Abstract
Arthrospira (Spirulina) platensis is a promising feedstock and host strain for bioproduction because of its high accumulation of glycogen and superior characteristics for industrial production. Metabolic simulation using a genome-scale metabolic model and flux balance analysis is a powerful method that can be used to design metabolic engineering strategies for the improvement of target molecule production. In this study, we constructed a genome-scale metabolic model of A. platensis NIES-39 including 746 metabolic reactions and 673 metabolites, and developed novel strategies to improve the production of valuable metabolites, such as glycogen and ethanol. The simulation results obtained using the metabolic model showed high consistency with experimental results for growth rates under several trophic conditions and growth capabilities on various organic substrates. The metabolic model was further applied to design a metabolic network to improve the autotrophic production of glycogen and ethanol. Decreased flux of reactions related to the TCA cycle and phosphoenolpyruvate reaction were found to improve glycogen production. Furthermore, in silico knockout simulation indicated that deletion of genes related to the respiratory chain, such as NAD(P)H dehydrogenase and cytochrome-c oxidase, could enhance ethanol production by using ammonium as a nitrogen source.
Introduction
In recent years, bioproduction of fuels and chemicals from biomass has been intensively investigated to achieve a sustainable society. Cyanobacteria have also attracted increasing attention as biomass feedstock and host organisms for bioproduction because they can produce biomass and biofuels from CO2 as a carbon source and light as an energy source through photosynthesis [1]. Moreover, cyanobacteria have some advantages compared with higher plants, e.g., higher photosynthesis efficiency and lack of competition with food and land resources. To date, various products have been successfully produced using cyanobacteria, such as ethanol [2, 3], lactate [4], isobutanol [5], glycogen [6], and fatty acids [7].
Arthrospira (Spirulina) platensis, a filamentous non-N2-fixing cyanobacterium, is promising for use as a biomass feedstock because of its capability to accumulate a large amount of glycogen, which is an excellent feedstock for biofuel production [8, 9]. Under optimal light intensity and nitrate concentration, the intracellular glycogen content was reported to be up to 70% of the dry cell weight in A. platensis [6, 10]. Moreover, A. platensis has been produced as a superior nutrient because of its high content of protein and carotenes [11], and it is the most industrially cultivated microalgal species [12]. Its superior characteristics such as high pH tolerance and high salt tolerance [11] could prevent contamination by other organisms in outdoor cultivation.
Recently, metabolic simulation using a genome-scale metabolic model has been widely used for rational metabolic design for the improvement of target molecule production [13, 14]. A genome-scale metabolic model is an in silico metabolic model that includes most of the metabolic reactions and metabolites of the target strain [15, 16]. Flux balance analysis (FBA) enables the simulation of the metabolic flux distribution in the whole metabolic reactions of the metabolic model with the following two assumptions: steady-state metabolism and maximization of cell growth [17, 18]. This method can be applied to analyze the effects of gene manipulation and culture conditions on the metabolic flux distribution, and various studies have reported successful improvement of target production [13, 14]. In cyanobacteria, several genome-scale metabolic models have been constructed, such as those for Synechocystis sp. PCC 6803 [19–22], Synechococcus sp. PCC 7002 [23], and Cyanothece sp. ATCC 51142 [24], thus providing a detailed understanding of cyanobacterial metabolism. In the case of A. platensis, two small metabolic models were constructed that include 22 metabolic reactions [25] and 121 metabolic reactions [26]. Recently, a genome-scale metabolic model of A. platensis C1 (PCC9438) including 875 reactions was constructed, and metabolic phenotypes and essential genes under various culture conditions, such as autotrophic and mixotrophic conditions, were analyzed [27]. However, metabolic simulations to identify the candidate genes to be manipulated for improvement of target production have not been conducted.
In this study, we constructed a genome-scale metabolic model of A. platensis NIES-39 and developed metabolic engineering strategies to improve production of valuable materials, such as glycogen and ethanol. A. platensis NIES-39 is a well-studied A. platensis strain, as the whole genome sequence [28], glycogen production [6], and metabolome [29] have been analyzed. After construction of the metabolic model, simulation results using the constructed model were evaluated by comparison with the experimental results with regard to growth rates and growth capabilities on various organic substrates. Moreover, metabolic engineering strategies to improve autotrophic production of glycogen and ethanol were developed using the metabolic simulation.
Materials and Methods
2.1. Construction of the genome-scale metabolic model of A. platensis NIES-39
A genome-scale metabolic model of A. platensis NIES-39 was constructed based on various information sources such as databases, the literature, and genome sequences. The annotation data were collected from CyanoBase [30], Kyoto Encyclopedia of Genes and Genomes (KEGG) [31], and the literature [28], and a draft metabolic model was constructed. To fill in the missing reactions of the draft model, candidate genes were identified from the genome sequence of A. platensis NIES-39 by comparison with the protein sequence of related cyanobacteria species such as Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7002 using BLASTP in KEGG (E-Value < 10−10) (http://www.genome.jp/tools/blast/). When the candidate genes for the synthesis reactions for the biomass components were not identified in the genome sequence, the reactions were added to the draft model based on the KEGG database. The cofactors of the reactions, such as NADPH and NADH, and transport reactions between the cytosol and periplasm and the periplasm and extracellular environment were added to the metabolic model based on previous genome-scale metabolic models of Synechocystis sp. PCC 6803 [29] and Escherichia coli [32].
The biomass components of A. platensis were collected from various previous reports. The macromolecular portion of the biomass was composed of 68% proteins, 16% carbohydrates, 0.88% DNA, 3.12% RNA, 11% lipids, and 1% chlorophyll, based on a previous study [26]. The amino acid composition in the proteins and the lipid composition were obtained from the literature [11, 33]. The carbohydrates were composed of glycogen [29], lipopolysaccharides [11], and polyhydroxybutyrate [34]. Carotenoids [35], which are categorized as hydrocarbons, were also included in the carbohydrates. The residual fraction of the carbohydrates consisted of peptidoglycans. The composition of nucleic acids in DNA and RNA was assumed to be proportional to the frequency in the whole genome sequence, e.g., 44.3% G + C content [28], following other study [20]. Because the ATP demand for biomass production in A. platensis NIES-39 was unknown, that of Synechocystis sp. PCC 6803 under autotrophic condition [36] was used in this model. The export reactions for intracellular metabolites, such as ethanol, lactate, acetate, pyruvate, and formate, were added to the metabolic model because the production of these metabolites was identified in the present study and also reported by previous studies [10, 37].
Finally, the metabolic model of A. platensis NIES-39 was constructed, which includes 673 unique metabolites and 746 metabolic reactions including 652 intracellular reactions, 60 transport reactions and 34 exchange reactions. The details of all reactions, metabolites, and biomass compositions are summarized in S1–S3 Tables.
2.2. Flux balance analysis
The metabolic flux distribution on the genome-scale metabolic model of A. platensis NIES-39 was calculated using FBA [18, 32]. Briefly, a pseudo-steady state of the metabolism was assumed, and the maximum and minimum ranges of the flux for each reaction were defined. These constraints provided a feasible space for the flux distribution in the metabolic model. To obtain the optimal flux distribution in the feasible space, an objective function was introduced and a linear programming technique was applied. This problem is represented by the following equation:
where S represents the stoichiometric matrix of metabolites in metabolic reactions, and v indicates a vector of the flux for each metabolic reaction. The values v min and v max represent the minimum and maximum constraints of the flux for each reaction, and c is a vector that represents the objective function to be maximized or minimized. In this study, biomass production was used as the objective function to be maximized, with the assumption that cellular metabolism is self-organized to maximize growth rate.
Because linear programming problems can present multiple solutions with an identical value for the objective function, the flux variability of each reaction was calculated [38]. In brief, after the maximum biomass production was determined by FBA, each flux was maximized and minimized under the condition of maximum biomass production.
All simulations were performed using Matlab (Mathworks Inc. Natick, MA) with a linear programming solver, GLPK (GNU Linear Programming Kit, http://glpkmex.sourceforge.net/).
For FBA simulation, the following external metabolites were assumed to be freely transported through the cell membrane: CO2, O2, H2O, sulfate, and phosphate. Nitrate and ammonium were used as the nitrogen source. Unless otherwise stated, the nitrate uptake rate was set to be free, and the ammonium uptake rate was set to zero. For the multiple knockout simulations to screen gene knockouts which are effective to increase target productions, we simply iterated FBA using metabolic networks with all possible combinatorial knockouts (e.g., three gene knockouts).
To simulate the metabolic flux distribution under autotrophic conditions, the photon uptake rate was set to a given value, and the glucose uptake rate was set to zero. For heterotrophic conditions, the uptake rate of one of the organic substrates, such as glucose, was set to a given value, and the photon uptake rate was set to zero. For simplification, we assumed that all photons are used for photosynthesis and neglected the effect of excess light energy to dissipate.
2.3. Culture experiment
The detailed culture conditions of A. platensis NIES-39 were described in a previous study [6]. The cells were grown in 500 mL Erlenmeyer flasks containing 250 mL of SOT medium under continuous illumination using white fluorescent bulbs (Life look HGX and NHG; NEC Corporation, Tokyo, Japan) with agitation at 100 rpm. The SOT medium consisted of 16.8 g/L NaHCO3, 0.5 g/L K2HPO4, 2.5 g/L NaNO3, 1.0 g/L K2SO4, 1.0 g/L NaCl, 0.2 g/L MgSO4 7H2O, 0.04 g/L CaCl2 2H2O, 0.01 g/L FeSO4 7H2O, 0.08 g/L Na2 EDTA, and 1.0 mL of A5 solution (2.86 g/L H3BO3, 2.5 g/L MnSO4 7H2O, 0.222 g/L ZnSO4 7H2O, 0.079 g/L CuSO4 5H2O, and 0.021 g/L Na2MoO4 2H2O). Pre-cultivated cells were grown under 50 μmol photons/m2/s at 30°C. In the log-growth phase, the pre-cultivated cells were harvested by filtration and inoculated into fresh SOT medium without nitrate at 0.15 g dry cell weight (DW)/L. The main culture was performed under 250 μmol photons/m2/s at 30°C. Before inoculation, cells were washed three times with nitrate-free SOT medium to prevent a change in nitrate concentration.
2.4. Analytical methods
The cell concentration was measured according to the optical density of the culture medium at 750 nm (OD750) by using a UVmini-1240 spectrophotometer (Shimadzu, Kyoto, Japan).
The glycogen concentration was measured following the method of a previous study [6]. Glycogen was extracted from the cells using a modification of a method from a previous study [39]. The culture broth was centrifuged and the cell pellet was hydrolyzed at 90°C for 90 min with 100 μL of 30% KOH (g/L). The sample was cooled on ice and mixed with 600 μL of cooled ethanol. After 1 h, the sample was centrifuged and mixed with 600 μL of cooled ethanol. This step was repeated three times. Then, the sample was dried at 80°C for 10 min and re-suspended in 100 μL of water. The supernatant of the centrifuged sample was subjected to glycogen measurement. Glycogen content was determined by high-performance liquid chromatography (HPLC) (Shimadzu, Kyoto, Japan) using a size exclusion HPLC column (OHpak SB-806 M HQ; Shodex, Tokyo, Japan) and a reflective index detector (RID-10A; Shimadzu, Kyoto, Japan). The column temperature was kept at 60°C, and the mobile phase, 0.1 M NaNO3, was run at 1 mL/min.
The organic acid concentrations were measured using a HPLC system (Shimadzu, Kyoto, Japan) equipped with an Aminex HPX-87H column (Bio-Rad, Hercules, CA, U.S.A.). During HPLC analysis, the column temperature was kept at 65°C; the mobile phase, 2 mM H2SO4, was run at 0.5 mL/min; and the eluent absorbance was detected at 210 nm.
The ethanol concentration was measured using a gas chromatograph (GC) system with a hydrogen flame ionization detector (7890A; Agilent Technologies, USA) and a Stabiliwax column (0.32-mm internal diameter, 60-m length, and 1-μm thickness; Restek Co., USA).
Results and Discussion
3.1. Construction of the genome-scale metabolic model of A. platensis NIES-39
A genome-scale metabolic model of A. platensis NIES-39 was constructed based on information from genome sequences, databases such as KEGG [31] and CyanoBase [30], and previously published material. The draft metabolic model was constructed based on the metabolic reactions collected from the KEGG database. Biomass compositions were collected from several previous reports [11, 26, 29, 33, 34]. The draft model was manually revised based on the literature and genome sequence information. For example, cyanobacteria have been reported to possess an incomplete TCA cycle because of a lack of α-ketoglutarate dehydrogenase, which converts α-ketoglutarate to succinyl-CoA [40, 41], and α-ketoglutarate dehydrogenase also has not been identified in A. platensis NIES-39. However, a recent study reported that a TCA bypass pathway complements the lack of the reaction [42] that converts α-ketoglutarate to succinate via the following two reactions: the conversion of α-ketoglutarate to succinic semialdehyde by α-ketoglutarate decarboxylase, and the conversion of succinic semialdehyde to succinate by succinic semialdehyde dehydrogenase. In A. platensis NIES-39, the gene encoding succinic semialdehyde dehydrogenase has been already annotated (NIES39_L06120), but the gene encoding α-ketoglutarate decarboxylase has not been identified. The candidate gene for α-ketoglutarate decarboxylase was searched for in the A. platensis NIES-39 genome sequence by BLASTP (http://www.genome.jp/tools/blast/) using the amino acid sequence of SynPCC7002_A2770, which was identified as α-ketoglutarate decarboxylase in Synechococcus sp. PCC 7002 [42], as a query sequence. As result, NIES39_L06130 was identified with high reliability (E-value = 0, identity = 79%). Thus, the reactions for the TCA bypass pathway were introduced into the metabolic model. In other cases, missing reactions that were not identified in the genome sequence of A. platensis NIES-39 but that are required for synthesis of biomass components, e.g., methionine, serine, and fatty acid biosynthesis reactions, were added based on the information of the KEGG database. Because A. platensis has been reported to produce some metabolites such as ethanol, formate, lactate, and acetate [10, 37], the export reactions of these metabolites were added to the metabolic model. Other reactions identified by BLASTP are summarized in S1 Table.
Finally, the metabolic model of A. platensis NIES-39, which included 746 reactions and 673 metabolites, was constructed (Table 1). The detailed information for the metabolic model is summarized in S1 and S2 Tables, and the functional classification of the metabolic reactions is described in S3 Table.
Table 1. Characteristics of the reconstructed metabolic model of A. platensis NIES-39.
The number of unique metabolites was calculated by considering the metabolites present in more than one compartment as a single metabolite.
| Features | Number | |
|---|---|---|
| Genomic information | ||
| Genome size (bp) | 6,788,435 | |
| No. of open reading frames (ORFs) | 6,631 | |
| No. of annotated genes | 2,542 | |
| Reconstructed metabolic model | ||
| Metabolites | 673 | |
| Reactions | 746 | |
| Annotated reactions | 644 | |
| Transport reactions | 60 | |
| Exchange reactions | 34 | |
| No. of ORFs including in the model | 620 | |
| ORF coverage | 9% | |
3.2. Evaluation of the metabolic simulation results using the constructed model
To evaluate the simulation results using the constructed model, the simulated specific growth rates under various conditions were compared with the experimental results. To analyze the growth rate under autotrophic conditions, the maximum specific growth rate of A. platensis NIES-39 was calculated to be 0.074 (/h) from culture results under various light intensity conditions [6]. The maximum CO2 uptake rate was estimated to be 3.1 mmol/gDW/h from the carbon composition in the biomass [43] and the maximum specific growth rate, with the assumption that the cells do not excrete any metabolites into the medium in the log growth phase. The specific growth rate under autotrophic conditions was simulated using the metabolic model with the following parameters: CO2 uptake rate, 3.1 mmol/gDW/h; photon uptake rate, free; glucose uptake rate, zero. The simulated specific growth rate (0.069/h) agreed well with the experimental results (0.074/h). The specific growth rate under heterotrophic conditions was also analyzed. The specific growth rate and specific glucose uptake rate were calculated from a previous experiment under heterotrophic conditions generated with glucose and without light illumination [37]. Metabolic simulation was performed to imitate the experimental conditions with the following parameters: glucose uptake, 0.16 mmol/gDW/h calculated from the experimental result [37]; photon uptake rate, zero. The simulated specific growth rate (0.11/h) was almost identical to the experimental result (0.10/h).
For further evaluation of the simulation results, growth capabilities on various organic substrates under heterotrophic conditions were analyzed. In a previous study, A. platensis NIES-39 was grown on 14 organic substrates, such as sugars, metabolites of the central metabolism, and amino acids, under heterotrophic conditions [44]. Note that A. platensis NIES-39 was described with its synonymous name, Spirulina platensis IAM M-135, in the previous study [44]. To simulate the growth capabilities of the metabolic model on these organic substrates, the uptake rate of each substrate was set to 10 mmol/gDW/h, and the photon uptake rate was set to zero. Because the uptake rate of each substrate was not measured in the experiment, the same uptake rate was set for each substrate for metabolic simulation, and only the growth capability was compared. The simulation results indicated that A. platensis could grow on all substrates and were consistent with the experimental results in 11 of 14 cases (Table 2). Inconsistencies between the simulation and experimental results were found in the case of α-ketoglutarate, succinate, and fumarate. To enable growth using α-ketoglutarate as a sole carbon source, a complete TCA cycle or glyoxylate shunt is required to supply the carbon for gluconeogenesis. However, the enzymes isocitrate lyase and malate synthase in the glyoxylate shunt and α-ketoglutarate dehydrogenase in the TCA cycle have not been identified in the genome sequence of A. platensis NIES-39. Recently, the TCA bypass pathway was determined to complement the incomplete TCA cycle in cyanobacteria [42]. A. platensis NIES-39 possesses enzymes highly homologous to those of the TCA bypass pathway and can grow on citrate and glutamate [44], which also requires a complete TCA cycle, thus suggesting the existence of a TCA bypass pathway. Therefore, it is possible that A. platensis NIES-39 lacks α-ketoglutarate uptake under the examined conditions because low uptake capability for α-ketoglutarate was reported in cyanobacteria [45]. Another possible reason for this result is that A. platensis NIES-39 does not possess a transporter for these metabolites because the transporter of α-ketoglutarate was annotated as a “putative” transporter (NIES39_G00660 gene) in A. platensis NIES-39. In fact, metabolic simulation without the α-ketoglutarate transport reaction predicted non-growth on α-ketoglutarate.
Table 2. Comparison of the growth capabilities on the various organic substrates.
| Substrate | Experimental [44] | Simulation |
|---|---|---|
| Glucose | + | + |
| Maltose | + | + |
| Sucrose | + | + |
| Maltotriose | + | + |
| Pyruvate | + | + |
| Lactate | + | + |
| Citrate | + | + |
| α-Ketoglutarate | − | + |
| Succinate | − | + |
| Fumarate | − | + |
| Malate | + | + |
| Oxaloacetate | + | + |
| Glutamate | + | + |
| Aspartate | + | + |
+ and − indicate growth or non-growth on the corresponding substrate, respectively, under heterotrophic conditions.
In the case of succinate and fumarate, the growth ability on citrate and glutamate suggests the existence of the activity of succinate dehydrogenase and fumarase required for utilization of succinate and fumarate in A. platensis NIES-39 (Table 2). The transporter of these metabolites was also annotated as being expressed from the same gene as the α-ketoglutarate transporter. Thus, this inconsistency may have resulted from a lack of existence of the transporter for these metabolites or a lack of their uptake under the examined conditions.
Overall, although further experimental validation and improvement of the metabolic model are required to eliminate these inconsistencies, the simulation results for the growth rate and growth capability are consistent with the experimental results, which suggests that the constructed metabolic model is reliable and could be applied for metabolic simulations of A. platensis NIES-39.
3.3. Glycogen production under nitrogen limitation conditions
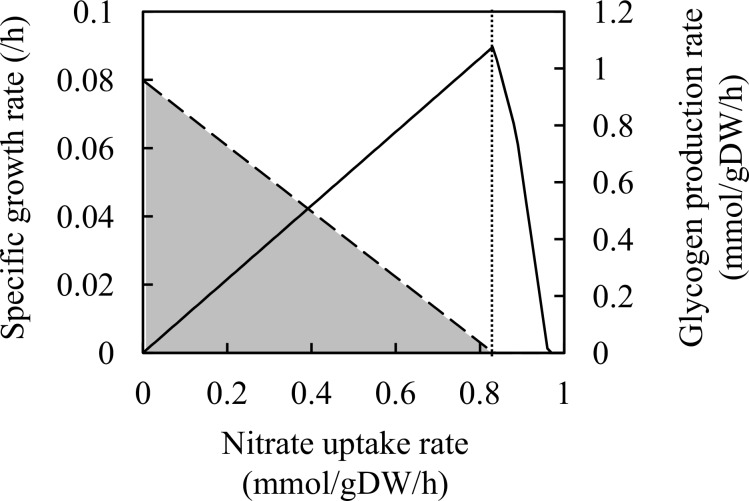
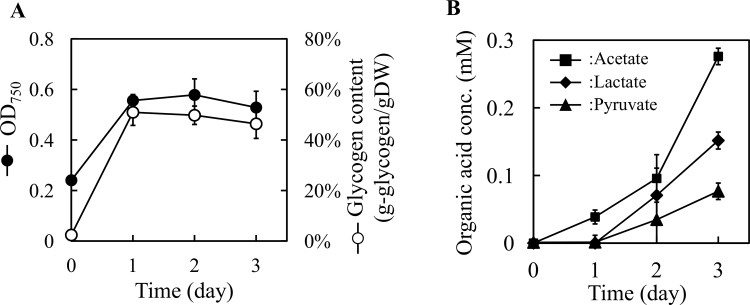
Glycogen accumulation by nitrogen limitation was simulated using the metabolic model. A. platensis is a promising host strain for use as a biomass feedstock because of its ability to accumulate a large amount of glycogen [6]. Previous studies reported a drastic increase of intracellular glycogen contents by nitrogen depletion of the culture medium [6, 10, 29]. The effects of nitrogen limitation on metabolism under autotrophic conditions were analyzed using the metabolic model. A virtual reaction for transport of glycogen from the cytosol to the extracellular compartment was added to the metabolic model to simulate glycogen production. For simplification, glycogen is treated as a monomer (C6H10O5, MW = 162.141) in this model, where one mole of glycogen is generated from one mole of glucose-1-phosphate. Metabolic simulations were performed with the following parameters: photon uptake rate, 50 mmol/gDW/h; glucose uptake rate, zero. The effect of nitrogen limitation was analyzed by changing the nitrogen uptake rate. The biomass and glycogen production rates as functions of the nitrate uptake rate are shown in Fig 1. When the nitrate uptake rate was lower than the optimal uptake rate for biomass production, the biomass production rate decreased and the glycogen production rate increased as observed for the experimental results [6]. The mechanism underlying the increase of glycogen production is as follows. Limitation of nitrate uptake rate decreased the growth rate, and the excess carbon caused by growth limitation was balanced with glycogen production. However, the increase of glycogen production was only one of the feasible solutions, and production of other metabolites such as ethanol, lactate, acetate, pyruvate, and formate was also one of the solutions under nitrate limitation. To confirm the production of these metabolites, the extracellular metabolites in the culture medium under nitrate depletion were experimentally analyzed. A. platensis NIES-39 was cultured with nitrate-supplemented medium, and cells were transferred to the fresh medium without nitrate. The glycogen content was increased by nitrate depletion, and production of acetate, lactate, and pyruvate was identified as estimated from the simulation results (Fig 2). Fumarate, succinate, and ethanol were not identified from HPLC and GC. Based on these results, deletion of genes related to the biosynthesis of acetate and lactate could be a strategy to improve glycogen production using A. platensis under nitrate depletion conditions.
Fig 1. Effect of nitrate uptake rate on growth and glycogen production.
The specific growth rate (solid line) and glycogen production rate (dashed line) at each nitrate uptake rate are shown. The dotted line indicates the optimal uptake rate for biomass production. The grey area represents the solution space of the glycogen production rate calculated by flux variability analysis [38], as the glycogen production rate was undetermined.
Fig 2. Culture profile under nitrogen depletion conditions.
Growth and intracellular glycogen (A) together with the concentrations of organic acids in the culture medium (B) were summarized. Closed circle, OD750; open circle, glycogen content; square, acetate; diamond, lactate; triangle, pyruvate. Error bars represent the standard deviation of triplicate experiments.
3.4. Design of a metabolic network for glycogen production
The metabolic model enabled us to develop metabolic engineering strategies for improvement of glycogen production. We performed an in silico multiple knockout simulation to identify the candidate genes to be deleted for improvement of glycogen production under autotrophic conditions with the following parameters: photon uptake rate, 50 mmol/gDW/h; glucose uptake rate, zero. All possible single, double, and triple gene knockout simulations were performed. Note that if several genes were assigned to one metabolic reaction in the model, knockout of one of these genes was used to knock out the reaction. Knockout simulations of more than four genes could not be performed because of the computational time required. However, a multiple knockout could not identify candidate genes to be deleted, possibly because glycogen production does not contribute to the improvement of biomass production, as glycogen biosynthesis is not accompanied by NAD(P)H production or consumption and requires ATP consumption, which reduces the growth rate.
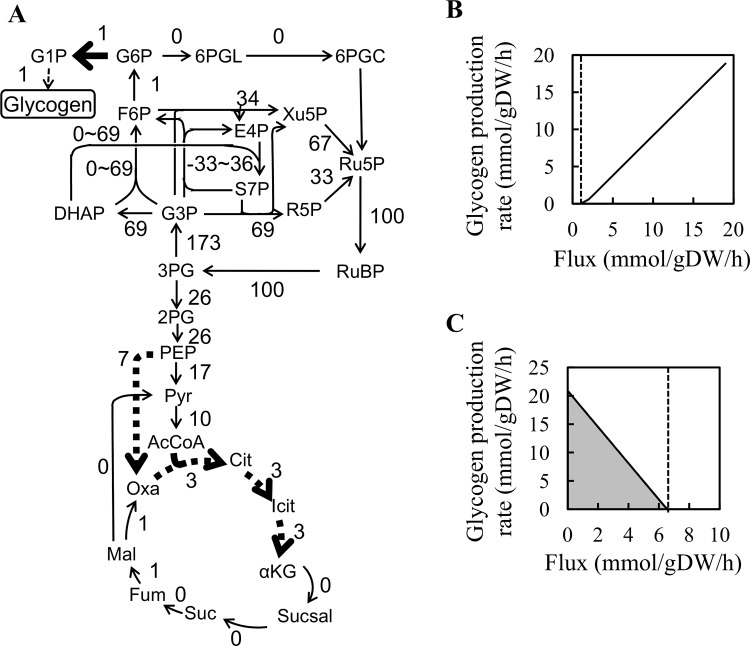
Next, we analyzed the effect of increasing or decreasing the flux of each reaction on glycogen production by flux response analysis (FRA) [14, 46]. In FRA, the changes of the target flux (glycogen production) are analyzed in response to artificial repression or activation of the flux of each reaction. FRA was performed with the following parameters: photon uptake rate, 50 mmol/gDW/h; glucose uptake rate, zero. Then, FBA was performed by changing the flux of each reaction related to the central metabolism, such as glycolysis, TCA cycle, pentose phosphate pathway (PPP), and the Calvin cycle. As expected, activation of the phosphoglucomutase reaction (G6P → G1P, abbreviations are described in figure legend of Fig 3), which produces a precursor of glycogen, enhanced glycogen production (Fig 3). Repression of reactions such as those mediated by phosphoenolpyruvate carboxylase (PEP → Oxa), citrate synthase (Oxa + AcCoA → Cit), aconitate hydratase (Cit → Icit), and isocitrate dehydrogenase (Icit → aKG), enhanced glycogen production, as decreased flux of these reactions repressed biomass production, and excess carbon was used for glycogen production.
Fig 3. Flux response analysis for glycogen production.
The simulated metabolic flux distribution under autotrophic conditions is shown (A). The values indicate the metabolic flux of each reaction. The flux was normalized to that of the reaction catalyzed by ribulose 1,5-bisphosphate carboxylase/oxygenase. Bold and dashed-bold arrows indicate the reactions whose activation or repression increased glycogen production, respectively, identified by flux response analysis. The reactions “DHAP + E4P ↔ S7P”, “DHAP + G3P ↔ F6P” and “G3P + S7P ↔ E4P + F6P” were undetermined, and the flux range of these reactions was calculated by flux variability analysis [38]. FRA results of the phosphoglucomutase reaction (G6P → G1P) (B) and phosphoenolpyruvate carboxylase reaction (PEP → Oxa) (C) are shown. The X-axis indicates the flux of the corresponding reaction and the Y-axis indicates the glycogen production rate excluding the flux for biomass production, as glycogen is a component of the biomass. The dotted line indicates the optimal flux of the corresponding reaction. The grey area in Fig 3(C) represents the solution space calculated by flux variability analysis. 2PG, glycerate-2-phosphate; 3PG, 3-phosphoglycerate; 6PGC, 6-phospho-gluconate; 6PGL, 6-phospho-glucono-1,5-lactone; AcCoA, acetyl-CoA; αKG, α-ketoglutarate; Cit, citrate; DHAP, dihydroxyacetone phosphate; E4P, erythrose-4-phosphate; F6P, fructose-6-phosphate; Fum, fumarate; G1P, glucose-1-phosphate; G3P, glyceraldehyde-3-phosphate; G6P, glucose-6-phosphate; Icit, isocitrate; Mal, malate; Oxa, oxalate; PEP, phosphoenolpyruvate; Pyr, pyruvate; R5P, ribose-5-phosphate; Ru5P, ribulose-5-phosphate; RuBP, ribulose-1,5-bisphosphate; S7P, sedoheptulose-7-phosphate; Suc, succinate; Sucsal, succinyl semialdehyde; Xu5P, xylulose-5-phosphate.
An increase of the precursor supply is an important strategy to improve target molecule production. However, FRA estimated that glycogen production was not enhanced by increased flux of the phosphoglucose isomerase reaction (F6P → G6P). When the flux of phosphoglucose isomerase was increased, the flux into oxidative PPP was increased because increase of oxidative PPP flux contributes to biomass production rather than increase of glycogen production. When zwf encoding glucose-6-phosphate dehydrogenase was deleted, glycogen production was predicted to be increased by activation of the phosphoglucose isomerase reaction (data not shown). Under the nitrate depletion condition in Synechocystis sp. PCC 6803, in which the flux toward glycogen was increased via phosphoglucose isomerase, the intracellular concentration of 6-phosphogluconate belonging to oxidative PPP was increased, suggesting that the flux through PPP was increased [47]. Therefore, deletion of genes related to the oxidative PPP, such as zwf, is a possible strategy to improve glycogen production under nitrogen limitation conditions as predicted by metabolic simulation.
3.5. In silico knockout simulation for ethanol production
The metabolic model was further applied to design the metabolic network to improve the production of another useful product, ethanol, which has the potential to replace fossil fuels. A. platensis NIES-39 is a natural producer of ethanol [10]. Both NADH and NADPH dependent alcohol dehydrogenase reactions were included in the metabolic model, since the specificity of alcohol dehydrogenase was unknown. First, the triple gene knockout simulation was performed to identify the candidate genes to be deleted for improvement of ethanol production with following parameters: photon uptake rate, 50 mmol/gDW/h; glucose uptake rate, zero. Nitrate was used as the nitrogen source. In this analysis, all flux estimations were performed under the autotrophic condition, the purpose of this screening was to improve the production of ethanol from CO2 as a carbon source and light as an energy source, for bio-engineering purposes. As results of this screening, two sets of knockouts were identified (Tables 3 and 4). The details, such as the identified genes and reactions, are summarized in S4 Table. The mechanisms underlying the increased ethanol production in these strains are not clear, but the excess NAD(P)H might be generated by these knockouts and was balanced with ethanol production, i.e., through the alcohol dehydrogenase reaction. Although the candidates were identified, their estimated carbon-molar yield (C-mol%), which was calculated as the carbon ratio between ethanol production to fixed CO2, were very low (approximately 2 C-mol%). When nitrate was used as a nitrogen source, a large amount of excess NAD(P)H could not be produced because a great amount of NADPH was used to reduce the oxidized ferredoxin generated by ferredoxin-nitrate reductase, which reduced nitrate to ammonium. Therefore, we speculated that using ammonium rather than nitrate could reduce the oxidization of NADPH by nitrate reduction and generate excess NADPH.
Table 3. In silico knockout simulation for ethanol production with nitrate as nitrogen source.
Detailed information is summarized in S4 Table.
| Relative growth rate to wild type model. | Ethanol yield (C-mol%) | Knockout targets |
|---|---|---|
| 96% | 2% | NADH dehydrogenase |
| NADPH dehydrogenase | ||
| L-Valine:pyruvate aminotransferase | ||
| Malic enzyme | ||
| 97% | 1% | NADH dehydrogenase |
| NADPH dehydrogenase | ||
| L-Valine:pyruvate aminotransferase | ||
| Malate dehydrogenase |
Table 4. In silico knockout simulation for ethanol production with ammonium as nitrogen source.
Detailed information is summarized in S4 Table.
| Relative growth rate to wild type model. | Ethanol yield (C-mol%) | Knockout targets |
|---|---|---|
| 18% | 55% | NADH dehydrogenase |
| NADPH dehydrogenase | ||
| Cytochrome-c oxidase | ||
| Phosphoglycerate mutase | ||
| 18% | 55% | NADH dehydrogenase |
| NADPH dehydrogenase | ||
| Cytochrome-c oxidase | ||
| Enolase | ||
| 20% | 54% | NADH dehydrogenase |
| NADPH dehydrogenase | ||
| Cytochrome-c oxidase | ||
| Phosphoenolpyruvate synthase | ||
| 37% | 43% | NADH dehydrogenase |
| NADPH dehydrogenase | ||
| Cytochrome-c oxidase |
The triple gene knockout simulation was performed using ammonium as a nitrogen source by changing the nitrate uptake rate to zero and the ammonium uptake rate to free. The knockout simulations identified several sets of knockout candidates whose deletion could improve ethanol production (Table 4 and S4 Table). The reactions of NADH dehydrogenase, NADPH dehydrogenase, and cytochrome-c oxidase, which belong to the respiratory chain, were commonly included among the candidates. NADH dehydrogenase and NADPH dehydrogenase are encoded by genes from NIES39_E02860, NIES39_A01530, NIES39_A08120, NIES39_K04850, NIES39_O01600, NIES39_O01620, NIES39_A06520, NIES39_K04840, NIES39_A06510, NIES39_O01610, NIES39_E02840, NIES39_O07110, NIES39_E02850, NIES39_A08140, NIES39_A08130, NIES39_F00440, NIES39_J01870, NIES39_H00330, NIES39_L04070, NIES39_L04060, and NIES39_L04040. Cytochrome-c oxidase is encoded by NIES39_O03120, NIES39_O03080, NIES39_O03090, NIES39_O03100, NIES39_A01920, and NIES39_A01910. The ethanol yield was estimated to be zero in the original metabolic model and 43 C-mol% in this knockout model. Because NAD(P)H was oxidized through the respiratory chain, excess NADPH was generated in the deletion model of these reactions. Generally, in non-photosynthetic microorganisms, ethanol production is performed under anaerobic conditions to generate excess intracellular NADH by reducing respiratory chain activity. However, in cyanobacteria, oxygen is produced by photosynthesis under autotrophic conditions, which makes it difficult to generate anaerobic conditions. Thus, disruption of the respiratory chain may be effective to increase ethanol production rather than anaerobic cultivation in cyanobacteria. In this case, the specificity of alcohol dehydrogenase for NADH and NADPH did not affect the results because transhydrogenase converts NADPH to NADH in our metabolic model. The disruption of respiratory chain also resulted in increasing flux on Photosystem I (PSI) and decreasing flux on Photosystem II (PSII). This change in PSI and PSII fluxes was probably due to compensating the decrease in proton pumping flux by the disruption of respiratory chain. Although the biomass production significantly decreased by the disruption of respiratory chain, still the cells can produce biomass. In the disrupted network, the proton pumping activity is maintained by the process of photosynthesis, which in turn results in producing ATP.
Moreover, additional deletions of some genes in the knockout of the respiratory chain were predicted to further increase ethanol production (Table 4), i.e. genes related to phosphoglycerate mutase, enolase and pyruvate kinase. Additional knockout of phosphoglycerate mutase encoded by NIES39_D07330, which converts 3-phosphoglycerate to 2-phosphoglycerate, resulted in a 55 C-mol% of ethanol yield. This knockout prevented the flux from 3-phosphoglycerate downstream of glycolysis, as 3-phosphoglycerate was converted to pyruvate via the serine biosynthesis pathway rather than glycolysis. In this strain, ethanol production was increased by balancing the excess NAD(P)H generated by following mechanisms: (1) NADH was generated by the serine biosynthesis pathway, and (2) the decreased growth rate reduced the usage of NAD(P)H for biomass formation because knockout of 3-phosphoglycerate prevented ATP production by pyruvate kinase. As for the deletion of enolase encoded by NIES39_B01040 and NIES39_C00710, the mechanism was the same as that in the knockout of phosphoglycerate mutase. In the case of the deletion of pyruvate kinase encoded by NIES39_M02310 and NIES39_P00240, the reduced ATP production resulting from knockout of pyruvate kinase decreased the growth rate, resulting in excess NAD(P)H.
Concluding Remarks
In this study, we constructed a genome-scale metabolic model of A. platensis NIES-39 and developed novel metabolic engineering strategies for glycogen and ethanol production. The metabolic model was constructed based on information from databases and the literature, and the simulation results using the metabolic model were evaluated by comparing them with the experimental results for growth rates and growth capabilities on various organic carbon sources. The high consistency between the simulation and experimental results indicates the availability of the constructed metabolic model for metabolic simulation. The metabolic simulation was further applied to demonstrate glycogen production under nitrogen limitation conditions and to design metabolic engineering strategies to increase glycogen production by FRA. Moreover, in silico knockout simulation successfully identified candidate genes to be deleted for improvement of ethanol production.
A. platensis is expected to be a suitable feedstock and host strain for bioproduction because of its superior characteristics for industrial bioproduction processes. The genome-scale metabolic model constructed in this study will provide a detailed metabolic understanding and valuable information to achieve industrial bioproduction using A. platensis.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, JAPAN.
References
- 1. Angermayr S.A., Hellingwerf K.J., Lindblad P., de Mattos M.J., Energy biotechnology with cyanobacteria, Curr. Opin. Biotechnol., 20 (2009) 257–263. 10.1016/j.copbio.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 2. Gao Z.X., Zhao H., Li Z.M., Tan X.M., Lu X.F., Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria, Energy & Environmental Science, 5 (2012) 9857–9865. [Google Scholar]
- 3. Deng M.D., Coleman J.R., Ethanol synthesis by genetic engineering in cyanobacteria, Appl Environ Microbiol, 65 (1999) 523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joseph A., Aikawa S., Sasaki K., Tsuge Y., Matsuda F., Tanaka T., Kondo A., Utilization of lactic acid bacterial genes in Synechocystis sp. PCC 6803 in the production of lactic acid, Biosci Biotechnol Biochem, 77 (2013) 966–970. [DOI] [PubMed] [Google Scholar]
- 5. Atsumi S., Higashide W., Liao J.C., Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde, Nat. Biotechnol., 27 (2009) 1177–1180. 10.1038/nbt.1586 [DOI] [PubMed] [Google Scholar]
- 6. Aikawa S., Izumi Y., Matsuda F., Hasunuma T., Chang J.S., Kondo A., Synergistic enhancement of glycogen production in Arthrospira platensis by optimization of light intensity and nitrate supply, Bioresour Technol, 108 (2012) 211–215. 10.1016/j.biortech.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 7. Liu X., Sheng J., Curtiss R. 3rd, Fatty acid production in genetically modified cyanobacteria, Proc Natl Acad Sci U S A, 108 (2011) 6899–6904. 10.1073/pnas.1103014108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aikawa S., Joseph A., Yamada R., Izumi Y., Yamagishi T., Matsuda F., Kawai H., Chang J.-S., Hasunuma T., Kondo A., Direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis processes, Energy & Environmental Science, 6 (2013) 1844. [Google Scholar]
- 9. Ueno Y., Kurano N., Miyachi S., Ethanol production by dark fermentation in the marine green alga, Chlorococcum littorale , J. Ferment. Bioeng., 86 (1998) 38–43. [Google Scholar]
- 10. Aoyama K., Uemura I., Miyake J., Asada Y., Fermentative metabolism to produce hydrogen gas and organic compounds in a cyanobacterium, Spirulina platensis , J. Ferment. Bioeng., 83 (1997) 17–20. [Google Scholar]
- 11. Vonshak A., Spirulina platensis (Arthrospira): Physiology, Cell-biology and Biotechnology, (2002). [Google Scholar]
- 12. Pulz O., Gross W., Valuable products from biotechnology of microalgae, Appl Microbiol Biotechnol, 65 (2004) 635–648. [DOI] [PubMed] [Google Scholar]
- 13. Tokuyama K., Ohno S., Yoshikawa K., Hirasawa T., Tanaka S., Furusawa C., Shimizu H., Increased 3-hydroxypropionic acid production from glycerol, by modification of central metabolism in Escherichia coli , Microb. Cell. Fac., 13 (2014) 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee K.H., Park J.H., Kim T.Y., Kim H.U., Lee S.Y., Systems metabolic engineering of Escherichia coli for L-threonine production, Mol Syst Biol, 3 (2007) 149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feist A.M., Palsson B.O., The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli , Nat. Biotechnol., 26 (2008) 659–667. 10.1038/nbt1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oberhardt M.A., Palsson B.O., Papin J.A., Applications of genome-scale metabolic reconstructions, Mol. Syst. Biol., 5 (2009) 320 10.1038/msb.2009.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Orth J.D., Thiele I., Palsson B.O., What is flux balance analysis?, Nat Biotechnol, 28 (2010) 245–248. 10.1038/nbt.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schilling C.H., Schuster S., Palsson B.O., Heinrich R., Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era, Biotechnol. Prog., 15 (1999) 296–303. [DOI] [PubMed] [Google Scholar]
- 19. Yoshikawa K., Kojima Y., Nakajima T., Furusawa C., Hirasawa T., Shimizu H., Reconstruction and verification of a genome-scale metabolic model for Synechocystis sp. PCC6803, Appl Microbiol Biotechnol, 92 (2011) 347–358. 10.1007/s00253-011-3559-x [DOI] [PubMed] [Google Scholar]
- 20. Nogales J., Gudmundsson S., Knight E.M., Palsson B.O., Thiele I., Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis, Proc Natl Acad Sci U S A, 109 (2012) 2678–2683. 10.1073/pnas.1117907109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knoop H., Zilliges Y., Lockau W., Steuer R., The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth, Plant Physiol., 154 (2010) 410–422. 10.1104/pp.110.157198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Montagud A., Navarro E., de Cordoba P. Fernandez, Urchueguia J.F., Patil K.R., Reconstruction and analysis of genome-scale metabolic model of a photosynthetic bacterium, BMC Systems Biology, 4 (2010) 156 10.1186/1752-0509-4-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vu T.T., Hill E.A., Kucek L.A., Konopka A.E., Beliaev A.S., Reed J.L., Computational evaluation of Synechococcus sp. PCC 7002 metabolism for chemical production, Biotechnol J, 8 (2013) 619–630. 10.1002/biot.201200315 [DOI] [PubMed] [Google Scholar]
- 24. Segrè D., Vu T.T., Stolyar S.M., Pinchuk G.E., Hill E.A., Kucek L.A., Brown R.N., Lipton M.S., Osterman A., Fredrickson J.K., Konopka A.E., Beliaev A.S., Reed J.L., Genome-Scale Modeling of Light-Driven Reductant Partitioning and Carbon Fluxes in Diazotrophic Unicellular Cyanobacterium Cyanothece sp. ATCC 51142, PLoS Comp. Biol., 8 (2012) e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meechai A., Pongakarakun S., Deshnium P., Cheevadhanarak S., Bhumiratana S., Metabolic flux distribution for gamma-linolenic acid synthetic pathways in Spirulina platensis , Biotechnol Bioproc E, 9 (2004) 506–513. [Google Scholar]
- 26. Cogne G., Gros J.B., Dussap C.G., Identification of a metabolic network structure representative of Arthrospira (spirulina) platensis metabolism, Biotechnol Bioeng, 84 (2003) 667–676. [DOI] [PubMed] [Google Scholar]
- 27. Klanchui A., Khannapho C., Phodee A., Cheevadhanarak S., Meechai A., iAK692: A genome-scale metabolic model of Spirulina platensis C1, BMC Systems Biology, 6 (2012) 71 10.1186/1752-0509-6-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fujisawa T., Narikawa R., Okamoto S., Ehira S., Yoshimura H., Suzuki I., Masuda T., Mochimaru M., Takaichi S., Awai K., Sekine M., Horikawa H., Yashiro I., Omata S., Takarada H., Katano Y., Kosugi H., Tanikawa S., Ohmori K., Sato N., Ikeuchi M., Fujita N., Ohmori M., Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39, DNA Res, 17 (2010) 85–103. 10.1093/dnares/dsq004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hasunuma T., Kikuyama F., Matsuda M., Aikawa S., Izumi Y., Kondo A., Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion, J Exp Bot, 64 (2013) 2943–2954. 10.1093/jxb/ert134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura Y., Kaneko T., Hirosawa M., Miyajima N., Tabata S., CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803, Nucleic Acids Res., 26 (1998) 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanehisa M., Goto S., KEGG: kyoto encyclopedia of genes and genomes, Nucleic Acids Res., 28 (2000) 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feist A.M., Herrgard M.J., Thiele I., Reed J.L., Palsson B.O., Reconstruction of biochemical networks in microorganisms, Nat. Rev. Microbiol., 7 (2009) 129–143. 10.1038/nrmicro1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quoc K.P., Dubacq J.P., Justin A.M., Demandre C., Mazliak P., Biosynthesis of eukaryotic lipid molecular species by the cyanobacterium Spirulina platensis , Biochim Biophys Acta, 1168 (1993) 94–99. [DOI] [PubMed] [Google Scholar]
- 34. Vincenzini M., Sili C., de Philippis R., Ena A., Materassi R., Occurrence of poly-beta-hydroxybutyrate in Spirulina species, J Bacteriol, 172 (1990) 2791–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miki W., Yamaguchi K., Konosu S., Carotenoid composition of Spirulina maxima , Bull Jpn Soc Sci Fish, 52 (1986) 1225–1227. [Google Scholar]
- 36. Shastri A.A., Morgan J.A., Flux balance analysis of photoautotrophic metabolism, Biotechnol. Prog., 21 (2005) 1617–1626. [DOI] [PubMed] [Google Scholar]
- 37. Marquez F.J., Sasaki K., Kakizono T., Nishio N., Nagai S., Growth characteristics of Spirulina platensis in mixotrophic and heterotrophic conditions, J. Ferment. Bioeng., 76 (1993) 408–410. [Google Scholar]
- 38. Mahadevan R., Schilling C.H., The effects of alternate optimal solutions in constraint-based genome-scale metabolic models, Metab Eng, 5 (2003) 264–276. [DOI] [PubMed] [Google Scholar]
- 39. Ernst A., Kirschenlohr H., Diez J., Boger P., Glycogen content and nitrogenase activity in Anabaena variabilis , Arch. Microbiol., 140 (1984) 120–125. [Google Scholar]
- 40. Pearce J., Leach C.K., Carr N.G., The incomplete tricarboxylic acid cycle in the blue-green alga Anabaena variabilis , J Gen Microbiol, 55 (1969) 371–378. [DOI] [PubMed] [Google Scholar]
- 41. Smith A.J., London J., Stanier R.Y., Biochemical basis of obligate autotrophy in blue-green algae and thiobacilli, J Bacteriol, 94 (1967) 972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S., Bryant D.A., The tricarboxylic acid cycle in cyanobacteria, Science, 334 (2011) 1551–1553. 10.1126/science.1210858 [DOI] [PubMed] [Google Scholar]
- 43. Sydney E.B., Sturm W., de Carvalho J.C., Thomaz-Soccol V., Larroche C., Pandey A., Soccol C.R., Potential carbon dioxide fixation by industrially important microalgae, Bioresour. Technol., 101 (2010) 5892–5896. 10.1016/j.biortech.2010.02.088 [DOI] [PubMed] [Google Scholar]
- 44. Ohira Y., Fukuchi W., Shimadzu M., Obata E., Effect of organic substrates on heterotrophic culture of Spirulina platensis , Kagaku Kogaku Ronbunshu, 35 (2009) 553–556. [Google Scholar]
- 45. Vazquez-Bermudez M.F., Herrero A., Flores E., Uptake of 2-oxoglutarate in Synechococcus strains transformed with the Escherichia coli kgtP gene, J Bacteriol, 182 (2000) 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park J.H., Kim T.Y., Lee K.H., Lee S.Y., Fed-batch culture of Escherichia coli for L-valine production based on in silico flux response analysis, Biotechnol Bioeng, 108 (2011) 934–946. 10.1002/bit.22995 [DOI] [PubMed] [Google Scholar]
- 47. Osanai T., Oikawa A., Shirai T., Kuwahara A., Iijima H., Tanaka K., Ikeuchi M., Kondo A., Saito K., Hirai M.Y., Capillary electrophoresis-mass spectrometry reveals the distribution of carbon metabolites during nitrogen starvation in Synechocystis sp. PCC 6803, Environ Microbiol, 16 (2014) 512–524. 10.1111/1462-2920.12170 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.