Abstract
The known subunits of yeast mitochondrial cytochrome c oxidase are reviewed. The structures of all eleven of its subunits are explored by building homology models based on the published structures of the homologous bovine subunits and similarities and differences are highlighted, particularly of the core functional subunit I. Yeast genetic techniques to enable introduction of mutations into the three core mitochondrially-encoded subunits are reviewed. This article is part of a Special Issue entitled: Respiratory Oxidases.
Keywords: Mitochondria, Cytochrome c oxidase, Complex IV, Yeast, Mutants, Supernumerary subunits
1. Introduction
In eukaryotes, the phosphorylating electron transfer chain is located in the inner membrane of their mitochondria and uses oxygen as the terminal sink for electrons. In mammalian respiratory chains, the sole enzyme that catalyses the transfer of electrons to oxygen to form water is cytochrome c oxidase (CcO) [1]. A second terminal oxidase, the alternative ubiquinol oxidase, can also be present in mitochondria of fungi, yeasts, algae and plants. However, genes for this enzyme (and those for complex I) have been lost in the yeast Saccharomyces cerevisiae which, like mammalian systems, relies entirely on CcO for aerobic respiration. CcO receives electrons from cytochrome c that resides in the intermembrane space. In both mammalian and yeast mitochondria the cytochrome c is reduced by complex III, the cytochrome bc1 complex. However, in yeast mitochondria, cytochrome c can also be reduced by lactate via L(+)lactate–cytochrome c oxidoreductase (flavocytochrome b2), a soluble flavin (FMN)- and haem B-containing enzyme that is co-localised in the intermembrane space [2].
Electrons from four successive cytochrome c oxidations are required, together with four protons from the mitochondrial matrix, to reduce molecular oxygen to two water molecules. In CcO, the intra-protein transfer of these four electrons to the catalytic oxygen reduction site is also linked to the translocation of four protons from the matrix into the intermembrane space [3]. This electron transfer-coupled proton transfer greatly increases the efficiency of the conversion of energy released by the exergonic chemistry of oxygen reduction into stored energy in the transmembrane protonmotive force. The chemistry of oxygen reduction is complex; details and questions of the intermediates involved have been covered in several reviews [4–6].
2. Bovine CcO: structure and mechanism
Bovine mitochondrial CcO is a homodimer with each half composed of the three mtDNA-encoded subunits (I, II and III), together with ten smaller, nuclear-encoded ‘supernumerary’ polypeptides (IV, Va, Vb, VIa, VIb, VIc, VIIa, VIIb, VIIc, VIII; Kadenbach nomenclature [7]). Subunits I, II and III form a catalytic core that is conserved throughout the large superfamily of homologous eukaryotic and prokaryotic oxidases but homologues of the additional subunits are not found in prokaryotic forms of CcO. Atomic structures of several bacterial aa3-type CcOs [8,9] and related superfamily members such as cytochrome bo [10], ba3 [11] and cbb3 [12] are known and the structures of their common core subunits are remarkably similar to those of bovine CcO, the only mitochondrial CcO structure that has been solved to date [13].
2.1. Core structure and mechanism
The core subunit structures and catalytic mechanism have been reviewed extensively and will not be covered in detail here. Subunit II consists of a transmembrane domain of two α-helices, together with a globular domain within the intermembrane space. This latter domain forms part of the docking site for cytochrome c and houses a dinuclear CuA metal centre that receive its electrons and passes them to haem a within subunit I. Subunit III has no cofactor but is nevertheless widely conserved (though absent from some of the more distant superfamily members, for example Thermus thermophilus c552-oxidising cytochrome ba3) [11,14]. It has seven transmembrane helices with short interhelical links. A V-shaped cleft in the folded helical structure may provide a channel through which substrate oxygen diffuses from the membrane into the catalytic binuclear centre (BNC) [8,9,13].
Subunit I has twelve membrane-spanning α-helices when viewed perpendicular to the membrane surface, which are arranged in three groups of four to form a ‘cloverleaf’ pattern [8]. It contains a bis-histidine coordinated low spin haem a, together with the BNC that is formed by a high spin histidine-ligated haem a3 and a copper atom, CuB, that is ligated by further three histidines. One of these, H240, is covalently linked through its Nτ nitrogen to a ring meta carbon of adjacent tyrosine, Y244 [13], which is thought to act as an electron and proton donor during catalysis by formation of a neutral tyrosyl radical [4–6]. The covalent linkage also creates a pentameric ring of amino acids (H240PEVY244), the central glutamic acid (E242 in bovine) of which is an essential component in internal proton transfer. Several arrays of relatively hydrophilic amino acids and water molecules provide possible pathways for the protons required for water formation and for those translocated across the membrane [8,13]. The ‘K’ channel appears to connect the Negative (N) aqueous phase to the BNC. A second, the ‘D’ channel, leads from the N aqueous phase to the E242 residue that is deeply buried and close to the edges of both haems. These structures are clearly evident in most (but not all) bacterial homologues and in bovine CcO. The consensus view for bacterial CcOs is that the K channel provides a route for 1–2 of the four substrate protons and that the D channel is the route of the remaining 3–2 protons required for water formation as well as providing part of the pathway for all four translocated protons [15]. However, a third structure, the ‘H’ channel, has been described in bovine CcO. This spans the entire subunit I close to haem a and has been proposed to provide the route for translocated protons in bovine CcO [16]. It is also evident in part in bacterial CcOs, but mutagenesis studies argue against a similar role in bacterial CcOs [17] although this is still under debate [16]. Current models of coupling between electron transfer and proton translocation include electrostatically-driven movement protons from E242 to a temporary ‘trap’ site [4,18], conformationally-retained strain that is released for proton translocation after the oxygen chemistry has occurred [19] or gated proton movement through elements of the H channel [16].
2.2. Supernumerary subunits
Bovine CcO has 10 additional supernumerary subunits. They do not share sequence or structural similarities and so have not arisen by gene duplication [20]. The absence of homologues in α-proteobacteria or in the mitochondrial genome of the protozoan Reclinomonas americana (which has the largest number of genes retained in its mtDNA) further suggests that they did not evolve from the original promitochondria [20]. They are not centrally involved in the core catalytic mechanism. In general, their specific functions are poorly understood. However, clear homologues of most of them are found in mitochondrial CcOs of quite distant species [20] and several have isoforms in some organisms that are differentially-expressed dependent on growth conditions, tissue type or stage of development.
For example, the heart isoform of subunit VIa (isoform 2, COX6-A2_BOVIN in Table 1) is expressed in heart and skeletal muscle but not in smooth muscle [21]. In human foetal heart and skeletal muscle, expression of both subunits VIa and VIIa switches from liver to heart isoforms after birth [22]. Furthermore, the N-terminal domain of the heart isoform (but not the liver isoform) of bovine subunit VIa can bind ATP or ADP [23]; at high intramitochondrial phosphate potential in a purified reconstituted system, this resulted in a 50% decrease of the H+/e– stoichiometry, an effect suggested to be physiologically-significant [24]. A binding site for ADP involving residues Arg14 and Arg17 of the heart isoform of subunit VIa was also suggested from the crystal structure of bovine CcO [13]. Other studies have suggested additional ATP binding sites on both cytosolic and matrix domains of subunit IV; its matrix-facing ATP site has been reported to be modulated itself by factors binding to the nearby domain of subunit Va [25]. Subunit Vb has a binding site for zinc that is formed by four cysteine residues. It has also been implicated in mitochondrial dysfunction in spinobulbar muscular atrophy through an interaction with human androgen receptor [26]. Specific functions of the remaining supernumerary subunits are not well understood but further roles of some subunits in assembly, stability, monomer/dimer or super-complex formation seem likely.
Table 1.
Sequence identities of subunits of yeast and bovine cytochrome c oxidases. Subunit sequences were obtained from the SwissProt DataBank and aligned automatically without manual corrections. Subunits in the heart form of bovine CcO whose structures were used for homology modelling are highlighted in yellow.
| Yeast subunit (length) | UniProt identifier | Bovine subunita (length) | UniProt identifier | Sequence Identity % (vs shortest sequence) |
|---|---|---|---|---|
| Cox1 (534) | COX1_YEAST | I (514) | COX1_BOVIN | 59% (303/514) |
| Cox2 (236) | COX2_YEAST | II (227) | COX2_BOVIN | 43% (98/227) |
| Cox3 (269) | COX3_YEAST | III (261) | COX3_BOVIN | 47% (122/261) |
| Cox4 (130) | COX4_YEAST | Vb (98) | COX5B_BOVIN | 35% (34/98) |
| Cox5A (133)b | COX5A_YEAST | IV isoform1 (147) | COX41_BOVIN | 20% (26/133) |
| IV isoform2 (171) | E1B8K7_BOVINc | 20% (26/133) | ||
| Cox5B (134)b | COX5B_YEAST | IV isoform1 (147) | COX41_BOVIN | 16% (22/134) |
| IV isoform2 (171) | E1B8K7_BOVINc | 20% (27/134) | ||
| Cox6 (108) | COX6_YEAST | Va (109) | COX5A_BOVIN | 37% (40/108) |
| Cox7 (59) | COX7_YEAST | VIIa isoform 1(59) | CX7A1_BOVIN | 22% (13/59) |
| VIIa isoform 2 (60) | CX7A2_BOVIN | 25% (15/59) | ||
| Cox8 (47) | COX8_YEAST | VIIc (47) | COX7C_BOVIN | 38% (18/47) |
| Cox9 (55)d | COX9_YEAST | VIc (73) | COX6C_B0VIN | 18% (10/55) |
| VIIb (56) | COX7B_BOVIN | 16% (9/55) | ||
| VIII isoform 1 (44) | COX8A_BOVIN | 9% (4/44) | ||
| VIII isoform 2(46) | COX8B_BOVIN | 11% (5/46) | ||
| Cox12 (82) | COX12_YEAST | VIb isoform 1 (85) | CX6B1_BOVIN | 43% (35/82) |
| VIb isoform 2 (88) | CX6B2_BOVIN | 39% (32/82) | ||
| Cox13 (120) | COX13_YEAST | VIa isoform 1 (85) | CX6A1_BOVIN | 33% (28/85) |
| VIa isoform 2 (85) | CX6A2_BOVIN | 35% (30/85) |
Kadenbach nomenclature [7].
Subunits 5A and 5B, which have 68% sequence identity, are expressed under normoxic and hypoxic conditions respectively [58]. The homology model in Fig. 1 was built using Cox5A.
Cox9 has been aligned to subunits VIc, VIIb and VIII for comparison. Although sequence identities with VIc and VIIb are similar, as discussed in Section 5 it is actually distantly homologous only with subunit VIc.
In the case of bovine subunit IV, COX_41_BOVIN is isoform 1 and is present in the heart enzyme. E1B8K7_BOVIN is clearly a second isoform – its full length, unprocessed length of 171 is cited though this probably includes a ~20 amino acid transit peptide.
3. Yeast CcO: genes, genetics and assembly
The genes encoding the subunits of yeast and bovine CcO are distributed in the same way between the mitochondrial and nuclear genomes. The three core subunits, Cox1, Cox2 and Cox3, are encoded by the mitochondrial genome. Mitochondrial genes are transcribed as polycistronic RNAs that are then processed and translated on mitochondrial ribosomes and immediately inserted into the membrane (see Ref. [27] for a review on mitochondrial gene expression). All these steps are practically simultaneous as transcription and translation are coupled and co-localised near the inner mitochondrial membrane [28]. The supernumerary subunits are encoded by nuclear genes, translated within the cytoplasm and imported into mitochondria via the TOM/TIM machineries [29]. Assembly of the subunits proceeds immediately to form the full complex. In addition, supermolecular associations between different mitochondrial complexes, or with the TIM import machinery and the ADP/ATP carrier, have been reported [30–33].
CcO biogenesis has been studied in detail in yeast. The nuclear genes encoding CcO components and many of the extrinsic factors required for the complex biogenesis have been identified by characterising collections of ‘petite’ mutants (pet mutants), mutants unable to grow on respiratory medium because of a defective mitochondrial function due to a nuclear mutation. A large proportion of the PET genes were found to be involved specifically in the biogenesis of CcO, acting at different steps of the process: processing of mitochondrial pre-mRNAs, translation of the mature mRNAs, and post-translational events, mainly assembly of the subunits and cofactors [34,35]. More than 30 such factors have been identified to date; homologues have been identified in humans, some of which have been linked to diseases [36,37].
Assembly of CcO is a rather complicated process and appears to be highly regulated. The majority of information has come from studies of S. cerevisiae though many features appear to be shared by mammalian systems. The mtDNA-encoded Cox1 is the seed or scaffold around which the other subunits are sequentially assembled. Several assembly intermediates have been characterised, the first of which is formed with subunit IV in both human and S. cerevisiae forms of CcO [38]. In S. cerevisiae, unassembled subunits are rapidly degraded by AAA protease [39,40]. In addition, it has been reported that synthesis of Cox1 is decreased when other Cox subunits are unavailable for assembly. Similarly, Cox1 synthesis was severely attenuated in mutant cells that could not bind haem to Cox1 [41]. The regulation of Cox1 translation should prevent the accumulation both of unassembled proteins that might perturb the mitochondrial membrane and also of possible stalled pre-complexes that could contribute to the production of pro-oxidant intermediates [42]. Mss51 seems to play a crucial role in this regulation. This auxiliary factor has a dual function: it is involved in Cox1 translation and also accompanies the newly formed Cox1 in the early steps of assembly [43,44]. In S. cerevisiae, CcO assembly is also regulated by other members of the OXPHOS system as the presence of cytochrome c and of ATP synthase is required for proper CcO assembly; in humans, disease-related defects in ATP synthase can also be linked to CcO deficiencies (reviewed in Ref. [43]).
4. Yeast as a model system for mutation of mitochondrial CcO subunits
Mutant forms of bacterial CcOs have been particularly useful for addressing specific questions of mechanism and roles of specific amino acids [5,45]. However, equivalent information on mutant forms of mitochondrial CcOs is sparse. Particularly problematic is the generation of mutations in core subunits I, II and III because they are usually encoded in mtDNA, the genetic manipulation of which is technically difficult. For mammalian CcO, this has been achieved only in an immortal cell line system in which mutated forms of bovine subunit I were incorporated into human CcO to produce chimeric bovine/human forms [16].
The yeast S. cerevisiae offers a flexible eukaryotic unicellular model system for the study of the biogenesis and catalytic mechanism of respiratory complexes. It is able to rely exclusively on glucose fermentation for its energy requirement, and so can survive without oxidative phosphorylation. This means that it is possible to analyse the consequences of severe defects in respiratory complexes, which in other organisms would be lethal. More importantly, both nuclear and mitochondrial genomes are amenable to genetic transformation. Hence, it is possible to introduce specific mutations into both nuclear- and mtDNA-encoded subunits of the respiratory complexes.
Furthermore, the subunits of yeast and mammalian forms of CcO share extensive sequence similarities [20,46–48]. Hence, any insights into structure/mechanism gained from studies of mutations should also apply to other mitochondrial forms of CcO. As expected, core subunits I, II and III exhibit strong homologies with sequence identities of 59, 43 and 47%, respectively. Table 1 provides a comparative overview of all yeast and bovine subunits. Five of the eight additional supernumerary subunits of yeast CcO have sequence identities of 35% or more with bovine CcO subunits. The lowest sequence identities were those between yeast/bovine Cox5A/IV, Cox7/VIIa and Cox9/VIc; these are discussed in more detail in Section 5.
The yeast system has already provided valuable insights into roles of the supernumerary subunits, in particular from the effects of gene deletions [47]. Deletion mutations in any of COX4, COX5A + COX5B, COX6, COX7, and COX9 genes resulted in the loss of detectable CcO or its activity. Hence, their corresponding subunits must be essential at some stage between expression and stable assembly of the functional enzyme. Null mutants of COX8 resulted in 80% of WT CcO activity levels and null mutants of COX12 resulted in much more severely diminished levels of CcO and its activity. In contrast, null mutants of COX13 expressed CcO at normal levels, though the CcO had different responses to ionic strength and adenine nucleotides. It has been suggested that both Cox8 and Cox13 subunits might be involved in enzyme activity regulation [47]. A suggestion of a regulatory function of Cox13 is strengthened by the fact that it is a homologue of bovine subunit VIa which has been implicated in allosteric control by binding of ATP/ADP [24].
5. A model of yeast CcO based on homology modelling with bovine CcO
A comparative model of the yeast CcO complex was built using Modeller 9v8 [49] with the structure of fully oxidised bovine CcO at 1.8 Å resolution as the template (PDB ID: 1V54, [50]). Sequence alignments between the yeast subunits and the bovine sequences used in the 1V54 structure were prepared as inputs to Modeller. Where the sequence identity between yeast and corresponding bovine subunit was high, MAFFT v6.857 multiple alignment was performed [51] of the sequences of yeast and bovine subunits together with those of mutual orthologues hand selected from UniProt [52] (yeast subunits 1, 2, 3, 6, 8, and 12). Distant homology between Cox5A and bovine subunit IV, Cox7 and bovine subunit VIIa, and Cox9 and bovine subunit VIc was confirmed using the HHpred server (http://toolkit.tuebingen.mpg.de/hhpred) to search the pdb70_9Jul11 library of hidden Markov models [53]. Chains D, J and I of PDB ID: 1V54 were the top matches respectively, with E-values of 0, 1.2e-26 and 0.0081. Where sequence identity between the yeast and corresponding bovine subunit was low, the HHpred alignment was inspected and considered to be superior to that produced by MAFFT (yeast subunits 4, 5A, 7, 9, and 13). No template was available in 1V54 for two substantial N-terminal regions: residues 26–50 of yeast Cox4 and 10–35 of yeast Cox13. Secondary structure predictions for these regions were performed using the PSIPRED v3.0 server (http://bioinf.cs.ucl.ac.uk/psipred/) and a single short α-helix was predicted in each region [54]. Restraints were applied in Modeller to make these regions of the model alpha helical. However, because their spatial positions are uncertain, these parts of the sequences of Cox4 and Cox13 have been omitted from Fig. 1. DOPE-based loop refinement was also applied in Modeller to explore the conformational space available to the remaining loop regions with no template. The N-termini of these loop regions were initially pinned, using the sequence alignments, to the N-termini of the bovine chains to promote compactness of the resulting model. The loop model with the best (lowest) DOPE score was selected. All relevant ligands in the bovine complex were modelled in the yeast complex as rigid bodies. Residue numbers in each chain of the model were changed so that they correspond to positions in the UniProt sequences. Model chain to subunit correspondences are A = 1, B = 2, C = 3, D = 4, E = 5A, F = 6, G = 7, H = 8, I = 9, J = 12, and K = 13. The alignments of all subunits used by Modeller, together with a Table of their sequence identities, are provided in Supplementary information. In several cases, short gaps are present in these alignments. It is not unusual to see such gaps in alignments between pairs of functionally equivalent homologues though detailed functional homology should not be assumed.
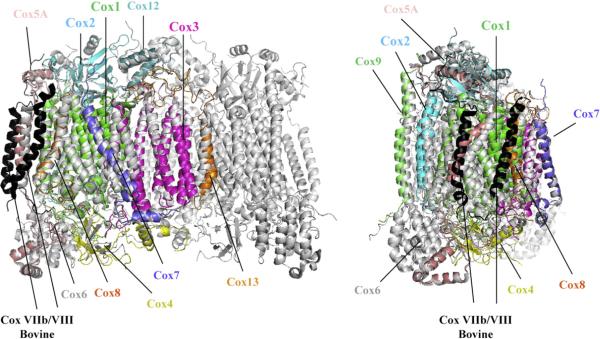
Fig. 1.
Superposition of homology models of the 11 subunits of yeast CcO on the dimeric bovine CcO structure. The whole, dimeric structure of bovine CcO (drawn from PDB ID: 1V54 [16]) is shown in light grey. Each of the eleven subunits of yeast CcO is modelled on its counterpart in one half of the bovine dimer only and is colour-coded to aid recognition. The two bovine subunits without homologues in yeast CcO are shown in black.
The resulting model of the whole monomeric yeast structure (11 subunits with Cox5A isoform) is available for download in PDB format in Supplementary information. It was overlaid on the dimeric bovine heart CcO structure (13 subunits in each monomer, heart isoforms subunits shaded in yellow in Table 1). Two aspects of this homology model are shown in Fig. 1.
5.1. Core subunits and metal binding sites
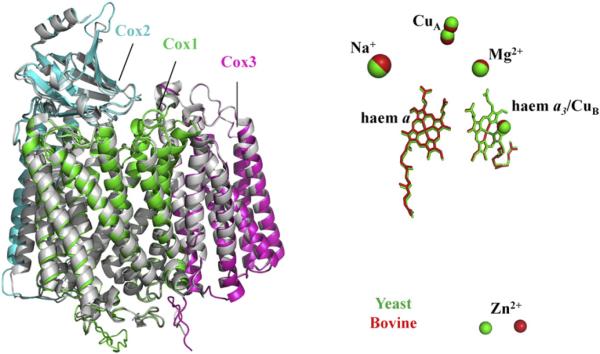
As expected for the common core that is conserved across eukaryotes and prokaryotic haem–copper oxidase, the three core subunits of yeast CcO are of similar lengths and have high sequence identities with their bovine homologues. The same number of membrane-spanning helices (12, 2 and 7, respectively), arranged in roughly the same positions along the primary sequences, are predicted. An overlay of homology-modelled structures of Cox1 and Cox2 on the structures of the bovine subunits has already been presented by Burke and Poyton in relation to Cox5A/B isoforms [55]. An overlay of all three core subunits is shown in Fig. 2A. Since all ligands to the redox centres are conserved and predicted to be located in roughly the same positions, the redox cofactors themselves are modelled in similar positions (Fig. 2B). CcOs also have binding sites for Mg2+ (formed in bovine CcO from H268/D369 of subunit I, E198 of subunit II and waters) and Ca2+/Na+ (formed in bovine CcO from E40 and backbones of Q43, G45 and S441 of subunit I). In yeast CcO, the amino acids that form this Mg2+ binding site are conserved. However, whereas E40 of the bovine Ca2+/Na+ site is conserved, the Q43 and S441 positions are different (A42 and P441); hence, the presence of an equivalent Ca2+/Na+ site in yeast CcO remains more questionable.
Fig. 2.
Superposition of models of the core subunits of yeast CcO on bovine CcO subunits. Colour-coding and source information are as for Fig. 1. The figure on the right shows an overlay of the positions of metal centres (see Section 5.3.1).
5.2. Core subunits and hydrophilic channels
Of particular interest in relation to proton coupling mechanism are the possible hydrophilic channels that provide the necessary routes for intraprotein proton movements. Superposition of the predicted yeast and published bovine subunit I structures suggests that the buried hydrophilic residues in yeast Cox1 map closely onto the K, D and H hydrophilic channel areas of bovine CcO (Fig. 3).
Fig. 3.
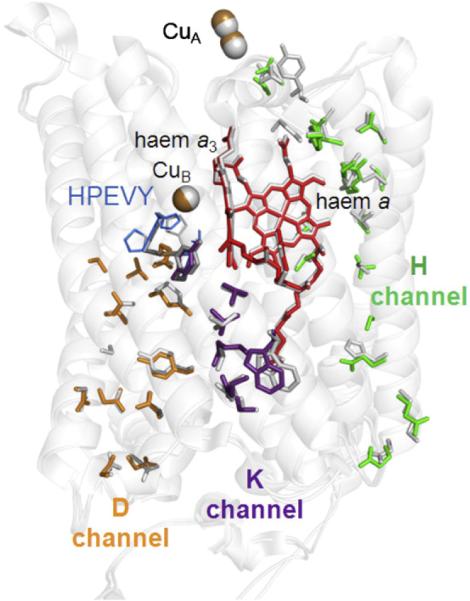
Buried hydrophilic amino acids in Cox1 and 2 of yeast CcO in comparison to the K, D and H channel residues in bovine subunit I. The yeast homology model (in colour) was superimposed onto bovine CcO (PDB ID: 1V54, white). Residues of the K channel (in purple) include: S256, Y245, T316 and K319 of Cox1 together with E82, W85 and T86 of Cox2. Residues of the D channel (in orange) include: N10, Y18, M72, N81, D92, N99, T154, S155, S157, S158, N164 and E243, all in Cox1. Residues of the H channel (in green) include: E407, Q411, Q413, S464, T462, S382, S458, S33, M428, S455, R37, N451, Y371, N56, S52 and N51 of Cox1.
All residues that form the K pathway in bovine CcO are conserved in yeast, from E82 of subunit II at its entrance to Y244 at the catalytic site. The covalent linkage between H240 and Y244 was not imposed on the yeast structure and, as a result, the N(H240)–C(Y244) distance is longer in the homology model (2.78 Å vs. 1.37 Å in bovine). Nevertheless, the overall arrangement of the amino acids 241–245 in a pentameric ring was retained. Hydrophilic residues in the D channel are also conserved, from the D92/N99 pair at its entrance to E243. The only exception is bovine residue S101, facing tyrosine Y19 (Y18 in yeast), which is replaced by an A102 in yeast. The corresponding mutation in Paracoccus denitrificans showed no effect on enzymatic activity [45], suggesting that the functioning of the D channel is unaffected by this substitution [56]. Hence, both the K and the D channels (in purple and orange in Fig. 3, respectively) are clearly evident in yeast CcO.
Many hydrophilic residues can also be identified in yeast CcO in the region of the bovine H channel (in green in Fig. 3). These form in yeast a clear hydrophilic channel that spans the entire subunit I, though there are significant differences in comparison to the bovine structure. In bovine CcO, the matrix-facing entrance is formed by H413, which is H-bonded with two buried water molecules [50] in yeast, this is replaced by a glutamine (Q413). The mutation of the corresponding residue (H448) of P. denitrificans to leucine affected both oxygen reduction and proton pumping activities [45], suggesting a need for a hydrophilic group at this location, though its purpose is still under debate. For instance, in the bovine enzyme, the double mutation M390W/V386L was introduced in order to destabilise and block the H channel waters above H413, and this resulted in loss of coupled proton pumping (but not oxygen reduction) [16]. In yeast CcO, the model suggests that bulkier residues F390 and I386 occupy this region (similar to P. denitrificans CcO, where these residues are F and V). The H channel region closer to haem a appears to be more similar to the bovine structure. It includes the haem a-ligating H378, S382 (a key residue in the proposed gated mechanism for proton movement in the H channel), together with S33 (H-bonded to haem a farnesyl) and R37 (H-bonded to haem a formyl) [50]. In bovine CcO, the top of the H channel is close to the Ca2+/Na+ binding site and also to a nearby sequence (G49–N55) that contains an aspartic acid, D51, that undergoes a dramatic redox-linked conformational change [57]. This region is not well conserved in yeast CcO: in particular, the D51 and a tyrosine, Y54, suggested to be a possible proton donor to D51, are altered (to S and F, respectively). The route for the proton from Y54 to D51 is proposed to involve a tautomerisation of the amide bond between Y440 and S441 with the S441 headgroup also playing a crucial H-bonding role [16,57]. In yeast CcO, the sequence 432–447 is fully conserved except, notably, residues Y440 and S441 which are replaced by I440 and P441.
Hence, in summary, it appears that yeast CcO has K and D channel structures that closely resemble those in other superfamily members. It also has a series of hydrophilic residues that map into much of the region proposed to form the H channel of bovine CcO. However, the model suggests that any H channel in yeast CcO will differ significantly in some parts of the water-containing regions below haem a and at the ‘top’ of the H channel where the Ca2+/Na+ site and the redox-sensitive region that contains D51 are located in bovine CcO. Nevertheless, as pointed out by Shimokata et al. [16], it is possible that such structural variations could still allow a protonmotive function and further experimental data will be required to resolve this issue of H channel function.
5.3. Supernumerary subunits
5.3.1. Cox4
Cox4 was modelled on matrix-located bovine subunit Vb, which ligates a zinc atom with four cysteine residues. Three of these cysteines are conserved in yeast Cox4 but the fourth is substituted by glycine. However, in the predicted folded structure, a methionine (M122) is in close proximity and could provide a fourth ligand. Hence, it seems likely that Cox4 may have retained this zinc site, though its predicted position is shifted slightly in relation to the positions of the core subunits and redox groups (Fig. 2). Cox4 has a substantial N-terminal extension (residues 26–50) that is absent from bovine Vb and is not included in the homology model of Fig. 1. It is predicted to contain α-helical structure. No specific functions of this subunit or its bound Zn are known.
5.3.2. Cox5A/5B
Cox5A and Cox5B are the only examples of isoforms of a subunit of yeast CcO. They share 68% sequence identity and both are homologous to bovine subunit IV. Bovine subunit IV also has two isoforms, though there is no obvious additional similarity between specific yeast/bovine isoforms pairs. In any case, these isoform pairs must have different functional and control details. In mammals, isoform 1 is expressed at high levels only in lung whereas isoform 2 is expressed in all tissues [58]. In contrast, Cox5A and Cox5B are differentially expressed under normoxic and hypoxic conditions, respectively [59] and it has been reported that CcO with the Cox5B subunit has 3–4 times greater kcat than that with Cox5A [60]. A model of these subunits, based on their homology with bovine subunit IV, has been proposed previously [55]. It was concluded that the effect on kcat is mediated through an interaction of the single predicted membrane-spanning α-helix of these subunits with transmembrane helix 12 of Cox1, causing an alteration in the rate of internal haem–haem electron transfer. In the model of Fig. 1, the helical region of subunit 5A lies close to both helix 12 and helix 11 of subunit I. These two helices contain the majority of the hydrophilic residues that appear to form the equivalent of the bovine CcO H channel; these residues face inwards, away from Cox5A/B, to form an internal hydrophilic channel that runs close to haem a. Hence, the intriguing possibility arises that effects on kcat might be mediated through modulation of the properties of the H channel.
5.3.3. Cox6
Cox6 is homologous to bovine subunit Va (37% sequence identity), a subunit that is wholly matrix-located and primarily composed of 5 short α-helices. It interacts closely with part of the large N-terminal matrix domain of subunit IV and has been suggested to influence one of its nucleotide binding sites. Cox6 is predicted to have the same α-helical secondary structure and so had been modelled with virtually the same topology and location (Fig. 1).
5.3.4. Cox7 and Cox8
Cox7 and Cox8 are homologous to bovine subunits VIIa and VIIc, respectively. Sequence identity between Cox7 and VIIa is low but distant homology was confirmed using the HHpred server. In contrast, Cox8 and bovine subunit VIIc share a high sequence identity of 38%. In bovine CcO, these two relatively short polypeptides are both composed primarily of a matrix-located N-terminal domain attached to a single transmembrane α-helix. This domain structure appears to be conserved in yeast Cox7 and Cox8. Despite the clear structural conservation, no specific function of either of these subunits has been identified.
5.3.5. Cox9
Cox9 has been suggested previously to share homology with bovine subunit VIc [20,46–48]. However, sequence identity is low with matched residues spread along the sequences. Distant homology between Cox9 and bovine subunit VIc was however confirmed using the HHpred server even though sequence identity when building the homology model was only 15% (Supplementary Table 1). Cox9 is predicted to have the same domain structure as bovine VIc, namely an N-terminal transmembrane α-helix, together with a small C-terminal domain. Hence, it has been included in the homology model in Fig. 1 based on the bovine VIc structure. No specific functions have been identified.
5.3.6. Cox12 and Cox13
Yeast Cox12/VIb shows the highest sequence identity (43%) of any of the homologous pairs of supernumerary subunits listed in Table 1. VIb faces the IMS and interacts with the VIb subunit of the opposite monomer via (presumably ion pairing of) an antiparallel alignment of a K46GGD sequence located between two short α-helices. When modelled on bovine VIb, Cox12 is predicted to form the same pair of α-helices with the bridging K41GxD sequence between them. It also retains the four cysteine residues that form two disulphide bridges in bovine VIb.
Yeast Cox13 is homologous in part with bovine VIa and is predicted to retain the hydrophobic helix that is presumably similarly positioned between the monomers. Bovine subunit VIa is also thought to house an allosteric nucleotide binding site close to its matrix-facing N-terminus and involving Arg14 and Arg17. However, these residues are not conserved in yeast Cox13. It does however have an additional 37 amino acid domain extension on its matrix-facing N-terminus that is not present in VIa. This domain was therefore omitted from the model shown in Fig. 1. Secondary structure prediction suggests that it contains a single short α-helix but no ligand binding motif could be found. It is possible that this domain may provide the yeast CcO with an as-yet unidentified allosteric regulatory site.
In the bovine structure, subunits VIa and VIb make direct connections with both halves of the homodimeric structure. Subunits VIb of each monomer bind to each other, as described in Section 2.2. Subunit VIa is positioned between helices 4 and 5 of subunit III of one monomer and helix 2 of subunit II and helices 7 and 8 of subunit I of the other monomer. Hence, it seems likely that both of these subunits will contribute to stability of the dimeric structure [13]. Given the striking similarities with bovine VIb and VIa, a major function of yeast Cox12 and Cox13 is predicted to also be dimer formation/stability. Early yeast CcO preparations [61] retained activity but lacked these two subunits and they were established only in later preparations as bone fide components [62,63]. From the above considerations, it seems likely that these earlier preparations may have become monomeric during purification, causing (or due to) loss of Cox12 and Cox13.
5.3.7. Missing subunits
The two bovine subunits that do not appear to have homologues in the 11-subunit yeast CcO are subunits VIIb and VIII. The positions of these two supernumerary subunits in bovine CcO are shown in black in Fig. 1. It is notable that these are rather peripheral and surface-located in the bovine CcO structure and hence should not disrupt other subunit interactions if absent.
6. Yeast mutations in the core subunits of CcO
Point mutations have been introduced into the core subunits of CcO with two parallel approaches: (1) screening of random mutants based on the phenotype; (2) introduction of site-directed mutations in the mitochondrial genes COX1, 2 and 3, with mutations chosen on the basis of the available atomic structures, homology models and catalytic mechanism.
6.1. Screening of random mutants with mutated CcO
Mutants with defective CcO due to random point mutations in the core mitochondrial subunits, Cox1, Cox2 and Cox3, were generated with a genetic screening protocol. This procedure was based on the use of a modified yeast strain in which any deleterious mutations in the mtDNA are counter-selected, except for those located in the CYTB (cytochrome b) or COX genes. Positive mutants were then screened with a genetic test that identified and mapped those with mutations in the COX1, 2 or 3 genes [64].
Using this random mutagenesis approach, 15 mutants with amino acid substitutions in Cox1 were obtained. Nine mutations severely impaired the assembly of the complex as judged by the absence of spectroscopically-detectable CcO. Six mutations resulted in an assembled but dysfunctional complex: I67N [65], T316K [66], G352D, V380M, G384D and D445E [67]. For those mutants with a defective CcO that resulted in a severe respiratory growth defect, revertants could then be selected with secondary mutations that compensated the respiratory defect induced by the primary mutation [67]. With this approach, functional interactions between residues could be identified in pairs of residues that could be spatially close or quite distant in the 3D structure.
6.2. Site-directed mutagenesis by mitochondrial transformation
Mitochondrial biolistic transformation by microprojectile bombardment, adapted and developed from Ref. [68], was used to introduce chosen mutations into the yeast COX gene [69]. Briefly, the mutated DNA coated on tungsten particles is bombarded into recipient cells devoid of mitochondrial DNA (rho0) and a genetic screen is used to identify mitochondrial transformants. These are then crossed with a second recipient strain containing a complete mitochondrial genome (rho+). The highly active homologous recombination machinery in yeast mitochondria allows the integration of the mutated gene at its locus. After a few generations, the cells are homoplasmic i.e. they harbour only one population of mitochondrial DNA, either parental or recombinant. A further genetic screen allows the identification of cells with a rho+ recombinant mitochondrial DNA.
Defined cox mutants have already been produced with this technique, including several mutations reported to be related to human diseases (Table 2). Three of the five Cox3 mutations had no impact in yeast: F251L, on the basis of the structure was not expected to hinder the complex assembly or function; G78S and A200T were also analysed in the bacterial model Rhodobacter sphaeroides and showed no effect on CcO activity. The data thus suggested that these changes might not be the primary cause of mitochondrial dysfunction in the patients. In contrast, two of the mutations, ΔF94–F98 and W249Stop, had severe effects and caused a dramatic decrease in CcO levels in yeast cells, which con-firmed their likely primary pathogenicity in patients.
Table 2.
Disease-related mutations introduced into yeast CcO. See Ref. [73] and MITOMAP: A Human Mitochondrial Genome Database http://www.mitomap.org, 2011 for more extensive details.
| Human CcO mutation (Residue in WT Yeast) | Related disease | Effect on yeast growth in respiratory medium |
|---|---|---|
| Cox1 | ||
| L196I (L197) | Epilepsy | No effect |
| A223S (S224) | Multisystem disorder/polymorphism | No effect |
| M273T (M273) | AISA | Mildly deleterious |
| I280T (I280) | AISA | Mildly deleterious |
| G317S (G317) | Polymorphism | No effect |
| Cox3 | ||
| G78S (G86) | LHON | No effect |
| A200T (G208) | LHON | No effect |
| F251L (F259) | MELAS | No effect |
| ΔF94–F98 (F102–F106) | Myoglobinuria | Severely deleterious |
| W249Stop (W257) | Encephalopathy | Severely deleterious |
Of five reported disease-linked mutations in human Cox1, three were without effect in yeast CcO. G317S occurs as a natural polymorphism and, as therefore expected, the change had no effect on yeast respiratory function. The pathogenicity of A223S is similarly controversial as this change also occurs as a natural polymorphism (MITOMAP: A Human Mitochondrial Genome Database. http://www.mitomap.org, 2011) and indeed in yeast WT this is naturally a serine. The third mutation, L196I, which is close to the D channel, occurs frequently in asymptomatic relatives of patients suffering from epilepsy [70]. Therefore, it is likely that this change has at most a weak effect on the respiratory function, and may not have been detected when monitoring respiratory growth of the equivalent yeast mutant. In contrast, subunit I mutations M273T and I280T, both of which are close to the K channel, have been observed in patients suffering from sideroblastic anaemia (AISA). When introduced into yeast, they caused a twofold decrease in CcO activity [71], which confirmed their likely primary pathogenicity in humans [72].
More recently, amino acid substitutions have been introduced into regions of Cox1 to explore specific structure/function questions, with residues to be mutated chosen on the basis of the homology model presented in Section 5. In order to facilitate the large scale purification of CcO required for detailed biochemical/biophysical analyses of mutated forms, a six-histidine tag has also been added to the nuclear-encoded Cox13 subunit (Meunier et al., submitted). The tag did not affect CcO assembly or activity. To date, mutations have been introduced successfully into all three channel structures, including multiple mutations, and their characterisation is in progress.
7. Conclusions
Comparison of the 11 subunit yeast CcO with its bovine mitochondrial counterpart highlights the remarkable overall structural and functional similarities of both core and supernumerary subunits. The structural homology model further highlights the likelihood that many of the atomic structural features of bovine subunit I appear to be conserved in yeast Cox1, including the presence of a possible H channel equivalent. Crystallisation and structure determination of the yeast CcO will now be required in order to confirm and refine this model and to locate the buried water molecules and conformational states that are crucial for coupled electron/proton transfer. The model nevertheless already provides a clear guide as to which residues should be functionally important and, therefore, should be targeted by mutagenesis. The ability to prepare purified yeast CcO with such mutations in core and/or supernumerary subunits provides a facile system for the study of structure/function aspects of mitochondrial forms of the CcO superfamily.
Supplementary Material
Acknowledgements
This work was supported by the Agence Nationale de la Recherche (grant code ANR-07-BLAN-0360-02 to BM) and the Biotechnology and Biological Sciences Research Council UK (grant code BB/H000097/1 to PR). DL is funded by the NIH as part of the Protein Structure Initiative.
Abbreviations
- CcO
cytochrome c oxidase
- BNC
Binuclear centre
Footnotes
This article is part of a Special Issue entitled: Respiratory Oxidases.
Supplementary materials related to this article can be found online at doi:10.1016/j.bbabio.2011.08.011.
References
- 1.Rich PR, Maréchal A. The mitochondrial respiratory chain. Essays Biochem. 2010;47:1–23. doi: 10.1042/bse0470001. [DOI] [PubMed] [Google Scholar]
- 2.Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 3.Wikström M. Proton pump coupled to cytochrome c oxidase in mitochondria. Nature. 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- 4.Kaila VRI, Verkhovsky MI, Wikström M. Proton-coupled electron transfer in cytochrome oxidase. Chem. Rev. 2010;110:7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 5.Brzezinski P, Gennis RB. Cytochrome c oxidase: exciting progress and remaining mysteries. J. Bioenerg. Biomembr. 2008;40:521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: proton transfer through the respiratory complexes. Annu. Rev. Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadenbach B, Jarausch J, Hartmann R, Merle P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecylsulfate-gel electrophoretic procedure. Analytical Biochemistry. 1983;129:517–521. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- 8.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 9.Svensson-Ek M, Abramson J, Larsson G, Törnroth S, Brzezinski P, Iwata S. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 10.Abramson J, Riistama S, Larrson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikström M. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 11.Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 13.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 14.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem–copper oxygen reductases. Biochim. Biophys. Acta Bioenerg. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 15.Zaslavsky D, Gennis R. Proton pumping by cytochrome oxidase: progress, problems and postulates. Biochim. Biophys. Acta. 2000;1458:164–179. doi: 10.1016/s0005-2728(00)00066-9. [DOI] [PubMed] [Google Scholar]
- 16.Shimokata K, Katayama Y, Murayama H, Suematsu M, Tsukihara T, Muramoto K, Aoyama H, Yoshikawa S, Shimada H. The proton pumping pathway of bovine heart cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4200–4205. doi: 10.1073/pnas.0611627104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H-M, Das TK, Rousseau DL, Mills D, Ferguson-Miller S, Gennis RB. Mutations in the putative H-channel in the cytochrome c oxidase from Rhodobacter sphaeroides show that this channel is not important for proton conduction but reveal modulation of the properties of heme a. Biochemistry. 2000;39:2989–2996. doi: 10.1021/bi9924821. [DOI] [PubMed] [Google Scholar]
- 18.Rich PR. Towards an understanding of the chemistry of oxygen reduction and proton translocation in the iron–copper respiratory oxidases. Aust. J. Plant. Physiol. 1995;22:479–486. [Google Scholar]
- 19.Faxén K, Gilderson G, Ädelroth P, Brzezinski P. A mechanistic principle for proton pumping by cytochrome c oxidase. Nature. 2005;437:286–289. doi: 10.1038/nature03921. [DOI] [PubMed] [Google Scholar]
- 20.Das J, Miller ST, Stern DL. Comparisons of diverse protein sequences of the nuclear-encoded subunits of cytochrome c oxidase suggests conservation of structure underlies evolving functional sites. Mol. Biol. Evol. 2004;21:1572–1582. doi: 10.1093/molbev/msh161. [DOI] [PubMed] [Google Scholar]
- 21.Anthony G, Stroh A, Lottspeich F, Kadenbach B. Different isozymes of cytochrome c oxidase are expressed in bovine smooth muscle and skeletal or heart muscle. FEBS Lett. 1990;277:97–100. doi: 10.1016/0014-5793(90)80817-3. [DOI] [PubMed] [Google Scholar]
- 22.Bonne G, Seibel P, Possekel S, Marsac C, Kadenbach B. Expression of human cytochrome c oxidase subunits during fetal development. Eur. J. Biochem. 1993;217:1099–1107. doi: 10.1111/j.1432-1033.1993.tb18342.x. [DOI] [PubMed] [Google Scholar]
- 23.Anthony G, Reimann A, Kadenbach B. Tissue-specific regulation of bovine heart cytochrome-c oxidase activity by ADP via interaction with subunit VIa. Proc. Natl. Acad. Sci. U. S. A. 1993;90:1652–1656. doi: 10.1073/pnas.90.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank V, Kadenbach B. Regulation of the H+/e– stoichiometry of cytochrome c oxidase from bovine heart by intramitochondrial ATP/ADP ratios. FEBS Lett. 1996;382:121–124. doi: 10.1016/0014-5793(96)00096-8. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig B, Bender E, Arnold S, Hüttemann M, Lee I, Kadenbach B. Cytochrome c oxidase and the regulation of oxidative phosphorylation. Chem. Biochem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 26.Beauchemin AMJ, Gottlieb B, Beitel LK, Elhaji YA, Pinsky L, Trifiro MA. Cytochrome c oxidase subunit Vb interacts with human androgen receptor: a potential mechanism for neurotoxicity in spinobulbar muscular atrophy. Brain Res. Bull. 2001;56:285–297. doi: 10.1016/s0361-9230(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 27.Lipinski KA, Kania-Golik A, Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome — from genetics to evolution and systems biology. Biochim. Biophys. Acta. 2010;1797:1088–1098. doi: 10.1016/j.bbabio.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Naithani S, Saracco SA, Butler CA, Fox TD. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 30.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1773–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- 32.Saddar S, Dienhart MK, Stuart RA. The F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J. Biol. Chem. 2008;283:6677–8666. doi: 10.1074/jbc.M708440200. [DOI] [PubMed] [Google Scholar]
- 33.Dienhart MK, Stuart RA. The yeast Aac2 protein exists in physical association with the cytochrome bc1–COX supercomplex and the TIM23 machinery. Mol. Biol. Cell. 2008;19:3934–3943. doi: 10.1091/mbc.E08-04-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pel HJ, Tzagoloff A, Grivell LA. The identification of 18 nuclear genes required for the expression of the yeast mitochondrial gene encoding cytochrome c oxidase subunit 1. Curr. Genet. 1992;21:139–146. doi: 10.1007/BF00318473. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann JM, Funes S. Biogenesis of cytochrome oxidase — sophisticated assembly lines in the mitochondrial inner membrane. Gene. 2005;354:43–52. doi: 10.1016/j.gene.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- 38.Ugalde C, Coenen MJH, Farhoud MH, Gilinsky S, Koopman WJH, van den Heuvel LP, Smeitink JAM, Nijtmans LGJ. New perspectives on the assembly process of mitochondrial respiratory chain complex cytochrome c oxidase. Mitochondrion. 2002;2:117–128. doi: 10.1016/s1567-7249(02)00012-0. [DOI] [PubMed] [Google Scholar]
- 39.Langer T. AAA proteases: cellular machines for degrading membrane proteins. Trends Biochem. Sci. 2000;25:247–251. doi: 10.1016/s0968-0004(99)01541-8. [DOI] [PubMed] [Google Scholar]
- 40.Tatsuta T, Langer T. AAA proteases in mitochondria: diverse functions of membrane-bound proteolytic machines. Res. Microbiol. 2009;120:711–717. doi: 10.1016/j.resmic.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 2010;30:1004–1017. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khalimonchuk O, Bird A, Winge DR. Evidence for pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- 43.Fontanesi F, Soto IC, Barrientos A. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life. 2008;60:557–568. doi: 10.1002/iub.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfitzner U, Odenwald A, Ostermann T, Weingard L, Ludwig B, Richter O-MH. Cytochrome c oxidase (heme aa3) from Paracoccus denitrificans: analysis of mutations in putative proton channels of subunit I. J. Bioenerg. Biomembr. 1998;30:89–97. doi: 10.1023/a:1020515713103. [DOI] [PubMed] [Google Scholar]
- 46.Capaldi RA, González-Halphen D, Takamiya S. Sequence homologies and structural similarities between the polypeptides of yeast and beef heart cytochrome c oxidase. FEBS Lett. 1986;207:11–17. doi: 10.1016/0014-5793(86)80004-7. [DOI] [PubMed] [Google Scholar]
- 47.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu. Rev. Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 48.Barrientos A, Gouget K, Horn D, Soto IC, Fontanesi F. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta. 2009;1793:97–107. doi: 10.1016/j.bbamcr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 50.Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinozawa-Itoh K, Yamashita E, Yao M, Ishimura Y, Yoshikawa S. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The UnitProt Consortium Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burke PV, Poyton RO. Structure/function of oxygen-regulated isoforms in cytochrome c oxidase. J. Exp. Bot. 1998;201:1163–1175. doi: 10.1242/jeb.201.8.1163. [DOI] [PubMed] [Google Scholar]
- 56.Pfitzner U, Hoffmeier K, Harenga A, Kannt A, Michel H, Bamberg E, Richter O-MH, Ludwig B. Tracing the D-pathway in reconstituted site-directed mutants of cytochome c oxidase from Paracoccus denitrificans. Biochemistry. 2000;39:6756–6762. doi: 10.1021/bi992235x. [DOI] [PubMed] [Google Scholar]
- 57.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, Yamaguchi H, Tomizaki T, Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 58.Hüttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267:111–123. doi: 10.1016/s0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- 59.Burke PV, Raitt DC, Allen LA, Kellog EA, Poyton RO. Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J. Biol. Chem. 1997;272:14705–14712. doi: 10.1074/jbc.272.23.14705. [DOI] [PubMed] [Google Scholar]
- 60.Allen LA, Zhao X-J, Caughey W, Poyton RO. Isoforms of yeast cytochrome c oxidase subunit V affect the binuclear reaction center and alter the kinetics of interaction with the isoforms of yeast cytochrome c. J. Biol. Chem. 1995;270:110–118. doi: 10.1074/jbc.270.1.110. [DOI] [PubMed] [Google Scholar]
- 61.Power SD, Lochrie MA, Sevarino KA, Patterson TE, Poyton RO. The nuclear-coded subunits of yeast cytochrome c oxidase. I. Fractionation of the holoenzyme into chemically pure polypeptides and the identification of two new subunits using solvent extraction and reversed phase high performance liquid chromatography. J. Biol. Chem. 1984;259:6564–6570. [PubMed] [Google Scholar]
- 62.Taanman J-W, Capaldi RA. Purification of yeast cytochrome c oxidase with a subunit composition resembling the mammalian enzyme. J. Biol. Chem. 1992;267:22481–22485. [PubMed] [Google Scholar]
- 63.Geier BM, Schägger H, Ortwein C, Link TA, Hagen WR, Brandt U, von Jagow G. Kinetic properties and ligand binding of the eleven subunit cytochrome c oxidase from Saccharomyces cerevisiae isolated with a novel large scale purification method. Eur. J. Biochem. 1995;227:296–302. doi: 10.1111/j.1432-1033.1995.tb20388.x. [DOI] [PubMed] [Google Scholar]
- 64.Meunier B, Lemarre P, Colson AM. Genetic screening in Saccharomyces cerevisiae for large numbers of mitochondrial point mutations which affect structure and function of catalytic subunits of cytochrome-c oxidase. Eur. J. Biochem. 1993;213:129–135. doi: 10.1111/j.1432-1033.1993.tb17742.x. [DOI] [PubMed] [Google Scholar]
- 65.Meunier B, Ortwein C, Brandt U, Rich PR. Effect of mutation of residue I67 in yeast cytochrome c oxidase on redox-linked protonation processes. Biochem. J. 1998;330:1197–1200. doi: 10.1042/bj3301197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ortwein C, Link TA, Meunier B, Colson A-M, Rich PR, Brandt U. Structural and functional analysis of deficient mutants in subunit I of cytochrome c oxidase from Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1997;1321:79–92. doi: 10.1016/s0005-2728(97)00035-2. [DOI] [PubMed] [Google Scholar]
- 67.Meunier B, Rich PR. Second-site reversion analysis is not a reliable method to determine distance in membrane proteins: an assessment using mutations in yeast cytochrome c oxidase subunits I and II. J. Mol. Biol. 1998;283:727–730. doi: 10.1006/jmbi.1998.2132. [DOI] [PubMed] [Google Scholar]
- 68.Bonnefoy N, Fox TD. Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol. 2001;65:381–396. doi: 10.1016/s0091-679x(01)65022-2. [DOI] [PubMed] [Google Scholar]
- 69.Meunier B, Taanman J-W. Mutations of cytochrome c oxidase subunits 1 and 3 in Saccharomyces cerevisiae: assembly defect and compensation. Biochim. Biophys. Acta. 2002;1554:101–107. doi: 10.1016/s0005-2728(02)00217-7. [DOI] [PubMed] [Google Scholar]
- 70.Varlamov DA, Kudin AP, Vielhaber S, Schröder R, Sassen R, Becker A, Kunz D, Haug K, Rebstock J, Heils A, Elger CE, Kunz WS. Metabolic consequences of a novel missense mutation of the mtDNA CO I gene. Hum. Mol. Genet. 2002;11:1–9. doi: 10.1093/hmg/11.16.1797. [DOI] [PubMed] [Google Scholar]
- 71.Meunier B. Site-direct mutations in the mitochondrially-encoded subunits I and III of yeast cytochrome oxidase. Biochem. J. 2001;354:407–412. doi: 10.1042/0264-6021:3540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bröker S, Meunier B, Rich PR, Gattermann N, Hofhaus G. MtDNA mutations associated with sideroblastic anaemia cause a defect of mitochondrial cytochrome c oxidase. Eur. J. Biochem. 1998;258:132–138. doi: 10.1046/j.1432-1327.1998.2580132.x. [DOI] [PubMed] [Google Scholar]
- 73.Bratton M, Mills D, Castleden CK, Hosler J, Meunier B. Disease-related mutations in cytochrome c oxidase studied in yeast and bacterial models. Eur. J. Biochem. 2003;270:1–9. doi: 10.1046/j.1432-1033.2003.03482.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.