Abstract
The Luminex xTAG® respiratory viral panel (RVP) kit simultaneously detects and identifies multiple respiratory viruses including several subtypes of influenza A using a multiplex nucleic acid amplification test assay platform. The emitted fluorescence signal from the RVP assay provides qualitative information on the presence of a particular viral species in respiratory specimens. However, a quantitative assessment is preferred when monitoring environmental samples for respiratory viruses. In this study, we explored the potential use of the RVP kit as a semi-quantitative screening assay for influenza virus detection. The concentration- response of the RVP assay was modeled using four-parameter logistic (4-PL) fits of mean fluorescence intensity (MFI) versus dilute ranges of the influenza A matrix gene, seasonal influenza vaccine, and 2009 H1N1 influenza vaccine. The goodness of fit of the 4-PL model was evaluated by comparing the copy number determined with the fitted model (observed copy number) with the copy number calculated from the dilution of the matrix DNA or vaccine (expected copy number). For the matrix DNA and 2009 H1N1 vaccine, the 4-PL model provided good fit for the influenza A RVP assay response over factors of 103 to 104. For seasonal influenza vaccine, the model provided good fit for RVP assay response to influenza A, influenza B, H1, and H3.
Keywords: Influenza virus, polymerase chain reaction, semi-quantitative multiplex measurement
Introduction
Taqman® real-time polymerase chain reaction (real-time PCR)-based techniques can be used to produce qualitative or quantitative results depending on the application. Diagnosis of influenza virus infection can be done using real-time PCR which can provide a qualitative identification of the strain of influenza A subtype H1N1 virus causing the infection (World Health Organization, 2009). Techniques employing quantitative PCR (qPCR) have been developed for applications such as determination of viral contamination in water samples (Karim et al. 2009). For the determination of the relative contribution of various routes of exposure to influenza virus infection, quantitative assessment of surface contamination and air concentration in the environment can be done with qPCR using calibration with influenza virus vaccine (Blachere et al. 2007) or calibration with influenza matrix gene DNA (Lindsley et al. 2010). These qPCR techniques have been utilized to assess airborne concentrations in healthcare facilities (Blachere et al. 2009) or from cough samples (Lindsley et al. 2010).
The Luminex xTAG® respiratory viral panel (RVP) kit was developed to diagnose multiple respiratory viral infections simultaneously using a multiplex microbead-based technique (Krunic et al. 2007; Mahony 2007a; Mahony et al. 2007b; Merante et al. 2007). It has also been useful for recognizing the presence of unknown strains of the H1 subtype of influenza A (Ginocchio and St. George 2009a; Ginocchio et al. 2009b), where response to influenza A but no response to H1 or H3 subtypes showed the possible presence of a novel strain of H1 influenza A. The RVP kit has been used in a qualitative manner in applications where response above a threshold is used to indicate the presence of a given viral species. In this proof of concept study, we measured the response of the RVP kit over a range of concentrations to explore its use for semi-quantitative or quantitative applications such as environmental monitoring. There was a limited amount of valid response data produced from this study since there were few RVP kits available. We therefore had to limit our evaluation to determining the response of the kits for a semi-quantitative interpretation of the data and areas for more work are given in the discussion.
Methods and materials
Reagents
The Luminex xTAG® RVP kits (TDAS RVP-1 version 1.11 Lot IK019C-0012) were manufactured by Luminex Molecular Diagnostics (Toronto, Ontario). The kits have all reagents needed to perform the assay. The RNase-free water was obtained from Invitrogen (Carlsbad, CA, USA).
Vaccine samples and cloned DNA
The influenza vaccines used in the production of response curves were 2005–2006 seasonal FluMist and 2009 H1N1 FluMist. The 2005–2006 seasonal FluMist was purchased from MedImmune Vaccine, Inc. (Gaithersburg, MD, USA). The 2005–2006 seasonal FluMist was a live, trivalent vaccine, composed of the A/New Caledonia/20/99 (H1N1), A/California/7/2004 (H3N2), and B/Jiangsu/10/2003 (B/Shanghai/361 /2002-like) strains. These strains were genetically altered to attenuated, cold-adapted, and temperature-sensitive phenotypes, which limits viral replication to the nasal pharynx. Each 0.5 mL dose has been formulated to contain approximately 107 TCID50 (106.5–107.5 median tissue culture infectious dose) per viral strain. In calculating concentrations, it was assumed that there are 2 × 107 copies of each virus in 1 mL of the undiluted vaccine so that H1 subtype, H3 subtype, and influenza B were present at 2 × 107 copies/mL in the undiluted vaccine. However, the influenza A matrix gene (the influenza A response of the RVP kit) was present for both strains of influenza A so it was assumed to be 4 × 107 copies/mL in the undiluted vaccine. The 2009 H1N1 FluMist vaccine was purchased from MedImmune and was monovalent (A/California/7/2009 (H1N1) strain) with a 0.2 mL dose containing 107 TCID50 (106.5–107.5 median tissue culture infectious dose). In calculations, it was assumed that the undiluted vaccine had 5 × 107 viral copies/mL.
The cloned influenza matrix gene M1 used in this pilot study was kindly provided by Francoise Blachere, NIOSH (Morgantown, WV, USA). The matrix DNA was diluted by factors of 10 from 10−4 to 10−12 with RNase-free water before running with the RVP kit and the concentration in the diluted samples was determined by multiplying the stock concentration by the dilution factor.
RNA extraction
The 2005–2006 seasonal and 2009 H1N1 vaccines were diluted by factors of 10 from 10−2 to 10−8 with RNase-free water before RNA extraction and the concentration in the diluted samples was determined by multiplying the undiluted vaccine concentration by the dilution factor. The RNA was extracted from each diluted vaccine sample using a MagNA Pure Compact (MPC) (Roche Diagnostics, Indianapolis, IN, USA) with an RNA isolation kit (Roche 04 802 993 001) and a QIAamp MinElute Virus Spin kit (VSK) (product number 57704, Qiagen, Valencia, CA, USA).
The MPC instrument performs the lysis, binding via magnetic beads to adsorb the RNA, washing, and elution of the RNA steps automatically. For the MPC extraction procedure, 159 μL of the diluted vaccine was combined with 15.9 μL of phage MS2 internal control from the RVP kit and 175 μL of lysis buffer from the RNA isolation kit. The treated samples were loaded into the instrument and run with RNA Tissue-V3-1 program with the sample volume set at 350 μL. All reagents needed for the extraction were contained in kit cartridges that were loaded into the instrument. The final elution volume was set at 50 μL.
Manual isolation of viral RNA was performed using the VSK according to manufacturer instructions. For the VSK procedure, 182 μL of diluted vaccine was combined with 18.2 μL of the phage MS2 internal control from the RVP kit, 25 μL protease, and 200 μL lysis buffer and incubated for 15 min at 56°C. Ethanol was added to the treated sample and the RNA was adsorbed onto a membrane cartridge where it was washed several times with different kit buffers and ethanol and finally eluted into 50 μL. In calculating the concentration of recovered RNA from the MPC- and VSK-extracted samples, it was assumed that all the RNA from the diluted vaccines was extracted and was concentrated in the extraction procedure by a factor (initial sample volume/final elution volume) (159/50 for the MPC and 182/50 for the VSK).
RNA and DNA determination using the RVP kit
The xTAG RVP kit was used in accordance to the manufacturer instructions. The thermal cycler employed was a model PTC-100, Programmable Thermal Controller (MJ Research, Inc., Watertown, MA, USA). Detection of positive and negative bead signals was performed using a BioRad Bioplex model 100 system (BioRad, Hercules, CA, USA). Data collection with the Bioplex system used Luminex IS 100 2.3 software (Luminex Inc., Austin, TX, USA) with the xTAG RVP T-A kit software template. In addition to the extracted RNA from the diluted vaccines and the diluted matrix DNA samples, three negative controls (RNase-free water) and a run control (Lambda DNA supplied with the kit) were included in each run as specified by the kit instructions.
Data analysis
The batch file containing the data from the experiment was used to obtain mean fluorescence intensity (MFI) values for the diluted vaccine and matrix DNA samples. The dilution calculated copy number for each sample was calculated by multiplying the concentration as determined above by the volume of sample used in the reverse transcription PCR (RT-PCR) step of the assay, which was 5 μL. The MFI values for the diluted H1N1 and seasonal vaccine samples were normalized by calculating the average response from the internal control (phage MS2) for the vaccine samples. The MFI value for each individual diluted vaccine sample was multiplied by the ratio: average internal control MFI/individual sample internal control MFI. This helped to correct the differences in RNA extraction and well to well variations in the assay. The MFI versus dilution calculated copy number curve was fitted using four-parameter logistic model (4-PL, SigmaPlot, SPSS, Chicago, IL, USA). Assessment of the “goodness of fit” and the dynamic ranges of the assays were investigated by evaluating the fit of the standards data to the 4-PL model by “standards recovery” (Nix and Wild 2001), calculated by evaluating interpolated results from each 4-PL fit (observed copy number) and comparing it to the calculated copy number derived from dilution of the vaccine or matrix DNA (the expected copy number). The resultant data were analyzed for linearity by linear regression.
Results
Influenza A matrix gene DNA
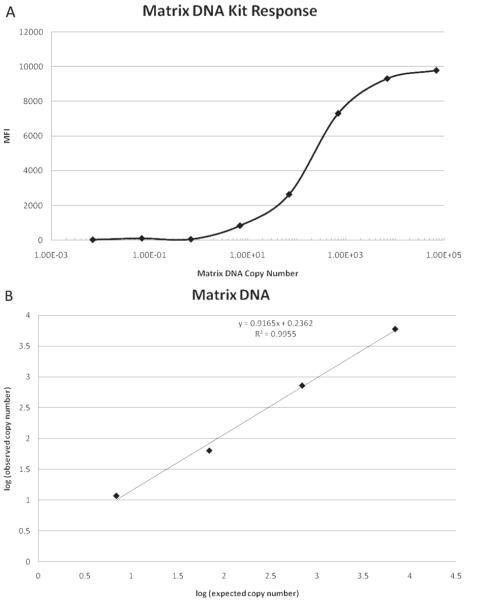
Figure 1A shows the influenza A MFI response of the RVP kit as a function of the dilution calculated matrix gene DNA copy number. The assay response begins to rise when the dilution calculated copy number becomes greater than 1 (at 6.93 copies) and levels off at 6.93 × 103 copies. The response curve was fitted with the 4-PL model from 6.93 × 10−2 copies up to 6.93 × 104 copies and a fit with R2 > 0.999 was obtained. Observed copy number was calculated with the 4-PL model and was correlated with expected copy number from 6.93 to 6.93 × 103 copies as shown in Figure 1B.
Figure 1.
(A) Influenza A response of Luminex xTAG RVP kit versus cloned influenza A matrix gene DNA dilution calculated copy number. The response is given as MFI as measured by the BioPlex reader. The copy number was calculated by multiplying the concentration of the diluted stock solution by the volume used in the RT-PCR step of the assay, which was 5 μL. The concentration of the diluted DNA was determined by multiplying the stock influenza A matrix gene concentration by the dilution factor.(B). Observed copy number versus expected copy number for the cloned influenza A matrix gene DNA. The observed copy number was calculated from the 4-PL fit of the influenza A response of the RVP kit for the cloned influenza A matrix gene DNA. The expected copy number was calculated from dilution of the stock influenza A matrix gene DNA.
2009 H1N1 vaccine
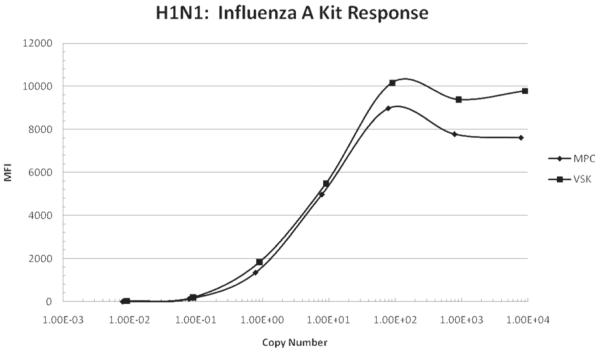
Figure 2 shows influenza A MFI response of the RVP kit as a function of calculated influenza A virus copies for the 2009 H1N1 influenza vaccine. The RNA in the diluted virus samples was extracted using both the MPC and VSK kits as described above. The assay response begins to rise before the dilution calculated virus copy number reaches 1 copy (at about 8 × 10−2 copies) and levels off at about 8 × 101 dilution calculated copies. Again this data was fitted with the 4-PL model with R2 > 0.999 obtained for both MPC and VSK curves and the observed copy number calculated with the model correlated well with the expected copy number (R2 close to 1) over the range 8 × 10−2 to 8 × 101 dilution calculated copies. Also note that the MPC and VSK kits gave similar recoveries.
Figure 2.
Influenza A response of Luminex xTAG RVP kit versus influenza A dilution calculated copy number for the 2009 H1N1 vaccine. The RNA from the samples was extracted with the VSK and MPC. The response is given as MFI as measured by the BioPlex reader. The copy number was calculated by multiplying the concentration of the diluted vaccine by the volume of sample used in the RT-PCR step of the assay, which was 5 μL. The concentration of the diluted vaccine was determined by multiplying the undiluted vaccine concentration by the dilution factor.
Seasonal influenza vaccine
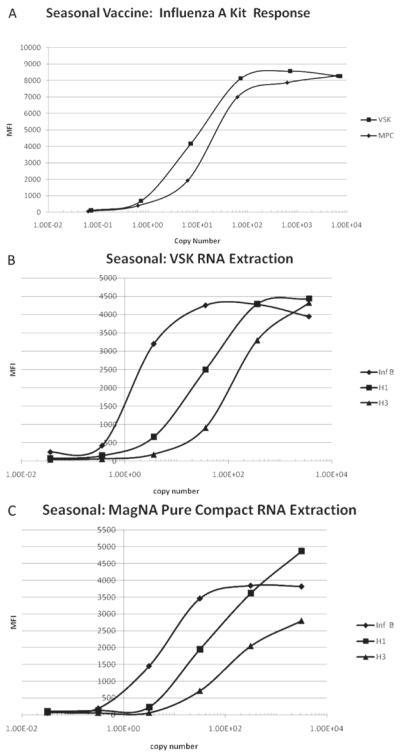
Figure 3A shows influenza A MFI response of the RVP kit as a function of influenza A virus copies calculated from dilution for the 2005–2006 seasonal influenza vaccine. In a similar manner to the 2009 H1N1 vaccine, the influenza A response begins to rise before the calculated virus copy number reaches 1 copy (at about 8 × 10−2 copies) and levels off at about 8 × 101 calculated copies. This data was fitted with the 4-PL model (R2 > 0.999) and the copy number calculated with the model correlated well with the expected copy number (R2 close to 1). Both the MPC and VSK kits gave similar RNA recoveries.
Figure 3.
(A) Influenza A response of the RVP kit versus influenza A dilution calculated copy number for the 2005–2006 seasonal influenza vaccine. The RNA from the samples was extracted with the VSK and MPC. The response is given as MFI as measured by the BioPlex reader. The copy number was calculated by multiplying the concentration of the diluted vaccine by the volume of sample used in the RT-PCR step of the assay, which was 5 μL. The concentration of the diluted vaccine was determined by multiplying the undiluted vaccine concentration by the dilution factor. (B). Influenza B, H1 influenza A subtype, and H3 influenza A subtype response of the RVP kit versus dilution calculated copy number for the 2005–2006 seasonal influenza vaccine for samples extracted with VSK. The response is given as MFI as measured by the BioPlex reader. The copy number was calculated by multiplying the concentration of the diluted vaccine by the volume of sample used in the RT-PCR step of the assay, which was 5 μL. The concentration of the diluted vaccine was determined by multiplying the undiluted vaccine concentration by the dilution factor. (C). Influenza B, H1 influenza A subtype, and H3 influenza A subtype response of Luminex xTAG RVP kit versus dilution calculated copy number for the 2005–2006 seasonal influenza vaccine for samples extracted with MPC. The response is given as MFI as measured by the BioPlex reader. The copy number was calculated by multiplying the concentration of the diluted vaccine by the volume of sample used in the RT-PCR step of the assay, which was 5 μL. The concentration of the diluted vaccine was determined by multiplying the undiluted vaccine concentration by the dilution factor.
Figure 3B and 3C show the influenza B, H1 influenza A subtype, and H3 influenza A subtype MFI response of the RVP kit as a function of influenza B, H1, and H3 dilution calculated copies for the 2005–2006 seasonal influenza vaccine extracted with the VSK and MPC kits, respectively. The influenza B MFI response of the RVP kit for both the VSK and MPC extracted samples began to increase at about 3 × 10−2 dilution calculated copies and leveled off at about 3 × 101 dilution calculated copies and the observed copy number calculated with the 4-PL model correlated well with the expected copy number calculated from dilution over this range. For H1 subtype, the MFI response of the RVP kit began increasing at about 3 × 10−1 dilution calculated copies and leveled off at 3 × 102 dilution calculated copies and the 4-PL model again provided good fit to this data. The H3 subtype response began to increase at about 3 × 100 dilution calculated copy number and had not completely leveled at 3 × 103, the highest dilution calculated copy number studied. Again the 4-PL model fit the data with high correlation of observed copy number with expected copy number.
Discussion
For a number of types of applications, such as environmental sampling, it is desirable to obtain semi-quantitative or quantitative information about the concentration of multiple viral species present in samples. The xTAG RVP assay is able to detect and identify multiple viral species, but the analysis software provides only qualitative interpretation of the measurements. In this pilot study, we have examined the response of the RVP kit to cloned influenza A matrix gene DNA and it was found that the influenza A response of the kit could be modeled with a 4-PL fit over a range of dilutions. The matrix gene DNA provided the most accurate measure of response since the concentration in the stock solution was calculated using UV spectroscopy. The RVP kit response from the matrix gene DNA did not increase until after the calculated copy number was greater than 1. For both vaccine samples, the influenza A response of the RVP kit response began to increase before the dilution calculated copy number reached 1 so the assumption that one tissue culture infectious dose (TCID) represents one viral genome copy apparently underestimates the actual copy number. However the 4-PL model provided a good fit of the influenza A RVP kit response for influenza A dilution calculated copy number for both the vaccines. Additionally, the 4-PL model provided a good fit of influenza B, H1, and H3 RVP kit response to calculated copy number for the 2005–2006 seasonal vaccine samples. The RVP kit has no significant H1 response to 2009 H1N1 virus (Ginocchio and St. George, 2009a; Ginocchio et al. 2009b) so only influenza A response was observed for the 2009 H1N1 vaccine in this study. The 4-PL fit is used for many immunoassays and is present in instrument software for immunoassay instruments such as BioPlex and Luminex instruments which can use Luminex IS 100 2.3 software. Several aspects of RVP kit performance could not be addressed in this study due to the limited availability of the RVP kits. The reproducibility of the response curves from the kits for the vaccine and matrix DNA samples could not be addressed because of limited data. It would be useful to estimate the limit of detection which would require more response data near the limit of detection in addition to the reproducibility data. In addition, the use of the RVP kit could be evaluated with environmental samples which were also evaluated with qPCR to compare the results from the two methods. These are areas for further work which could evaluate the ability of the RVP kits to produce quantitative data. The MPC automated and VSK manual RNA extraction procedures gave similar recoveries in this study but a more detailed comparison of the extraction procedures would require additional data. However, either procedure should provide useful data concerning the relative contribution of various sources of viral contamination.
One advantage of multiplexed methods is that multiple controls can be used with each sample. In the present study using the RVP kit, only the phage MS2 internal control is added to every extracted sample before RNA extraction. This provides a useful assessment of the extraction process. However, the addition of more controls could be useful if semi-quantitative or quantitative results are desired. If the run control were added to every sample at the RT-PCR step, it could provide a response control in the midrange of the assay. Several other nonviral DNA sequences could be added at a lower concentration and a higher concentration than the run control. The RT-PCR primers and target specific primer extension primers would have to be added to the primer mix for these additional sequences along with appropriate bead sets for these additional controls. In this way, a relative calibration curve could be generated in every sample well and used to compensate for differences in assay response for different samples. This concept has been exploited previously in multiplex clinical immunoassays (Inverness Medical).
Conclusion
Our data suggest that the RVP kit has utility as a semi-quantitative assessment tool for determination of influenza A, influenza B, and the H1 and H3 subtypes of influenza A in environmental samples. It may also be useful for semi-quantitative determination of other respiratory viruses in these samples.
Acknowledgments
The authors alone are responsible for the content and writing of the paper. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health. Mention of company or product names does not imply endorsement by the National Institute for Occupational Safety and Health.
Footnotes
Declaration of interest
The authors report no conflicts of interest.
References
- Blachere FM, Lindsley WG, Pearce TA, Anderson SE, Fisher M, Khakoo R, Meade BJ, Lander O, Davis S, Thewlis RE, Celik I, Chen BT, Beezhold DH. Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis. 2009;48:438–440. doi: 10.1086/596478. [DOI] [PubMed] [Google Scholar]
- Blachere FM, Lindsley WG, Slaven JE, Green BJ, Anderson SE, Chen BT, Beezhold DH. Bioaerosol sampling for the detection of aerosolized influenza virus. Influenza Other Respi Viruses. 2007;1:113–120. doi: 10.1111/j.1750-2659.2007.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginocchio CC, St George K. Likelihood that an unsubtypeable influenza A virus result obtained with the Luminex xTAG respiratory virus panel is indicative of infection with novel A/H1N1 (swine-like) influenza virus. J Clin Microbiol. 2009a;47:2347–2348. doi: 10.1128/JCM.01027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginocchio CC, Zhang F, Manji R, Arora S, Bornfreund M, Falk L, Lotlikar M, Kowerska M, Becker G, Korologos D, de Geronimo M, Crawford JM. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Virol. 2009b;45:191–195. doi: 10.1016/j.jcv.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inverness Medical AtheNA Multi-Lyte® Test System, Intra-Well Calibration Technology®
- Karim MR, Rhodes ER, Brinkman N, Wymer L, Fout GS. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl Environ Microbiol. 2009;75:2393–2399. doi: 10.1128/AEM.00922-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krunic N, Yager TD, Himsworth D, Merante F, Yaghoubian S, Janeczko R. xTAG RVP assay: Analytical and clinical performance. J Clin Virol. 2007;40:39–46. doi: 10.1016/S1386-6532(07)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley WG, Blachere FM, Thewlis RE, Vishnu A, Davis KA, Cao G, Palmer JE, Clark KE, Fisher MA, Khakoo R, Beezhold DH. Measurements of airborne influenza virus in aerosol particles from human coughs. PLoS ONE. 2010;5:e15100. doi: 10.1371/journal.pone.0015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony JB. The clinical need for the RVP test. J Clin Virol. 2007a;40:36–38. doi: 10.1016/S1386-6532(07)70008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. Development of a respiratory virus panel test for detection of 20 human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007b;45:2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merante F, Yaghoubian S, Janeczko R. Principles of the xTAG respiratory viral panel assay (RVP Assay) J Clin Virol. 2007;40:31–35. doi: 10.1016/S1386-6532(07)70007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix B, Wild D. In: The immunoassay handbook. Wild D, editor. Nature; New York: 2001. pp. 198–210. [Google Scholar]
- World Health Organization CDC Protocol of Realtime RT-PCR for Influenza A(H1N1) 2009.