Abstract
Seed storability in rice (Oryza sativa L.) is an important agronomic trait. Two segregating populations with N22 (indica) as a common parent, viz. a set of 122 backcross-inbred lines (BILs) derived from the backcross Nanjing35 (japonica)/N22//Nanjing35 and another population comprising 189 recombinant inbred lines (RILs) from the cross of USSR5 (japonica) and N22, were studied to detect quantitative trait loci (QTL) controlling seed storability. Germination percentage (GP) was used to evaluate seed storability after aging treated under three different conditions, viz. natural, artificial and combined aging treatments. A total of seven QTLs were identified on chromosomes 1, 2, 5, 6 and 9. Among them, a major QTL, qSSn-9, was common in the two populations. In contrast, four QTLs (qSSnj-2-1, qSSn-2-2, qSSn-5 and qSSn-6) were detected in BILs and the QTL qSSn-1 was identified in RILs, which was a new QTL for seed storability. The N22-derived alleles increased the seed storability at all the loci except qSSnj-2-1. We also investigated the effect of QTLs using five selected lines with high storability from BILs and verified qSSn-5 with a near-isogenic line (NIL). These results provide an opportunity for pyramiding or map-based cloning major QTLs for seed storability in rice.
Keywords: rice (Oryza sativa L.), seed storability, backcross inbred lines (BILs), recombinant inbred lines (RILs), quantitative trait locus (QTL), artificial aging, natural aging
Introduction
Seed storability in rice is an important trait for both ecological and agronomic aspects due to its correlation with high seed germination and seedling vigor (Yamauchi and Winn 1996). During warehousing period, seeds begin to deteriorate, lose vigor, and as a result, become more sensitive to stresses during germination, and ultimately die. Generally storability is evaluated by seed germination ratio (Miura et al. 2002, Sasaki et al. 2005, Xue et al. 2008). Strong storability reduces the risk of rapid seed deterioration, which is a serious problem for rice production in tropical Asia.
So far, quantitative trait loci (QTL) controlling seed storability have been identified by using either natural or artificial aging in rice (Jiang et al. 2011, Li et al. 2012, Miura et al. 2002, Sasaki et al. 2005, Xue et al. 2008, Zeng et al. 2006). Natural aging is intrinsically time-consuming (always more than one year), thus not suitable for rapid screening of a large amount of samples and germplasm. Instead, artificial aging with elevated ambient temperature and relative humidity (RH) was utilized to rapidly assess seed storability of rice (Miura et al. 2002, Xue et al. 2008, Zeng et al. 2006). Using a backcross population of Nipponbare/Kasalath, Miura et al. (2002) identified three QTLs related to seed longevity on chromosome 2, 4, and 9, through artificial aging method. Later, three QTLs for seed storability were mapped on chromosome 9, 11, and 12 using doubled-haploid population derived from the cross ZYQ8/JX17 (Zeng et al. 2006). Besides, Xue et al. (2008) detected three QTLs associated with seed storability on chromosomes 1, 3, and 9 using recombinant inbred lines (RILs) derived from the cross IR24/Asominori.
Under natural storage conditions, other QTLs for seed storability were detected in diverse genetic backgrounds (Jiang et al. 2011, Li et al. 2012, Sasaki et al. 2005). After keeping seeds for 1, 2, and 3 years, Sasaki et al. (2005) detected four QTL (RC7, RC9-1, RC9-2, and RC9-3) for seed longevity in 191 RILs (F7 generations) derived from Milyang 23/Akihikari. Employing similar aging treatments, Jiang et al. (2011) found seven QTL associated with seed storability using two sets of RILs; one derived from Milyang 23/Tong 88-7, the other from Dasanbyeo/TR22183. Thereafter, Li et al. (2012) identified six QTLs affecting seed storability on chromosomes 2, 3, 4, 6, 9, and 11 in a backcross population of Koshihikari/Kasalath after natural aging for 32 and 48 months. Although these QTL data enhanced our understanding to molecular mechanism of seed storability, QTLs detected under natural and artificial aging treatments were not entirely consistent and currently available information has not been applied by breeders.
N22 is a traditional indica cultivar from India. After stored at room temperature (<30°C) and 40–60% RH for 42 months, N22 still displayed >90% germination capability. However, the genetic factors controlling seed storability for N22 have not been identified. Nanjing35 and USSR5 (all japonica) with weak aging resistance (the former was much weaker than the latter), were used as other parents to construct mapping population (Li et al. 2013, Xie et al. 2011). In the present study, our objectives were: 1) to map QTL for seed storability using two populations with N22 as a common parent; 2) to estimate the genetic effect of QTL under different aging conditions; 3) to explore the possibility to select breeding materials with good seed storability by pyramiding QTL through marker-assisted selection.
Materials and Methods
Plant materials
A BIL (BC1F8) population of 122 individuals from the backcross Nanjing35/N22//Nanjing35 and a RIL (F2:7) population comprising 189 recombinant inbred lines from the cross of USSR5 and N22 were both developed by single seed descent (Li et al. 2013, Xie et al. 2011). A near-isogenic line (NIL), which contained a single genomic fragment from N22 that included the region between markers RM178 and RM3170 on chromosome 5 introgressed into the Nanjing35 genetic background (Supplemental Fig. 1), was developed using phenotype (strong seed dormancy) and marker-assisted selection by Lu et al. (2011). Further investigation of this NIL was used to verify the QTL of seed storability presently identified in this region of chromosome 5.
Parental accessions (Nanjing35, USSR5, and N22) were planted in 2009, 2010, 2011, and 2012, at Nanjing Agricultural University Rice Station (Tuqiao, Nanjing, China). BILs were planted in 2009, and 2010, whereas RILs in 2009, 2011, and 2012. Two rows of each BILs and RILs were planted and the distance of each plant was 15 cm. The heading date was recorded, and seeds were harvested 35 days after flowering. Then, the sample seeds were kept in an oven at 50°C for seven days to fully break dormancy and dried under well-ventilated conditions to a constant weight (11–13% seed moisture content).
Different treatment methods before seed storability measurement
Natural aging treatment was performed as described by Li et al. (2012). BILs and RILs harvested in 2009 were both stored at room temperature (<30°C) and 40–60% RH in Nanjing for 32 months.
Artificial aging treatment was based on an improved method originally proposed by Zeng et al. (2006). Initially, to investigate the seed storability of parental accessions and determine appropriate evaluation periods of artificial aging, a preliminary experiment was conducted for comparing the performance of three parents (Nanjing35, USSR5, and N22). Using a LH-150 (Jiangsu, China) thermostatic moisture regulator, seeds of Nanjing35 and N22 harvested in 2010 were treated at 40°C and 80% RH for 25, 35, and 45 days, for a total of 3 treatments. Seeds of USSR5 and N22 harvested in 2012 were treated at 40°C and 85% RH for 32, 35, 38, and 41 days. After optimization, 80% RH and storage periods of 35 days were used for testing BIL-2010 population. The RILs harvested in 2012 were treated at 40°C and 85% RH for 38 days.
Combined aging treatment was performed according to a previous study (Miura et al. 2002) with minor modifications. After being stored for 32 months at indoor temperature (<30°C) and 40–60% RH in Nanjing, BILs harvested in 2009 were treated at 40°C and 80% RH for 10 days. For RILs, the sample seeds were stored for 12 months and then kept at 40°C and 80% RH for 35 days. All populations harvested in 2009–2012 with different aging treatments were summarized in Table 1. ‘B’ and ‘R’ indicate BIL and RIL population, respectively. ‘A’, ‘N’, and ‘C’ denote artificial, natural, and combined aging treatment, respectively.
Table 1.
Different treatment methods before seed storability measurement for Nanjing35/N22//Nanjing35 BILs and USSR5/N22 RILs
| Code | Population | Aging methods | Year | Treated conditions |
|---|---|---|---|---|
| B-A-10 | BIL | Artificial aging | 2010 | 40°C and 80% RH for 35 days |
| R-A-12 | RIL | 2012 | 40°C and 85% RH for 38 days | |
| B-N-09 | BIL | Natural aging | 2009 | Room temperature (<30°C) and 40–60% RH in Nanjing for 32 months |
| R-N-09 | RIL | 2009 | Room temperature (<30°C) and 40–60% RH in Nanjing for 32 months | |
| B-C-09 | BIL | Combined aging | 2009 | 40°C and 80% RH for 10 days based on stored for 32 months |
| R-C-11 | RIL | 2011 | 40°C and 80% RH for 35 days based on stored for 12 months |
Evaluation of seed storability
Germination percentage (GP) was used to determine the degrees of seed storability. Three replications of 50 treated seeds of each line were placed on doubled sheets of moistened filter paper in 9-cm Petri dishes maintained at 30°C for 7 days. The emergence of the radicle and plumule denoted the seed with high storability. The average GP of each line was arcsin-transformed for the purpose of QTL analysis (Li et al. 2012, Miura et al. 2002).
QTL and statistical analyses
Linkage maps were constructed using MapMaker/EXP 3.0 (Li et al. 2013, Xie et al. 2011). The composite interval mapping procedure in Windows QTL Cartgrapher v2.5 (Wang et al. 2007) was used for QTL mapping. The permutation method (Churchill and Doerge 1994) was used to obtain the empirical thresholds for claiming main-effect QTL of the experiment based on 1000 runs of randomly shuffling the trait values, which were 3.0, 3.8, and 3.0 for B-A-10, B-N-09, and B-C-09, respectively, and 3.0, 2.9, and 3.1 for R-A-12, R-N-09, and R-C-11, respectively. Thus, these LOD thresholds were used for claiming significant main-effect QTL in this study. Confidence intervals (CI) were set at one LOD unit at either side of the peak. Adjacent QTLs with overlapping CIs were treated as a single QTL. QTL nomenclature followed the guideline of McCouch et al. (1997). For example, qSSn-2-1 denoted the first Seed Storability QTL on chromosome 2 and ‘n’ indicated additive effect enhancing seed storability was conferred by the allele from N22.
Seed viability assay
Seeds of Nanjing35 and five lines selected from BILs harvested in 2011 were stored under disgusting-ventilated conditions (poor ventilation and wide variation of humidity accompanied with seasonal alternation) for 18 months, while seeds of Nanjing35 and the NIL harvested in 2012 were stored at room temperature (<30°C) and 40–60% RH in Nanjing for 20 months. Then the seeds were dehulled and imbibed in tap water for 24 hour at 30°C incubator. Subsequently the seeds were soaked in a 1% solution of 2,3,5-triphenyltetrazolium chloride (TTC) at 35°C for 2 hour in the dark, and then rinsed with distilled water until the water ran clear. Embryos were considered as viable if they were completely stained into pink or red, and uncolored embryos were considered as non-viable. Deeper staining was assumed to reflect greater viability (Towill and Mazur 1975).
Results
Seed storability of parent lines
To investigate the seed storability of three parents (N22, USSR5, and Nanjing35) and determining appropriate evaluation periods of artificial aging in BILs and RILs, a time-response experiment was initially conducted.
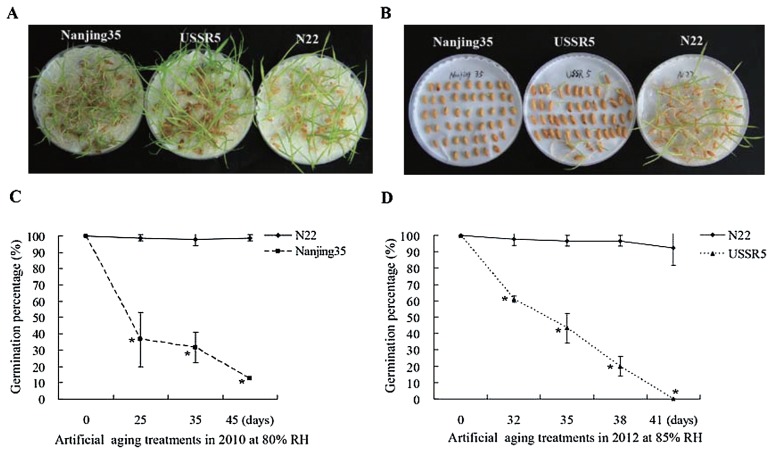
All parental accessions exhibited normal germination (near 100%) in the absence of aging treatments, indicating a high viability of the untreated seeds (Fig. 1A). As indicated in Fig. 1B and 1C, the GPs of Nanjing35 decreased faster than those of N22 after aging treatment at 80% RH, suggesting Nanjing35 showed the lower storability. The difference of storability between Nanjing35 and N22 became pronounced (p < 0.05) after artificial aging for 35 days, suggesting this period was favorable to evaluate storability in Nanjing35/N22//Nanjing35 BILs (Fig. 1C).
Fig. 1.
Response of parental accessions to different artificial aging treatment times (days). (A) Germination of Nanjing35 (left), USSR5 (middle), and N22 (right) without aging treatments. (B) Germination of Nanjing35 (left), USSR5 (middle), and N22 (right) after artificial aging treatment for 38 days in 2012. (C) Response of Nanjing35 and N22 to different artificial aging treatment times (days) in 2010. (D) Response of USSR5 and N22 to different artificial aging days in 2012. Solid, short broken and long broken lines indicate GPs for N22, USSR5, and Nanjing35, respectively. Error bars indicate the standard deviations. Asterisk means significant at the 0.05 probability level.
Similarly, the GPs of USSR5 after artificial aging treatments at an increased RH (85%) displayed a dramatic loss, whereas only slight decrease was observed in N22. Remarkably, after treatment for 41 days, the GP of USSR5 dropped to 0%, in contrast to 92.2 ± 10.7% of N22 (Fig. 1D). Hence aging treatment for 38 days was selected for USSR5/N22 RILs.
GPs in BILs and RILs after different aging treatments
Germination tests were performed on seeds of Nanjing35, USSR5, N22, and their derived BIL and RIL populations after three different aging methods.
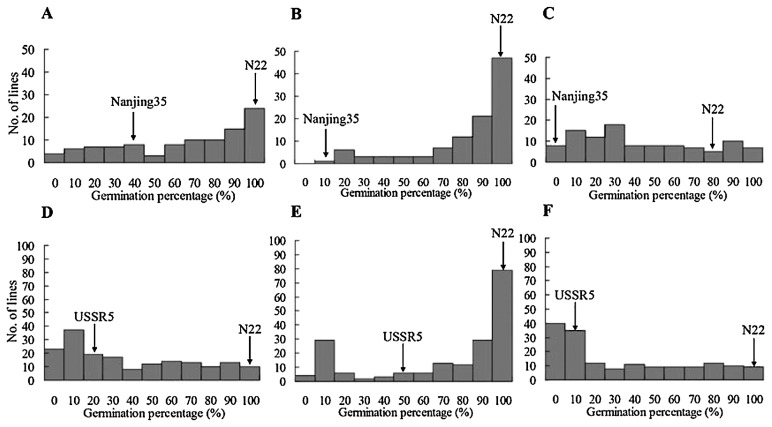
As indicated in Fig. 2A–2C, the GPs of Nanjing35 were 31.6 ± 9.4%, 6.7 ± 1.9%, and 0.0% after three different aging treatments B-A-10, B-N-09, and B-C-09, respectively, while the GPs of N22 were 97.8 ± 3.9%, 100%, and 77.8 ± 7.7%. The differences of GPs between Nanjing35 and N22 were significant (p < 0.05) and variations of BILs ranged from 0% to 100% in three treatment environments. The distribution of GPs of the BILs in treatment B-N-09 was highly skewed with nearly half of the lines above 90% (Fig. 2B). The populations showed a wide distribution in GPs after aging treatments B-A-10 (Fig. 2A) and B-C-09 (Fig. 2C).
Fig. 2.
Frequency distributions of GPs after aging treatments in Nanjing35/N22//Nanjing35 BILs and USSR5/N22 RILs. (A–C) represent frequency distributions of GPs in Nanjing35/N22//Nanjing35 BILs after artificial aging for 35 days in 2010, natural aging for 32 months in 2009, and combined aging treatment in 2009, abbreviated as treatment B-A-10, B-N-09, and B-C-09, respectively. (D–F) represent frequency distributions of GPs in USSR5/N22 RILs after artificial aging for 38 days in 2012, natural aging for 32 months in 2009, and combined aging treatment in 2011, abbreviated as treatment R-A-12, R-N-09, and R-C-11, respectively. GPs of the parents are indicated by arrows.
As indicated in Fig. 2D–2F, the GPs of USSR5 were 20.0 ± 5.8%, 44.4 ± 3.8%, and 6.7 ± 0.0% after three different aging treatments R-A-12, R-N-09, and R-C-11, respectively, while the GPs of N22 were 96.7 ± 3.3%, 100%, and 97.8 ± 1.9%. The differences of GPs between USSR5 and N22 were also significant (p < 0.05) and GPs of the RILs ranged from 0% to 100% with continuous variation after three aging treatments. Comparing with wide distributions after treatments R-A-12 and R-C-11 (Fig. 2D, 2F), the distribution of GPs in RILs after the treatment R-N-09 was highly skewed with nearly half of the lines above 90% (Fig. 2E).
QTL detection in BILs and RILs after different aging treatments
A total of five putative QTLs were detected in BILs and they were on chromosome 2, 5, 6, and 9 (Table 2 and Supplemental Fig. 2). Apart from the Nanjing35 allele at qSSnj-2-1, N22 alleles at other QTLs increased seed storability.
Table 2.
Location and genetic effects of QTL for seed storability in Nanjing35/N22//Nanjing35 BILs and USSR5/N22 RILs after aging treatments
| Population | Aging treatmentsa | QTL nameb | Chromosome | Marker interval | LOD scorec | PVE (%)d | Additive effect |
|---|---|---|---|---|---|---|---|
| BIL | B-A-10 | qSSnj-2-1 | 2 | RM324-RM262 | 4.1 | 12.3 | −9.3 |
| qSSn-2-2 | 2 | RM526-RM240 | 3.8 | 11.4 | 8.8 | ||
| qSSn-9 | 9 | RM7038-RM105 | 3.7 | 12.2 | 10.4 | ||
| B-N-09 | qSSn-5 | 5 | RM421-RM26 | 5.3 | 15.0 | 9.0 | |
| B-C-09 | qSSn-5 | 5 | RM421-RM26 | 5.5 | 14.4 | 10.4 | |
| qSSn-6 | 6 | RM588-RM217 | 4.1 | 13.6 | 9.5 | ||
| qSSn-9 | 9 | RM444-RM105 | 4.3 | 13.1 | 10.2 | ||
| RIL | R-A-12 | qSSn-1-1 | 1 | RM443-RM128 | 3.6 | 10.8 | 7.9 |
| qSSn-9 | 9 | RM1328-RM105 | 4.9 | 11.1 | 7.9 | ||
| R-N-09 | qSSn-1 | 1 | RM237-RM128 | 4.9 | 11.3 | 9.8 | |
| qSSn-9 | 9 | RM1328-RM105 | 6.8 | 13.7 | 10.1 | ||
| R-C-11 | qSSn-1-1 | 1 | RM443-RM128 | 4.2 | 9.9 | 8.4 | |
| qSSn-1-2 | 1 | RM237 | 3.1 | 7.3 | 7.4 | ||
| qSSn-9 | 9 | RM1328-RM105 | 10.2 | 21.5 | 12.0 |
Aging treatments: ‘B’ and ‘R’ indicate BIL and RIL population, respectively. ‘A’, ‘N’, and ‘C’ denote artificial, natural, and combined aging treatment, respectively, ‘09’, ‘10’, ‘11’, and ‘12’ indicate 2009, 2010, 2011, and 2012, respectively.
QTL name: the letter ‘q’ stands for QTL and ‘SS’ for seed storability; the letter ‘n’ and ‘nj’ for rice cultivar N22 and Nanjing35, respectively; the first number denotes the chromosomal location; the second number denotes different QTL located on the same chromosome.
LOD score: logarithm of odds.
PVE (%): percentage of total phenotypic variance explained by the QTL.
The QTL qSSn-9 was simultaneously detected in the same region located between markers RM7038 and RM105 on chromosome 9 after treatments B-A-10 and B-C-09, respectively, with relatively high LOD values of 3.7–4.3 and explained phenotypic variations of 12.2–13.1% (Table 2 and Supplemental Fig. 2). qSSn-5, flanked by markers RM421 and RM26 on chromosome 5, was detected repeatedly in treatments B-N-09 and B-C-09, and explained the highest phenotypic variations ranging from 14.4 to 15.0% with the highest LOD scores of 5.3–5.5. Other three QTLs, viz. qSSnj-2-1, qSSn-2-2, and qSSn-6, were detected only once on chromosome 2 and 6 (Table 2 and Supplemental Fig. 2).
Three QTLs, qSSn-1-1, qSSn-1-2, and qSSn-9 were detected on chromosomes 1 and 9 in RILs after three aging treatments (Table 2 and Supplemental Fig. 3). N22 alleles of all QTLs contributed to increased seed storability. qSSn-1-1 and qSSn-1-2 were tightly linked and only qSSn-1-1 was detected repeatedly in three treatments. The closest SSR loci to qSSn-1-1 and qSSn-1-2 were RM128 and RM237, respectively (Table 2 and Supplemental Fig. 3).
Five selected lines with high seed storability from Nanjing35/N22//Nanjing35 BILs
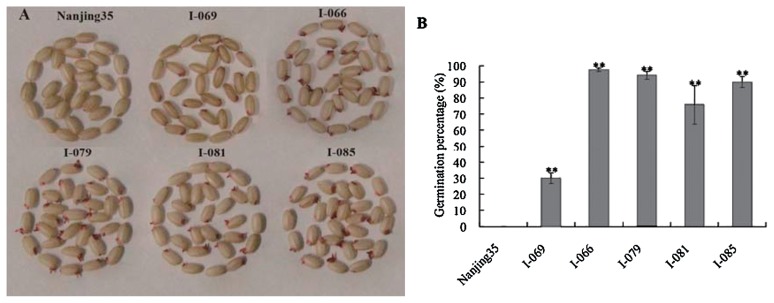
Five lines harboured different N22-derived alleles at QTLs for seed storability were selected in BILs (Table 3). A line (named I-069) carrying N22 alleles at qSSnj-2-1 and qSSn-9, exhibited less GP than background Nanjing35 after treatment B-A-10 (p < 0.05). However, the GP of I-069 increased significantly after treatment B-N-09, suggesting that the N22 allele at qSSnj-2-1 reduced seed storability but the N22 allele of qSSn-9 exerts strong favorable effect on storability and overpowers qSSnj-2-1 in natural aging conditions (Table 3). Additionally, 4 lines, named I-066, I-079, I-081, and I-085, possessed different QTLs and exhibited similar or equal GPs to N22, regardless of aging treatments (Table 3).
Table 3.
Genotypes and phenotypes of five lines with different numbers of QTLs in Nanjing35/N22//Nanjing35 BILs
| Line name | Genotypes | Germination percentages | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| qSSnj-2-1 | qSSn-2-2 | qSSn-5 | qSSn-5 | qSSn-6 | qSSn-9 | qSSn-9 | ||||
|
| ||||||||||
| RM341 | RM525 | RM480 | RM26 | RM588 | RM7038 | RM105 | B-A-10a | B-N-09 | B-C-09 | |
| Nanjing35 | 1b | 1 | 1 | 1 | 1 | 1 | 1 | 31.6 ± 9.4 | 6.7 ± 1.9 | 0 |
| N22 | 2c | 2 | 2 | 2 | 2 | 2 | 2 | 97.8 ± 3.9 | 100 | 77.8 ± 7.7 |
| I-069 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 6.7 ± 0.0 | 71.1 ± 8.4 | 1.3 ± 1.5 |
| I-066 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 100 | 100 | 98.9 ± 1.9 |
| I-079 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 91.0 ± 6.9 | 97.8 ± 3.9 | 62.0 ± 34.1 |
| I-081 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 96.7 ± 5.8 | 97.5 ± 3.5 | 90.0 ± 0.0 |
| I-085 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 86.67 ± 0.0 | 98.9 ± 1.9 | 98.0 ± 2.0 |
‘B’ indicate the BIL population. ‘A’, ‘N’, and ‘C’ denote artificial, natural, and combined aging treatment, respectively, ‘09’ and ‘10’ indicate 2009 and 2010, respectively.
1 denotes genotypes of Nanjing35.
2 denotes genotypes of N22.
Seeds of the selected five lines from BILs harvested after maturity were stored under disgusting-ventilated conditions (poor ventilation and wide variation of humidity accompanied with seasonal alternation) for 18 months and then those seeds stained with TTC to determine seed viability. As shown in Fig. 3A, seed batches of Nanjing35 exhibited maximum mortality due to its high percentage of uncolored embryos. Seed batches of I-069 showed moderate mortality while those of I-066, I-079, I-081, and I-085 exhibited the strongest viability because almost all seed embryos were stained into deep red. The differences in seed viability by TTC staining were consistent with those on seed GPs (Fig. 3B).
Fig. 3.
Tetrazolium assay and GPs of Nanjing35 and five lines from BILs after stored under disgusting-ventilated conditions (poor ventilation and wide variation of humidity accompanied with seasonal alternation) for 18 months. Error bars indicate the standard deviations. ** mean significant at the 0.01 probability level. (A) represent tetrazolium assay of Nanjing35 and five lines from BILs. (B) represent GPs of Nanjing35 and five lines from BILs.
Confirmation of qSSn-5 with a NIL and TTC test
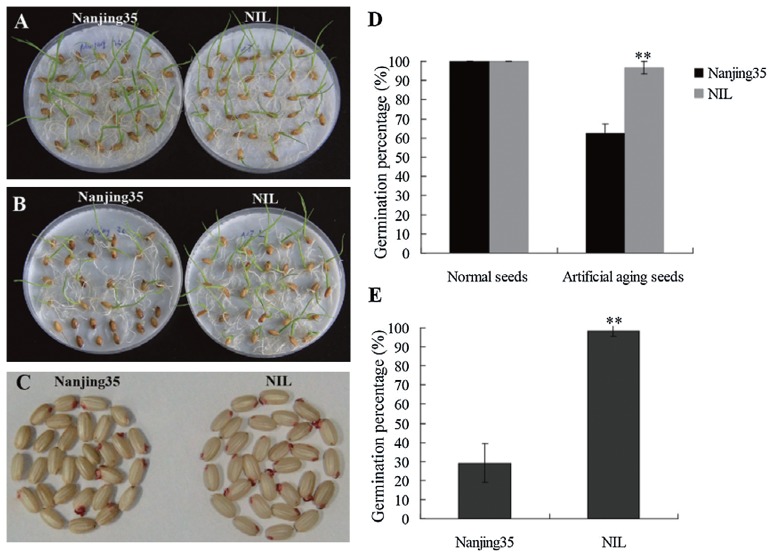
A NIL selected from a previous study (Lu et al. 2011), which contained a single genomic fragment from N22 that included the qSSn-5 region (length between markers RM178 and RM3170) introgressed into the Nanjing35 genetic background (Supplemental Fig. 1), was used to determine whether the N22 allele at qSSn-5 enhancing seed storability or not. The germination performances of Nanjing35 and the NIL were analyzed after artificial aging treated at 40°C and 85% RH for 20 days. As indicated in Fig. 4B and 4D, the GP of Nanjing35 was significantly less than that of the NIL (p < 0.01), though both Nanjing35 and NIL exhibited normal germination capability after harvest (Fig. 4A).
Fig. 4.
Response of Nanjing35 and the NIL to artificial aging and natural aging treatments. (A and B) represent germination of Nanjing35 (left) and the NIL (right) without aging treatment and after artificial aging for 20 days at 40°C and 85% RH, respectively. (C and E) represent tetrazolium assay and GPs of Nanjing35 and the NIL after natural aging for 20 months, respectively. (D) represents GPs of Nanjing35 and the NIL after artificial aging for 20 days at 40°C and 85% RH. Error bars indicate the standard deviations. ** mean significant at the 0.01 probability level.
As shown by the tetrazolium salt staining in vivo (Fig. 4C), seed batches of Nanjing35 after natural aging for 20 months exhibited a higher extent of seed mortality. This was illustrated by the detection of a substantially higher number of uncolored embryos in Nanjing35 than those in the NIL. The difference in seed viability was consistent with the observed deleterious effect of natural aging treatment on seed germination performance (Fig. 4E). Altogether these data supported that the N22 allele at qSSn-5 exerts an increasing effect on seed storability.
Discussion
Recent QTL analysis for seed storability in rice suggests that it is generally governed by multi-genes and is vulnerably influenced by growing environments (Jiang et al. 2011, Li et al. 2012, Miura et al. 2002, Sasaki et al. 2005, Xue et al. 2008, Zeng et al. 2006). In this study, we mainly focused on genetic dissection of the seed storability trait in cultivated rice N22, which exhibits high levels of both dormancy (Lu et al. 2011, Wan et al. 2006, Xie et al. 2011) and storability. A total of five QTLs associated with the seed storability were detected in BILs from Nanjing35/N22//Nanjing35 and only three in RILs from USSR5/N22 (Table 2 and Supplemental Figs. 2, 3). It could be explained that Nanjing35 has weaker seed storability than USSR5 (Supplemental Fig. 4), suggesting that USSR5 might contain the same alleles as N22 at some loci for seed storability, resulting in fewer loci detected in RILs from USSR5/N22 than BILs from Nanjing35/N22//Nanjing35. Besides, various factors, such as population size, population type, number of molecular markers, environmental conditions, methods, and critical standards of the germination test, may have affected the mapping results (Wan et al. 2006, Yano et al. 1997). The N22-derived QTL, qSSn-9, was simultaneously detected in the almost same region in the two populations, suggesting that this QTL is robustly expressed across distinct genetic backgrounds and deserves to be investigated in the future.
Artificial aging does not completely mimic natural aging
Natural aging and artificial aging methods are widely used in seed storability (Li et al. 2012, Miura et al. 2002, Sasaki et al. 2005, Xue et al. 2008, Zeng et al. 2006). Sensitivity of seeds to artificial aging has been used successfully to rapidly evaluate and predict seed storability (Miura et al. 2002, Xue et al. 2008, Zeng et al. 2006). High storability seeds germinate normally after being subjected to artificial aging treatments, but low storability seeds produce abnormal seedlings or die. Several physiological and biochemical processes have been identified that occur during artificial aging of seeds, similar to natural aging (Han et al. 2014, Rajjou et al. 2008). In this study, qSSn-1 and qSSn-9 was detected both under natural aging and artificial aging conditions in RIL populations (Table 2 and Supplemental Fig. 3). However, in the results from identification of QTL for seed storability in BILs, two QTLs (qSSnj-2-1 and qSSn-2-2) were detected only after artificial aging treatment, and other one, qSSn-6, only after combined aging treatment, while qSSn-5 was detected in both B-N-09 and B-C-09 environments (Table 2 and Supplemental Fig. 2), indicating artificial and combined aging treatments do not completely mimic deterioration process in conventional storage conditions.
The five selected lines with high storability could be used for further study and breeding
Except the line I-069, four lines all showed high GPs after different aging treatments, even higher than N22 in B-C-09, though they did not harbour all QTLs of seed storability. One possible explanation is that the interaction between different QTLs might have important effects on seed storability, especially relationship of qSSnj-2-1 and the other QTLs, in which negative effects of N22 locus might be exceptional among N22 genotype QTL. In 2009 after combined aging treatment B-C-09, the line I-079 had the lowest GP value of 62.0 ± 34.1% (Table 3), while in 2011 after storage under poor-ventilated conditions for 18 months, the line I-081 had the lowest GP value of 76.0 ± 12.0% (Fig. 3B), among these four lines. This might be the consequence of different planted years and aging treatments, since it is known that growing environments strongly affect seed quality (Contreras et al. 2008). The five selected lines showed significantly higher storability than Nanjing35 (p < 0.01) (Fig. 3), suggesting that these N22-derived alleles could increase the seed storability. Therefore, we think that high storability was achieved by the additive effect of two or more QTLs. Based on these five lines, additional advanced backcross lines will be developed to isolate single QTL, and analyze the effect of each QTL on seed storability.
Comparison of QTLs identified in present and previous studies
Based on common markers, qSSn-9 is the same locus as either qLG-9 or qSS-9 that was detected on chromosome 9 for seed storability (Li et al. 2012, Miura et al. 2002). Using an F2 population from a cross between Nipponbare and SL36, which the target qSS-9 region on chromosome 9 was substituted with a segment from Kasalath in the genetic background of Nipponbare, the qSS-9 locus was located between the Indel markers Y10 and Y13 reported by Lin et al. (2015), corresponded to a delimited region of 147 kb in the Nipponbare genome. The QTL qSSn-2-2 was located between markers RM526 and RM240, and explained 11.4% of the phenotypic variation with the LOD score of 3.8, corresponding to qSS-2 flanked by the region R1826-R3393 reported by Li et al. (2012), which explained 17.3% of the phenotypic variation with the LOD score of 5.6. Meanwhile, qSSn-5, located between markers RM421 and RM26 on chromosome 5, and explained 14.4–15.0% of phenotypic variations with the LOD scores of 5.3–5.5, was similar to qMT-SGC5.1 reported by Jiang et al. (2011) with the LOD scores of 3.2–6.0 explaining 7.9–12.5% of total phenotypic variances. In addition, the interval overlapping the region G200-C214 containing qSS-6, which was found on chromosome 6, and explained 7.6–14.5% of the phenotypic variations with the LOD scores of 4.2–6.7 reported by Li et al. (2012), included qSSn-6, which explained 13.6% of the phenotypic variation with the LOD score of 4.1. Due to the lack of common markers and a long interval, it was difficult to confirm whether qSSn-2-2, qSSn-5, and qSSn-6 were the same loci of qSS-2, qMT-SGC5.1, and qSS-6, respectively. This requires further study. Nevertheless, qSSn-1 in the present study, located in the region between markers RM237 and RM128 on chromosome 1, was a new QTL for seed storability, and was resolved into the two separate QTLs, qSSn-1-1 and qSSn-1-2 in RILs after R-C-11. It is necessary to estimate and confirm whether there are actually two QTLs by developing near-isogenic lines carrying either or both of the two QTLs.
Colocation of QTL related to seed storability and seed dormancy in N22
Seed dormancy, which defined as a temporal failure of a living seed to germinate under favor conditions for germination (Bewley 1997), is also a very important seed trait as well as storability. So far, there are three different viewpoints about the relationship of seed storability and seed dormancy. Comparing QTLs for seed dormancy with those for seed longevity detected in backcross population of Nipponbare/Kasalath, Miura et al. (2002) suggested that seed dormancy and seed longevity were controlled by different genetic factors in Kasalath, supporting no association between the seed dormancy and longevity reported in several earlier studies (Juliano et al. 1990, Roberts 1963). In contrast, based not only on the performance of mutants in the key regulators of seed maturation such as leafy cotyledon1 (lec1), abscisic acid intensitive3 (abi3) mutants (Ooms et al. 1993, Sugliani et al. 2009) and testa-defective mutants such as tt, ats (Debeaujon et al. 2000) but also on the loss-of-function mutant in the DOG1 gene (Bentsink et al. 2006), the current idea is that seed dormancy and seed longevity are positively correlated. All these mutants had a reduced dormancy level consistent with reduced seed longevity. However, Nguyen et al. (2012) firstly reported that the natural variation for seed longevity and seed dormancy were negatively correlated in Arabidopsis.
Previous studies indicated that N22 showed very strong dormancy and two major QTLs for seed dormancy in N22 (qSdn-1 and qSdn-5) were identified (Lu et al. 2011, Wan et al. 2006, Xie et al. 2011). In order to exclude the interference of seed dormancy on evaluation of seed storability, sampled seeds were treated at 50°C for 7 days after harvest to fully break dormancy, so that seeds exhibited normal germination (near 100%) before aging treatments (data not shown). A total of seven QTLs for seed storability were identified after three different aging treatments. Among them, qSSn-1 and qSSn-5 simultaneously resided in the near region of qSdn-1 and qSdn-5, respectively (Supplemental Fig. 5). Additionally, qSSn-2-2 was at the same locus as another minor QTL about dormancy viz. qSd-2 (Xie et al. 2011). A NIL, which substituted with a segment from N22 that included the qSSn-5 region in the genetic background of Nanjing35 (Supplemental Fig. 1), was showed strong seed dormancy (Lu et al. 2011). After natural aging for 20 months or artificial aging for 20 days, the GPs of the NIL were significantly higher than those of Nanjing35 (p < 0.01). These results indicated that the allele of qSSn-5 in N22 can enhance both seed storability and dormancy, suggesting that two traits may be positively correlated, consistent with previous studies in Arabidopsis (Bentsink et al. 2006, Debeaujon et al. 2000, Sugliani et al. 2009) but contrary to the report by Nguyen et al. (2012). However it was difficult to confirm whether qSSn-1/qSdn-1 and qSSn-2-2/qSd-2 were also responsible for both seed storability and dormancy or not, due to a lack of corresponding NIL.
Understanding of gene action and environmental effects for a trait will help plant breeders to determine appropriate strategy to improve the trait in breeding programs. Based on the present study, N22 possesses a few major genes together with many minor genes and could be used as a donor genotype for the improvement of seed storability through marker-assisted selection (MAS) and for further research in relationship of seed storability and seed dormancy in rice.
Supplementary Material
Acknowledgments
This research was supported by the Yangtze River Valley Hybrid Rice Collaboration Innovation Center and Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in Mid-lower Yangtze River, Ministry of Agriculture, P. R. China, and supported by grants from the National Transform Science and Technology Program (2013ZX08001-006), High Technology Program from NDRC ([2012]1961), Jiangsu Cultivar Development Program (projects BE2014394), Jiangsu Independence Innovation Project (CX(12)1003) and Qing Lan Project.
Literature Cited
- Bentsink, L., Jowett, J., Hanhart, C.J. and Koornneef, M. (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G.A. and Doerge, R.W. (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras, S., Bennett, M.A., Metzger, J.D. and Tay, D. (2008) Maternal light environment during seed development affects lettuce seed weight, germinability, and storability. HortScience 43: 845–852. [Google Scholar]
- Debeaujon, I., Léon-Kloosterziel, K.M. and Koornneef, M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122: 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Z., Ku, L., Zhang, Z., Zhang, J., Guo, S., Liu, H., Zhao, R., Ren, Z., Zhang, L., Su, H.et al. (2014) QTLs for seed vigor-related traits identified in maize seeds germinated under artificial aging conditions. PLoS ONE 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Lee, J., Jin, Y.M., Qiao, Y., Piao, R., Jang, S.M., Woo, M.O., Kwon, S.W., Liu, X., Pan, H.Y.et al. (2011) Identification of QTLs for seed germination capability after various storage periods using two RIL populations in rice. Mol. Cells 31: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano, B.O., Perez, C.M. and Chang, T.T. (1990) Varietal differences in longevity of tropical rough rice stored under ambient conditions. Seed Sci. Technol. 18: 361–369. [Google Scholar]
- Li, L.F., Lin, Q.Y., Liu, S.J., Liu, X., Wang, W.Y., Hang, N.T., Liu, F., Zhao, Z.G., Jiang, L. and Wan, J. (2012) Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.). Plant Breed. 131: 739–743. [Google Scholar]
- Li, L., Liu, X., Xie, K., Wang, Y., Liu, F., Lin, Q., Wang, W., Yang, C., Liu, B., Liu, S.et al. (2013) qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor. Appl. Genet. 126: 2313–2322. [DOI] [PubMed] [Google Scholar]
- Lin, Q.Y., Jiang, Y.M., Sun, A.L., Cao, P.H., Li, L.F., Liu, X., Tian, Y.L., He, J., Liu, S.J., Chen, L.M.et al. (2015) Fine mapping of qSS-9, a major and stable quantitative trait locus, for seed storability in rice (Oryza sativa L.). Plant Breed. 134: 293–299. [Google Scholar]
- Lu, B., Xie, K., Yang, C., Wang, S., Liu, X., Zhang, L., Jiang, L. and Wan, J. (2011) Mapping two major effect grain dormancy QTL in rice. Mol. Breed. 28: 453–462. [Google Scholar]
- McCouch, S.R., Cho, Y.G., Yano, M., Paul, E. and Blinstrub, M. (1997) Report on QTL nomenclature. Rice Genet. Newsl. 14: 11–13. [Google Scholar]
- Miura, K., Lin, S.Y., Yano, M. and Nagamine, T. (2002) Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 104: 981–986. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.P., Keizer, P., van Eeuwijk, F., Smeekens, S. and Bentsink, L. (2012) Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis. Plant Physiol. 160: 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms, J.J.J., Leon-Kloosterziel, K.M., Bartels, D., Koornneef, M. and Karssen, C.M. (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (A comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol. 102: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou, L., Lovigny, Y., Groot, S.P.C., Belghazi, M., Job, C. and Job, D. (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol. 148: 620–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, E.H. (1963) An investigation of inter-vatietal differences in dormancy and viability of rice seed. Ann. Bot. 27: 365–369. [Google Scholar]
- Sasaki, K., Fukuta, Y. and Sato, T. (2005) Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage. Plant Breed. 124: 361–366. [Google Scholar]
- Sugliani, M., Rajjou, L., Clerkx, E.J.M., Koornneef, M. and Soppe, W.J.J. (2009) Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive 3–5 (abi3-5) and leafy cotyledon 1-3 (lec1-3). New Phytol. 184: 898–908. [DOI] [PubMed] [Google Scholar]
- Towill, L.E. and Mazur, P. (1975) Studies on the reduction of 2,3,5-triphenyltetrazolium chloride as a viability assay for plant tissue cultures. Can. J. Bot. 53: 1097–1102. [Google Scholar]
- Wan, J.M., Jiang, L., Tang, J.Y., Wang, C.M., Hou, M.Y., Jiang, W. and Zhang, L.X. (2006) Genetic dissection of the seed dormancy trait in cultivated rice (Oryza sativa L.). Plant Sci. 170: 786–792. [Google Scholar]
- Wang, S., Basten, C.J. and Zeng, Z.B. (2007) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. [Google Scholar]
- Xie, K., Jiang, L., Lu, B.Y., Yang, C.Y., Li, L.F., Liu, X., Zhang, L., Zhao, Z.G. and Wan, J.M. (2011) Identification of QTLs for seed dormancy in rice (Oryza sativa L.). Plant Breed. 130: 328–332. [Google Scholar]
- Xue, Y., Zhang, S.Q., Yao, Q.H., Peng, R.H., Xiong, A.S., Li, X., Zhu, W.M., Zhu, Y.Y. and Zha, D.S. (2008) Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.). Euphytica 164: 739–744. [Google Scholar]
- Yamauchi, M. and Winn, T. (1996) Rice seed vigor and seedling establishment in anaerobic soil. Crop Sci. 36: 680–686. [Google Scholar]
- Yano, M., Harushima, Y., Nagamura, Y., Kurata, N., Minobe, Y. and Sasaki, T. (1997) Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor. Appl. Genet. 95: 1025–1032. [Google Scholar]
- Zeng, D.L., Guo, L.B., Xu, Y.B., Yasukumi, K., Zhu, L.H. and Qian, Q. (2006) QTL analysis of seed storability in rice. Plant Breed. 125: 57–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.