Summary
Error-free repair of DNA double-strand breaks (DSB) is achieved by homologous recombination (HR), and BRCA1 is an important factor for this repair pathway1. In the absence of BRCA1-mediated HR, administration of PARP inhibitors induces synthetic lethality of tumor cells of patients with breast or ovarian cancers2,3. Despite the benefit of this tailored therapy, drug resistance can occur by HR restoration4. Genetic reversion of BRCA1-inactivating mutations can be the underlying mechanism of drug resistance, but this does not explain resistance in all cases5. In particular, little is known about BRCA1-independent restoration of HR. Here, we show that loss of REV7 (also known as MAD2L2) re-establishes CtIP-dependent end resection of DSBs in BRCA1-deficient cells, leading to HR restoration and PARP inhibitor resistance, reversed by ATM kinase inhibition. REV7 is recruited to DSBs in a manner dependent on the H2AX-MDC1-RNF8-RNF168-53BP1 chromatin pathway, and appears to block HR and promote end joining in addition to its regulatory role in DNA damage tolerance6. Finally, we establish that REV7 blocks DSB resection to promote non-homologous end-joining (NHEJ) during immunoglobulin class switch recombination. Our results reveal an unexpected critical function of REV7 downstream of 53BP1 in coordinating pathological DSB repair pathway choices in BRCA1-deficient cells.
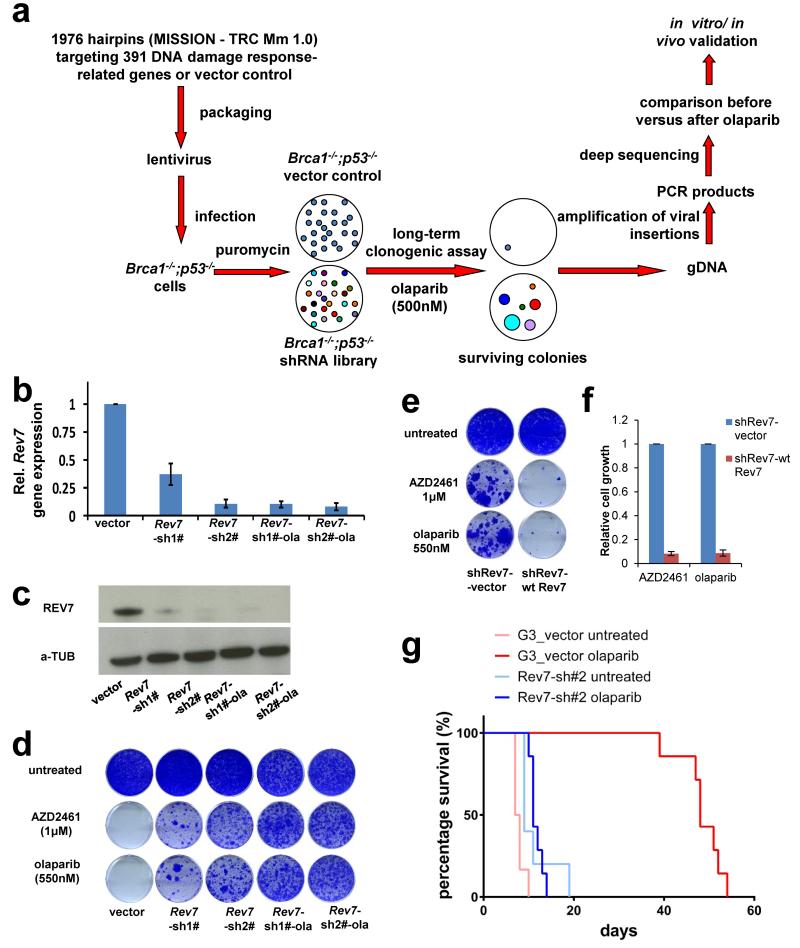
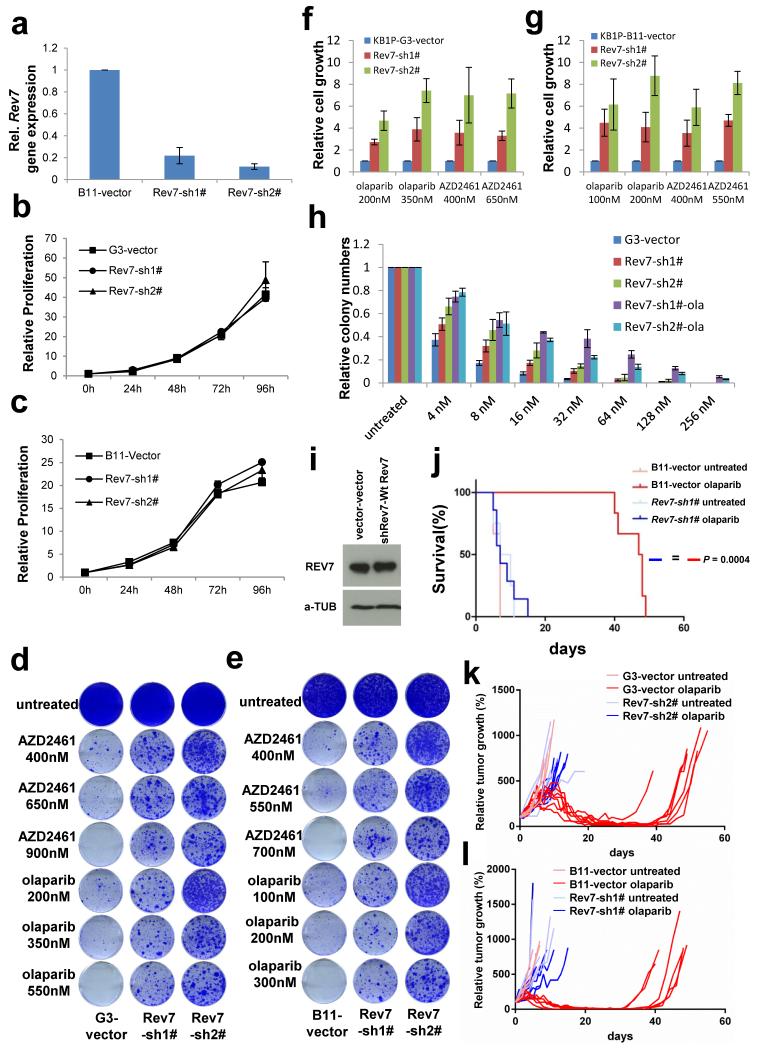
To identify mechanisms of BRCA1-independent restoration of the homologous recombination (HR) pathway, we carried out a loss-of-function shRNA screen using the KB1P-B11 and KB1P-G3 cell lines that we previously derived from Brca1−/−;p53−/− mouse mammary tumors7 (Fig. 1a and Supplementary Table 1). Cells with HR restoration were selected with a high concentration of olaparib (500nM, about 100-fold the IC50), which killed cells of the empty vector control. Sequencing of the olaparib-surviving colonies revealed a reproducible enrichment of various individual hairpins targeting Rev7 or 53bp1. To validate the Rev7 hit, we introduced 2 different hairpins into the B11 and G3 cell lines that resulted in a substantial inhibition of Rev7 expression (Fig. 1b, c and Extended Data Fig. 1a). Despite the role of REV7 in metaphase-to-anaphase transition8, the level of Rev7 inhibition in these cells did not affect proliferation (Extended Data Fig. 1b, c), allowing long-term clonogenic survival assays. We confirmed that loss of Rev7 resulted in increased resistance to the PARP inhibitors (PARPi) olaparib and AZD24617 in both cell lines (Fig. 1d and Extended Data Fig. 1d-g). Resistant cells that survived olaparib treatment (Rev7-sh#-ola) yielded even lower REV7 expression levels and increased numbers of colonies after PARPi treatment (Fig. 1b-d and Extended Data Fig. 1h). When we reconstituted the Rev7-depleted cells with shRNA-resistant Rev7 cDNA resulting in similar REV7 protein levels (Extended Data Fig. 1i), we successfully re-sensitized the tumor cells to PARPi (Fig. 1e, f).
Figure 1. Identification of loss of Rev7 in PARPi-resistant Brca1−/−; p53−/− mammary tumor cells.
a, Design of the functional shRNA screen. b, c, Quantification of Rev7 transcript (b) or protein (c) levels in KB1P-G3 cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± SD. d, e, Long-term clonogenic assay using KB1P-G3 cells transduced with the indicated constructs (wt Rev7 stands for pLenti6-wt Rev7) and treatments. f, Quantification of the clonogenic assay (e) by determining the absorbance of crystal violet at 590nm. All the groups were normalized by the absorbance of the vector control. The data represent the mean ± SD. g, Overall survival of mice with KB1P-G3-derived Rev7-depleted or control tumors treated with one regimen of 50 mg olaparib per kg daily for 28 days or left untreated. The P value was calculated using the log-rank test.
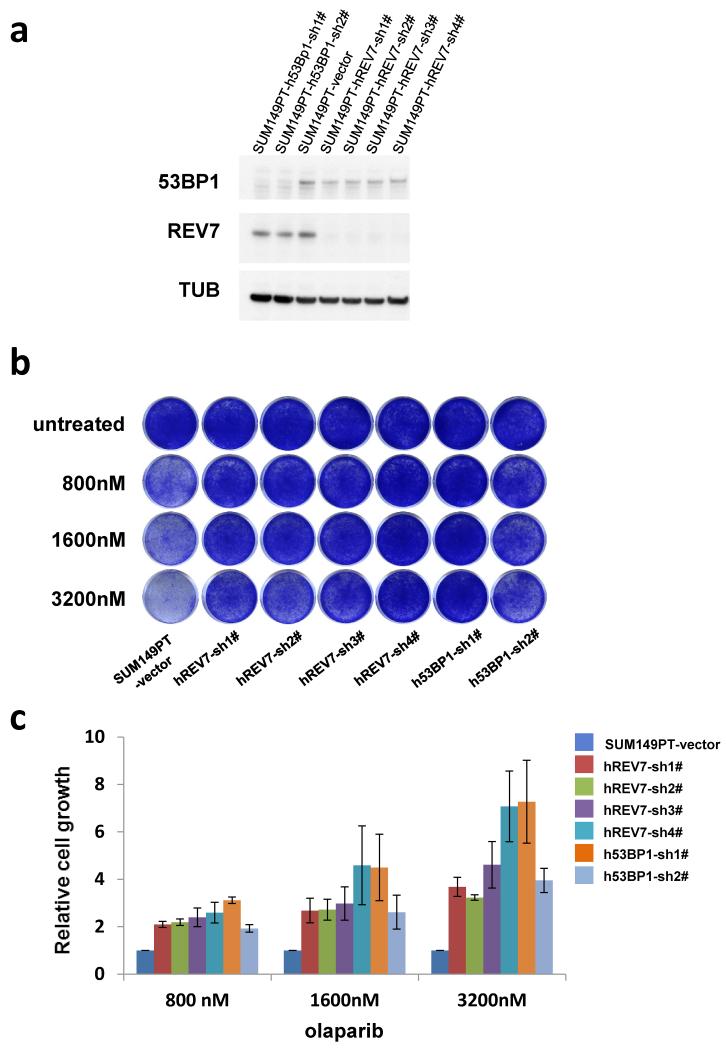
Tumors derived from the Brca1−/−;p53−/− cells with stable Rev7 inhibition also showed olaparib resistance in vivo, in contrast to the empty vector controls (Fig.1g and Extended Data Fig. 1j-l). In addition, we found that Rev7 loss explains some cases of in vivo acquired PARPi resistance in BRCA1-deficient mouse mammary tumors (data not shown). REV7 depletion also resulted in PARPi resistance of the human BRCA1-deficient cell line SUM149PT (Extended Data Fig. 2). Together, these data strongly indicate that inhibition of Rev7 confers PARPi resistance in BRCA1-deficient tumor cells.
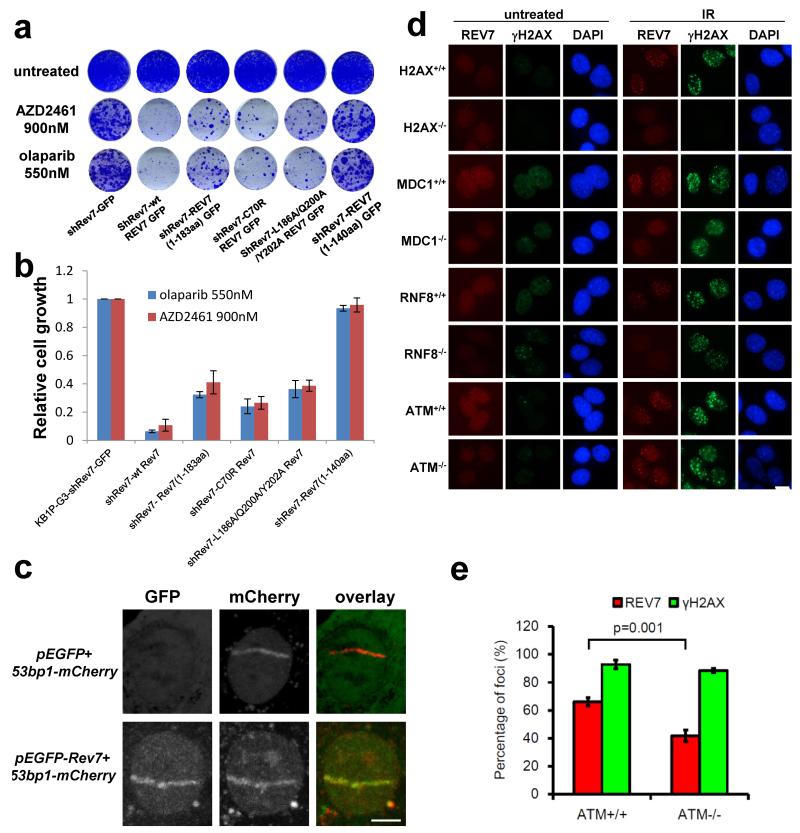
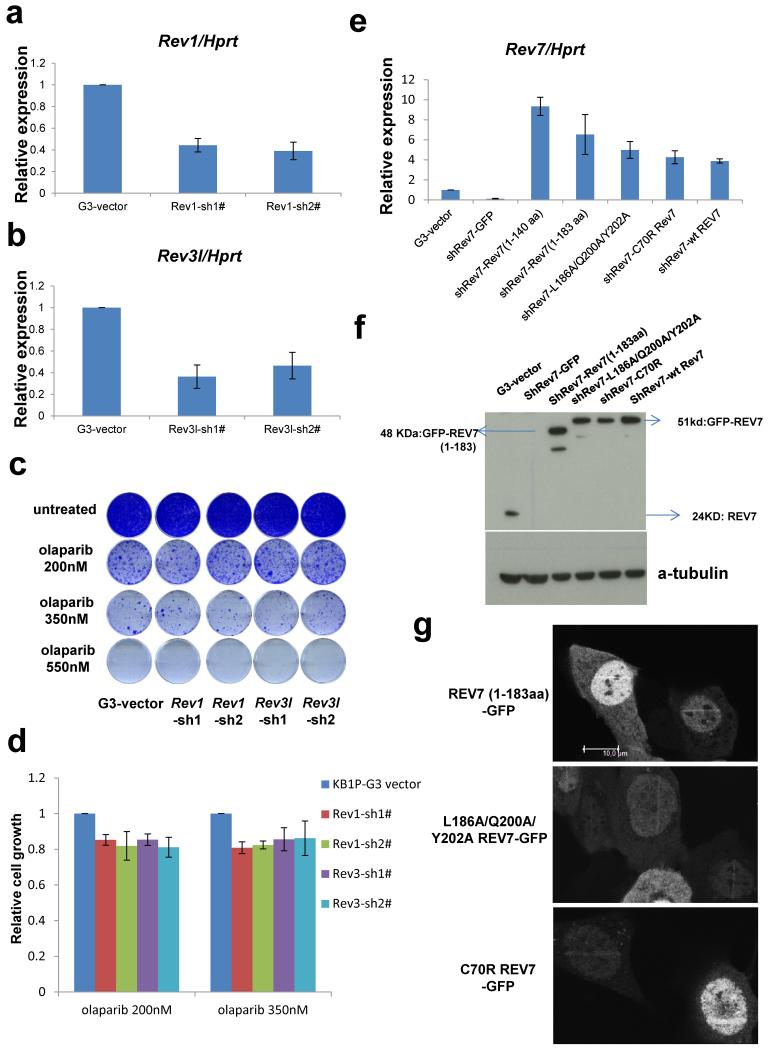
REV7 is known to form the TLS polymerase ζ together with the catalytic subunit REV3, and it interacts with REV19. We therefore investigated whether REV1 or REV3 loss also confers PARPi resistance in Brca1−/−;p53−/− cells. A 60% inhibition of Rev1 or Rev3 transcripts did not cause olaparib resistance (Extended Data Fig. 3a-d). Moreover, we studied various shRNA-resistant REV7 mutants that lack REV1 (L186A/Q200A/Y202A and 1-183aa) or REV3 (C70R) binding sites10,11. In contrast to the truncated 1-140aa REV7 protein, these mutants are recruited to DNA damage sites (Extended Data Fig. 3e-g) and their expression in the KB1P-B11-shRev7 and KB1P-G3-shRev7 cells significantly restored the sensitivity to PARPi to a degree approaching that of wild-type REV7 (Fig. 2a, b; P<0.001, t-test). The remaining differences of the L186A/Q200A/Y202A or C70R mutants with wild-type REV7 may be explained by unequal expression levels (Extended Data Fig. 3f). Together, these data suggest that the REV1 or REV3 interaction is not absolutely required for the REV7-mediated function in this context.
Figure 2. Dissection of REV7 function and its dependent factors.
a, b, Long-term clonogenic assay (a) and quantification (b) using KB1P-G3 cells transduced with the indicated constructs (wt Rev7 stands for pMSCV-GFP-wt Rev7) and treatments. All the groups were normalized by the absorbance of the shRev7-GFP control. The data represent the mean ± SD. c, GFP-REV7 recruitment to sites of DNA damage (visualized by 53BP1-mCherry) was observed 5 min after 405 nm laser exposure (0.99mW, 60% laser power, 50 seconds) in KB1P-B11 cells. Scale bar, 5μm. d, REV7 foci formation in H2AX−/−, ATM−/−, MDC1−/− and RNF8−/− MEF cells and their corresponding controls before and 4h post IR (10 Gy). Scale bar, 10μm. e, Quantification of REV7 foci formation (>8 foci per cell) in ATM−/− and ATM+/+ MEF cells. The quantification of foci-positive cells was performed by counting a total of 100 cells per sample. Data are presented as mean ± SD from 3 different experiments. The P value was calculated using the t-test.
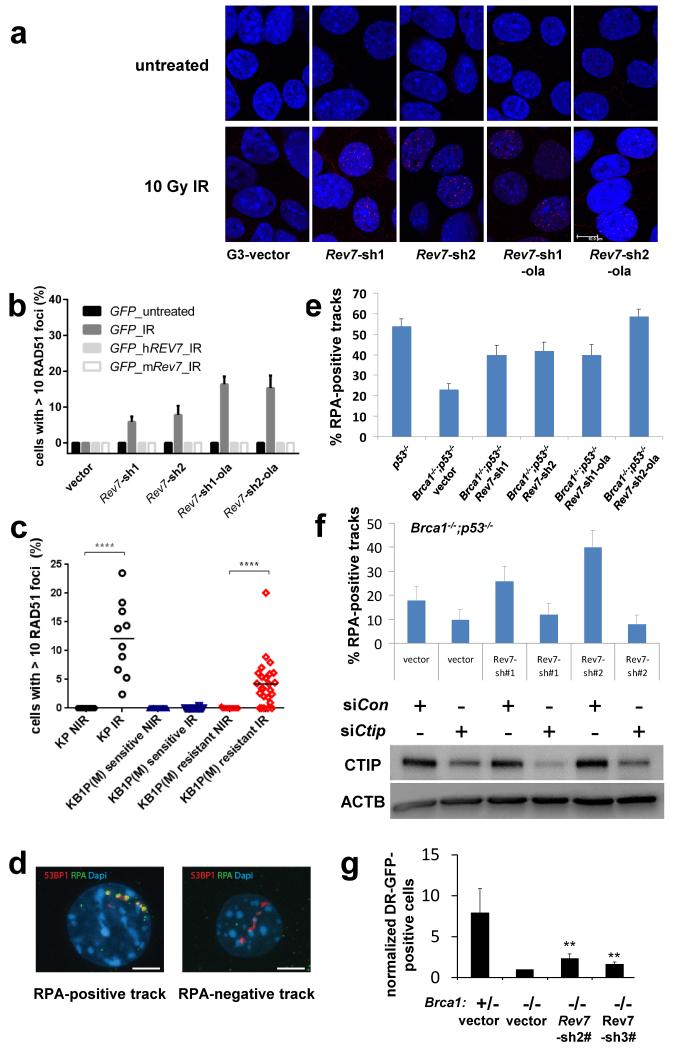
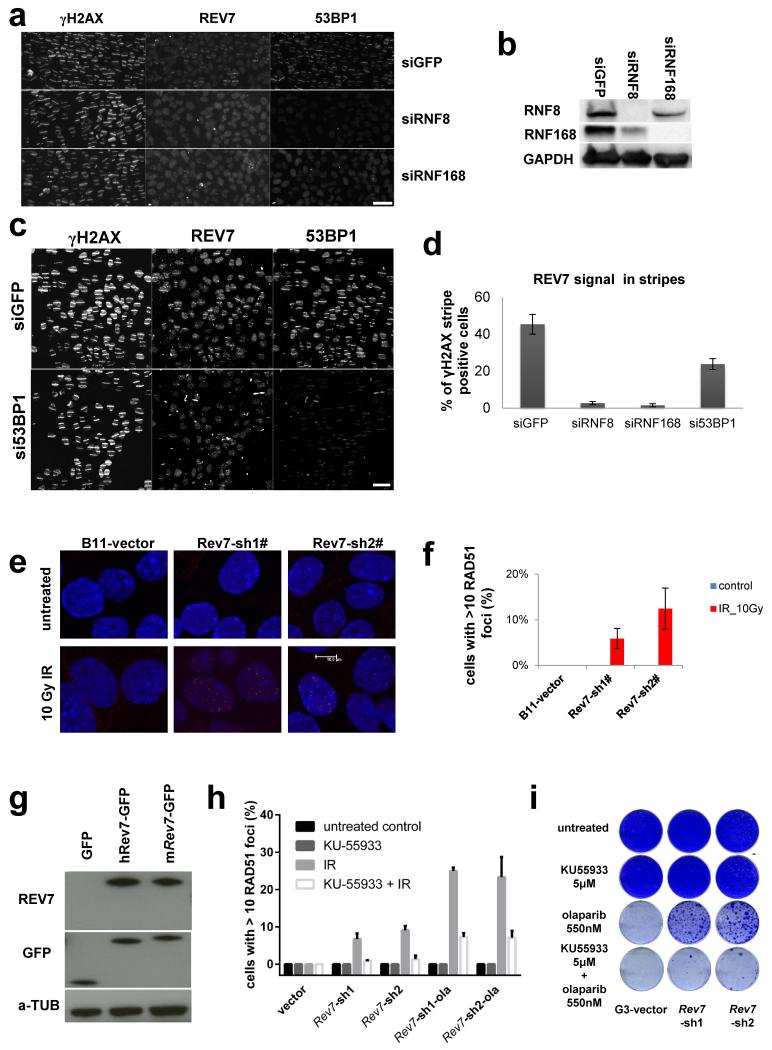
Next, using GFP-tagged REV7, we observed that REV7 co-localizes with 53BP1 shortly after DNA damage induction, suggesting that REV7 acts directly at the site of DNA damage (Fig. 2c). This REV7 recruitment depends on H2AX, MDC1, RNF8, RNF168, and partly ATM, and we observed the requirement of this canonical pathway in both mouse and human cells (Fig. 2d, e and Extended Data Fig.4a-d). To examine whether PARPi resistance in Rev7-depleted Brca1−/−;p53−/− tumor cells is due to HR restoration, we investigated RAD51 focus formation 5h post 10Gy IR. As shown in Fig. 3a, b and Extended Data Fig. 4e, f we observed Rev7 loss to result in the restoration of RAD51 foci formed following DNA damage. To exclude potential off-target effects of the hairpins, we reconstituted shRev7#1+2 cells with shRNA-resistant mouse or human REV7-GFP fusion proteins (Extended Data Fig. 4g). REV7 re-expression abolished RAD51 focus formation upon DNA damage in GFP-positive cells (Fig. 3b). As shown in Fig. 3c, we confirmed the re-appearance of RAD51 foci upon tumor irradiation in vivo using CT-guided high precision cone beam irradiation of animals carrying PARPi-resistant KB1P(M) tumors with low Rev7 gene expression.
Figure 3. The effect of REV7 inhibition on RAD51 and RPA focus formation of Brca1−/−; p53−/− cells.
a, RAD51 focus (red) formation in KB1P-G3 cells before and 5h post IR (10 Gy). Scale bar, 10μm. b, Quantification of RAD51 foci in KB1P-G3 cells (with or without REV7 depletion) transfected with an empty vector (GFP) or vectors containing mouse or human Rev7/REV7. At least 150 GFP-positive cells were analyzed per group in 3 independent experiments each. The data represent the mean ± SD. IR= 5h post 10 Gy. c, In situ analysis of RAD51 foci in PARPi-resistant KB1P(M) tumors with a low Rev7 gene expression. IR= 2h post 15Gy, NIR= no IR, **** P value < 0.0001 (Mann-Whitney test). d, e, Representative examples of images (d) of 53BP1-labelled alpha tracks in cells positive or negative for RPA and quantification of RPA-positive tracks 2h post IR (e). KP (p53−/−) or KB1P-G3 cells with or without Rev7-targeting shRNAs were tested. Scale bar, 5μm. f, Quantification of RPA and 53BP1-positive alpha tracks in KB1P-G3 cells transfected with non-targeting control siRNAs or siRNAs against CtIP. CTIP protein expression of the indicated groups was checked by Western blotting. g, Quantification of HR using the DR-GFP reporter assay. GFP-positive cells normalized to the vector-transduced Brca1−/−; shp53; cells are shown. The data represent the mean ± SD. The P values (** P<0.01) were calculated using the t-test (2-tailed).
We then tested whether the processing of broken DNA ends requires ATM in Rev7-depleted cells, and found that inhibition of ATM using KU55933 efficiently suppresses DNA damage-induced RAD51 foci and increases olaparib sensitivity (Extended Data Fig. 4h, i). Hence, the partial restoration of RAD51 focus formation in Brca1-deficient mammary tumor cells upon DNA damage by inhibition of Rev7 is ATM-dependent.
In contrast to the results with BRCA1-deficient cells, Rev7 depletion in BRCA2-deficient cells did not result in PARPi resistance (Extended Data Fig. 5a-f). Furthermore, we did not observe increased PARPi resistance following Rev7 inhibition in the BRCA1/2-proficient p53−/− tumor cell line KP3.33 (Extended Data Fig. 5g-i). This indicates that REV7 works upstream of BRCA2 and is antagonized by BRCA1. We therefore tested whether DNA end resection is altered in the absence of Rev7 in BRCA1 deficient cells. Accumulation of the single strand binding protein RPA was employed as a marker for the generation of single stranded DNA. Cells were exposed to alpha particles12, and BRCA1 deficiency resulted in a marked decrease in RPA-positive alpha tracks compared to BRCA1-proficient cells (Fig. 3d, e). REV7 depletion in the Brca1−/−;p53−/− cells largely suppressed this defect in both KB1P-G3 and KB1P-B11 cells (Fig. 3e and Extended Data Fig. 6a). In addition to coating resected DNA ends, RPA also interacts with ssDNA gaps (e.g. during replication)13. To exclude that the observed RPA accumulation reflected interaction with internal ssDNA gaps, we prevented DNA end resection (without influencing replication) by knocking down CtIP14. This eliminated the increase in RPA-positive tracks induced by Rev7 knockdown in Brca1−/−;p53−/− cell lines (Fig. 3f and Extended Data Fig. 6b), without affecting cell cycle distribution (Extended Data Fig. 6c). We therefore conclude that increased resection and not binding to ssDNA gaps is responsible for RPA accumulation.
As Rev7 loss could restore end resection in BRCA1-deficient cells, we analyzed whether its depletion could restore full HR proficiency in this context. Using mES cells with a Brca1 selectable conditional knockout allele15, we observed that Rev7 loss indeed prevented cell death of mES cells upon BRCA1 deletion and restored RAD51 focus formation upon DNA damage (Extended Data Fig. 6d-h). Moreover, we reproducibly observed a partial restoration of HR function in the DR-GFP reporter assay for homologous recombination16 when Rev7 was depleted in BRCA1-deficient mES cells (Fig. 3g).
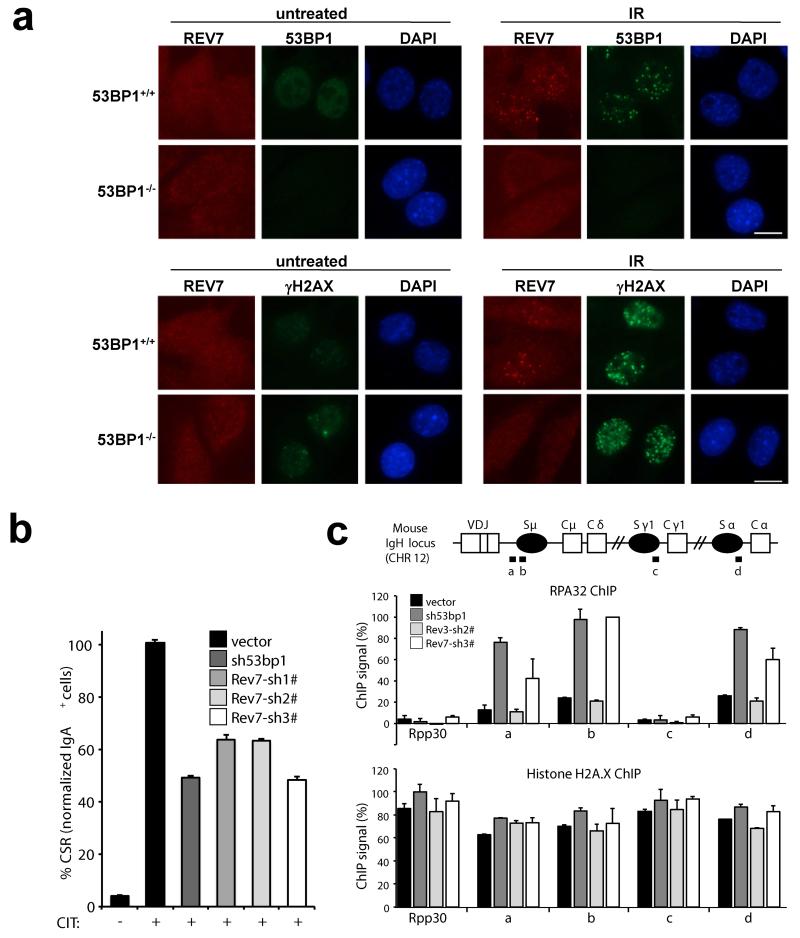
Our data for REV7 are reminiscent of previous findings that 53BP1-loss occurs in subsets of human breast carcinomas15, and can also restore HR to BRCA1-deficient cells7,15,17. Like for 53BP1, we found a frequent aberrant reduction or loss of the REV7 protein in human triple-negative breast carcinomas (Extended Data Fig. 7). Addressing the relationship between REV7, 53BP1 and 53BP1’s NHEJ effector protein RIF118, we found that Rev7 deficiency did not compromise the formation of endogenous 53BP1 or RIF1 foci (Extended Data Fig. 8a-d). In contrast, we observed endogenous REV7 foci or laser-induced stripes to be absent in 53BP1-depleted mouse and human cells (Figure 4a and Extended Data Fig. 4c, d), strongly suggesting that REV7 acts downstream of 53BP1. This is also consistent with our results that PARP inhibitor resistance is not increased when both Rev7 and 53bp1 were depleted (data not shown). Despite such strong evidence for a cooperative role for REV7 and 53BP1, we did not detect REV7 in 53BP1 immunocomplexes isolated from untreated cell lysates, or ATM-phosphorylated 53BP1 immunocomplexes containing RIF1 that were induced by DNA damage19 (Extended Data Fig. 8e and data not shown). Although the intricacies of the interactions remain to be determined, REV7 recruitment to DNA damage sites by 53BP1 may result from indirect interactions or an activity elicited by 53BP1 protein complexes in chromatin at DSB sites.
Figure 4. REV7 is a downstream effector of 53BP1 on inhibiting end resection and promoting CSR.
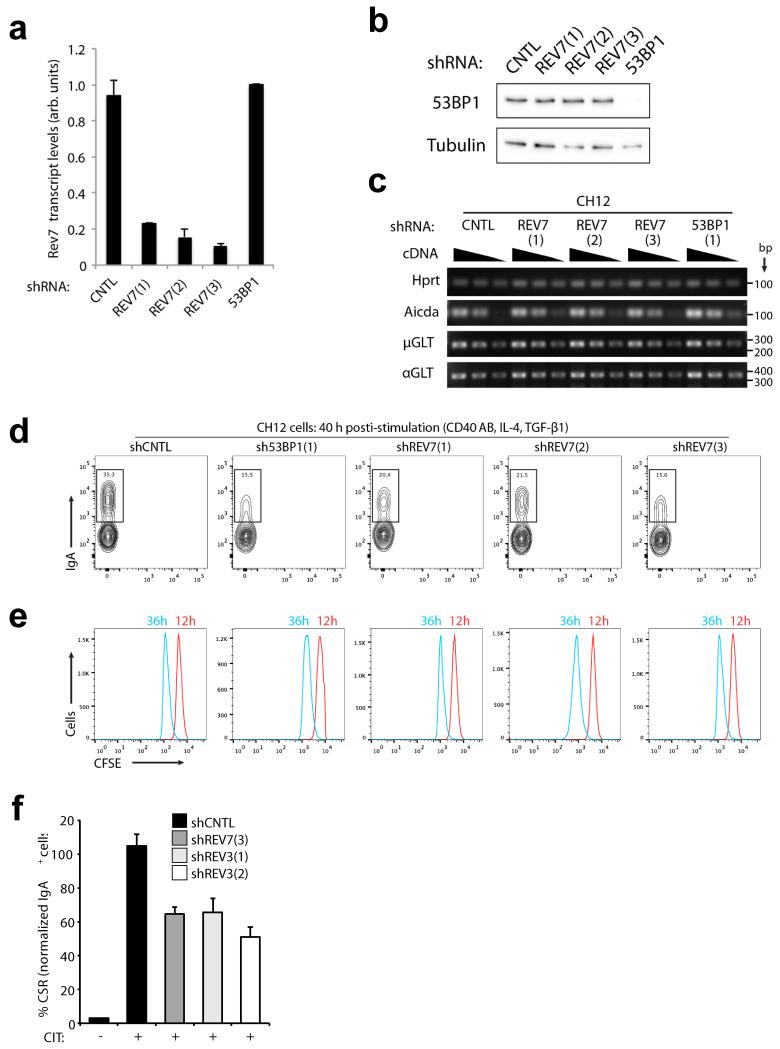
a, REV7 foci formation in 53bp1−/− and 53bp1+/+ MEF cells before and 4h post IR (10 Gy). Scale bar, 10μm. b, Quantification of CSR to IgA of shRNA-transduced CH12 cells 40h post stimulation (CD40Ab, IL-4, TGFβ-1; CIT). Data represent the mean ± SD from two independent experiments performed in triplicate. c, Schematic of IgH locus shows relative positions of qPCR amplicons used in ChIP experiments. A control non- IgH locus (Rpp30) was also examined. Indicated CH12 cell-lines stimulated for 30 h with CIT were subjected to ChIP with IgG (control), histone H2A.X, and RPA32 monoclonal antisera. Following background subtraction, values were normalized to the DNA input signals, followed by the maximum value in each data set. Mean signals, two replicates ± SEM.
To examine whether REV7, like 53BP118, also promotes non-homologous end joining (NHEJ) of DSBs during class switch recombination CSR, we depleted Rev7 transcripts in the murine CH12 B-cell line, which upon stimulation undergo CSR from IgM to IgA at a high rate20. Indeed, efficient Rev7 knockdown was achieved using multiple shRNAs, reducing CSR efficiency to levels comparable with 53BP1-depleted cells when compared to control-depleted cells (Fig. 4b and Extended Data Fig. 9a, b). Moreover, these defects were not accompanied by defects in cell proliferation, AID (Aicda) or germline transcript (μGLT/αGLT) expression (Extended Data Fig. 9c-e). Conditional REV3-ablation has been reported to reduce CSR efficiency in B-lymphocytes21, suggesting that distinct from 53BP1, REV7 might participate in CSR through Polζ function. This considered, we reasoned we might separate the function of REV7 from that of Polζ during CSR, at the level of DSB resection inhibition. To this end, RPA enrichment was measured by chromatin immunoprecipitation (ChIP) at control and multiple IgH loci in control, 53BP1, REV3 or REV7-depleted CH12-lines stimulated to undergo CSR. Consistent with 53BP1’s known role in resection inhibition, 53BP1-depletion was accompanied by 3-5 fold enrichments of RPA ChIP signal specifically in donor (Sμ) and acceptor (Sα) IgH switch (S) regions where DSBs occur during IgM to IgA CSR, but not at an IgH Sγ1 locus or a control non-IgH Rpp30 locus (Fig. 4c). Strikingly, these defects were closely mimicked upon REV7-depletion, while shRNA mediated REV3-depletion yielded no detectable increase in RPA IgH S-region enrichment, despite diminishing CSR efficiency (Extended Data Fig. 9f) as expected21. Importantly, at each locus equivalent ChIP signals for total histone were obtained between cell-lines. Thus, our data support a Polζ-independent function of REV7 in inhibiting the nucleolytic processing of DSBs generated during CSR.
In summary, our data uncover a critical role for REV7 in regulating DSB repair. REV7 depletion restores homology-directed DNA repair of BRCA1-deficient cells resulting in PARPi resistance. We attribute this result to the inhibitory effect of REV7 on DNA end resection. Like 53BP1 together with RIF1 and PTIP18, REV7 may function as a NHEJ factor that performs a regulatory role in DSB repair pathway choice. This provides new insight into the versatile functions of REV7 in addition to its role in translesion synthesis9,22, and helps to explain how HR can be partially restored in the absence of BRCA1.
METHODS
Cell culture and reagents
KB1P_B11 (B11) and KB1P_G3 (G3) cell lines were derived from a Brca1−/−;p53−/− mouse mammary tumor as described7. KB2P_1.21 and KB2P_3.4 cell lines originate from a Brca2−/−;p53−/− mouse mammary tumor and KP3.33 cell line from a p53−/− mouse mammary23. These cell lines were cultured in DMEM/F-12 (Life Technologies) supplemented with 10% FCS, 50 units/mL penicillin, 50ng/ml streptomycin, 5μg/mL insulin (Sigma), 5ng/mL epidermal growth factor (Life Technologies) and 5ng/mL cholera toxin (Gentaur) under low oxygen conditions (3% O2, 5% CO2, 37°C). SUM149PT cells were grown in RPMI1640 (Life Technologies) supplemented with 10% FCS, under normal oxygen conditions (21% O2, 5% CO2, 37°C). U2OS, phoenix, 293T cells were cultured in DMEM (Life Technologies) supplemented with 10% FCS, under normal oxygen conditions (21% O2, 5% CO2, 37°C). Mouse ES cells with a selectable conditional BRCA1 deletion (R26CreERT2/wt;Brca1SCo/Δ)15 were cultured on gelatin-coated plates in 60% buffalo red liver (BRL) cell conditioned medium supplied with 10% fetal calf serum, 0.1 mM β-mercaptoethanol (Merck) and 103U/ml ESGRO LIF (Millipore) under normal oxygen conditions (21% O2, 5% CO2, 37°C). CH12 cells (CH12F3-2) were cultured in RPMI-1640 medium (Sigma) supplemented with 10% FBS, 5% NCTC 109 medium (Sigma), 50μM β-mercaptoethanol, 50 units/mL penicillin and 50ng/ml streptomycin under normal oxygen conditions.
Olaparib and AZD2461 were provided from AstraZeneca, KU-55933 (KuDOS) was bought from Selleckchem (S1092).
Lentivirus-based transduction of cells with shRNA
Glycerol stocks of shRNA hairpins were obtained from the Sigma Mission library (TRC 1.0) and isolation of plasmids was carried out with the high pure plasmid Mini Kit or Genopure maxi kit (Roche). 293T cells were seeded 16 hrs prior to transfection. For each 10cm dish, 0.5ml 2× HBS (8.18 g NaCl, 0.2g Na2HPO4-7H2O, 5.95g HEPES in 500ml MilliQ water and pH7.01) was added into a sterile falcon tube. In another sterile falcon tube, 6μg of plasmid DNA of interest, 2μg of pHCMV-G envelope vector (pMD.G), 2μg of pRSV-Rev, 2μg of packaging vector pMDLg/pRRE, 250μl of 0.5M CaCl2 and distilled water were added to bring up to 0.5ml. The CaCl2/plasmid DNA mix was added to the 2× HBS and incubated for 20 min and then added to the cells. Medium was refreshed after 6h and another 18h respectively. The supernatant of 293T cells containing lentivirus was collected after 24h to infect cells with polybrene (6μg/ml) for 12h. The medium was refreshed after lentivirus infection and the cells were selected with puromycin.
Individual shRNA vectors used were collected from the TRC library.
Mouse Rev7:
Sh1#: TRCN0000012844_ CCAGTGGAGAAGTTTGTCTTT;
Sh2#: TRCN0000012846_ CATCTTCCAGAAGCGCAAGAA;
Sh3#: TRCN0000012847_ GATACAGGTCATCAAGGACTT;
Human REV7:
Sh1#:TRCN0000006569_CCCTGATTCCAAGTGCTCTTA;
Sh2#:TRCN0000006570_CCCGGAGCTGAATCAGTATAT;
Sh3#:TRCN0000006571_CCCAGTGGAGAAATTCGTCTT;
Sh4#:TRCN0000006573_CATCTTCCAGAAACGCAAGAA;
Mouse 53bp1:
(puromycin) sh: TRCN0000081778_GCTATTGTGGAGATTGTGTTT;
(Neomycin) sh: same sequences as above;
Human 53BP1:
Sh1#:TRCN0000018866_CCAGTGTGATTAGTATTGATT;
Sh2#:TRCN0000018865_GATACTTGGTCTTACTGGTTT;
Mouse p53:
(Neomycin) TRCN0000054551_AGAGTATTTCACCCTCAAGAT;
Mouse Rev1:
Sh1#: TRCN0000120298_ GCCGAGATCAACTATGGAATA;
Sh2#: TRCN0000120297_ CAGCAGTGCTTGTGAGGTATT;
Mouse Rev3:
Sh1#: TRCN0000119969_ CCGTCACATTAGTGAGACTAT;
Sh2#: TRCN0000119970_ GCCCACATACACTTTCTTCTT;
Mouse Rif1: (only to referee 3) related to the IR studies
Sh1#: TRCN0000071339_CCCTCTATGATCCGAGAAATA;
Sh2#: TRCN0000071342_CCAGCATATCAGGTTGCTAAT.
Loss-of-function screen
1976 lentiviral hairpins (pLKO.1) from the Sigma Mission library (TRC Mm 1.0) which target 391 mouse genes involved in the DNA damage response were selected (see Supplementary Table 1). This library was used to generate pools of lentiviral shRNA in 293T cells to infect target cells. After infection, the cells stably expressing integrated shRNA were selected with puromycin. Cells with HR restoration were selected with a high concentration of olaparib (500nM, about 100-fold the IC50) which killed cells of the empty vector group. Surviving cells were pooled and genomic DNA was extracted using the Gentra Puregene kit according to the manufacturer’s protocol (Qiagen). ShRNA inserts were retrieved from 50ng genomic DNA by PCR amplification (PCR1 and PCR2) using the following conditions: (1) 95 °C, 5min; (2)95 °C, 30s; (3) 60°C, 30s; (4)72°C, 1min; (5)Go to step (2), 20 cycles; (6)72°C, 5min; (7) 4°C. The PCR reaction system were as follows: 0.6 ul DMSO, 4.0 ul phusion HF buffer 5×, 0.4 ul dNTPs, 1.0 ul primer f (10 mM), 1.0 ul primer r (10 mM), 11.8 ul mQ, 0.2 ul phusion,1.0 ul gDNA for PCR1 or 1ul PCR1 products for PCR2.Adaptors and Indexes for deep sequencing (Illumina HiSeq 2000) were incorporated into PCR primers as follows:
PCR1 forward:
PCR1_01_PLKO1_f_Integration determination_1, ACACTCTTTCCCTACACGACGCTCTTCCGATCT CGTGAT CTTGTGGAAAGGACGAAACACCGG;
PCR1_02_PLKO1_f_Integration determination_2, ACACTCTTTCCCTACACGACGCTCTTCCGATCT GTAGCC CTTGTGGAAAGGACGAAACACCGG;
PCR1_03_PLKO1_f_untreated_1, ACACTCTTTCCCTACACGACGCTCTTCCGATCT CACTGT CTTGTGGAAAGGACGAAACACCGG;
PCR1_04_PLKO1_f_untreated_2, ACACTCTTTCCCTACACGACGCTCTTCCGATCT ATTGGC CTTGTGGAAAGGACGAAACACCGG;
PCR1_05_PLKO1_f_olaparib_1, ACACTCTTTCCCTACACGACGCTCTTCCGATCT GATCTG CTTGTGGAAAGGACGAAACACCGG;
PCR1_06_PLKO1_f_olaparib_2, ACACTCTTTCCCTACACGACGCTCTTCCGATCT TCAAGT CTTGTGGAAAGGACGAAACACCGG;
PCR1_07_PLKO1_f_olaparib_3, ACACTCTTTCCCTACACGACGCTCTTCCGATCT CTGATC CTTGTGGAAAGGACGAAACACCGG;
PCR1_08_PLKO1_f_olaparib_4, ACACTCTTTCCCTACACGACGCTCTTCCGATCT AAGCTA CTTGTGGAAAGGACGAAACACCGG;,
PCR1 reverse: P7_pLKO1_r, CAAGCAGAAGACGGCATACGAGAT TTCTTTCCCCTGCACTGTACCC;
PCR2 forward: P5_IlluSeq, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGAT CT;
PCR2 reverse: P7, CAAGCAGAAGACGGCATACGAGAT.
PCR2 products were purified using the PCR purification Kit from Qiagen. The shRNA stem sequence was segregated and aligned to the TRC library. The reads of different hairpins were counted and the following criteria were used to select the top hits for further validation: first, hairpins targeting the same gene in survival clones should have at least104 reads (total ~6*106 reads); second, at least 2 different hairpins targeting the same gene should be present; third, hairpins in resistant clones should be highly enriched (>8 fold) in cells after olaparib selection; fourth, hairpins should be present in 4/4 independent screens.
PARPi treatment study
Long term clonogenic assay: on d0, 1.5×104 (B11) or 1×104 (G3, KB2P_1.21 or KB2P_3.4) or 6×103 (KP3.33) cells were seeded per well with PARPi (or untreated control) into 6-well plates. The medium of the PARPi treatment groups was refreshed with PARPi every 4 days. On d5, the untreated control group was stopped and the PARPi treatment groups were stopped after another 2~3 weeks and stained with 0.1% crystal violet. Using the SUM149PT cells, 4 ×104 cells were seeded per well with olaparib (or untreated control) into 12-well plates. The medium of the PARPi treatment groups was refreshed with olaparib every 4 days. On d6, the untreated control group was stopped and the olaparib treatment groups were stopped on d8 and stained with 0.1% crystal violet. Quantification of the clonogenic assay was done by determining the absorbance of crystal violet at 590nm.
Short term clonogenic assay: On d0, 4×102 (G3) cells were seeded per well with olaparib (or untreated control) into 6-well plates. On d4, the medium was refreshed with olaparib or untreated control. On d8, all the groups were stained with Leishman dye and quantification was done by the relative colony numbers.
ATMi and PARPi combination study
On d0, 1 ×104 (G3) cells were seeded per well into 6-well plates and then ATMi or olaparib or their combination was added. The medium was refreshed every 3 days with the different drugs. For the combination therapy groups, ATMi was applied for 6 days. On d5, ATMi alone and untreated control groups were stopped and the other groups were stopped on d12 and stained with 0.1% crystal violet.
Constructs
Human REV7 was amplified by PCR using Platinum Taq polymerase (Invitrogen) from U2OS cDNA using the following primers 5′-ATAGAATTCAATGACCACGCTCACACGACAAGAC-3′ and 5′-ATATGGTACCATCAGCTGCCTTTATGAGCGCGC-3′. Mouse Rev7 was amplified from mouse lung cDNA in two parts to introduce silent mutations, making it resistant to Rev7-sh2# (mRev7R). The following primers were used for part A: [IB11m] 5′-ATATGAATTCGATGACCACCCTCACGCGC-3′ [IB14m] 5′-TACTTCTTCCGTTTCTGAAAGATGCCCACCGGGTA-3′ and part B: [IB12m] 5′-ATATGGTACC-AT-GCTGTTCTTATGCGCTCGCT-3′ [IB13m] 5′-GGGCATCTTTCAGAAACGGAAGAAGTACAACGTGC-3′. Equal parts of both PCR reactions were mixed and used for a PCR using IB11m and IB12m to create the complete mouse Rev7 sequence including the silent mutations.
The PCR product and pEGFP-C1 vector were digested using EcoRI and KpnI and ligated using the T4 DNA Ligase (Roche) to generate pEGFP-hRev7 and pEGFP-mRev7R.
Using pEGFP-mRev7R as a template, pEGFP-C1 based Rev7 truncated constructs was amplified by PCR using the following primers:
Forward primer: 5′-ATAT-GAATTC-G-ATGACCACCCTCACGCGC-3′
Reverse primers:
mRev7R (1-55aa): 5′-ATAT-GGTACC-GGACATCTGAACCGGCAC -3′
mRev7R (1-81aa): 5′-ATAT-GGTACC-CTCCACATCGTTCTTCTCCAGG -3′
mRev7R (1-110aa): 5′-ATAT-GGTACC-GATGGACAGCAAGGGAGGC-3′
mRev7R (1-140aa): 5′-ATAT-GGTACC-GTTGTGATCCAGGACAGC -3′
mrev7R (1-183aa): 5′-ATAT-GGTACC-GTCGTGCATGTGGACATCCTG-3′
pEGFP-Rev7 mutants (shRNA resistant) were ordered as gBlocks (IDT) and cloned into the pEGFP-C1 vector using EcoRI and KpnI restriction enzymes and quick ligase (NEB). The DNA sequences that were ordered as gBlocks are:
mREV7_shRNAresistant_wildtype: CGCCGCGAATTCCGCCACCatgaccaccctcacgcgccaagacctcaactttggccaagtggtggctgacg tgctctccgagttcctggaggtggccgtgcacctgattctctatgtgcgcgaggtctacccggtgggCATCTTTCAGA AACGGAAGAAgtacaacgtgccggttcagatgtcctgtcacccggagctgaaccagtacatccaggacacactcc actgcgtcaaacctctcctggagaagaacgatgtggagaaggtggtggtggtgattttggataaggaacaccgcccagtg gagaagtttgtctttgagatcactcagcctcccttgctgtccatcaattcagactccctcctgtctcatgtggagcagctgcttcg agccttcatccttaagattagtgtgtgtgatgctgtcctggatcacaaccctccaggctgcacatttacagtcctcgtgcacac aagagaagctgctactcgaaacatggagaagatacaggtcatcaaggacttcccatggatcctggcagatgaacagga tgtccacatgcacgacccccgcttgatacccctaaaaaccatgacgtcggacattttaaagatgcagctctacgttgaaga gcgagcgcataagaacagctgaGGTACCCCGGG;
mREV7_shRNAresistant_L186A/Q200A/Y202A: CGCCGCGAATTCCGCCACCatgaccaccctcacgcgccaagacctcaactttggccaagtggtggctgacg tgctctccgagttcctggaggtggccgtgcacctgattctctatgtgcgcgaggtctacccggtgggCATCTTTCAGA AACGGAAGAAgtacaacgtgccggttcagatgtcctgtcacccggagctgaaccagtacatcc aggacacactccactgcgtcaaacctctcctggagaagaacgatgtggagaaggtggtggtggtgattttggataaggaacaccgcccagtg gagaagtttgtctttgagatcactcagcctcccttgctgtccatcaattcagactccctcctgtctcatgtggagcagctgcttcg agccttcatccttaagattagtgtgtgtgatgctgtcctggatcacaaccctccaggctgcacatttacagtcctcgtgcacac aagagaagctgctactcgaaacatggagaagatacaggtcatcaaggacttcccatggatcctggcagatgaacagga tgtccacatgcacgacccccgcgctatacccctaaaaaccatgacgtcggacattttaaagatggctctcgctgttgaag agcgagcgcataagaacagctgaGGTACCCCGGG;
mREV7_ shRNAresistant _C70R: CGCCGCGAATTCCGCCACCatgaccaccctcacgcgccaagacctcaactttggccaagtggtggctgacg tgctctccgagttcctggaggtggccgtgcacctgattctctatgtgcgcgaggtctacccggtgggCATCTTTCAGA AACGGAAGAAgtacaacgtgccggttcagatgtcctgtcacccggagctgaaccagtacatccaggacacactcc accgcgtcaaacctctcctggagaagaacgatgtggagaaggtggtggtggtgattttggataaggaacaccgcccagt ggagaagtttgtctttgagatcactcagcctcccttgctgtccatcaattcagactccctcctgtctcatgtggagcagctgcttc gagccttcatccttaagattagtgtgtgtgatgctgtcctggatcacaaccctccaggctgcacatttacagtcctcgtgcaca caagagaagctgctactcgaaacatggagaagatacaggtcatcaaggacttcccatggatcctggcagatgaacagg atgtccacatgcacgacccccgcttgatacccctaaaaaccatgacgtcggacattttaaagatgcagctctacgttgaag agcgagcgcataagaacagctgaGGTACCCCGGG.
mREV7_shRNAresistant_wildtype: CGCCGCGAATTCCGCCACCatgaccaccctcacgcgccaagacctcaactttggccaagtggtggctgacg tgctctccgagttcctggaggtggccgtgcacctgattctctatgtgcgcgaggtctacccggtgggCATCTTTCAGA AACGGAAGAAgtacaacgtgccggttcagatgtcctgtcacccggagctgaaccagtacatccaggacacactcc actgcgtcaaacctctcctggagaagaacgatgtggagaaggtggtggtggtgattttggataaggaacaccgcccagtg gagaagtttgtctttgagatcactcagcctcccttgctgtccatcaattcagactccctcctgtctcatgtggagcagctgcttcg agccttcatccttaagattagtgtgtgtgatgctgtcctggatcacaaccctccaggctgcacatttacagtcctcgtgcacac aagagaagctgctactcgaaacatggagaagatacaggtcatcaaggacttcccatggatcctggcagatgaacagga tgtccacatgcacgacccccgcttgatacccctaaaaaccatgacgtcggacattttaaagatgcagctctacgttgaaga gcgagcgcataagaacagctgaGGTACCCCGGG
mREV7_shRNAresistant_L186A/Q200A/Y202A: CGCCGCGAATTCCGCCACCatgaccaccctcacgcgccaagacctcaactttggccaagtggtggctgacg tgctctccgagttcctggaggtggccgtgcacctgattctctatgtgcgcgaggtctacccggtgggCATCTTTCAGA AACGGAAGAAgtacaacgtgccggttcagatgtcctgtcacccggagctgaaccagtacatccaggacacactcc actgcgtcaaacctctcctggagaagaacgatgtggagaaggtggtggtggtgattttggataaggaacaccgcccagtg gagaagtttgtctttgagatcactcagcctcccttgctgtccatcaattcagactccctcctgtctcatgtggagcagctgcttcg agccttcatccttaagattagtgtgtgtgatgctgtcctggatcacaaccctccaggctgcacatttacagtcctcgtgcacac aagagaagctgctactcgaaacatggagaagatacaggtcatcaaggacttcccatggatcctggcagatgaacagga tgtccacatgcacgacccccgcgctatacccctaaaaaccatgacgtcggacattttaaagatggctctcgctgttgaaga gcgagcgcataagaacagctgaGGTACCCCGGG
mREV7_shRNAr_C70R: CGCCGCGAATTCCGCCACCatgaccaccctcacgcgccaagacctcaactttggccaagtggtggctgacg tgctctccgagttcctggaggtggccgtgcacctgattctctatgtgcgcgaggtctacccggtgggCATCTTTCAGA AACGGAAGAAgtacaacgtgccggttcagatgtcctgtcacccggagctgaaccagtacatccaggacacactcc accgcgtcaaacctctcctggagaagaacgatgtggagaaggtggtggtggtgattttggataaggaacaccgcccagt ggagaagtttgtctttgagatcactcagcctcccttgctgtccatcaattcagactccctcctgtctcatgtggagcagctgcttc gagccttcatccttaagattagtgtgtgtgatgctgtcctggatcacaaccctccaggctgcacatttacagtcctcgtgcaca caagagaagctgctactcgaaacatggagaagatacaggtcatcaaggacttcccatggatcctggcagatgaacagg atgtccacatgcacgacccccgcttgatacccctaaaaaccatgacgtcggacattttaaagatgcagctctacgttgaag agcgagcgcataagaacagctgaGGTACCCCGGG
Using pEGFP-mRev7R, C70R and L186A/Q200A/Y202A mutants as templates, Gateway compatible pMSCV-GFP or Plenti6-UBC (Invitrogen) based Rev7 truncated or Rev7 mutated constructs were amplified by PCR using the following primers:
Forward primer: 5′-gggg aca act ttg tac aaa aaa gtt ggc ATG acc acc ctc acg cgc caa-3′;
Reverse primers:
mrev7R (full length for wt REV7, C70R REV7, L186A/Q200A/Y202A REV7): 5′- gggg aca act ttg tac aag aaa gtt ggg tat tca gct gtt ctt atg cgc tcg ctc -3′;
mRev7R (1-55aa): 5′- gggg aca act ttg tac aag aaa gtt ggg tat TCA GGA CAT CTG AAC CGG CAC -3′;
mRev7R (1-81aa): 5′- gggg aca act ttg tac aag aaa gtt ggg tat TCA CTC CAC ATC GTT CTT CTC CAG G -3′;
mRev7R (1-110aa): 5′- gggg aca act ttg tac aag aaa gtt ggg tat TCA GAT GGA CAG CAA GGG AGG C -3′;
mRev7R (1-140aa): 5′- gggg aca act ttg tac aag aaa gtt ggg tat TCA GTT GTG ATC CAG GAC AGC -3′;
mrev7R (1-183aa): 5′- gggg aca act ttg tac aag aaa gtt ggg tat TCA GTC GTG CAT GTG GAC ATC CTG -3′. PCR products were purified using PCR purification kit (Roche) and then subjected to BP (BP-clonase II, Invitrogen) and LR (LR-clonase II, Invitrogen) reaction according to manufacturer’s instructions.
Wt 53BP1 and 53BP120AQ constructs were previously described19.
All the constructs were verified by sequencing.
siRNA, cDNA transfection and cDNA transduction
SMARTpool siRNAs targeting mouse CtIP (siGENOME: M-055713-02, Thermo Scientific) and non-targeting control were transfected into cells using Dharmacon 1 transfection reagent (Thermo Scientific). 48h later, cells were subjected to western blot and Alpha track assay. siRNAs against 53BP1, RNF8 or RNF168 were transfected into cells using Lipofectamine RNAiMax according to the manufacturer’s instruction. The sequences of si53BP1: 5′-GAGAGCAGAUGAUCCUUUA-3′24; siRNF8: 5′-GGACAAUUAUGGACAACAATT-3′25; siRNF168: 5′-GGCGAAGAGCGAUGGAGGAtt-3′26, siGFP (5′-GGCUACGUCCAGGAGCGCACCTT-3′. Immunofluorescence and Western blotting analysis were done 64 hours after transfection.
For pEGFP-based constructs, transient transfection was done using X-treme GENE HP DNA transfection reagent (Roche) according to the manufacturer’s manual. GFP-positive cells were sorted by flow cytometry and subjected to Western blotting and immunofluorescence staining.
For pMSCV-GFP (retrovirus) based constructs, transient transfection was done using X-treme GENE HP DNA transfection reagent in phoenix cells and the medium was refreshed after 24h. Retroviruses were collected 48h after transfection and then infected target cells for 24h 2 times with an interval of 12h. The medium was refreshed after retroviruses infection and the cells were selected with blasticidin.
For pLenti6-UBC (lentivirus) based constructs, together with the packaging plasmids p59, p60 and p61, transient transfection was done using X-treme GENE HP DNA transfection reagent in 293T cells and the medium was refreshed after 24h. Lentiviruses were collected 48h after transfection and then infected target cells for 24h 2 times with an interval of 12h. The medium was refreshed after lentivirus infection and the cells were selected with blasticidin.
Mice, generation of PARPi resistant mouse mammary tumors
All mouse experiments were approved by the Animal Experiments Review Board of the Netherlands Cancer Institute (Amsterdam), complying with Dutch legislation. Olaparib-resistant KB1PM and AZD2461-resistant KB1P mouse mammary tumors were generated as described7. In this study we analyzed a total of 55 PARPi-resistant and 52 PARPi-sensitive tumors derived from 13 individual KB1PM and 10 individual KB1P donor tumors.
To generate mouse mammary tumors from cell lines, cells (5×105) were orthotopically transplanted into 6-week old female wild-type FVB/N_Ola129 mice as reported previously7. Mice were randomized to the PARPi or untreated control groups.
RT-qPCR
Total RNA was isolated from cells using the high pure RNA isolation kit (Roche). cDNA was made from 1ug RNA with the GoScript™ Reverse Transcription system (Promega). For the quantitative PCR cDNA, Primer Fw&Rv (400nM) and Lightcycler 480 SYBR Green I Master (Roche) were applied in a Lightcycler 480 96-well plate (Roche). The SYBR green signals were measured with Lightcycler 480 II (Roche). The Cp value of the gene of interest was subtracted from the housekeeping gene. This value was put in the power of 2 and this was also done for the SD. The primer sequences used in this study are as follows:
Mouse Hprt forward: CTGGTGAAAAGGACCTCTCG;
Mouse Hprt reverse: TGAAGTACTCATTATAGTCAAGGGCA;
Mouse Rev7 forward: ACACTCCACTGCGTCAAACC;
Mouse Rev7 reverse: AAAGACAAACTTCTCCACTGGGC;
Mouse Rev1 forward: ACAGGATTGCTTGGTGCCTGTG;
Mouse Rev1 reverse: TGAAGTCCGCGTTGCTCTTCTC;
Mouse Rev3 forward: AAGAGATGTCACAGACAGGCCC;
Mouse Rev3 reverse: AGTTAGACAGCCGCTGTTGTGC;
Mouse αGLT forward: GACATGATCACAGGCACAGG;
Mouse αGLT reverse: TTCCCCAGGTCACATTCATCGT;
Mouse μGLT forward: TAGTAAGCGAGGCTCTAAAAAGCAT;
Mouse μGLT reverse: AGAACAGTCCAGTGTAGGCAGTAGA;
Mouse AID forward: GAAAGTCACGCTGGAGACCG;
Mouse AID reverse: TCTCATGCCGTCCCTTGG.
Human Hprt-P1 (primers pair 1) forward: gcagactttgctttccttgg;
Human Hprt-P1 reverse: acacttcgtggggtcctttt;
Human Hprt-P2 forward: tgctcgagatgtgatgaagg;
Human Hprt-P2 reverse: aatccagcaggtcagcaaag;
Human Rev7-P1 forward: tgctgtccatcagctcagac;
Human Rev7-P1 reverse: tcttctccatgttgcgagtg;
Human Rev7-P2 forward: gctcacacgacaagacctca;
Human Rev7-P2 reverse: gaccggcacgttgtacttct;
Mouse 53bp1 forward: TCAGCCAAACAGGACAAGCA
Mouse 53bp1 reverse: GCAGAATCTTCAGCAGCAAGG
Western blot
Cells were washed with ice-cold PBS and lysed on ice for 30 minutes with RIPA lysis buffer supplemented with 3 protease inhibitors (P8340, P5726, P0044, Sigma-Aldrich). Protein concentration was determined by the Bradford Protein Assay Kit (Bio-Rad) and a calibration standard curve created from the BSA. The samples were prepared for loading by adding 4× sample buffer (Invitrogen) and heating the samples at 70°C for 10 min. Total proteins were separated by SDS-PAGE on 3-8% Tris-Acetate (for 53BP, RIF1) or 4-12% Bis-Tris gradient gels (all others). Next, proteins in the gel were electrophoretically transferred to PVDF membrane (Millipore) (for REV7, ACTB, alpha-Tubulin) or to NC membranes (Invitrogen) (all others) and then the membrane was blocked in 5% milk with Tris-buffered saline Triton X-100 buffer (100 mM Tris, pH 7.4, 500 mM NaCl, 0.1% Triton X-100) (TBS-T0.1%). Membranes were incubated with primary antibodies in 5% milk in TBS-T0.1% overnight at 4°C. HRP-conjugated secondary antibody incubation was performed for 1 hour at room temperature in 5% milk in TBS-T0.1% and signals were visualized by ECL. Primary antibodies used in this study were as follows: mouse anti-REV7 (612266, BD Biosciences), 1:5,000 dilution; rabbit anti-53BP1 (NB100-304, Novus), 1:1,000 dilution; rabbit anti-53BP1 (A300-272A, Bethyl), 1:5,000 dilution; rabbit anti-CtIP (ab70163, Abcam), 1:1,000 dilution; mouse anti-alpha-tubulin (T6074, Sigma), 1:5,000 dilution; mouse anti-ACTB (MAB1501R, Millipore), 1:5,000 dilution; rabbit anti-mouse Rif1 (SK1316)19, 1:2,000 dilution; mouse anti-RNF8 (B-2, Santa Cruz), 1:1,000 dilution; rabbit anti-RNF16827, 1:5000 dilution; mouse anti-GAPDH (1D4, GeneTex) 1:1000 dilution. Secondary antibodies used in this study were as follows: polyclonal rabbit anti-mouse immunoglobulins/HRP (P0161, Dako), 1:10,000 dilution; polyclonal swine anti-rabbit immunoglobulins/HRP (P0217, Dako), 1:10,000 dilution.
Immunofluorescence
Cells were grown on glass coverslips (12mm) in 24-well plates. To induce Ionizing Radiation-Induced Foci (IRIF), cells γ-irradiated (10Gy) and compared to non-irradiated controls 5 hours post IR. For this purpose the cells were pre-extracted using cold CSK buffer (10 mM Hepes KOH pH 7.9, 100 mM NaCl, 300 mM Sucrose, 3 mM MgCl2, 1 mM EGTA and 0.5% v/v Triton X100) on ice for 5 min and then cold CSS buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% v/v Tween and 0,5% w/v Sodium Deoxycholate) on ice for 5 min. Cells were washed with PBS++ (PBS with 1mM CaCl2 and 0.5mM MgCl2) and fixed using 2% PFA/PBS++ for 20’ at room temperature (RT). Fixed cells were washed 3× with PBS++ and stored at 4°C. The cells were incubated 20’ in 0.2% Triton X-100/PBS++ to be permeabilized. Then the cells were washed 3× in staining buffer (PBS++, BSA (1%), glycine (0.15%), Triton X-100 (0.1%)), incubated 30’ in staining buffer at RT, incubated with the 1st antibody for 2hrs at RT in staining buffer, washed 3× in staining buffer, incubated with the 2nd antibody for 1hr at RT in staining buffer and washed 3× in staining buffer. Next, the cells were counter-stained with DAPI for 5 minutes, washed in staining buffer, washed in PBS++, mounted in Vectashield and sealed with nail polish. Primary antibodies used in this study were as follows: rabbit anti-RAD51 (70-001, BioAcademia), 1:20,000 dilution; rabbit anti-53BP1 (A300-272A, Bethyl), 1:4000 dilution; Secondary antibodies used in this study were as follows: Alexa fluor 568 F(ab’)2 Fragment goat anti-rabbit (A21069, Invitrogen), 1:1,000 dilution; Alexa fluor 488 anti-mouse antibody (A11001, Invitrogen), 1:1,000 dilution.
For REV7 staining in MEF cells, cells cultured on coverslips were treated with IR (10 Gy) and allowed to recover for 4 hours. Cells were then washed with PBS, pre-extracted with 0.5% triton solution for 3 min and fixed with 3% paraformaldehyde for 12 minutes. Coverslips were washed with PBS and then immunostained with rev7 antibody (612266, BD Biosciences, 1:200 dilution) and anti-γH2AX or anti-53bp1 in 5% goat serum for 60 min at RT. Coverslips were washed and incubated with secondary antibodies conjugated with Rhodemine or FITC for 30 minutes at RT. Cells were then stained with DAPI to visualize nuclear DNA. The coverslips were mounted onto glass slides with anti-fade solution. For RIF1 staining in MEF cells, wild-type MEFs stably transduced with indicated shRNA-expressing lentiviruses were examined for RIF1 foci following neocarzinostatin (NCS) treatment. Automated quantification of RIF1 foci following mock and NCS treatment was performed using Cell-Profiler software (Broad Institute).
Laser irradiation of human cells and Immunofluorescence staining
Cells were grown on plastic disks (17mm in diameter) which were cut using CNC cutter from the bottom of standard 10cm cultivation dish (TPP, Switzerland) UV-sterilized and placed inside the wells of a 12-well plate. BrDU (0.5 μM) was added into siRNA transfected cells 40 hours after the transfection to pre-sensitize cells towards UV-A wavelength. 24 hours after BrDU addition the plastic disk with cells were removed and covered by a coverslip and immediately placed inside Zeiss AxioObserver Z.1 inverted microscope combined with LSM 780 confocal module. Cells were irradiated at 20°C via 40× water immersion objective (Zeiss C-Apo 40×/1.2W DICIII,) using 355nm 65mW laser set on 100% power. The total laser dose which can be further manipulated by the amount of irradiation cycles was empirically set to 6 irradiation cycles. Laser track was pre-defined to cover all the cells within the acquisition area with at least one stripe across the nucleus. After the irradiation process the coverslip was gently removed and plastic disk was quickly placed back into the same well of the 12-well plate and incubated for another 45minutes at standard cultivation conditions. The plastic disks with laser-irradiated cells were first processed by pre-extraction at 4°C. It involves washing by PBS (4°C), equilibration for 2min in sucrose Buffer 1 (10mM PIPES pH 6.8, 100mM NaCl, 1.5mM MgCl2, 300mM sucrose) on ice and then pre-extraction for 15 minutes on ice, on slow-moving shaker using sucrose Buffer 2 (10mM PIPES pH 6.8, 100mM NaCl, 1.5mM MgCl2, 300mM sucrose, 0.5% Triton X, 5ug/ml Leupeptin, 2ug/ml Aprotinin, 0.1mM PMSF,1mM DTT). After the pre-extraction cells were washed by PBS (4°C) and fixed by 4% PAF for 15min at RT. PAF was washed out three times by PBS (RT). The disks were further processed as standard coverslips. It means, blocking in blocking solution (D-MEM + 20% Fetal serum) for 1hour RT followed by incubation with primary antibodies involving REV7 (BD bioscience, Mouse, 1:200), pS139-H2AX (Cell Signaling, 20E3, Rabbit, 1:300) and 53BP1 (Santa Cruz, H-300, Rabbit 1:400) for 2hours at RT and with appropriate secondary antibodies coupled to AlexaFluor 488 and AlexaFlour 568 fluorophores (dilution 1:1000) (Life Technologies). Both primary and secondary antibodies were dissolved in the blocking solution. After washes in PBS, the disks were incubated in 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in dH2O at RT for 5min and air dried. Dried disks were placed on a standard microscopy glass (layer of cells face up) and anchored by two rubber bands laced over the glass. Stained cells were mounted using VectaShield (Vector Labs) mounting medium and covered by coverslip. The samples were examined using Zeiss AxioObserver Z.1 inverted microscope combined with LSM 780 confocal module using 40× oil objective (Zeiss EC PlnN 40×/1.3 Oil DICII). It means that after the first acquisition the plastic disk and the microscopy glass was marked by diamond cutter (to ensure same positioning of the disk in the future), the coverslip was gently removed and disk was washed three times by PBS/Tween 0.5% to remove the mounting medium. Next, the disk was incubated at RT in the 1× Re-Blot solution (Re-Blot Plus Mild, Millipore) for 30minutes on a slow moving shaker. The solution was washed out three times by PBS (RT). Such sample was ready for new staining procedure involving new set of primary and secondary antibodies following the same protocol as described above.
In situ analysis of RAD51 foci formation
5 matched PARPi-resistant and -sensitive KB1P(M) tumors were orthotopically transplanted into wild-type FVB/N recipient mice. When tumors reached about 500 mm3 in volume the mice were randomized to be either irradiated (IR dose: 15 Gy) using a CT-guided high precision cone beam micro-irradiator (X-RAD 225Cx) or to be left untreated. As a positive control a BRCA1-proficient KP tumor was taken along. 2h after the irradiation tumors were taken out and fixed in 4% formalin. IF staining was performed on FFPE slides. RAD51 foci were detected using a non-commercial Ab provided by Roland Kanaar (Erasmus MC) in a dilution of 1:5000. 53BP1 foci were detected using rabbit anti-53BP1 (A330-272A, Bethyl), diluted 1:500. As a secondary Ab for both stainings goat-anti rabbit-AlexaFluor568 (Invitrogen) was used in a dilution of 1:1000 (2ug/ml). Images were taken by a “blinded” investigator using a confocal microscope (Leica SP5, Leica Microsystems GmbH), equipped with a 100× objective. For each tumor 5 random areas (246×246 μm) were imaged. Image stacks (~4 slices) were analyzed in ImageJ, using an in-house developed macro to automatically and objectively evaluate the RAD51 foci. Briefly, nuclei were segmented by thresholding the (median-filtered) DAPI signal, followed by a watershed operation to separate touching nuclei. For every z-stack the maximum-intensity projection of the foci signal was background-subtracted using a Difference of Gaussians method. Next, for each nucleus, foci candidates were identified as locations where the resulting pixel values exceeded the background by a factor (typically 10-fold) times the median standard deviation of all nuclei in the image. In combination with additional filters discriminating for foci size and absolute brightness this procedure yielded a robust and reliable foci count for all nuclei. Results were validated by visual inspection.
REV7 recruitment to local laser-induced DNA damage sites
pEGFP-Rev7 or pEGFP and 53BP1-mCherry were co-transfected into Brca1−/−;p53−/− cells using X-treme GENE HP DNA Transfection Reagent according to the manufacturer’s manual. GFP and mCherry double positive cells were sorted by Flow cytometry and seeded onto coverslips. Cells were sensitized by pre-incubation with Hoechst33342 and were subsequently irradiated using a 405 nm diode laser (63× objective, 0.99mW, 60% laser power, 50 seconds) on a Leica SP5 confocal microscope equipped for live-cell imaging. EGFP-REV7 and 53BP1-mCherry recruitment in living cells was monitored by time-lapse imaging.
Alpha track assay
Cells were seeded in dishes with a mylar surface as previously described28, allowing α-particle irradiation through the bottom of the dish. 1h or 2h post irradiation for 3× 30 sec with a 241Americium source, cells were washed once in ice-cold PBS. Subsequently, cells were extracted with cold CSK buffer (10 mM Hepes KOH pH 7.9, 100 mM NaCl, 300 mM Sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% v/v Triton X100) and cold CSS buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% v/v Tween20, 0.5% w/v Sodium Deoxycholate) for 5 min each before fixation in 4% PFA in PBS for 20 min at room temperature. Fixed cells were washed 5 times in PBS+0.1% Triton X100 and washed once in blocking solution (0.5 %BSA + 0.15 % Glycine in PBS). Primary antibodies were diluted in blocking solution and cells were incubated overnight at 4°C. After incubation, cells were washed 5 times with PBS+0.1% TritonX100 and washed once in blocking solution. Secondary antibodies were diluted in blocking solution and cells were incubated at room temperature for at least 1 hour. Afterwards, cells were washed 5 times in PBS+0.1% TritonX100 and once in PBS. Finally, mylar films were glued on glass slides and cells were mounted using Vectashield with DAPI. For quantification, at least 100 53BP1 or Mre11 positive tracks were scored for the presence of RPA. Primary antibodies used in this study were as follows: Rabbit anti-53BP1 (NB100-304, Novus), 1:1000 dilution; mouse anti-RPA2 (Ab2175, Abcam), 1:500 dilution; MRE11 antibody (de Jager M, et al. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res 29:1317-1325, 2001), 1:200 dilution. Secondary antibodies used in this study were as follows: Alexa fluor 594 Goat anti-Rabbit IgG (A 31631, Invitrogen), Alexa fluor 488 Goat anti-Mouse IgG (A11001, Invitrogen).
BrdU-PI cell cycle assay
Cells were seeded in 6 cm dishes and attached overnight. Next day, cells were incubated for 15 min with 5 μM BrdU in growth medium, trypsinized, washed in PBS and fixed in 70% ethanol overnight. Fixed cells were washed in PBS, resuspended in pepsin solution (5 mg pepsin in 10 ml 0.1N HCl) and incubated for 20’ at room temperature. Subsequently blocking solution (0.5% Tween, 0.1% BSA in PBS) was added to wash. Next, cell were resuspended in 2N HCl and incubated 12’ at 37 °C. To neutralize, 100 mM Borate buffer pH 8.5 was added and cells were pelleted. Anti BrdU-FITC antibody (347583, BD Bioscience) was diluted in blocking solution, added to cells and incubated for 2h on ice. Stained cells were washed once with blocking solution and resuspended in 500 μl PBS + 12.5 μl RNAseA + 1 μl Propidium Iodide (P3566, Invitrogen). Cell cycle analysis was performed the next day.
Survival assay of mES
R26CreERT2/wt; Brca1SCo/Δ mES15 were infected with hairpins targeting Rev7 or the vector control and selected with puromycin. Expression of mouse Brca1 were switched off by overnight incubation with 0.5 μM 4-OHT. Four days after switching, 5,000 cells of the indicated groups were seeded per well into 6-well plates and assayed for growth. Surviving colonies were fixed in formalin and stained with crystal violet.
DR-GFP for HR assay
R26CreERT2/wt; Brca1SCo/Δ mES were targeted with a modified version of the p59X DR-GFP construct as described29. To allow experiments on a p53-deficient background, cells were infected with a lentiviral p53 shRNA (5′-AGAGTATTTCACCCTCAAGAT-3′) using a pLKO1 vector provided with a neomycin resistance marker. A G418 selected p53 deficient clone was subsequently infected with hairpins targeting Rev7 or the vector control and selected with puromycin. Expression of mouse Brca1 was switched off by overnight incubation with 0.5 μM 4-OHT to measure the effect of Rev7 loss on HR. HR reporter assays were performed by Lipofectamine 2000 (Invitrogen) transfection of ISceI-mCherry plasmid which was generated by providing the cBas I-SceI expression plasmid with CMV-mCherry (Clontech). 48 hours after transfection mCherry/GFP double positive cells were monitored by flow cytometry as described15.
53BP1 pull-down
53BP1 pull-downs were performed as described in19. Flag pull-downs were performed from 2mg lysate prepared from MEFs following mock or NCS treatment (2 h at 250 ng/μl). Control, 53BP1, and 53BP120AQ immunoprecipitates were then treated with sequential low salt (150mM) and high salt (500mM) RIPA buffer washes, before re-equilibration in nuclear extract buffer (20 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% Glycerol, 0.5 mM PMSF, 0.5 mM DTT, protease inhibitors (Roche, Complete)). Following incubation in Hela Nuclear Extract (HNE, 2mg), control, 53BP1, and 53BP120AQ complexes were washed 4× in nuclear extract buffer, then eluted with triple-Flag peptide (Sigma)
Immunoglobulin class switch recombination
CH12 cells were either mock treated or stimulated with agonist anti-CD40 antibody (0.5 μg/ml; eBioscience; HM40-3), mouse IL-4 (5 ng/μl; R & D Systems) and TGF-β1 (1.25 ng/μl; R & D systems). Cell-surface IgA expression was determined by flow cytometry, immunostaining with biotinylated anti-mouse IgA antibody (eBioscience; 13-5994), and Alexa488-streptavidin conjugate (Life Technologies).
CFSE assay
Cell proliferation was assessed in stimulated CH12 cells using CFSE according to manufacturer’s instructions (CellTrace™; Life Technologies).
Chromatin Immunoprecipitation
Each ChIP was performed from chromatin prepared from ~107 CH12 cells stimulated for 30 h with agonist CD40 antibody, IL-4, and TGF-β essentially as described19. For each individual ChIP, 4μg of RPA34-20 (Ab-3, Calbiochem), 2μg H2a.X (3522-1, Epitomics), or 4μg control mouse anti-IgG (sc-2025; Santa Cruz) coupled to 25μl Protein-G Dynabeads® (Life Technologies, 10003D) was used. Quantities of immunoprecipitated chromatin were calculated relative to total input chromatin by quantitative PCR in duplicate on an CFX96 Real-Time Analyzer (Biorad) with the use of iQ SYBR® Green (Biorad) for each primer pair. The following sequences were used:
Rpp30 (target Rpp30, Fwd: TCCAGTGTGCAAGAAAGCTAAATG, Rev: GGCAGTGCGTGGAGACTCA);
A (IgH Sμ, Fwd: CAATGTGGTTTAATGAATTTGAAGTTGCCA, Rev: TCTCACACTCACCTTGGATCTAAGCACTGT);
B (IgH Sμ, Fwd: GCTAAACTGAGGTGATTACTCTGAGGTAAG, Rev: GTTTAGCTTAGCGGCCCAGCTCATTCCAGT);
C (IgH Sy1, Fwd: AGTGTGGGAACCCAGTCAAA, Rev: GTACTCTCACCGGGATCAGC);
D (IgH Sα, Fwd: TGAAAAGACTTTGGATGAAATGTGAACCAA, Rev: GATACTAGGTTGCATGGCTCCATTCACACA);
Immunohistochemistry on paraffin sections
The panel of formalin-fixed, paraffin-embedded archival specimens from a series of 50 human primary breast carcinomas was examined, surgically resected before any chemotherapy or radiotherapy. In parallel, sections from 10 specimens of formalin-fixed, paraffin-embedded histologically normal human breast tissues were used as normal tissue controls, for immunohistochemical analysis of REV7 expression patterns. The breast carcinomas were all of the triple-negative type, defined in this cohort as having fewer than 1% tumor cells positive in standard immunohistochemical staining for estrogen receptor and progesterone receptor proteins, and lacking amplification of the HER2 gene. For the immunohistochemical analysis, 4-micrometer thick sections were cut from representative blocks of the tumour tissues, the sections were deparaffinized and processed for sensitive immunoperoxidase staining with the primary mouse monoclonal antibody to REV7 (BD Biosciences, # 612266, diluted 1:100). The staining procedure was essentially as described15, with antigen unmasking in citrate buffer, pH 6,0 for 15-20 min in microwave oven, and overnight incubation with the primary antibody, followed by the Vectastain Elite kit (Vector Laboratories) and nickel sulphate enhancement without nuclear counterstaining30. Mouse normal serum served as negative, and antibody to gH2AX as positive controls, respectively. Evaluation of the staining patterns was performed by two independent observers (with very similar outcome) including a senior oncopathologist with over 20 years of experience with breast cancer pathology. The results were scored in the following categories, based on comparison of cancer cells with the series of normal breast tissue controls, and also the normal cells present within each of the tumour sections. As normal breast epithelium showed reproducible positivity for REV7 protein in over 90% of cells, we considered as aberrantly decreased expression in a tumour when fewer than 70% of cancer cells were positive. In addition to percentage of stained tumour cells, staining intensity was classified as either normal (comparable with the intensity of normal cells present on each section) or aberrantly low (clearly below the intensity seen in adjacent normal cells, and up to undetectable in some cases). Overall, while 6 of 47 informative cases showed concomitantly aberrant fraction of REV7-stained cells and reduced intensity of staining, 12 additional cases showed less pronounced defects limited to either staining intensity or reduced percentage of cancer cells, respectively. As the primary goal of these analyses was to establish a detection assay for REV7 on archival tissue specimens and to assess the frequency of potentially REV7-deficient breast tumours, correlation analyses with clinical parameters including treatment outcome remain to be performed on larger cohorts of patients in the future.
Extended Data
Extended Data Figure 1. Loss of Rev7 causes PARPi resistance in vitro and in vivo.
a, Quantification of Rev7 transcript levels in KB1P-B11 cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± SD. b-c, Cell proliferation rates in KB1P-G3 (b) or –B11 (c) cells analyzed using the MTT assay. d-g, Long-term clonogenic survival assays and quantification of KB1P-G3 (d, f) or –B11 (e, g) cells transduced with the indicated constructs and treatments. All the groups were normalized by the absorbance of the vector control. The data represent the mean ± SD. h, Quantification of the real colony numbers from the short term clonogenic survival assay of KB1P-G3 cells with or without Rev7 loss exposed to olaparib. i, REV7 protein levels were determined by Western Blotting of lysates derived from KB1P-G3 cells transduced with the indicated constructs. j, Overall survival of mice with KB1P-B11-derived Rev7-depleted or control tumors treated with one regimen of 50 mg/kg olaparib daily for 28 days or left untreated. The P value was calculated using the log-rank test. k, l, relative tumor growth of individual KB1P-G3 (k) and KB1P-B11 (l)-derived Rev7-depleted or control tumors treated with one regimen of 50 mg/kg olaparib daily for 28 days or left untreated.
Extended Data Figure 2. Loss of REV7 causes olaparib resistance in BRCA1-deficient SUM149PT cells.
a, Western Blotting analysis of REV7 or 53BP1 expression in SUM149PT cells transduced with REV7- or 53BP1-targeting hairpins or the vector control. b, Example of a long-term clonogenic survival assay using the indicated hairpins and olaparib concentrations. c, Quantification of the clonogenic assays using absorbance of crystal violet at 590nm. The data represent the mean ± SD. All the groups were normalized by the absorbance of the vector control and showed significant differences to the control (P< 0.01, t-test).
Extended Data Figure 3. Rev1 or Rev3 inhibition and PARPi sensitivity of Brca1−/−; p53−/− mammary tumor cells.
a-b, Quantification of Rev1 (a) or Rev3 (b) transcript levels in KB1P-G3 cells transduced with Rev1-or Rev3-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± SD. c, Long-term clonogenic survival assays of KB1P-G3 cells exposed to the indicated PARP inhibitors. d, Quantification of the clonogenic assay by determining the absorbance of crystal violet at 590 nm. All the groups were normalized by the absorbance of the vector control. The data represent the mean ± SD. e-f, Quantification of Rev7 transcript (e) or protein (f) levels in KB1P-G3 cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± SD. g, GFP-tagged REV7 mutants recruitment to sites of DNA damage was observed 5 min after 405 nm laser exposure (0.99mW, 60% laser power, 50 seconds) in KB1P-B11 cells. Scale bar, 10μm.
Extended Data Figure 4. REV7 recruitment to the DNA damage sites in human cells.
a-d, Human REV7 recruitment to sites of laser-induced DNA damage were analyzed in siRNF8, siRNF168 (a) si53BP1 (c) and siGFP U2OS cells. RNF8 and RNF168 protein levels were determined by Western Blotting (b) using lysates derived from U2OS cells transfecting with the indicated siRNAs. For the quantification of the REV7 signal within laser-induced DNA damage stripes (d), a minimum of 100 striped cells (meaning yH2AX positive) were analyzed for the presence of the REV7 signal in two independent experiments. White scale bars = 50 μm. e, RAD51 focus (red) formation in KB1P-B11 cells before and 5h post IR (10 Gy). Scale bar, 10 μm. f, Quantification of RAD51 foci in KB1P-B11 cells in the presence or absence of REV7 depletion. At least 150 cells were analyzed per group in 3 independent experiments each. Error bars indicate SD; IR= 5h post 10 Gy. g, Western Blotting analysis of REV7-depleted KB1P-G3 cells transfected with human REV7-GFP or shRev7 resistant mouse Rev7-GFP fusion proteins. h, Same assay as in (e, f) using the ATM inhibitor KU55933 with or without IR (5h post 10 Gy). i, Long-term clonogenic survival assay of KB1P-G3 cells exposed to olaparib in the presence or absence of KU55933 pretreatment.
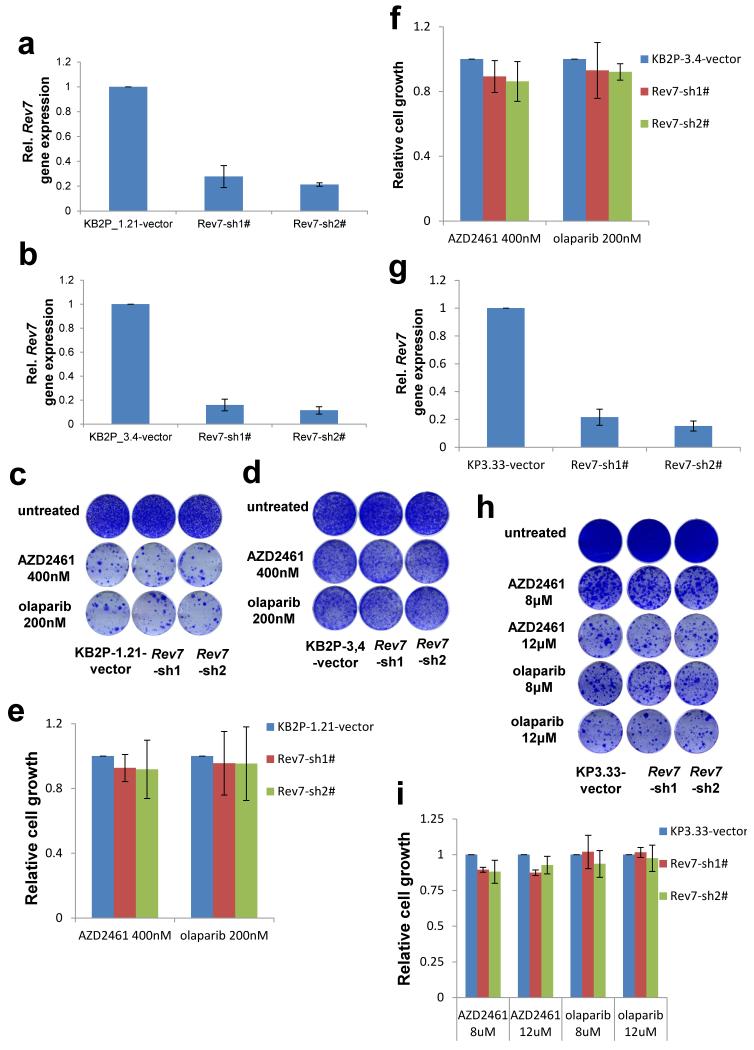
Extended Data Figure 5. Loss of Rev7 does not cause PARPi resistance in Brca2−/−; p53−/− or p53−/− mammary tumor cells in vitro.
a, b, Quantification of Rev7 transcript levels in Brca2−/−;p53−/− (KB2P_1.21 or KB2P_3.4) cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± SD. c-f, Long-term clonogenic survival assays and quantification of KB2P_1.21 or KB2P_3.4 cells with or without Rev7 depletion exposed to the indicated treatments. All the groups were normalized by the absorbance of the vector control. The data represent the mean ± SD. g, Quantification of Rev7 transcript levels in p53−/− (KP3.33) cells transduced with the indicated constructs. Hprt was used as a control for transcript expression and the data represent the mean ± SD. h, i, Long-term clonogenic survival assays and quantification of KP3.33 cells exposed to the indicated treatments. The data represent the mean ± SD.
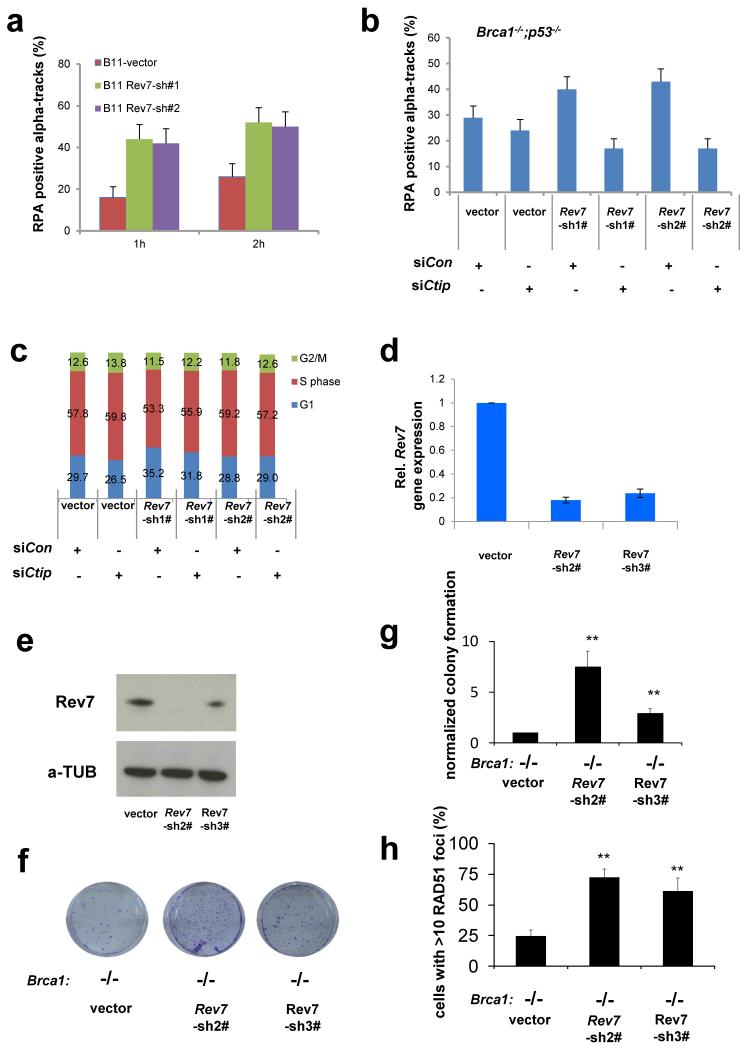
Extended Data Figure 6. Rev7 loss promotes end resection at DSBs in BRCA1-deficient cells after IR.
a, Quantification of RPA positive alpha tracks in KB1P-B11 cells 1h or 2h after irradiation with alpha particles. b, Quantification of RPA and 53BP1-positive alpha tracks in KB1P-B11 cells transfected with non-targeting control siRNAs or siRNAs against CtIP. c, Cell cycle analysis (BrdU incorporation and propidium iodine labeling) of KB1P-B11 cells transduced with the indicated constructs and siRNAs. . d, e, Quantification of Rev7 transcript (d) or protein (e) levels in BRCA1-deficient mES cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± SD. f, Representative images of surviving colonies of Brca1−/− mES cells transduced with an empty vector control or Rev7-targeting shRNAs. g, Quantification of colony formation normalized to the vector control. h, Quantification of RAD51 foci in Brca1−/− mES cells that were transduced with the indicated constructs.
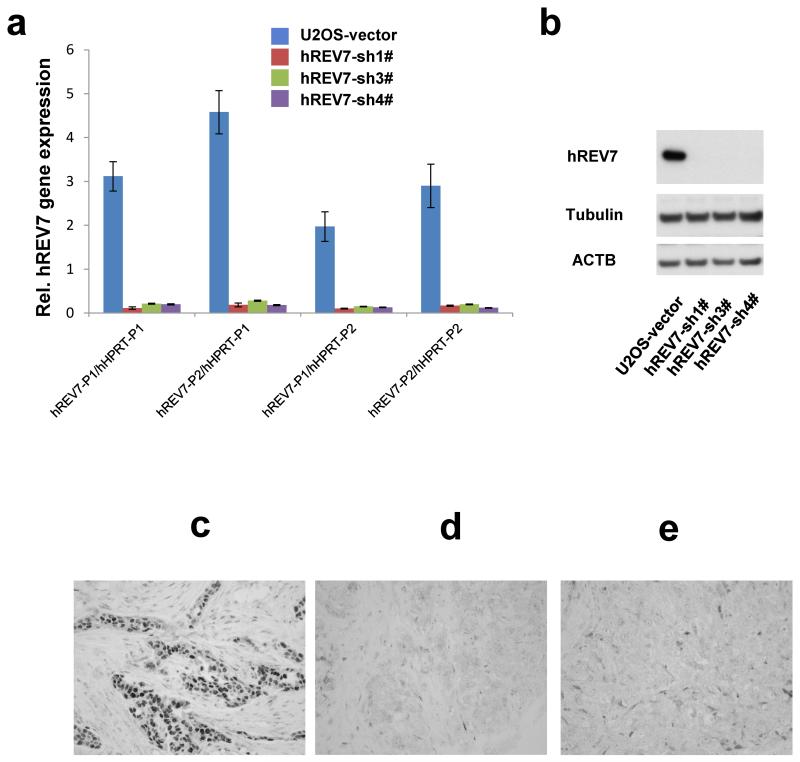
Extended Data Figure 7. REV7 loss frequently occurs in triple-negative breast cancer.
a, b, Quantification of hREV7 transcript levels (a) and protein levels (b) in U2OS cells transduced with indicated constructs. 2 different pairs of primers for hREV7 or hHPRT were used for the quantification of hREV7 transcript levels. c-e, Examples of aberrantly reduced REV7 protein expression in triple-negative human breast carcinomas. Immunohistochemical detection of REV7 in human breast carcinomas shows moderate to high nuclear expression in normal human breast tissue (data not shown) and the majority of invasive breast tumors (c). Aberrant reduction of REV7 with less than 70% of cancer cells that show nuclear positivity (d, e) was observed in 18/50 cases.
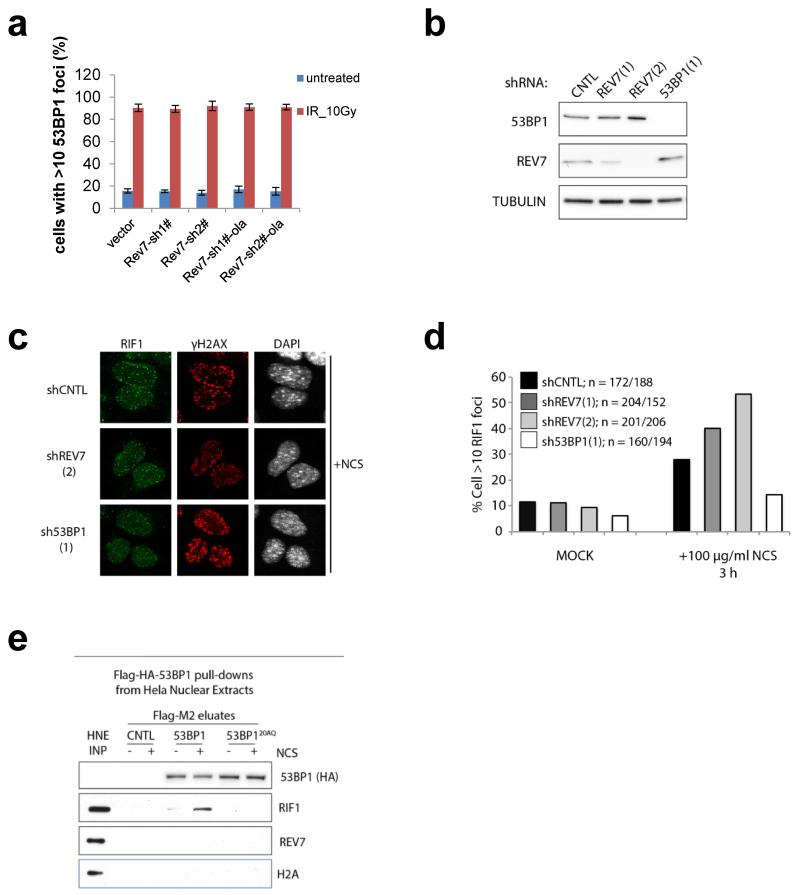
Extended Data Figure 8. REV7 is a downstream effector of 53BP1.
a, Quantification of 53BP1 focus in KB1P-G3 cells in the presence or absence of REV7 depletion. At least 100 cells were analyzed per group in 3 independent experiments each. Error bars indicate SD; IR= 5h post 10 Gy. b, REV7 or 53BP1 protein levels were determined by Western Blotting of lysates derived from MEF cells transduced with the indicated constructs. c, d, RIF1 foci formation (c) following neocarzinostatin treatment and quantification of RIF1 focus (d) in MEF cells in the presence or absence of REV7 or 53BP1 depletion. e, Flag-pulldowns were performed from 2 mg lysate prepared from 53BP1−/−, 53BP1−/−+53BP1 and 53BP1−/−+53BP120AQ MEFs following mock or NCS treatment. Control, 53BP1, and 53BP120AQ immunoprecipitates were incubated in Hela Nuclear Extract (HNE, 2mg) and then eluted with triple-Flag peptide.
Extended Data Figure 9. The effect of REV7 inhibition on CSR after antigenic stimulation of CH12 cells.
a, Rev7 mRNA levels determined by qRT-PCR were normalized against β-actin transcripts in the indicated shRNA-transduced CH12 cell-lines. The data represent the mean ± SEM from two primer sets specific for Rev7 transcript. b, 53BP1 protein of each group normalized to vector-transduced cells (CH12) was analyzed by Western Blotting. c, IgH μ and α germline transcripts and AID (Aicda) mRNA were estimated by semi-quantitative RT-PCR using 2-fold serial dilutions of cDNA made from indicated CH12 cell-lines 40 h following stimulation. Hprt was used as a control for transcript expression. d, Representative flow cytometric profiles of shRNA-transduced CH12 B-cells stained with anti-IgA antibody 40 h following stimulation with indicated cytokines. e, Cells (CH12) were labeled with CFSE immediately before cytokine stimulation as in figure 4d, and cell proliferation was assessed by flow cytometry at indicated time points. f, Quantification of CSR to IgA of shRNA-transduced CH12 cells 40h post stimulation with CD40Ab, IL-4, TGFβ-1 (CIT). Data represent the mean ± SD from 2 independent experiments performed in triplicate.
Supplementary Material
Acknowledgements
We thank Bram Gerritsen, Pasi Halonen and Ben Morris (NKI Robotics and Screening Center), Thanos Halazonetis (University of Geneva) and Olli Kallioniemi (Institute for Molecular Medicine, Helsinki) for advice on the DDR shRNA library, Alessia Gasparini and Gerben Borst (NKI) for their assistance with the cone beam micro-irradiator, Roland Kanaar (Erasmus MC) for his RAD51 antibody, Jacqueline Jacobs (NKI) for the pMSCV-GFP vector, and Mark O’Connor (AstraZeneca) for olaparib and AZD2461. This work was supported by the Netherlands Organization for Scientific Research (NWO-Toptalent to J.E.J. and NWO-VIDI (91711302) to S.R., the Dutch Cancer Society (NKI 2011-5220), CTMM Breast Care, the Swiss National Science Foundation, and the European Union (EU) FP6 Integrated Project CHEMORES and FP7 Project DDResponse. Work in J.R.C.’s group is funded by the Wellcome Trust (090532/Z/09/Z). The work in the J.B’s laboratories was funded by the Danish Cancer Society, the Danish Council for Independent Research, the Lundbeck Foundation and the Czech National Program of Sustainability. S.J.B. is funded by Cancer Research UK and an ERC Advanced Investigator Grant (RecMitMei) and is a Royal Society Wolfson Research Merit Award Holder.
Footnotes
Online content: Additional Methods and Extended Data
The authors declare no competing financial interests.
References
- 1.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J-M, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO. 2013 doi: 10.1093/annonc/mdt384. doi:10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 4.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 2013;19:1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 5.Ang JE, et al. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: a multi-institutional study. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013;19:5485–5493. doi: 10.1158/1078-0432.CCR-13-1262. [DOI] [PubMed] [Google Scholar]
- 6.Chun AC-S, Kok K-H, Jin D-Y. REV7 is required for anaphase-promoting complex-dependent ubiquitination and degradation of translesion DNA polymerase REV1. Cell Cycle Georget. Tex. 2013;12:365–378. doi: 10.4161/cc.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaspers JE, et al. Loss of 53BP1 Causes PARP Inhibitor Resistance in Brca1-Mutated Mouse Mammary Tumors. Cancer Discov. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Listovsky T, Sale JE. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J. Cell Biol. 2013;203:87–100. doi: 10.1083/jcb.201302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan GN, Wittschieben JP, Wittschieben BØ, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 10.Hara K, et al. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J. Biol. Chem. 2010;285:12299–12307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalaj M, et al. A missense mutation in Rev7 disrupts formation of Polζ, impairing mouse development and repair of genotoxic agent-induced DNA lesions. J. Biol. Chem. 2014;289:3811–3824. doi: 10.1074/jbc.M113.514752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stap J, et al. Induction of linear tracks of DNA double-strand breaks by alpha-particle irradiation of cells. Nat. Methods. 2008;5:261–266. doi: 10.1038/nmeth.f.206. [DOI] [PubMed] [Google Scholar]
- 13.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 21.Schenten D, et al. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J. Exp. Med. 2009;206:477–490. doi: 10.1084/jem.20080669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi S, Hara K, Shimizu T, Sato M, Hashimoto H. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J. Biol. Chem. 2012;287:33847–33852. doi: 10.1074/jbc.M112.396838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evers B, et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin. Cancer Res. 2008;14:3916–3925. doi: 10.1158/1078-0432.CCR-07-4953. [DOI] [PubMed] [Google Scholar]
- 24.Fradet-Turcotte A, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe S, et al. JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1-mediated chromatin response to DNA breaks. Nat. Struct. Mol. Biol. 2013;20:1425–1433. doi: 10.1038/nsmb.2702. [DOI] [PubMed] [Google Scholar]
- 26.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 27.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 28.Stap J, et al. Induction of linear tracks of DNA double-strand breaks by alpha-particle irradiation of cells. Nat. Methods. 2008;5:261–266. doi: 10.1038/nmeth.f.206. [DOI] [PubMed] [Google Scholar]
- 29.Bouwman P, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;3:1142–1155. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- 30.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.