Nanoscale particles that undergo reversible and defined changes in morphology in response to stimuli are expected to have broad utility in a range of applications, including targeted drug delivery, detection strategies, soft templates, and self-healing materials. To date, programmable materials with these properties have not been reported, despite the many elegant examples of stimuli-responsive soft nanoparticles and micelles.[1–11] Inspired by the utility of DNA as an informational molecule in nanotechnology,[12–20] we report herein DNA-encoded polymeric materials that are capable of in situ controlled, selective, reversible, and user-defined shifts in morphology. The design is based on polymeric micelles formed from a novel set of amphiphilic DNA-brush copolymers (Figure 1).[16,21] By utilizing the sequence-selective recognition properties of DNA,[22] and its performance as a substrate for selective enzymatic cleavage,[23,24] information stored in the micelle shell may be read and manipulated in several modes, causing dramatic changes in morphology and particle size.
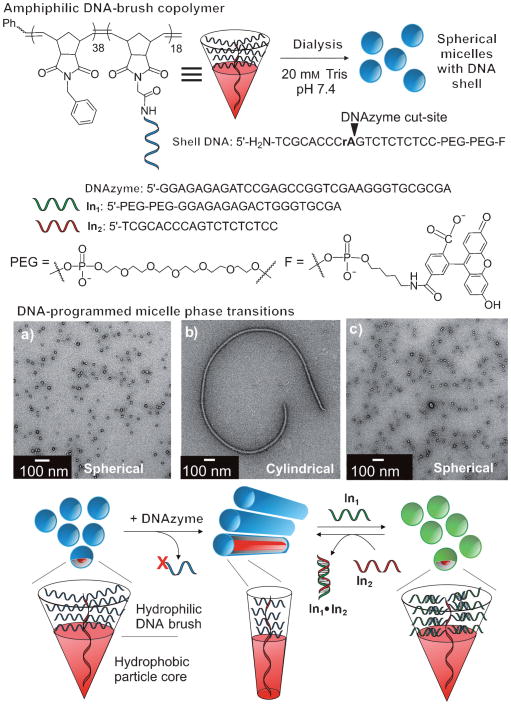
Figure 1.
Assembly of DNA-brush copolymers into micelles with spherical or cylindrical morphologies. Amphiphile structures are represented as cones for each respective morphology, with the hydrophobic domain highlighted in red. TEM images of a) 25 nm spherical micelles assembled from initial DNA-brush copolymers; b) cylindrical morphology formed following DNAzyme addition to spheres; c) spherical micelles (green) formed after addition of In1 to cylinders. See also Figure 3S in the Supporting Information.
The design rationale for DNA-programmed micelle morphology is based on rules that govern the aggregation of amphiphilic block copolymers.[25,26] Briefly, the phase (shape, size, overall morphology) of assembled amphiphiles is controlled by their geometric structure and electrostatics.[27] Therefore, it was hypothesized that DNA-brush copolymer amphiphiles would assemble into micelles with morphologies that are governed by sequence-selective interactions, which allow manipulation of the magnitude of steric and electrostatic repulsions in the micelle shells. Changes in geometric structure of the amphiphile are shown in Figure 1; larger cone angles give higher surface curvature aggregates (i.e., spheres). To demonstrate the concept of DNA-programmed micelle phase transition, three types of sequence-selective interactions were chosen: 1) enzymatic cleavage, 2) isothermal hybridization of complementary single-stranded DNA (ssDNA), and 3) thermal melting and annealing of DNA duplexes.
The DNA-brush copolymer amphiphiles assemble into spherical micelles (Figure 1, blue spheres) approximately 25 nm in diameter, as characterized by transmission electron microscopy (TEM), atomic force microscopy (AFM), and dynamic light scattering (DLS; see Figure 1S in the Supporting Information for DLS and AFM studies). The DNA-brush copolymer amphiphiles contain a RNA base (rA) as an enzymatic cleavage site, two 18-membered ethylene glycol moieties that increase steric bulk of the hydrophilic block, and a fluorescein tag to allow monitoring of reactions that occur at the particle shell. To facilitate a sphere-to-cylinder phase transition, the spherical micelles were mixed with a DNA-based phosphodiesterase (DNAzyme[24]), which was conveniently synthesized to recognize a given DNA sequence and cut at a RNA base. This process resulted in complete, rapid catalytic turnover of the DNA substrate that formed the bulk of the hydrophilic block and generated a truncated ssDNA sequence. A subsequent sphere-to-cylinder phase transition occurs as the “new” surfactants reorganize and pack accordingly (see Figure 2S in the Supporting Information for data showing turnover of shell DNA). To facilitate a cylinder-to-sphere phase transition, a 19-base input DNA sequence (In1) was added. This sequence was designed to form a 9-base duplex with the truncated DNA in the cylinder shell. Subsequent cylinder-to-sphere transition occurs as the bulky, extended duplex is better accommodated in the spherical micelle phase. Therefore, the new structures (Figure 1, green spheres) contain a noncovalent DNA duplex that is amenable to sphere-to-cylinder phase transition utilizing DNA strand invasion.[28,29] This was achieved by addition of a perfectly complementary 19-base ssDNA (In2) designed to invade into the shorter 9-base duplex in the micelle shell. The more thermodynamically stable 19-base long duplex (In1·In2) departs, leaving the truncated ssDNA amphiphile to reassemble into the cylindrical phase.
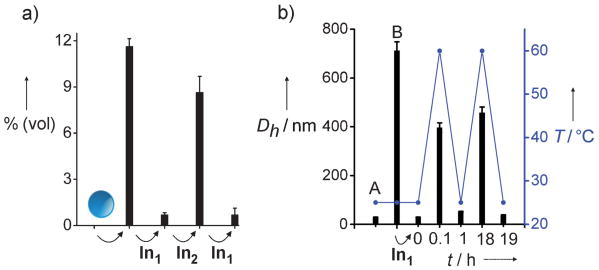
The reversibility of the phase transitions was examined in solution by DLS (Figure 2), with a confirmation of morphologies by TEM (Figure 3S in the Supporting Information). DLS data for isothermal DNA-directed phase transitions are shown as the percentage (volume distribution) of particles at the 396 nm size class, which result from various input additions after two-hour incubation periods (Figure 2). Initially, solutions of the spherical particles showed no observable scattering intensity for aggregates above 30 nm. Indeed, the particle diameter (reported as the hydrodynamic diameter Dh) was constant in the absence of DNAzyme over a time scale of many weeks. However, mixing of the spherical particles with the DNAzyme for two hours caused the expected increase in the Dh value (Figure 2a). Next, this solution was mixed with In1 showing the expected contraction in Dh. Subsequent addition of In2 results in duplex In1·In2, regenerating the truncated shell structure and causing another expansion of particle size. To complete the isothermal cycle, the particles were again treated with In1 resulting in regeneration of small aggregates. In addition, this isothermal phase-cycling process was monitored by the uptake and release of a small-molecule dye (Figure 4S in the Supporting Information).
Figure 2.
Reversible phase cycling experiments. a) Isothermal hybridization and invasion. The y axis shows the percentage (volume distribution) of species in Dh = 396 nm size class versus input; measurements were obtained by DLS 2 h after each input addition beginning with 25 nm spheres. The Dh value of 396 nm was chosen as it is the largest aggregate size class observed by DLS at 2 h (see Figure 3). b) Variable-temperature DLS. Plot shows Dh of largest aggregates in solution at given time points. Initial spheres (A) were treated with DNAzyme for 18 hrs (B) prior to addition of In1. DLS measurement at t = 0 min taken 1 h after addition of In1. Ramp rate: 25→60°C in 5 min. Instrument cooling time was 60 min; sample was cooled by placing in ice bath for 5 min, then rested at RT for 55 min. Second heating cycle conducted 18 h later with same ramp rate (25→60°C in 5 min) and cooling time (60 min). Conditions: particles (0.14 g L−1), DNAzyme (5 nm), In1 (50 nm), In2 (50 nm), 2-amino-2-hydroxymethyl-propane-1,3-diol (Tris; 20 mm, pH 7.4), MgCl2 (50 mm).
The hybridization state of complementary oligonucleotides is disrupted at elevated temperatures. The isothermal cycling experiments (Figure 2a) show a dependence of the morphology on the hybridization state of the DNA. Similarly, cycling of the solution temperature resulted in aggregate size changes, which were dependent on whether or not the DNA in the amphiphile was in duplex or single-stranded form (Figure 2b). These data show a correlation between the temperature of the solution and the aggregate size in solution, that is, the size increases above the melting temperature of the DNA duplex and decreases below the melting temperature. These data complement the isothermal cycling experiment with the same correlation with respect to the hybridization state of the amphiphile.
To further elucidate the mechanism and assess the selectivity of the DNAzyme-directed phase transition, the process was examined by TEM and DLS (Figure 3), and studied by fluorescence microscopy (Figure 4). TEM and DLS data show a solution populated with a decreasing concentration of intact 25 nm spheres (Figure 3, red curve) and growing cylindrical aggregates with time (Figure 3, blue curve; see also Figure 5S in the Supporting Information). The TEM data correlates with the increase in Dh observed by DLS (Figure 3, green curve). After one day, cylinders could be observed in high density, and TEM data showed the presence of well-defined cylinders (Figure 3d and inset). After two days in the presence of the DNAzyme, TEM data confirmed low concentrations of intact spherical structures (Figure 3e), and the presence of cylinders greater than 1 μm in length. This phase-transition process constitutes an approximate 100-fold increase in size and a dramatic change in morphology, which results in clear solutions and no precipitation. Insight into the mechanism of this process is provided by the observation that complete turnover of the shell DNA of the initial spheres is rapidly achieved (Figure 2S in the Supporting Information) prior to complete phase transition. Therefore, after DNAzyme addition, competing equilibria for monomeric amphiphile assembly are rapidly established and result in cylinder growth (amphiphile↔cylinder versus amphiphile↔sphere).
Figure 3.
DNA-directed size and phase change over time. Conditions: particles (0.14 g L−1), Tris (20 mm, pH 7.4), MgCl2 (50 mm). DNAzyme (5 nm) was mixed with particles at t = 0 min. a) t = 0 min, b) 2 min, c) 2 h, d) 1 day, e) 2 days. f) Particle size as a function of time following DNAzyme addition. Blue curve: average cylinder length (CL) measured by TEM. Green curve: Dh value of largest aggregates measured by DLS. Red curve: Sphere diameter (SD) measured by TEM. DLS data was taken at the time points shown following DNAzyme addition. Data points for CL and SD are averages of multiple measurements made from TEM images, with error bars indicating standard deviations (Figure 4S in the Supporting Information).
Figure 4.
Sequence-selective phase shifting observed by fluorescence microscopy. a) TEM image showing bundled cylinder structures analogous to optical images. Optical microscopy images show bright field, green and red fluorescence images taken after treatment with DNAzyme. b) D-1 recognizes only fluorescein labeled particles. c) D-2 recognizes only rhodamine labeled particles. d) D-1 and D-2 together cause fiber formation containing both labels. Optical image scale bars = 10 μm. Conditions: Micelles (0.14 g L−1), DNAzyme (5 nm), Tris (20 mm, pH 7.4), MgCl2 (50 mm). Shell DNA sequences are analogous to that shown in Figure 1, with an extra three bases at the 5′-terminus. The third base from the 5′-amide linkage to the polymer backbone is a dye-labeled thymidine residue (see the Supporting Information for DNA sequences used in these experiments).
A series of fluorescence microscopy experiments were conducted to test if a mixed population of particles encoded with different DNA sequences could respond independently and selectively in the presence of competing DNAzymes. Two fluorescent particles that contain two different DNA sequences were synthesized; one labeled with a fluorescein dye (through a fluorescein–thymidine (FlrT) phosphoramidite) and the other labeled with a rhodamine dye (through a rhodamine–thymidine (RhT) phosphoramidite; Figure 4 and Figure 6S in the Supporting Information). Conveniently, micrometer-sized fibers (bundles of cylinders) could be imaged by light microscopy; 25 nm spheres, which were below the resolution limit, contributed to diffuse background fluorescence that had much lower intensity than the fibers. The red and green fluorescent particles were mixed together and treated with two different DNAzymes (D-1 and D-2), each of which was complementary to one of the dye-labeled particles. Introduction of the DNAzyme selective for the sequence within the green fluorescent particle shell (D-1) resulted in the formation of green fluorescent cylinders, with no observable red fluorescence (Figure 4b). Conversely, only red fluorescent particles reacted with the DNAzyme selective for the sequence within their shell (D-2). This selectivity is confirmed by the absence of green fluorescence in the images of these cylinder structures and the appearance of red fluorescent cylinders (Figure 4c). Finally, when mixed with D-1 and D-2, both spheres in the mixture reacted, which caused the formation of structures containing both red and green fluorophores (Figure 4d). These studies provide evidence supporting the conclusion that truncation of the DNA is a necessary requirement for phase transition and is indeed sequence-selective. This experiment is possible because the two amphiphiles are chemically similar except for the information encoded in their respective DNA sequences. This characteristic makes each particle type in the mixed population independently addressable, a feature that is not easily accessible for systems designed to respond to non-informational stimuli such as temperature, light, or pH.
In summary, we have demonstrated the utility of DNA as an informational tool for morphology control in discrete, stimuli-responsive, nanoscale polymeric materials.[13] It is expected that this approach will be amenable for extension to a variety of stimuli given the versatility of nucleic acids as molecules that are capable of multiple modes of selective recognition. In a broader context, the development of generally applicable methods for predictably controlling size, shape, and functionality of soft materials at the nano-meter length scale will be critical for realizing their tremendous potential.[30–39]
Supplementary Material
Footnotes
We gratefully acknowledge the support of this work by the University of California and from a Henry and Camille Dreyfus Foundation New Faculty Award to N.C.G. We wish to thank Mahealani R. Bautista and Lila K. Habib for their technical assistance, and Norm Olson for his generous assistance. We acknowledge use of the UCSD Cryo-Electron Microscopy Facility, which is supported in part by NIH grant 1S10 RR020016, a gift from the Agouron Institute, UCSD funds provided to Dr. Timothy S. Baker, the Nano-3 facilities at UCSD for use of their AFM instrument, the National Center for Microscopy and Imaging Research at San Diego supported by NIH, and the support of the National Science Foundation under CHE-0741968.
Supporting information for this article is available on the WWW.under http://dx.doi.org/10.1002/anie.201000265.
References
- 1.Wang Y, Xu H, Zhang X. Adv Mater. 2009;21:2849. doi: 10.1002/adma.200901237. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Yu K, Eisenberg A. Science. 1996;272:1777. doi: 10.1126/science.272.5269.1777. [DOI] [PubMed] [Google Scholar]
- 3.Bütün V, Billingham NC, Armes SP. J Am Chem Soc. 1998;120:11818. [Google Scholar]
- 4.LaRue I, Adam M, Pitsikalis M, Hadjichristidis N, Rubinstein M, Sheiko SS. Macromolecules. 2006;39:309. [Google Scholar]
- 5.Liu X, Jiang M. Angew Chem. 2006;118:3930. [Google Scholar]; Chem Int Ed. 2006;45:3846. [Google Scholar]
- 6.Ishihara Y, Bazzi HS, Toader V, Godin F, Sleiman HF. Chem Eur J. 2007;13:4560. doi: 10.1002/chem.200601423. [DOI] [PubMed] [Google Scholar]
- 7.Kim JH, Lee S, Park K, Nam HY, Jang SY, Youn I, Kim K, Jeon H, Park RW, Kim IS, Choi K, Kwon IC. Angew Chem. 2007;119:5881–5884. doi: 10.1002/anie.200700767. [DOI] [PubMed] [Google Scholar]; Chem Int Ed. 2007;46:5779–5782. doi: 10.1002/anie.200700767. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Du W, Sun G, Wooley KL. Macromolecules. 2008;41:6605. doi: 10.1021/ma802057u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy D, Cambre JN, Sumerlin BS. Chem Commun. 2009:2106. doi: 10.1039/b900374f. [DOI] [PubMed] [Google Scholar]
- 10.Fernyhough C, Ryan AJ, Battaglia G. Soft Matter. 2009;5:1674. [Google Scholar]
- 11.Amir RJ, Zhong S, Pochan DJ, Hawker CJ. J Am Chem Soc. 2009;131:13949. doi: 10.1021/ja9060917. [DOI] [PubMed] [Google Scholar]
- 12.Seeman NC. Mol Biotechnol. 2007;37:246. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aldaye FA, Palmer AL, Sleiman HF. Science. 2008;321:1795. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 14.Gothelf KV, LaBean TH. Org Biomol Chem. 2005;3:4023. doi: 10.1039/b510551j. [DOI] [PubMed] [Google Scholar]
- 15.Storhoff JJ, Mirkin CA. Chem Rev. 1999;99:1849. doi: 10.1021/cr970071p. [DOI] [PubMed] [Google Scholar]
- 16.Alemdaroglu FE, Herrmann A. Org Biomol Chem. 2007;5:1311. doi: 10.1039/b617941j. [DOI] [PubMed] [Google Scholar]
- 17.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 18.Ofir Y, Samanta B, Rotello VM. Chem Soc Rev. 2008;37:1814. doi: 10.1039/b712689c. [DOI] [PubMed] [Google Scholar]
- 19.Venkataraman S, Dirks Robert M, Rothemund Paul WK, Winfree E, Pierce Niles A. Nat Nanotechnol. 2007;2:490. doi: 10.1038/nnano.2007.225. [DOI] [PubMed] [Google Scholar]
- 20.Adleman LM. Science. 1994;266:1021. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Zhang Y, Fullhart P, Mirkin CA. Nano Lett. 2004;4:1055. [Google Scholar]
- 22.Watson JD, Crick FH. Nature. 1953;171:737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 23.Kelly TJ, Jr, Smith HO. J Mol Biol. 1970;51:393. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- 24.Santoro SW, Joyce GF. Proc Natl Acad Sci USA. 1997;94:4262. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Israelachvili JN, Mitchell DJ, Ninham BW. J Chem Soc Faraday Trans 2. 1976;72:1525. [Google Scholar]
- 26.Jain S, Bates FS. Science. 2003;300:460. doi: 10.1126/science.1082193. [DOI] [PubMed] [Google Scholar]
- 27.Nagarajan R. Langmuir. 2002;18:31. [Google Scholar]
- 28.Hazarika P, Ceyhan B, Niemeyer CM. Angew Chem. 2004;116:6631. doi: 10.1002/anie.200461887. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2004;43:6469. [Google Scholar]
- 29.Zhang DY, Winfree E. J Am Chem Soc. 2009;131:17303. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 30.Smith D, Pentzer EB, Nguyen ST. Polym Rev. 2007;47:419. [Google Scholar]
- 31.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat Nanotechnol. 2007;2:249. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kishimura A, Koide A, Osada K, Yamasaki Y, Kataoka K. Angew Chem. 2007;119:6197. doi: 10.1002/anie.200701776. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2007;46:6085. [Google Scholar]
- 33.Kim KT, Cornelissen JJLM, Nolte RJM, van Hest JCM. Adv Mater. 2009;21:2787. [Google Scholar]
- 34.Xia Y, Olsen BD, Kornfield JA, Grubbs RH. J Am Chem Soc. 2009;131:18525. doi: 10.1021/ja908379q. [DOI] [PubMed] [Google Scholar]
- 35.MacKay JA, Chen M, McDaniel JR, Liu E, Simnick AJ, Chilkoti A. Nat Mater. 2009;8:993. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI. Proc Natl Acad Sci USA. 2010;107:3293. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 38.Haag R. Angew Chem. 2004;116:280. [Google Scholar]; Angew Chem Int Ed. 2004;43:278. [Google Scholar]
- 39.Nayak S, Lyon LA. Angew Chem. 2005;117:7862. doi: 10.1002/anie.200501321. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2005;44:7686. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.