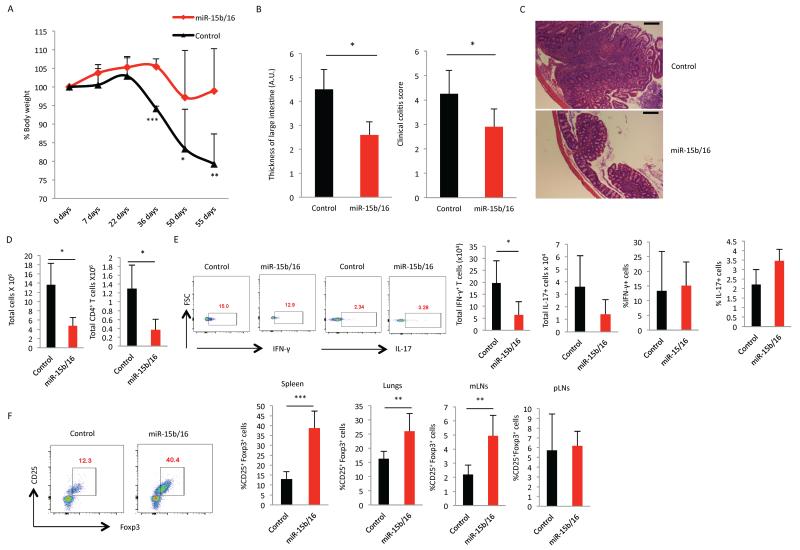
To test if overexpression of miR-15b/16 could also enhance the development of pTregs in vivo, Rag2−/− mice were reconstituted with conventional helper T cells (CD4+, CD25−) transduced with control or miR-15b/16-expressing retroviruses. The autoimmune reaction observed by this reconstitution was significantly inhibited by the overexpression of miR-15b/16, as measured by weight loss and histopathological review of the colon (Fig 2A, B, C). In addition, the total numbers of mononuclear cells populating the spleen was significantly reduced when miR15b/16 was overexpressed, as was the number of CD4+ T cells (Fig 2D). Consistent with the reduction in the total number of T cells, there were fewer IFN-γ-producing cells in the spleen when miR-15b/16 was overexpressed. Likewise, there appeared to be fewer IL-17-producing cells, but this apparent difference did not reach statistical significance. Interestingly, the percentage of IFN-γ and IL-17-producing cells was not statistically different so the overexpression of miR15b/16 did not appear to specifically impact the development of Th1 and Th17 cells over other Th subsets. Therefore, the reduction in the inflammatory response was most likely due to the overall reduction in T cell numbers. This could have been due to multiple effects of miR-15b/16 overexpression, but most importantly, there was a significant increase in the percentage of pTregs at the sites of inflammation (spleen, lungs, and mesenteric lymph nodes), but not away from the site of inflammation within peripheral lymph nodes (Fig 2F). Therefore, similar to the in vitro experiments, the overexpression of miR-15b/16 in CD4+ T cells enhanced the peripheral development of Tregs, and this was consistent with the reduced numbers of T cells and the attenuation of the autoimmune response. Alternative explanations for the decrease in autoimmunity might include suppression of T cell proliferation or activation of cell death because miR-15/16 can inhibit cell growth (26) and induce apoptosis (27). However, overexpression of miR-15b/16 did not affect T cell proliferation or apparent viability in vitro (Sup Fig 1A) so it is uncertain if these issues were important in vivo. Additionally, miR-15b/16 could also have impacted the suppressive function of Tregs, but our attempts to examine suppressive function of Tregs was hampered by the fact that ex vivo isolated Tregs transduced to overexpress miR-15b/16, much more stably maintained Foxp3 expression when cultured in vitro (Sup Fig 1B). Therefore, any effects on suppressive function would be difficult to interpret. Finally, to determine if the effect of miR-15b/16 on pTreg development was cell intrinsic or due to extrinsic factors created in the immune environment where T cells overexpressed miR-15b/16, unsorted miR-15b/16-transduced cells were adoptively transferred into Rag2−/− mice, and the percentage of pTregs was determined in both the GFP+ and GFP− populations of CD4+ T cells. Cells overexpressing miR-15b/16 had significantly more pTregs in both mesenteric lymph nodes and spleen (Sup Fig 2 A and B) suggesting that the effect of miR-15b/16 was intrinsic to the T cells overexpressing these miRNAs.
Figure 2. Overexpression of miR-15b/16 enhances the production of pTregs and decreases the autoimmune response in Rag2−/− reconstitution experiments.
Naïve CD4+ T cells were transduced with the control or miR-15b/16-expressing retroviruses. GFP+ cells were sorted and adoptively transferred into Rag2−/− mice, and the effect of miR-15b/16 overexpression was examined in the following assays: (A) Loss of body weight was followed over 55 days after reconstitution, and it was significantly less (day 36, ***, p=0.0003; day 50, *, p=0.05; day 55, **, p=0.01). (B) Thickness of colons and colitis score at the endpoint of analysis were measured using a scale of 1-5, and these were significantly less (colon thickness, *, p=0.01; colitis score, *, p=0.03). (C) Representative histological sections of the large intestine of mice demonstrated a large decrease in the inflammation. Scale bar represents 100μm. (D) Total numbers of mononuclear cells were significantly reduced (*, p=0.05) as were CD4+ T cells (GFP+ cells *, p=0.02). (E) Analysis of inflammatory IFN-γ and IL-17 producing cells. On the left are representative experiments, and on the right are the mean and standard deviation values. There was a significant reduction in IFN-γ-producing cells (*, p=0.03) but no significant change in IL-17-producing cells. Interestingly, the percentage of CD4+ T cells producing IFN-γ or IL-17 was not significantly different. (F) The development of pTregs was significantly increased. On the left is a representative experiment from spleen and on the right is the mean and standard deviations showing significant differences in the percentages of Tregs in the spleen (***, p=0.0001), lungs (**, p=0.007) and mesenteric lymph nodes (**, p=0.002), but not away from the site of inflammation at the peripheral lymph nodes. Data were derived from six control and five miR-15b/16 mice. Two independent experiments were performed with similar results.
Identification of relevant genes targeted by miR15b/16
Because miR-15b/16 had the greatest effect on iTreg induction in overexpression and blocking studies, we focused on finding relevant target genes for this miRNA family. As a first step, target prediction algorithms (28, 29) identified nearly a thousand genes with potential target sites for miR-15b/16. To narrow down this list, we decided to examine signal transduction pathways known to be important for Treg development and determine if any were altered in Dicer−/− CD4+ T cells. Candidate target genes within affected pathways could then be focused upon for analysis.
miRNA regulation of IFN-γ is not important for iTreg induction
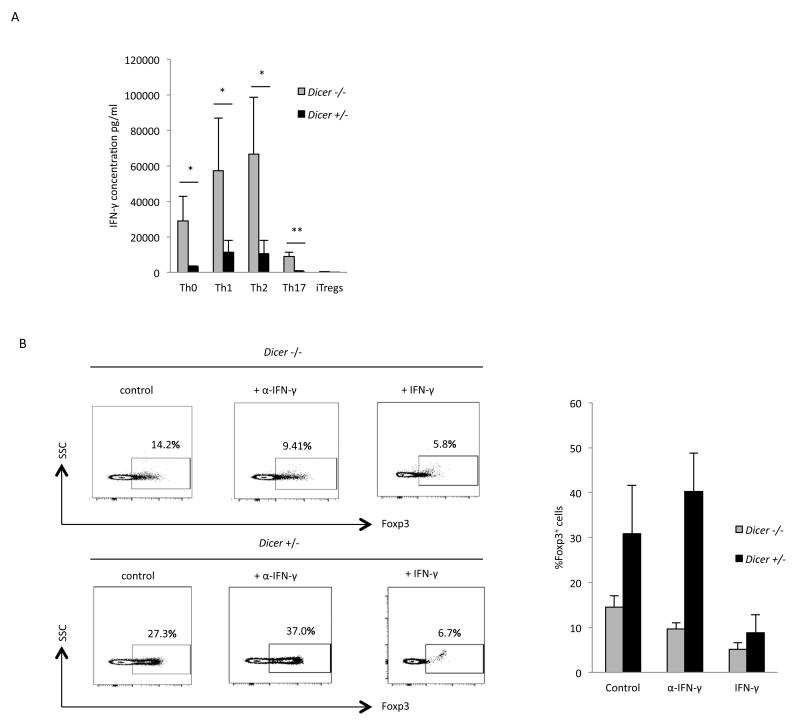
IFN-γ inhibits the induction of iTregs (30, 31), and Dicer−/− CD4+ T cells inherently produce IFN-γ when activated by co-stimulation of the T cell receptor and CD28 (32). Therefore, the importance of miRNAs in iTreg induction could be through their inhibition of IFN-γ expression. Previous studies have indicated that miR-29 regulates the expression of IFN-γ through its regulation of the transcription factors T-bet and Eomes that activate expression of IFN-γ (23, 33, 34). The gene encoding T-bet (Tbx21) contains a predicted target site for miR15b/16 in its 3′UTR in addition to the site targeted by miR-29 (29, 35). Therefore, the importance of these miRNAs in iTreg induction could be through their targeting of Tbx21 and thereby regulating IFN-γ expression. To test the importance of miRNAs in the regulation of IFN-γ expression, IFN-γ production was determined in Dicer+/− and Dicer−/− CD4+ T cells that were activated under different Th-polarizing conditions. Production of IFN-γ was significantly higher in Dicer−/− CD4+ T cells polarized towards Th0, Th1, Th2, and Th17 subtypes, but it was virtually absent in iTregs from both Dicer+/− and Dicer−/− CD4+ T cells (Fig 3A) (note: the production of IFN-γ in Th2 conditions in Dicer+/− cells was due to the lack of the addition of neutralizing antibody for IFN-γ, which is required for full polarization to the Th2 subset but would prevent the measurement of IFN-γ in the culture supernatants). Therefore, IFN-γ production does not appear to be responsible for the reduction in the induction of iTregsin Dicer−/− CD4+ T cells. Furthermore, the addition of aneutralizing antibody to the low amount of IFN-γ present in the conditions used in these studies did not significantly enhance the induction of iTregs in either Dicer+/− or Dicer−/− CD4+ T cells (Fig 3B) (in fact, induction was slightly inhibited in Dicer−/− CD4+ T cells). In contrast, the addition of IFN-γ did inhibit iTreg induction in both Dicer+/− and Dicer−/− CD4+ T cells. Therefore, miRNA regulation of IFN-γ expression does not appear to be important for iTreg induction.
Figure 3. miRNA regulation of IFN-γ expression is not important for induction of iTregs.
(A) IFN-γ expression in CD4+ T cells polarized towards the indicated Th cell subsets. IFN-γ levels were measured by a Flowcytomix-based assay, and the mean and standard deviation values from three independent experiments are shown. Expression was found to be significantly higher in Dicer−/− CD4+ T cells in Th0 (* p=0.03), Th1 (* p=0.05), Th2 (* p=0.04), and Th17 (* p=0.003) cell subsets. However, in iTregs IFN-γ production was virtually absent in both Dicer+/− and Dicer−/− CD4+ T cells. (B) iTreg induction in the presence of a neutralizing IFN-γ antibody or exogenously added IFN-γ (from culture supernatants of cells polarized under Th1 conditions). On the left is a representative experiment, and on the right are the mean and standard deviation values from three independent experiments, which show that the addition of an IFN-γ neutralizing antibody did not enhance iTreg induction in Dicer+/− or Dicer−/− CD4+ T cells; whereas exogenously added IFN-γ did inhibit induction in both Dicer+/− and Dicer−/− CD4+ T cells.
Proximal TGF-β signaling events are not regulated by miRNAs
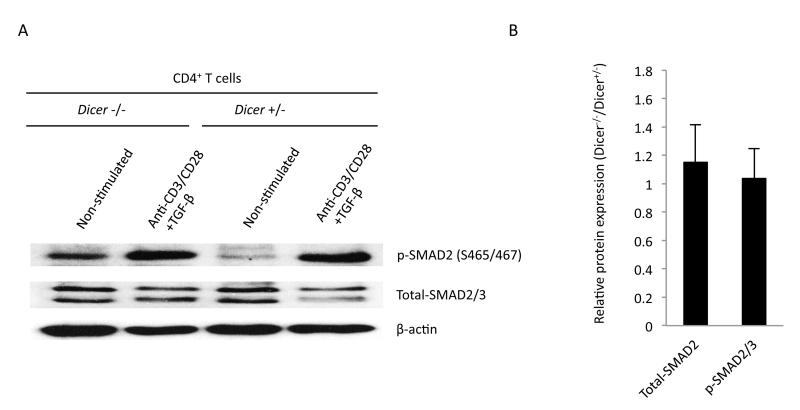
Since TGF-β signaling during T cell activation results in iTreg induction (36), miRNAs could potentially regulate the expression of key repressors of this pathway and provide a means to fine-tune its signaling. Therefore, miRNA regulation of proximal events in this pathway were analyzed by examining the phosphorylation of the downstream transcription factor, SMAD2, which is activated through the TGF-β receptor complex by phosphorylation on serines 465 and 467. As shown in Fig 4, Dicer+/− and Dicer−/− CD4+ T cells had no difference in phosphorylation of SMAD2 when activated by anti-CD3/CD28 and the addition of TGF-β. Therefore, miRNAs are not important for regulating TGF-β signaling up to the point of SMAD2 phosphorylation. However, it is possible that miRNAs could regulate TGF-β signaling downstream of this point. Target prediction algorithms found target sites of miR15b/16 in SMAD7, SMURF1/2 and WWP1, which are negative regulators of TGF-β signaling, but little is known about the importance of these factors in TGF-β signaling leading to iTreg induction so their analysis was not pursued further.
Figure 4. Proximal events in TGF-β signaling are not affected in Dicer−/− CD4+ T cells.
TGF-β signaling was examined in Western blots of phosphorylated SMAD2 at serines 465 and 467. (A) A representative experiment. (B) The mean and standard deviation values of the relative expression or phosphorylation levels in anti-CD3/CD28 + TGFβ stimulated cells from three separate experiments. Owing to the variability in the relative signal strength of the indicated protein compared to β-actin, we normalized the results in each experiment to β-actin, then determined the ratio between Dicer−/− and Dicer+/− CD4+ T cells and calculated the mean and standard deviation of this value for all experiments. No significant difference was observed in the levels of SMAD2/3 protein or the phosphorylation of SMAD2.
The mTOR-signaling pathway is regulated by miRNAs
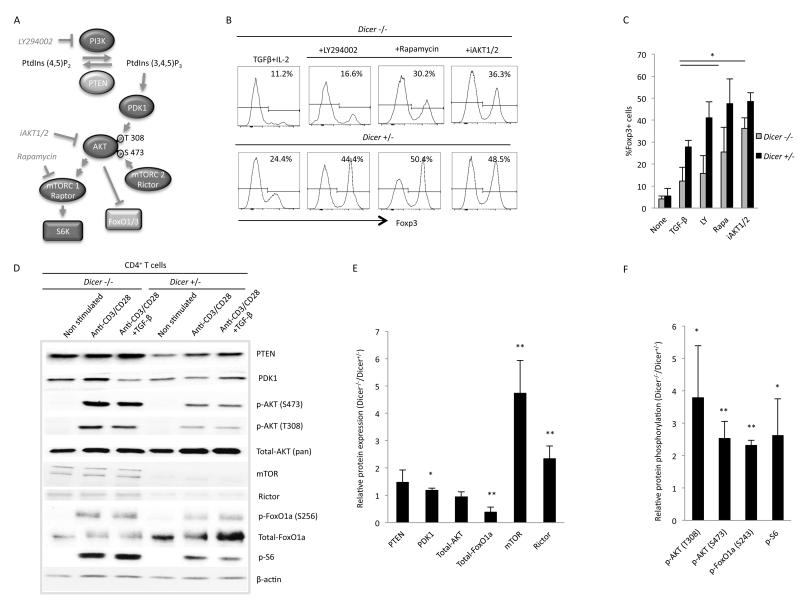
Induction of iTregs requires suppression of signaling through the mTOR pathway (18-20). A highly simplified diagram of the pathway is shown in Fig 5A. Briefly, stimulation of the T cell receptor and co-receptors leads to the activation of phosphoinositol 3 kinases (PI3K). These phosphorylate inositol lipids at the cell membrane (countered by the lipid phosphatase and tensin homolog (PTEN)), which then attract and activate phosphoinositide-dependent kinase-1 (PDK1). PDK1 then phosphorylates protein kinase B/AKT (at threonine 308) leading to its activation, which ultimately results in the activation of the mammalian target of rapamycin complex (mTORC). mTORC regulates multiple facets of cell metabolism and ultimately controls many steps in the determination of developmental fates (37). One key mediator is ribosomal protein S6 kinase, which is activated through its phosphorylation by mTOR and then phosphorylates many cellular proteins involved in cell growth, one being ribosomal protein S6. mTORC contains the mTOR kinase and other associated proteins. Two main complexes exist that differ primarily by the inclusion of Raptor (mTORC1) or Rictor (mTORC2). mTORC1 lies downstream and is activated via events mediated by AKT. However, mTORC2 is required for full activation of AKT through its phosphorylation of Serine 473. In addition to activating mTORC1, AKT also phosphorylates FoxO1a and FoxO3a proteins, which are important transcription factors required for the expression of Foxp3 and the development of Tregs (38, 39). Phosphorylation of FoxO proteins by AKT leads to their exclusion from the nucleus and subsequent degradation, which is one important mechanism through which mTOR signaling inhibits Treg development (40).
Figure 5. The mTOR signaling pathway is enhanced in Dicer−/− CD4+ T cells.
(A) A simplified diagram of the mTOR signaling pathway showing the targets of specific pharmacological inhibitors. See text for details. (B) A representative experiment and (C) the mean and standard deviation values for three independent experiments examining the effect of mTOR signaling pathway inhibitors on iTreg induction in Dicer−/− and Dicer+/− CD4+ T cells. The PI3K inhibitor LY294002 had minimal effect; whereas the mTORC1 inhibitor rapamycin (* p=0.05) and the AKT inhibitor iAKT1/2 (* p=0.02) significantly enhanced iTreg induction in Dicer−/− CD4+ T cells. (D) Levels of the indicated proteins are illustrated in a representative Western blot. (E and F) The mean and standard deviation values for the relative level of expression (E) or phospho-protein (F) of the indicated protein between Dicer−/− and Dicer+/− CD4+ T cells. Ratios were calculated as done in Fig 4 and were derived from three separate experiments. In Dicer−/− CD4+ T cells there was no significant change in PTEN or AKT expression, a slight increase in the level of PDK-1 (* p=0.02), a significant decrease in FoxO1a (** p=0.004), and a significant increase in mTOR (** p=0.005) and Rictor (** p=0.007). In addition, in Dicer−/− CD4+ T cells there was an increase in phosphorylated AKT at both Thr 308 (* p=0.03) and Ser 473 (** p=0.01) and an increase in phosphorylated FoxO1a (** p=0.002) and S6 (* p=0.03), indicating an enhanced response in the mTOR signaling pathway.
Pharmacological inhibitors of this pathway enhance the induction of iTregs. These include LY294002, rapamycin, and AKT1/2, which inhibit PI3K, mTORC1, and AKT respectively (20). To determine if miRNAs are important for suppressing the mTOR signaling pathway, the above inhibitors were tested for their effect on iTreg induction in Dicer−/− CD4+ T cells. As shown in Fig 5B and C, LY294002 enhanced the induction of iTregs in Dicer+/− CD4+ T cells, but it had no significant effect on Dicer−/− CD4+ T cells. In contrast, both rapamycin and iAKT1/2 significantly increased the induction of iTregs in both Dicer+/− and Dicer−/− CD4+ T cells. Therefore, miRNAs appear to be important for regulating the mTOR signaling pathway downstream of PI3K. Examining the levels of key proteins in this pathway (Fig 5D - F), PTEN appeared to be slightly upregulated in Dicer−/− CD4+ T cells, but upon further experiments, this difference was not apparent and revealed no statistically significant changes. Looking downstream, the level of PDK displayed a slight increase (10%) in Dicer−/− CD4+ T cells that was consistent in further experiments yielding statistically significant results. In contrast, the level of AKT was not affected by miRNAs. However, its phosphorylation at both Thr 308 and Ser 473 was significantly enhanced in Dicer−/− CD4+ T cells, indicating an upregulation of AKT activity. This correlated with an increase in the levels of mTOR and Rictor, which are part of mTORC2 that activates AKT through phosphorylation of Ser 473. Consistent with an increase in AKT activity was the downstream activation of mTORC1, as measured by phosphorylation of ribosomal protein S6. Likewise, the level of FoxO1a was significantly downregulated in Dicer−/− CD4+ T cells, and its relative phosphorylation was greatly enhanced. Because FoxO proteins are important regulators of Treg development through their activation of Foxp3 transcription (38, 39), we examined if overexpression of FoxO3a could enhance iTreg induction in Dicer−/− CD4+ T cells. Induction was enhanced by FoxO3a overexpression, and this was further enhanced by overexpression of a phosphorylation mutant of FoxO3a (A3A) that contains alanine residues at the three sites phosphorylated by AKT, thus making it resistant to degradation by AKT phosphorylation (Sup Fig 3) (41). A similar increase in induction was observed with overexpression of these FoxO3a proteins in Dicer+/− CD4+ T cells. However, the level of iTreg induction did not correlate with the absolute level of FoxO3a expression. Therefore, FoxO3a protein expression cannot by itself explain the effects on iTeg induction in Dicer−/− CD4+ T cells and its relative importance is unclear.
Rictor and mTOR are relevant targets of miR-15b/16 in iTreg induction
Target prediction algorithms identified potential target sites of miR-15b/16 in the 3′UTRs of several components of the mTOR signaling pathway (29, 35). These included Akt3, Sgk1, and Rictor. Of these, Akt3 expression was greater in Dicer−/− CD4+ T cells (as measured by RNA – our unpublished observations), but this was not thought to be relevant because as shown above, there was no increase in the total amount of AKT in Dicer−/− CD4+ T cells in which the AKT antibody used for the Western blot recognizes all three isoforms (AKT1, 2 and 3). Therefore, any changes in the level of AKT3 did not affect the overall level of AKT. Sgk1 encodes the serum glucocorticoid kinase (SGK1), which is related to AKT and can phosphorylate FoxO proteins at the same sites as AKT and similarly regulate their activity (42). SGK1 is important for Th17 development (43, 44) and also for Th1 and Th2 development (45). However, its role in Treg development is less clear. SGK1 levels (as measured by RNA) did not change significantly between conventionally activated T cells and iTregs, and overexpression of SGK1 had minimal effect on iTreg induction (our unpublished observations). Therefore, SGK1 was not thought to be a relevant target of miR-15b/16 in the induction of iTregs.
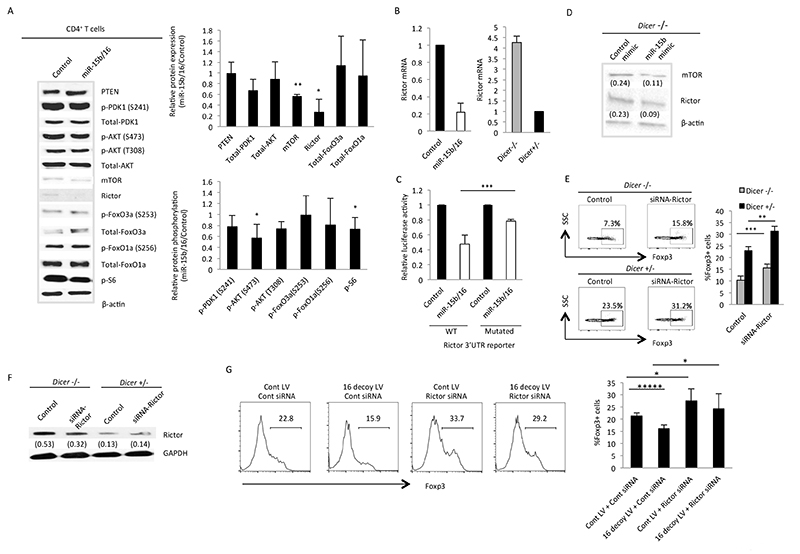
In contrast, Rictor did appear to be a relevant target of miR-15b/16. Its expression was reduced at both the protein and mRNA levels when miR-15b/16 was overexpressed (Fig 6A and B). The increase in protein level in Dicer−/− CD4+ T cells and the decrease in miR-15b/16 overexpressing cells was also observed at the mRNA level (Fig 6B). A luciferase reporter construct containing the 3′ UTR of Rictor was suppressed by overexpression of miR15b/16, and suppression was significantly reduced when the predicted target sequence was mutated (Fig 6C). Furthermore, the miR-15b miRNA mimic reduced the level of Rictor expression in Dicer−/− CD4+ T cells (Fig 6D). The relevance of the increased Rictor expression impacting iTreg induction in Dicer−/− CD4+ T cells was tested by siRNA knockdown of Rictor, which enhanced the induction of iTregs in Dicer−/− CD4+ T cells by about 1.5 fold (Fig 6E). This did not bring the level of induction back to that in Dicer+/− CD4+ T cells transfected with a control oligo, but the knockdown of Rictor in Dicer−/− CD4+ T cells did not reduce its level to what was observed in Dicer+/− CD4+ T cells. Therefore, the differences in iTreg induction between Rictor siRNA-transfected Dicer−/− and control-transfected Dicer+/− CD4+ T cells were consistent with the prevailing level of Rictor expression. Interestingly, there was not a significant knockdown of Rictor expression in Dicer+/− cells, but this could be due to its already low level of expression. Despite this caveat, the transfection of Rictor siRNAs still resulted in an increase in Foxp3 positive cells in these experiments of approximately 1.3-fold. Therefore, small changes of Rictor expression around its normal level could make significant differences in mTOR signaling and iTreg induction. Overexpression of miR-15b/16 did significantly reduce the level of Rictor in wild-type cells, increasing the percentage of Foxp3 cells on average by approximately 1.5-fold. However, these experiments were performed at different times, and the basal level of Foxp3-expressing cells was higher with the cells used in these experiments. This and the problems of comparing retroviral-transduced and siRNA-transfected experiments make it difficult to compare the effects of siRNA knockdown and miR-15b/16 expression on iTreg induction. Nevertheless, changes in Rictor levels over a broad range were important for iTreg induction, and most notably, the upregulation of Rictor in Dicer−/− CD4+ T cells appeared to play an important role in the reduced levels of iTreg induction.
Figure 6. miR-15b/16 regulates the expression of Rictor and mTOR in iTregs.
(A) Expression levels and phosphorylation of indicated proteins in control or miR-15b/16-expressing CD4+ T cells as measured by Western blot. On the left is a representative experiment, and on the right are the mean and standard deviation values for the relative level of expression or phospho-protein of the indicated protein between control or miR-15b/16-expressing CD4+ T cells. In mir-15b/16-overexpressing T cells, there was a reduction in the level of Rictor (* p=0.03) and mTOR (** p=0.002), and also the phosphorylation of AKT Ser 473 (* p=0.02) and S6 (* p=0.02). Values are derived from between three and six experiments. (B) The level of Rictor mRNA was similarly affected by miR-15b/16 overexpression, and it was increased in Dicer−/− CD4+ T cells. (C) Luciferase reporter constructs containing the 3′ UTR of Rictor were regulated by miR-15b/16 overexpression in HEK 293T cells, and this was significantly reduced when the predicted miR-15b/16 target site was mutated (*** p=0.0009). (D) An miRNA mimic for miR-15b decreased the expression of Rictor and mTOR in Dicer−/− CD4+ T cells as measured by Western blot. (E) siRNA knockdown of Rictor enhanced iTreg induction in both Dicer−/− and Dicer+/− CD4+ T cells. On the left is a representative experiment, and on the right are the mean and standard deviation values from two independent experiments, each performed in triplicate (Dicer−/−, *** p=0.0002; Dicer+/−, ** p=0.005). (F) Expression levels of Rictor in one of the two siRNA-knockdown experiments, with the relative level compared to GAPDH listed below each band. Similar results were obtained in each experiment. (G) The reduction in iTreg induction with the inhibition of miR15b/16 is counteracted by the knockdown of Rictor expression. CD4+ T cells were transduced with either a control (Cont LV) or miR-16 decoy expressing lentivirus (16 decoy LV). After 16-18 hours, cells were transfected with a control or Rictor siRNA then 48 hours later cells were harvested and analyzed for Foxp3 expression. A representative experiment is displayed on the left, and the mean and standard deviation values from four independent experiments are displayed on the right. Under these conditions the miR-15b decoy significantly inhibited iTreg induction (Cont LV + Cont siRNA vs 16 decoy + Cont siRNA) ***** p=0.000006. Likewise, the Rictor siRNA enhanced iTreg induction (Cont LV + Cont siRNA vs Cont LV + Rictor siRNA) *p=0.012. Most importantly, the Rictor siRNA counteracted the inhibition of iTreg induction by the miR-16 decoy (16 decoy LV + Cont siRNA vs 16 decoy LV - Rictor siRNA) *p=0.011.
To further address the relevance of miR15b/16 regulation of Rictor on iTreg induction, we found that the siRNA knockdown of Rictor expression in T cells reversed the inhibition of iTreg induction when miR-15b/16 were inhibited by the miR-15 decoy (Fig 6G). However, quantifying the importance of miR-15b/16 regulation of Rictor expression as apposed to other targets will require the gene mutation of the target site in the Rictor 3′UTR. Undoubtedly, other targets of miR-15b/16 will also play a role in iTreg induction, and in fact within the mTOR-signaling pathway, miR-15b/16 additionally regulated the expression of mTOR. Interestingly, target prediction algorithms did not identify sites for miR-15b/16 in its 3′UTR (29, 35), but like Rictor, its expression was reduced by overexpression of miR-15b/16 (Fig 6A), and an miRNA mimic for miR-15b decreased its expression in Dicer−/− CD4+ T cells (Fig 6D). Therefore, mTOR could have target sites for miR-15b/16 not recognized by the target prediction algorithms, or it may be regulated indirectly.
Changes in mTOR and Rictor levels were consistent with a decrease in AKT activation as measured by phosphorylation of Ser 473, but this was not reflected by any significant changes in FoxO1a or FoxO3a expression (Fig 6A). In contrast, mTORC1 activity was inhibited, as measured by phosphorylation of ribosomal protein S6. Therefore, miR-15b/16 appears to affect mTOR signaling downstream of mTORC1 but not by regulating expression or phosphorylation of FoxO proteins.
DISCUSSION
miRNAs are critical regulators of Treg development and function. This work has identified three individual miRNAs (miR-15b/16, miR-24, and miR-29a) that are important for the in vitro induction of iTregs, and it has also demonstrated the importance of miR-15b/16 in the in vivo generation of pTregs. These miRNAs were identified through overexpression and blocking experiments, but they could also reverse the defect in the induction of iTregs in Dicer−/− CD4+ T cells. Further support for the importance of miR-29 has come from the T cell specific deletion of the locus encoding it, which results in a significant reduction of Tregs in vivo. However, little difference in the induction of iTregs was found under the conditions utilized, which included the addition of retinoic acid (34). We have found that the influence of miRNAs is greatest under sub-optimal conditions for iTreg induction. Therefore, it would be interesting to determine if the deletion of miR-29a would affect the induction of iTregs in the conditions used in this report. No T-cell specific deletions of miR-15/16 or miR-24 have been characterized to the best of our knowledge. However, such experiments would be complicated because both miR-15/16 and miR-24 are encoded on two separate loci, and it is not known if one locus is preferentially expressed over the other in T cells or if the deletion of one locus would impact the expression of the other. Furthermore, miR-15/16 is part of a family of related miRNAs that are capable of targeting the same sequences in messages. These include miR-195, miR-322, miR-424, miR-497, and miR-1907 so it could be difficult to ensure lack of expression of all these miRNAs, which would make the interpretation of effects on gene expression challenging. Therefore, the overexpression and blocking approach offers the advantage of characterizing the effects of this whole family.
Because miR-15b/16 had the greatest effect in overexpression and blocking experiments, we focused on identifying relevant targets regulated by this miRNA family. Signaling through the mTOR pathway was enhanced in Dicer−/− CD4+ T cells, and the expression of the mTORC2 components Rictor as well as mTOR were found to be regulated by miR15b/16. It has been well established that a reduction in signaling through the mTOR pathway is required for Treg development. A T cell-specific deletion of mTOR results in the spontaneous induction of iTregs without the addition of TGF-β (18). The expression of a constitutively active AKT inhibits iTreg induction (19), and the pharmacological inhibition of the mTOR signaling pathway enhances iTreg induction (20). Therefore, regulation of this pathway by miRNAs is an obvious mechanism for fine-tuning its function in T-cell development.
The regulation of mTORC2 by miR-15b/16 affected mTOR signaling by reducing AKT activity and subsequent downstream events. This disruption in mTOR signaling is consistent with the T cell-specific deletion of Rictor, which diminishes AKT activation and enhances iTreg induction (46). It is also consistent with the effects observed in the T cell-specific deletion of mTOR (18). However, in contrast to the T cell-specific deletion of mTOR, iTreg induction in miR-15b/16-overexpressing cells still required the addition of TGF-β. In addition, the proximal events of TGF-β signaling were not affected in Dicer−/− CD4+ T cells as they were in Mtor−/− CD4+ T cells. Therefore, the effects on signaling by the reduction of mTOR levels do not entirely follow the complete loss of mTOR expression, suggesting that other signaling mechanisms involving the mTOR pathway exist that are only sensitive to large changes in mTORC activity. Interestingly, the reduced AKT activity in T cells overexpressing miR-15b/16 had no effect on FoxO protein levels or phosphorylation, suggesting that the change in AKT activation was not sufficient to regulate this part of the pathway. In contrast, it was sufficient to regulate mTORC1 activity, leading us to speculate that the reduction of both mTORC1 and mTORC2 levels from reduced mTOR expression amplified the effect of the reduced AKT activity. Alternatively, regulation of mTORC1 in T cells might be more sensitive to small changes in AKT activity.
The regulation of the mTOR signaling pathway is a major mechanism controlling Treg development, and miR-15b/16 affects this pathway by regulating the expression of Rictor and mTOR. However, there will undoubtedly be other important targets for this miRNA, as well as targets for miR-24 and miR-29a. Furthermore, the regulation of expression of these miRNAs will need to be differentially controlled in iTreg induction to enhance their levels over what is found in other Th subtypes. T-cell activation results in a global change in miRNA levels through their turnover due to the destabilization of Argonaute proteins; however, transcriptional regulation of miRNA transcripts and their posttranscriptional processing are also important (47). Therefore, it will be interesting to dissect the mechanisms controlling expression of these important miRNAs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Robert Unsell, Poorna Sharma, and Claudia Gale for technical assistance. We also thank Anna Morgunowicz, Grainne Mcgeever and Wendy Balderson for their technical help with animal experiments.
This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) grant (BB/H018573/1) and a BD Biosciences grant.
REFERENCES
- 1.Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150:29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational Repression and eIF4A2 Activity Are Critical for MicroRNA-Mediated Gene Regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 7.Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong MMW, Rasmussen JP, Rudensky AY, Rundensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liston A, Lu L-F, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu L-F, Thai T-H, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: The Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 13.Lu L-F, Boldin MP, Chaudhry A, Lin L-L, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyer M, Thabet Y, Müller R-U, Sadlon T, Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, Schönfeld EA, Böttcher J, Golovina T, Mayer CT, Hofmann A, Sommer D, Debey-Pascher S, Endl E, Limmer A, Hippen KL, Blazar BR, Balderas R, Quast T, Waha A, Mayer G, Famulok M, Knolle PA, Wickenhauser C, Kolanus W, Schermer B, Bluestone JA, Barry SC, Sparwasser T, Riley JL, Schultze JL. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Kouchkovsky D, Esensten JH, Rosenthal WL, Morar MM, Bluestone JA, Jeker LT. microRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J Immunol. 2013;191:1594–1605. doi: 10.4049/jimmunol.1203567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li Q-J. Molecular dissection of the miR-17-92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, Lund AH, Bluestone JA. MicroRNA 10a marks regulatory T cells. PLoS ONE. 2012;7:e36684–e36684. doi: 10.1371/journal.pone.0036684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O'Connor E, Shokat KM, Fisher AG, Merkenschlager M. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Research. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Meth. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CDC, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, Suar M, Van Kaer L, Bishai WR, Das G. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem. 2013;288:5056–5061. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patton DT, Garden OA, Pearce WP, Clough LE, Monk CR, Leung E, Rowan WC, Sancho S, Walker LSK, Vanhaesebroeck B, Okkenhaug K. Cutting edge: the phosphoinositide 3-kinase p110 delta is critical for the function of CD4+CD25+Foxp3+ regulatory T cells. J Immunol. 2006;177:6598–6602. doi: 10.4049/jimmunol.177.10.6598. [DOI] [PubMed] [Google Scholar]
- 26.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2012;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu C-G, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Research. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. microRNA.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei JJ, Duramad OO, Perng OAO, Reiner SLS, Liu Y-JY, Qin FX-FF. Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2007;104:18169–18174. doi: 10.1073/pnas.0703642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang J-HJ, Kim Y-JY, Han S-HS, Kang C-YC. IFN-gamma-STAT1 signal regulates the differentiation of inducible Treg: potential role for ROS-mediated apoptosis. Eur J Immunol. 2009;39:1241–1251. doi: 10.1002/eji.200838913. [DOI] [PubMed] [Google Scholar]
- 32.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 34.Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, Mavrikis Cox G, Satoskar AR, Croce CM, Racke MK, Lovett-Racke AE, Whitacre CC. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189:1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Jin W, Hardegen N, Lei K-J, Li L, Marinos N, Mcgrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouyang W, Beckett O, Ma Q, Paik J-H, Depinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 39.Kerdiles YM, Stone EL, Beisner DL, Mcgargill MA, Ch'en IL, Stockmann C, Katayama CD, Hedrick SM. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi H. Regulation and function ofmTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedrick SM, Michelini RH, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12:649–661. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Molecular and Cellular Biology. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, Hafler DA. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496:518–522. doi: 10.1038/nature11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–517. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heikamp EB, Patel CH, Collins S, Waickman A, Oh M-H, Sun I-H, Illei P, Sharma A, Fejes-Toth A. Naray, Fejes-Toth G, Misra-Sen J, Horton MR, Powell JD. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nat Immunol. 2014;15:457–464. doi: 10.1038/ni.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian Target of Rapamycin Protein Complex 2 Regulates Differentiation of Th1 and Th2 Cell Subsets via Distinct Signaling Pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, Rao A, Ansel KM. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–43. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.