Abstract
Sigmodontinae rodents represent one of the most diverse and complex components of the mammalian fauna of South America. Among them most species belongs to Oryzomyini and Akodontini tribes. The highly specific diversification observed in both tribes is characterized by diploid complements, which vary from 2n = 10 to 86. Given this diversity, a consistent hypothesis about the origin and evolution of chromosomes depends on the correct establishment of synteny analyzed in a suitable phylogenetic framework. The chromosome painting technique has been particularly useful for identifying chromosomal synteny. In order to extend our knowledge of the homeological relationships between Akodontini and Oryzomyini species, we analyzed the species Akodon montensis (2n = 24) and Thaptomys nigrita (2n = 52) both from the tribe Akodontini, with chromosome probes of Hylaeamys megacephalus (2n = 54) of the tribe Oryzomyini. The results indicate that at least 12 of the 26 autosomes of H. megacephalus show conserved synteny in A. montensis and 14 in T. nigrita. The karyotype of Akodon montensis, as well as some species of the Akodon cursor species group, results from many chromosomal fusions and therefore the syntenic associations observed probably represent synapomorphies. Our finding of a set of such associations revealed by H. megacephalus chromosome probes (6/21; 3/25; 11/16/17; and, 14/19) provides phylogenetic information for both tribes. An extension of these observations to other members of Akodontini and Oryzomyini tribes should improve our knowledge about chromosome evolution in both these groups.
Introduction
Sigmodontinae rodents represent one of the most diverse and complex components of the mammalian fauna of South America, currently compost by around 380 species belonging to 74 genera. With the exception of 11 incertae sedis genera, all species of the subfamily are grouped into nine tribes (Abrotrichini, Akodontini, Ichthyomyini, Oryzomyini, Phyllotini, Reithrodontini, Sigmodontini, Thomasoyini, and Wiedomyini) [1, 2]. About 60% of the Sigmodontinae belong to tribes Oryzomyini and Akodontini, which contain 120 and 106 recognized species, respectively [1].
The highly specific diversification observed in both tribes is characterized by a remarkable variability of diploid numbers, which vary from 2n = 16 to 86 in Oryzomyini and 2n = 10 to 54 in Akodontini [3, 4]. Given this diversity, a hypothesis about the origin and evolution of their chromosomes depends on the correct establishment of synteny analyzed in a suitable phylogenetic framework. Comparative analysis of G-banded chromosomes enabled the identification of numerous chromosomal alterations in Akodontini [5, 6], which has led to the elaboration of phylogenetic hypotheses that explain this chromosomal variability by the occurrence of centric fusions, tandem fusions and inversions. However, there are many chromosomal differences that cannot be explained solely by G-banding techniques and molecular cytogenetic techniques are required.
Chromosome painting has been particularly useful in solving problems of chromosome evolution in old world rodents (e.g. [7–10]). However, in new world rodents there have been only a few studies in Sigmodontinae. Among them, Swier et al. [11] used Sigmodon (Sigmodontini tribe) probes for cross-species FISH on nine species of the genus Sigmodon, which allowed them to postulate an ancestral Sigmodon karyotype and three diploid number reductions. Likewise, cross-species chromosome painting with Oligoryzomys moojeni chromosome probes allowed Di-Nizo et al. [12] to uncover the complex and extensive chromosome re-patterning involved in the diversification of seven Oligoryzomys species. Additionally, conventional and reverse chromosome painting experiments in four species of Akodon with different diploid numbers allowed Ventura et al. [13] to characterize a sequence of tandem fusion events involved in the chromosomal evolution of species in the Akodon cursor group with low diploid numbers, previously suggested by their G-banding patterns [14, 15].
Studies involving inter-generic comparison have uncovered several clues of conserved synteny among five Akodontini species (Akodon cursor, A. montensis, A. paranaensis, A. serrensis, Necromys lasiurus, and Thaptomys nigrita) and one Oryzomyini species (Oligoryzomys flavescens) using Mus musculus chromosome-specific probes [16, 17]. Finally, Nagamachi et al. [18] developed species-specific chromosome probes for Hylaeamys megacephalus that disclosed major karyotypic reconstruction in Cerradomys langguthi (Oryzomyini species). These studies demonstrate the value of chromosome painting in understanding chromosome evolution in new world rodents.
In order to extend our knowledge of the phylogenetic relationships between Akodontini and Oryzomyini species, we analyzed A. montensis (2n = 24) and T. nigrita (2n = 52) of the tribe Akodontini, with probes from H. megacephalus (2n = 54) of the tribe Oryzomyini. This approach allowed us to identify shared and unique syntenic associations in each of these species.
Material and Methods
Ethics Statement
Animals collected during this study were handled following procedures recommended by the American Society of Mammalogists [19], and were made under specific collecting licenses from the Ministerio de Ecologia y Recursos Naturales Renovables de la Provincia de Misiones, Argentina (9910-00136/14; headed by CL as principal investigator) and Instituto Chico Mendes de Conservação da Biodiversidade, Brazil (N°13248; headed by JCP as principal investigator).
JCP has a permanent field permit, number 13248 from “Instituto Chico Mendes de Conservação da Biodiversidade”. The Cytogenetics Laboratory from UFPa has permit number 19/2003 from the Ministry of Environment for sample transport and permit 52/2003 for using the samples for research. The Ethics Committee (Comitê de Ética Animal da Universidade Federal do Pará) approved this research. The rodents were maintained in the lab with food and water, free from stress, until their euthanasia by IP injection of buffered and diluted barbiturates after local anesthetic.
Sample and chromosomes
Akodon montensis male and female adults and Thaptomys nigrita female adults were collected in several localities of Misiones Province, Argentina (Iguazú S25°67’67”/W54°44’66”; San Ignacio S27°16’82”/O55°34’45”; Puerto Esperanza S25°59’20”/W54°38’35”; Uruguaí S25°51’22”/W54°10’; and, Candelaria S27°10’6”/W55°20’40”) using “Sherman” and “Tomahawk” live-traps. The skulls and skins were preserved and deposited in the Mastozoological collection housed in the Laboratorio de Genética Evolutiva, Instituto de Biología Subtropical, Posadas, Misiones, Argentina. Chromosome preparations were obtained from bone marrow following Ford and Hamerton [20], with some modifications. Gross chromosomal characterization was established by C-Banding [21], and G-Banding [22].
Probes and FISH
Each whole chromosome probe is the product of PCR amplification of copies of the same chromosome. The templates were chromosomes of H. megacephalus sorted at the University of Cambridge using a Dako flow cytometer [18]. The DNA sequence of the probes used for FISH is unknown and they were not molecularly characterized. Primary PCR products were labeled either with biotin-16-dUTP (Boehringer Mannheim), FITC-12-dUTP, or Cy3-dUTP (Amersham) by a second round of DOP-PCR with 6MW primers [23, 24]. The DNA fragment size of probes were checked on 1% agarose gel electrophoresis (100V, 40 min), and had an average size of 0,2–0,8 kb. As some H. megacephalus chromosome pairs could not be separated from others by cytometry (pairs 9/10, 13/22, and 16/17), the combined probes recognize homologous regions of more than one chromosome pair. Fluorescent In situ Hybridization (FISH) in metaphase chromosomes of A. montensis and T. nigrita with chromosome-specific probes was performed as previously described [25]. Briefly, slides were pretreated with pepsin solution (50 ug/mL in 0,01M HCl) at room temperature for 5 min, dehydrated progressively with an ethanol series (70%, 90%, and 100%) 4 min each, and incubated for at least two hours at 65°C. DNA of mitotic chromosomes was denatured at 62°C with 70% formamide/0,6xSSC for 60 seconds, and quickly stopped with 4°C 70% ethanol for 4 min, followed by another ethanol series of dehydration. About 300 ng of labeled DNA probe was dissolved in 15 uL of hybridization buffer (50% deionized formamide/2xSSC/12,5% dextran sulfate/50mM phosphate buffer solution), and heated at 75°C for 15 min before being applied to the slides. Hybridization reactions were performed at 77% stringency for 48-72h at 37°C in a humid chamber. Post-hybridizations washes were performed in 50% deionized formamide/1xSSC and 2xSSC for 10 min each at 42°C. Indirect detection of biotinylated probes was made with avidin-Cy3 or avidin-FITC antibodies. DAPI stained chromosomes and fluorescent hybridization signals were captured and digitalized using a cooled CCD camera (Axiocam MRm), coupled to a Zeiss Axiophot microscope with Axiovision software.
Results
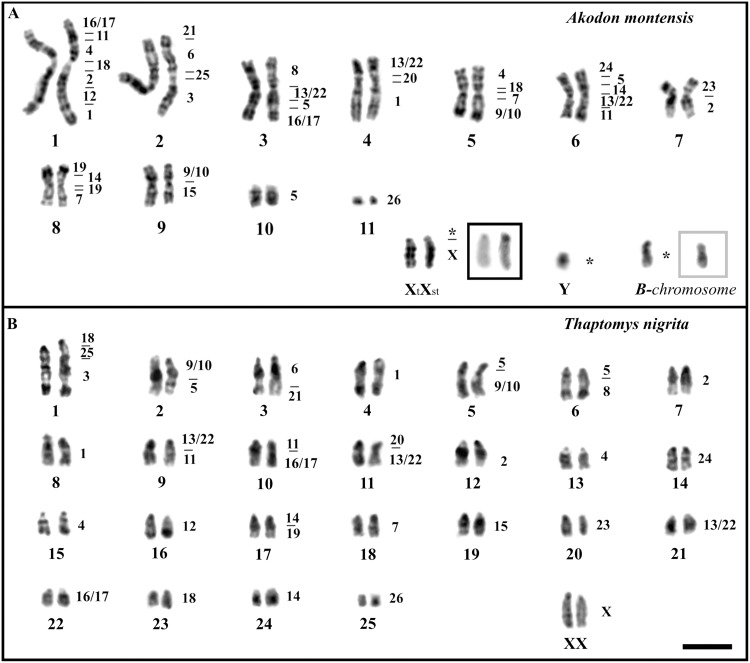
Akodon montensis has 2n = 24 (FN = 42), mostly metacentric chromosomes, except the submetacentric pair 2 and the acrocentric pair 10. X and Y sex chromosomes also showed acrocentric morphology, with the exception of a polymorphic subtelocentric X chromosome (Fig 1A). The B-chromosome observed in this species is submetacentric (Fig 1A). Thaptomys nigrita has 2n = 52 (FN = 52), the metacentric pair 25 being the only biarmed chromosome pair (Fig 1B). The C-banding technique shows that A. montensis chromosomes are characterized mainly by centromeric and telomeric constitutive heterochromatin (CH). The acrocentric X chromosome has centromeric CH and the subtelocentric X has an additional heterochromatic block in the short arm (Fig 1A). The B-chromosome in turn shows CH on the centromere and has an interstitial block on the long arm (Fig 1A). In T. nigrita the CH is restricted to centromeric regions.
Fig 1. Chromosome complements of Akodon montensis and Thaptomys nigrita with regions of homology to chromosome-specific probes of Hylaeamys megacephalus.
A-B: G-banded karyotypes. Black-square: C-banded X-chromosomes. Grey-square: C-banded B-chromosome. Bar = 10 μm.
All 24 whole chromosome probes of Hylaeamys megacephalus (1–8, 9/10, 11–12, 13/22, 14–15, 16/17, 18–21, 23–26, and X) were used to map chromosome homologies (Fig 1; S1 and S2 Figs) and positive and reproducible hybridization signals were observed in at least one chromosome region of both A. montensis and T nigrita (Fig 2). Table 1 summarizes these results. No hybridization signals were obtained for the Y chromosome (for which we have no H. megacephalus Y chromosome probe), for the short arm of the subtelocentric X chromosome of A. montensis, or for the B chromosome in the A. montensis female specimen (Fig 1).
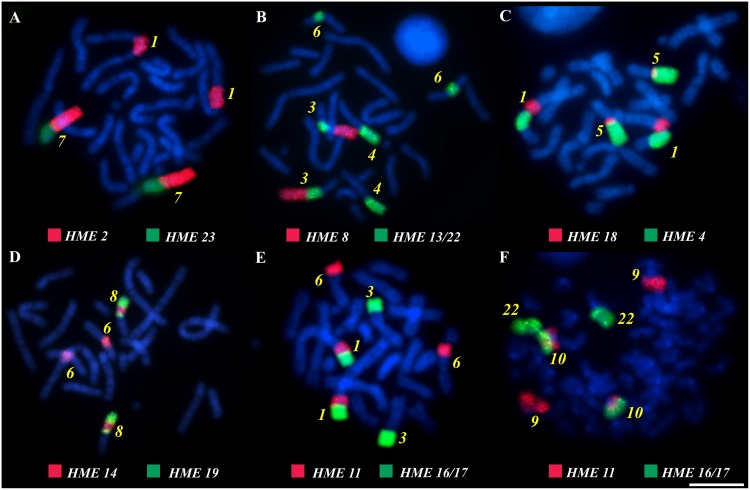
Fig 2. Hybridization in situ experiments with chromosome-specific probes of Hylaemys megacephalus hybridized to metaphase chromosomes of Akodon montensis (a, b, c, d and e) and Thaptomys nigrita (f) showing syntenic associations.
Yellow numbers indicating each matching chromosome pair on target species. Bar = 10 μm.
Table 1. Total number and location of FISH signals observed on Akodon montensis and Thaptomys nigrita with specific-chromosome probes of Hylaeamys megacephalus.
| Hylaeamys megacephalus Chromosome probes | Akodon montensis | Thaptomys nigrita | ||
|---|---|---|---|---|
| N° signals | Chromosome location | N° signals | Chromosome location | |
| 1 | 2 | 1q distal; 4q | 2 | 4; 8 |
| 2 | 2 | 1q interstitial; 7q | 2 | 7; 12 |
| 3 | 1 | 2q | 1 | 1 interstitial and distal |
| 4 | 2 | 1p proximal; 5p distal | 2 | 13; 15 |
| 5 | 3 | 3q interstitial;6p interstitial; 10 | 3 | 2 distal; 5 proximal; 6 proximal |
| 6 | 1 | 2p | 1 | 3 proximal and interstitial |
| 7 | 2 | 5q proximal; 8q | 1 | 18 |
| 8 | 1 | 3p | 1 | 6 distal |
| 9/10 | 2 | 5q; 9p | 2 | 2 proximal; 5 distal |
| 11 | 2 | 1p interstitial; 6q distal | 2 | 9 distal; 10 proximal |
| 12 | 1 | 1q interstitial | 1 | 16 |
| 13/22 | 3 | 3q proximal; 4p distal; 6q proximal | 3 | 9 proximal; 11 interstitial and distal; 21 |
| 14 | 2 | 6p proximal; 8p interstitial | 2 | 17 proximal; 24 |
| 15 | 1 | 9q | 1 | 19 |
| 16/17 | 2 | 1p distal; 3q distal | 2 | 10 distal; 22 |
| 18 | 2 | 1q proximal; 5p proximal | 2 | 1 proximal; 23 |
| 19 | 2 | 8p distal; 8q proximal | 1 | 17 distal |
| 20 | 1 | 4p proximal | 1 | 11 proximal |
| 21 | 1 | 2p distal | 1 | 3 distal |
| 23 | 1 | 7p | 1 | 20 |
| 24 | 1 | 6p distal | 1 | 14 |
| 25 | 1 | 2q proximal | 1 | 1 proximal |
| 26 | 1 | 11 | 1 | 25 |
| X | 1 | X (Xq*) | 1 | X |
* subtelocentric X chromosome.
Twelve of the 24 whole chromosome probes show conserved synteny in A. montensis (H. megacephalus chromosomes 3, 6, 8, 12, 15, 20, 21, 23–26, and X) and 14 in T. nigrita (H. megacephalus chromosomes 3, 6–8, 12, 15, 19–21, 23–26 and X). Of these, seven chromosomes of H. megacephalus are preserved as unique linkage groups in T. nigrita (five with the same morphology: 7, 12, 15, 26 and the X chromosome; and two with a different morphology in H. megacephalus: pairs 23 and 24). Only two chromosomes of H. megacephalus (pairs 26 and X) were preserved as a discrete unit in A. montensis (pairs 11 and X). Thus, only the smallest pair in each of the three karyotypes and the X chromosome are wholly conserved between the three species.
More than one signal was observed using the remaining whole chromosome probes. Chromosome probes of H. megacephalus pairs 5 and 13/22 gave the highest number of signals, complementary to three regions of three chromosome pairs in A. montensis (pairs 3, 6 and 10, and pairs 3, 4 and 6, respectively) and T. nigrita (pairs 2, 5 and 6, and pairs 9, 11 and 21, respectively). Finally, the remaining H. megacephalus chromosome probes shared homology with 2 chromosome regions in A. montensis (probes of pairs 1, 2, 4, 7, 9/10, 11, 14, 16/17, 18, and 19) and in T. nigrita (probes of pairs 1, 2, 4, 9/10, 11, 14, 16/17, and 18).
Both Akodontini species show the association of several probes of H. megacephalus in the same linkage group. Among them, probes 3 and 25, and probes 6 and 21 mapped to two chromosomes of T. nigrita respectively, but in only one chromosome of A. montensis, providing evidence of tandem fusion of chromosomes in A. montensis. Probes 5 and 8; 11 and 13/22; 11 and 16, 17; 20 and 13/22; 14 and 19 were also associated in both species. But as probes 5, 11, 13/22, 14 and 16/17 produce more than one signal in the karyotypes of T. nigrita and A. montensis, the synteny relationship of these chromosome fragments may need further confirmation. Table 1 and Fig 3 summarize the complementary relationships revealed by FISH experiments. The X chromosome is the only conserved chromosome both in the taxa studied here and in Mus musculus.
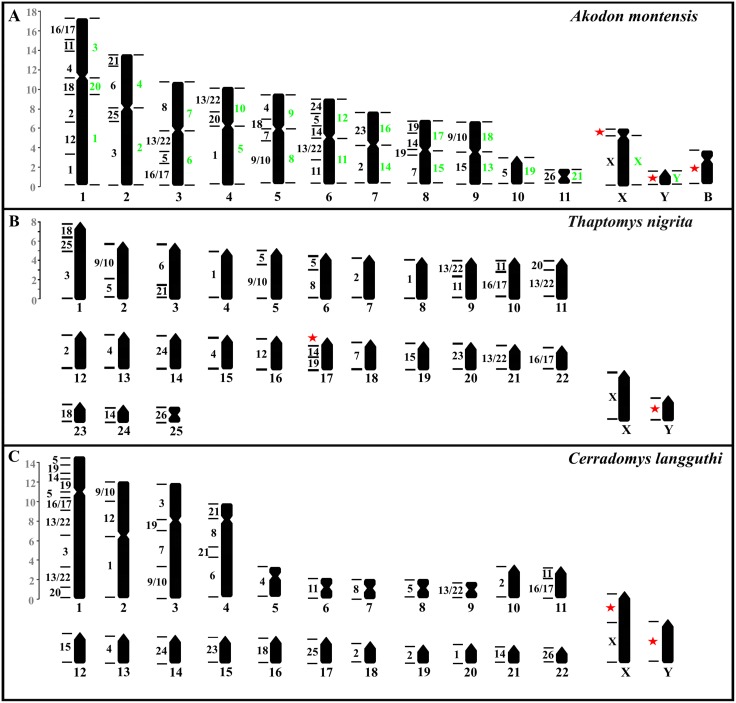
Fig 3. Ideograms of Akodon montensis (A), Thaptomys nigrita (B), and Cerradomys langguthi (C), showing chromosome homologies to Hylaeamys megacephalus (left black numbers) and Akodon paranaensis (right green numbers).
Red stars show regions without complementarity with the probes used. Scale shows the percent relative size of chromosome pairs. (Adapted from Ventura et al. [12], and Nagamachi et al. [17]).
Discussion
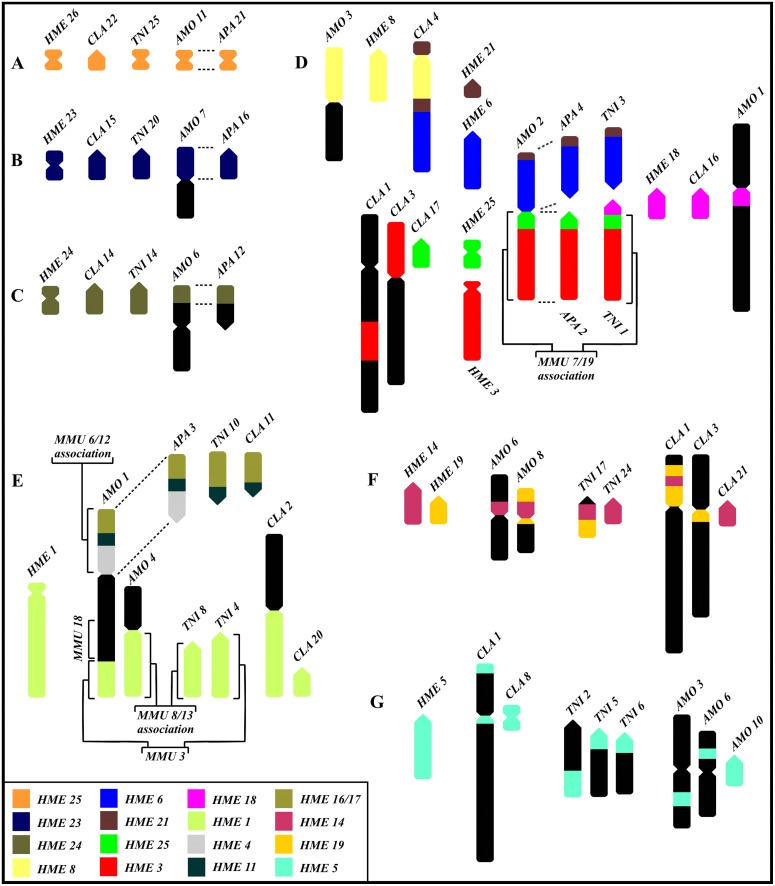
Bianchi et al. [26] have made the initial cytogenetic hypotheses on chromosome evolution within the Sigmodontinae. Despite methodological constraints, their phylogenetic approach using cytogenetic data group several sigmodontine species by the shared morphology of chromosome 1. They also placed the smallest metacentric pair as a shared characteristic of all karyotyped species. Recently, chromosome painting analyses of four Akodon species made by Ventura et al. [13] show the complete homeology of the smallest metacentric pair in A. cursor, A. montensis, A. paranaensis, and Akodon sp (2n = 10). The results presented here for A. montensis and T. nigrita, and the available data in Hylaeamys megacephalus and Cerradomys langguthi [18] indicate that this metacentric chromosome pair seems to be the only autosome entirely conserved as an independent linkage group among the Akodontini and Oryzomyini species that have been studied (Fig 4A). However, this chromosome is not completely conserved inasmuch as in C. langguthi it is telocentric, and this may be a derived condition, due to pericentric inversion or centromeric repositioning. Intriguingly, the same methodological approach, but with Mus musculus chromosome probes, fails to identify the homologies of this smallest pair in the sigmodontine species A. cursor, A. montensis, A. paranaensis, Oligoryzomys flavescens, T. nigrita, and Necromys lasiurus [16, 17]. These results may be due to technical problems related to its small size, and/or sequence divergence.
Fig 4. Syntenic relationships among several chromosome pairs of Hylaeamys megacephalus (HME), Akodon montensis (AMO), A. paranaensis (APA), Thaptomys nigrita (TNI), and Cerradomys langguthi (CLA) reveled by different chromosome-paint probes (Adapted from Hass et al [15, 16], Ventura et al. [12], and Nagamachi et al. [17]).
Dotted lines: indirectly inferred homology from Ventura et al. [12] data. See Discussion for details.
Some chromosomes pairs maintain their independent identity despite changes in morphology. Nagamachi et al. [18] describe the relationship among the bi-armed chromosome pairs 23, 24, and 25 of H. megacephalus (HME23, HME24 and HME25) and their telocentric homologues in C. langguthi pairs 15, 14 and 17, respectively (Fig 4B–4D). The results with H. megacephalus chromosome probes in A. montensis and T. nigrita, together with available data from four other Akodon species [13], suggest that the bi-armed HME23 and 24 are derived chromosomes, being homologous to the telocentric chromosomes in the two other species. Additionally, the metacentric HME25 is homeologous to the telocentric pair 17 of C. langguthi.
HME25 is a single pair in Oryzomyini species (H. megacephalus and C. langguthi), but is associated with HME3 in A. montensis (pair 2q) and T. nigrita (pair 1) (Fig 4D). The association HME25/3 is coincident with the association of Mus musculus chromosome pairs 7/19 reported for A. montensis and T. nigrita [16, 17], and this has been inferred as an ancestral muroid syntenic association, since it was found in other Cricetidae subfamilies, like Cricetinae [27, 8]. This association already existed before the split of Sigmodontinae and its absence is a derived condition. In C. langguthi this conserved region is tripartite, probably due to an additional fission of HME3 [18] (Fig 4D). A similar situation is found between HME6 and 21, which are associated in A. montensis (pair 2p), T. nigrita (pair 3), and C. langguthi (pair 4). In the latter there is an intercalary insertion of HME8 [18], splitting the HME21 signal (Fig 4D). In A. montensis and T. nigrita, these regions were found to be homeologous to M. musculus chromosome 2 [16, 17].
The association HME25/3/18 in T. nigrita could represent a putative autapomorphy (Fig 4D). Their close phylogenetic relationship with Akodon serrensis [28, 29], in which chromosome pair 1 is apparently derived from a chromosome fusion [6], needs to be more closely evaluated.
The Mus musculus association 8/13 was suggested as a putative synapomorphy for South American Cricetidae [16, 17]. Its presence in A. montensis [16] and T. nigrita [17] corresponds to one of the two signals observed with HME1 (Fig 4E). The second signal of this probe corresponds to the M. musculus chromosome probe of pair 3 in A. montensis (pair 1) and T. nigrita (pair 8), which strongly suggests that the H. megacephalus chromosome pair 1 is the result of a fusion event between the homeological regions of the highly conserved M. musculus chromosome pair 8/13 association and the M. musculus chromosome pair 3. This possibility had already emerged from a comparative G-banding pattern analysis between H. megacephalus, H. yunganus and Euryoryzomys nitidus (pairs 7, 4, and 8, respectively) [30].
The associations of chromosome pair 3/18 and 6/12 of M. musculus have been postulated as synapomorphies in Akodontini [16, 17]. The first association co-localized mainly with chromosome pair 1 of those Akodon species with karyotypes composed mainly of telocentric chromosomes (A. paranaensis and A. serrensis), the distal half of the long arm of pair 1 of A. montensis (Figs 3A and 4E), and on chromosome pair 5 of Necromys lasiurus, but is not found in T. nigrita [13, 17]. The second association seems to be restricted to chromosome 3 of A. paranaensis and A. serrensis [16], to their homeological regions of other Akodon cursor species group [13], and to pair 10 of N. lasiurus [17]. This association corresponds to the association of HME4/11/16/17 on the short arm of chromosome 1 of A. montensis (Fig 4E). However, in T. nigrita this is not fully preserved since a portion is co-localized on pair 10 associated with H. megacephalus chromosome pairs 11/16/17 (Fig 4E). Interestingly, this association (11/16/17) was also found in C. langguthi on chromosome pair 11 [18] (Figs 3C and 4E).
Hylaeamys megacephalus pair 14 probe paints two different chromosomes, one of which appears associated with HME19 in A. montensis, T. nigrita, and C. langguthi (Fig 4F) [18]. The fission seems to be a derived condition on H. megacephalus karyotype. In A. montensis and T. nigrita, the chromosome regions where FISH signals were observed for both probes are co-localized with M. musculus chromosome pair 11 [16, 17]. The additional signal observed for HME19 in A. montensis and Cerradomys langguthi could be explained by an inversion event, but it is necessary to corroborate whether this is one or two independent events.
The H. megacephalus chromosome probe 5 is the most fragmented chromosome pair in A. montensis, T. nigrita, and C. langguthi (Fig 4G). One interpretation of these results could be related to chromosomal fragmentation in the evolutionary pathway of Cerradomys, Thaptomys and Akodon species. However, a more parsimonious alternative is that it could be the outcome of the fusions of three independent syntenic units that derived from chromosome pair 5 of H. megacephalus.
The X chromosomes were the only syntenic units that are conserved as a unique and independent linkage group in all sigmodontine species studied and in Mus musculus. The high conservation of this sex chromosome in the rodent lineage, as in others mammals, was detected by several investigations [31]. However, some variations of its morphology were described. In A. montensis, the heteromorphism observed on the X chromosomes appears to be the result of the addition of heterochromatin in its short arm. This is supported by the absence of hybridization with the H. megacephalus probes and by the observation of a C positive band in its short arm (Fig 1A black square). The results presented by Nagamachi et al. [18] using the same probe of chromosome X of H. megacephalus has been useful also in distinguishing the additional constitutive heterochromatin on the X chromosome of Cerradomys langguthi.
A set of associations (H. megacephalus chromosome pairs 6/21, 11/16,17, and 14/19) has shown interesting synapomorphies among the Akodontini and Oryzomyini tribes. Extend these observations to other members of both tribes is necessary to confirm the phylogenetic assumptions presented here and to improve our knowledge of the chromosome evolution of these two important groups of Neotropical rodent fauna.
Supporting Information
(JPG)
(JPG)
Acknowledgments
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo à Pesquisa do Estado do Pará (FAPESPA) (Edital BIONORTE/CNPq Proc 552032/2010-7 and Edital BIONORTE/FAPESPA, ICAAF 007/2011) and the Banco Nacional de Desenvolvimento Economico e Social—BNDES (Operação 2.318.697.0001) for financial support on a project coordinated by CY Nagamachi; the FAPESPA for financial support (Edital Vale—Proc 2010/110447) on a project coordinated by JC Pieczarka, and the Welcome Trust for a grant to MA Ferguson-Smith. CL thanks PICT 2010/1095 and CONICET PIP 198. PS thanks CNPq by postdoctoral fellowship (159537/2011-8).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo à Pesquisa do Estado do Pará (FAPESPA) (Edital BIONORTE/CNPq Proc 552032/2010-7 and Edital BIONORTE/FAPESPA, ICAAF 007/2011) and the Banco Nacional de Desenvolvimento Economico e Social—BNDES (Operação 2.318.697.0001) for financial support on a project coordinated by CY Nagamachi; the FAPESPA for financial support (Edital Vale—Proc 2010/110447) on a project coordinated by JC Pieczarka, and the Welcome Trust for a grant to MA Ferguson-Smith. CL thanks PICT 2010/1095 and CONICET PIP 198. PS thanks CNPq by postdoctoral fellowship (159537/2011-8).
References
- 1. Musser G, Carleton M. Superfamily Muroidea In: Wilson D, Reeder D, editors. Mammal species of the world: a taxonomic and geographic reference 3rd ed. Baltimore, Maryland: Johns Hopkins University Press: 2005. pp. 894–1531. [Google Scholar]
- 2. D’Elía G, Pardiñas U, Teta P, Patton J. Definition and diagnosis of a new tribe of sigmodontine rodents (Cricetidae: Sigmodontinae), and a revised classification of the subfamily. Gayana. 2007; 71: 187–194. [Google Scholar]
- 3. Patton JL, da Silva MNF, Malcolm JR. Mammals of the rio Juruá and the evolutionary and ecological diversification of Amazonia. Bull Am Mus Nat Hist. 2000; 244:1–306. [DOI] [Google Scholar]
- 4. Silva MJJ, Patton JL, Yonenaga-Yassuda Y. Phylogenetic relationships and karyotype evolution in the Sigmodontine rodent Akodon (2n = 10 and 2n = 16) from Brazil. Genet Mol Biol. 2006, 3:469–474. 10.1590/S1415-47572006000300012 [DOI] [Google Scholar]
- 5. Sbalqueiro IJ, Nascimento AP. Occurrence of Akodon cursor (Rodentia, Cricetidae) with 14, 15 and 16 chromosome cytotypes in the same geographic area in Southern Brazil. Braz J Genet. 1996; 19: 565–569. 10.1590/S0100-84551996000400005 [DOI] [Google Scholar]
- 6. Geise L, Canavez FC, Seuánez HN. Comparative karyology in Akodon (Rodentia, Sigmodontinae) from Southeastern Brazil. Heredity 89: 158–163. 1998; 10.1093/jhered/89.2.158 [DOI] [PubMed] [Google Scholar]
- 7. Sitnikova NA, Romanenko SA, O’Brien PCM, Perelman PL, Fu B, Rubtsova NV, et al. Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). I. The genome homology of tundra vole, field vole, mouse and golden hamster revealed by comparative chromosome painting. Chromosome Res. 2007; 15: 447–456. 10.1007/s10577-007-1137-y [DOI] [PubMed] [Google Scholar]
- 8. Romanenko SA, Perelman PL, Serdukova NA, Trifonov VA, Biltueva LS, Wang J, et al. Reciprocal chromosome painting between three laboratory rodent species. Mamm Genome 17: 1183–1192. 2006; 10.1007/s00335-006-0081-z [DOI] [PubMed] [Google Scholar]
- 9. Romanenko SA, Volobouev VT, Perelman PL, Lebedev VS, Serdukova NA, Trifonov VA, et al. Karyotype evolution and phylogenetic relationships of hamsters (Cricetidae, Muroidea, Rodentia) inferred from chromosomal painting and banding comparison. Chromosome Res. 2007; 15: 283–297. 10.1007/s10577-007-1124-3 [DOI] [PubMed] [Google Scholar]
- 10. Romanenko SA, Perelman PL, Trifonov VA, Graphodatsky AS. Chromosomal evolution in Rodentia. Heredity. 2012; 108:4–16. 10.1038/hdy.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Swier VJ, Bradley RD, Rens W, Elder FFB, Baker RJ. Patterns of chromosomal evolution in Sigmodon, evidence from whole chromosome paints. Cytogenet Genome Res. 2009; 125:54–66. 10.1159/000218747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di-Nizo CB, Ventura K, Ferguson-Smith MA, O’Brien PCM, Yonenaga-Yassuda Y, Silva MJdJ. Comparative Chromosome Painting in Six Species of Oligoryzomys (Rodentia, Sigmodontinae) and the Karyotype Evolution of the Genus. PLoS ONE. 2015; 10(2): e0117579 10.1371/journal.pone.0117579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ventura K, O’Brien PCM, Yonenaga-Yassuda Y, Ferguson-Smith MA. Chromosome homologies of the highly rearranged karyotypes of four Akodon species (Rodentia, Cricetidae) resolved by reciprocal chromosome painting: the evolution of the lowest diploid number in rodents. Chromosome Res. 2009; 17:1063–1078. 10.1007/s10577-009-9083-5 [DOI] [PubMed] [Google Scholar]
- 14. Fagundes V, Vianna-Morgante AM, Yonenaga-Yassuda Y. Telomeric sequences localization and G-banding patterns in the identification of a polymorphic chromosomal rearrangement in the rodent Akodon cursor (2n = 14, 15 and 16). Chromosome Res. 1997; 5: 228–232. 10.1023/A:1018463401887 [DOI] [PubMed] [Google Scholar]
- 15. Fagundes V, Scalzi-Martin JM, Sims K, Hozier J, Yonenaga-Yassuda Y. ZOO-FISH of a microdissection DNA library and G-banding patterns reveal the homeology between the Brazilian rodents Akodon cursor and A. montensis . Cytogenet Cell Genet. 1997; 78: 224–228. 10.1159/000134662 [DOI] [PubMed] [Google Scholar]
- 16. Hass I, Sbalqueiro IJ, Müller S. Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetidae) from Southern Brazil established by Zoo-FISH using Mus musculus (Muridae) painting probes. Chromosome Res. 2008; 16:75–88. 10.1007/s10577-007-1211-5 [DOI] [PubMed] [Google Scholar]
- 17. Hass I, Müller S, Artoni RF, Sbalqueiro IJ. Comparative chromosome maps of neotropical rodents Necromys lasiurus and Thaptomys nigrita (Cricetidae) established by ZOO-FISH. Cytogenet Genome Res. 2011; 135:42–50. 10.1159/000330259 [DOI] [PubMed] [Google Scholar]
- 18. Nagamachi CY, Pieczarka JC, O’Brien PCM, Pinto JA, Malcher SM, Pereira AL, et al. FISH with whole chromosome and telomeric probes demonstrates huge karyotypic reorganization with ITS between two species of Oryzomyini (Sigmodontinae, Rodentia): Hylaeamys megacephalus probes on Cerradomys langguthi karyotypes. Chromosome Res. 2013; 21:107–119. 10.1007/s10577-013-9341-4 [DOI] [PubMed] [Google Scholar]
- 19. Sikes RS, Gannon WL. The Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal. 2011; 92:235–253. 10.1644/10-MAMM-F-355.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ford CE, Hamerton JL. A colchicine hypotonic citrate squash sequence for mammalian chromosome. Stain Technol. 1956; 3:247–251 [DOI] [PubMed] [Google Scholar]
- 21. Sumner AT. A simple technique for demonstrating centromere heterochromatin. Exp Cell Res. 1972; 75:304–306. [DOI] [PubMed] [Google Scholar]
- 22. Seabright M. A rapid banding technique for human chromosome. Lancet. 1971; 2:971–972. [DOI] [PubMed] [Google Scholar]
- 23. Telenius H, Carter NP, Bebb CE, Nordenskjo M, Ponder BAJ, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992; 13:718–725. [DOI] [PubMed] [Google Scholar]
- 24. Yang F, Carter NP, Shi L, Ferguson-Smith MA. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma. 1995; 103:642–652. 10.1007/BF00357691 [DOI] [PubMed] [Google Scholar]
- 25. Yang F, Graphodatsky AS. Animal Probes and ZOO-FISH In: Liehr T, editor. Fluorescence In Situ Hybridization (FISH)–Application Guide. Berlin: Springer-Verlag Berlin Heidelberg; 2009. pp. 323–346. 10.1007/978-3-540-70581-9_29 [DOI] [Google Scholar]
- 26. Bianchi NO, Reig OA, Molina OJ, Dulout FN. Cytogenetics of the south american Akodont rodents (Cricetidae). I. A progress report of argentinian and venezuelan forms. Evolution. 1971; 25: 724–736. [DOI] [PubMed] [Google Scholar]
- 27. Stanyon R, Yang F, Morescalchi AM, Galleni L. Chromosome painting in the longtailed field mouse provides insights into the ancestral murid karyotype. Cytogenet. Genome Res. 2004; 105: 406–411. 10.1159/000078213 [DOI] [PubMed] [Google Scholar]
- 28. Geise L, Smith MF, Patton JL. Diversification in the genus Akodon (Rodentia, Sigmodontinae) in southeastern South America: mitochondrial DNA sequence analysis. Journal of Mammal. 2001; 82: 92–101. [DOI] [Google Scholar]
- 29. D’Élia G. Phylogenetics of Sigmodontinae (Rodentia, Muroidea, Cricetidae), with special reference to the akodont group, and with additional comments on historical biogeography. Cladistics. 2003; 19: 307–323. [Google Scholar]
- 30. Volobouev VT, Aniskin VM. Comparative chromosome banding analysis of three South American species of rice rats of the genus Oryzomys (Rodentia, Sigmodontinae). Chromosome Res. 2000; 8: 295–304. 10.1023/A:1009223210737 [DOI] [PubMed] [Google Scholar]
- 31. Murphy WJ, Sun S, Chen ZQ, Pecon-Slattery J, O’Brien SJ. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 1999; 9: 1223–1230. 10.1101/gr.9.12.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPG)
(JPG)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.