Abstract
Limited guidance is available on practical approaches for maintaining genetic diversity in large NHP colonies that support biomedical research, despite the fact that reduced diversity in these colonies is likely to compromise the application of findings in NHP to human disease. In particular, constraints related to simultaneously housing, breeding, and providing ongoing veterinary care for thousands of animals with a highly complex social structure creates unique challenges for genetic management in these colonies. Because the composition of new breeding groups is a critical component of genetic management, here we outline a 3-stage protocol for forming new breeding groups of NHP that is aimed at maximizing genetic diversity in the face of frequent restrictions on age, sex, and numbers of animals per breeding group. As an example application of this protocol, we describe optimal combinations of rhesus macaques from an analysis of candidate animals available for breeding in July 2013, selected from among the approximately 4000 macaques maintained at the Oregon National Primate Research Center. In addition, a simulation study to explore the genetic diversity in breeding groups formed by using this protocol, indicated an approximate 10-fold higher genome uniqueness, 50% lower mean kinship, and an 84-fold lower mean inbreeding coefficient among potential offspring within groups, when compared with a suboptimal group design. We conclude that this protocol provides a practical and effective approach to breeding group design for colony managers who want to prevent the loss of genetic diversity in large, semiisolated NHP colonies.
Abbreviation: ONPRC, Oregon National Primate Research Center
The optimal pairing of animals to maximize genetic diversity is a major goal in animal colonies established for species conservation or public education, and resources and protocols that support the selection of animals for breeding in these and other small populations are readily available.4,16,25 However, similar protocols for much larger colonies of NHP are unavailable, despite the fact that these colonies support a wide range of biomedical and preclinical research that is expected to accurately reflect genetic effects on disease and drug response in humans.6,29 The need for practical genetic management protocols aimed at large NHP colonies is particularly relevant to the situation for one widely used NHP species, the Indian-origin rhesus macaque (Macaca mulatta), which can no longer be imported into the United States. This ban on importation effectively reduces US colonies to semiisolated populations in which genetic diversity will decline substantially over time without the careful selection of animals best suited to retain this diversity. In fact, population genetic theory predicts that the inappropriate assignment of animals for breeding can dramatically reduce standing genetic variation in just a few generations.1 The consequences of substantially reduced genetic diversity in macaque colonies are expected to be severe. In the short term, reduced genetic diversity can change the means and variability of important biomedical traits7 and might complicate or invalidate the interpretation of experimental findings in NHP to human disease. Over the long term, the loss of genetic diversity is expected to result in fewer viable offspring, increased morbidity and mortality in the colony, and spiraling costs for related clinical veterinary care.
As in the conservation setting, the primary goal of genetic management in large NHP colonies is the retention of genetic diversity present in founding colony members and in any more recent immigrants.3,9,30,32 However, due to large differences in colony size and the practical constraints of housing and caring for many thousands of animals, protocols developed for minimizing the loss of diversity in small colonies are not readily adapted to the needs of much larger NHP populations. The lack of practical genetic protocols for large NHP colonies is an important problem because, despite their apparent size, these colonies risk losing substantial genetic diversity if only a few animals are permitted to breed repeatedly over long periods of time.31 The highly complex social structure of many NHP species further compounds this problem, given that protocols need to take this characteristic into account to minimize animal stress and maladaptive behaviors that decrease the production of viable offspring.11 Here we present a 3-stage protocol for forming new multimale, multifemale breeding groups of Indian-origin rhesus macaques that we have developed over several years at the Oregon National Primate Research Center (ONPRC). As an example, we describe an analysis of the optimal combination of male and female adult macaques that were available at the ONPRC in July 2013 and that fulfilled specific criteria related to age, sex, and numbers of animals while retaining genetic diversity in the breeding colony as a whole.
Materials and Methods
Description of breeding groups.
The ONPRC is home to approximately 4000 Indian-origin rhesus macaques, most of which are housed in outdoor group enclosures containing from 10 to 260 animals. We aim for breeding groups to include: 1) 2 to 15 sexually mature male macaques that are unrelated both to each other and to the sexually mature females in the group; 2) 20 to 130 sexually mature female macaques of varied relatedness; and 3) any offspring of adult females that are below the age of sexual maturity. This social group structure mimics that found in wild rhesus macaques, in which adult females and immature female offspring are permanent members of lineages (or ‘matrilines’) within the group, unrelated adult male macaques immigrate into the group to breed, and males born in the group migrate out of the group at sexual maturity.21,22 At the ONPRC, the genetic composition of these multimale, multifemale groups is critically important, because the vast majority of breeding takes place opportunistically in these groups, with no further intervention after the assignment of animals to the group.
Overview of criteria and approach for example case.
In July 2013, the Division of Comparative Medicine at the ONPRC requested recommendations for 2 new breeding groups of rhesus macaques and provided a list of 204 candidate animals that were available for assignment to the new groups. In this case, candidate animals were those temporarily housed in indoor cages that were not assigned to research and animals belonging to social groups being considered for disbanding. This request for analysis further specified that each group should contain 18 to 20 female macaques 4 y of age or older, 3 or 4 males 6 y or older, and 1 to 30 juveniles between 1 and 4 y of age.
Breeding group design is based on 3 stages of analysis, outlined in detail in the following sections below. In the 1st stage, parentage analysis is conducted to place animals within the larger, comprehensive ONPRC pedigree (see the section ‘Parentage analysis and pedigree curation’). In the 2nd stage, an analysis of comprehensive pedigree information ranks the candidate list of animals individually by their genetic importance to the living colony according to several criteria (see ‘Genetic value analysis’). Within these rankings, a cutoff (or threshold) is drawn; animals below this cutoff are not recommended for further breeding. In the 3rd stage (see ‘Relatedness analysis and automated group formation’), by using all animals that rank above the threshold, several potential animal groups are characterized that fulfill the requested criteria.
Stage 1: Parentage analysis and pedigree curation.
The goal of parentage analysis is to correctly place all new offspring into the larger, comprehensive ONPRC pedigree. An accurate pedigree remains the most crucial resource for the appropriate selection of animals for breeding. Accurate pedigree data serve a function that is vital to both colony management and scientific research,3,30,32 by providing the baseline information required to maximize genetic diversity in the colony as a whole and by ensuring the independence of data collected on animals that are known to be unrelated when assigned to research. At the ONPRC, parentage analysis is conducted on an ongoing basis, as new offspring are recorded during the processing of enclosures for veterinary care and maintenance. Initial observations of parentage by animal care staff are recorded when possible, and parentage is verified later by using genetic typing. Parentage analysis at the ONPRC is conducted by testing Mendelian inheritance in offspring and all potential parents at 29 microsatellite or short tandem-repeat markers (Table 1). All potential parents are identified by using an automated query of the ONPRC animal records database (script available by request). Potential dams are identified as all breeding-age female animals (conservatively considered to be 2.5 y or older) that were present in the enclosure at offspring birth, regardless of their current location. Potential sires are identified as all breeding-age male macaques (2.5 y or older) that were cohoused with at least one of the potential dams during the conception window, regardless of their current location. We define the conception window as 155 to 175 d before the recorded offspring birthdate, according to the current estimated age, and a 164-d gestation time.8
Table 1.
Summary information on the 29 microsatellite markers used for parentage analysis at the Oregon National Primate Research Center, with markers ranked according to observed heterozygosity
| Marker | Chromosome no. (Rhesus) | Chromosome no. (Human) | No. of macaques typed | No. of alleles | Heterozygosity | Reference | |
| Observed | Expected | ||||||
| D12S67 | 11 | 12 | 7010 | 48 | 0.914 | 0.915 | 26 |
| D7S513 | 3 | 7 | 7949 | 51 | 0.902 | 0.908 | 2 |
| D15S823 | 7 | 15 | 6563 | 26 | 0.897 | 0.892 | 26 |
| D17S1300 | 16 | 17 | 7240 | 49 | 0.880 | 0.892 | 26 |
| MFGT21a | 8 | not applicable | 7783 | 22 | 0.870 | 0.875 | 2,15 |
| D3S1768 | 2 | 3 | 7950 | 25 | 0.864 | 0.864 | 2 |
| D2S1333 | 12 | 2 | 6476 | 15 | 0.856 | 0.860 | 26 |
| D8S1106 | unknown | 8 | 7896 | 17 | 0.844 | 0.851 | 2 |
| D22S685 | 13 | 22 | 743 | 18 | 0.843 | 0.863 | 23 |
| D13S765 | 16 | 13 | 7946 | 31 | 0.836 | 0.831 | 2,23 |
| D16S403 | 20 | 16 | 7900 | 21 | 0.817 | 0.826 | 2 |
| D4S413 | 4 | 4 | 6859 | 19 | 0.798 | 0.799 | 23 |
| D12S364 | 11 | 12 | 7507 | 18 | 0.783 | 0.792 | 26 |
| D11S925 | 14 | 11 | 7956 | 38 | 0.775 | 0.780 | 2 |
| D6S276 | 4 | 6 | 7894 | 17 | 0.773 | 0.775 | 2 |
| D9S921 | unknown | 9 | 6584 | 10 | 0.754 | 0.751 | unavailable |
| D6S1691 | 4 | 6 | 7897 | 22 | 0.745 | 0.742 | 2 |
| D6S501 | 4 | 6 | 6485 | 11 | 0.732 | 0.725 | 26 |
| D11S2002 | 11 | 11 | 7267 | 18 | 0.717 | 0.758 | 23 |
| D7S794 | 3 | 7 | 7995 | 14 | 0.704 | 0.703 | 2 |
| MFGT22a | unknown | not applicable | 7786 | 20 | 0.691 | 0.692 | 2 |
| D1S548 | 1 | 1 | 6580 | 11 | 0.631 | 0.627 | 26 |
| D5S1457 | 5 | 5 | 6579 | 11 | 0.631 | 0.624 | 23 |
| D18S72 | 18 | 18 | 7932 | 33 | 0.630 | 0.640 | 2 |
| D4S2365 | 5 | 4 | 7561 | 16 | 0.618 | 0.624 | 26 |
| D6S291 | 4 | 6 | 7895 | 12 | 0.617 | 0.616 | 2 |
| D18S537 | 18 | 18 | 6573 | 6 | 0.615 | 0.630 | 26 |
| D10S1412 | 9 | 10 | 7894 | 9 | 0.559 | 0.555 | 2 |
| DXS2506 | unknown | X | 6441 | 11 | 0.213 | 0.385 | unavailable |
These markers form the standard parentage marker panel used by the Veterinary Genetics Lab (University of California Davis, Davis, CA), which performed all genotyping services. When possible, a literature reference for the marker is indicated. Summary statistics were calculated by using CERVUS14 software (version 3.0.7).
MFGT markers developed from Macaca fuscata.
We use the method of exclusion to assign parentage to offspring.10,12,13 This approach seeks to exclude all incorrect parents, with the subject not excluded inferred as the correct parent. We exclude potential parents by the presence of apparent nonMendelian inheritance of alleles at 2 or more microsatellite loci that cannot be explained as genotyping error. One type of genotyping error that occurs frequently with microsatellite markers is single-nucleotide differences between offspring and potential parent, primarily due to technical problems in the assignment of alleles from raw data. However, differences of 2 basepairs or greater in allele size between offspring and potential parent are conservatively treated as cases of nonMendelian inheritance, despite the fact that an unknown number of these cases will represent true mutations between parent and offspring because of the high mutation rate of microsatellite markers. We apply this conservative rule because, in the absence of additional sequence information, we cannot distinguish between a mutation that is consistent with a true parent–offspring relationship and one that is not. In short, we prefer the risk of excluding all potential parents to the risk of miscalling a parent, because this single misattribution will create many more errors throughout the pedigree.
A second type of genotyping error that can occur with microsatellite markers is ‘null alleles,’ or sequences that fail to amplify for technical reasons, resulting in an apparent but incorrect homozygous genotype.5 A null allele is inferred when both the offspring and potential parent appear homozygous for different alleles at the same locus; these genotypes are removed from the offspring's genotype profile and are disregarded in the assignment of parentage. Note that X-linked markers are not useful for analyzing concordance of male offspring with potential sires because of the maternal origin of the X chromosome in male offspring. At X-linked loci, this phenomenon results in an apparent null allele between a potential sire and male offspring, because both the sire and male offspring will appear to be homozygous, usually for different alleles.
Offspring and any potential parent must show Mendelian inheritance of alleles at a minimum of 12 microsatellite markers before parentage is assigned; this low threshold allows us to accommodate the historical use of many fewer microsatellite loci for parentage analysis in previous generations of the colony. In addition, a maximum of one instance of non-Mendelian inheritance is allowed between offspring and parent, with this parentage assignment recorded as ‘provisional’ to indicate an acceptable level of uncertainty in the assignment. In the vast majority of cases, these decision rules result in a single dam and a single sire that are fully concordant with an offspring at all loci genotyped, with all other potential parents excluded. In rare cases in which multiple sets of potential parents satisfy all of the described criteria, the potential parents that share the greatest number of markers that are fully concordant with the offspring are assigned as the true parents. These decision rules have been honed over many years of parentage analysis based on microsatellite markers, with the goal of balancing the need for accuracy in parentage assignment against the twin limitations of budget and availability of historical samples. According to these parentage assignments, animals are added to the comprehensive ONPRC pedigree. The placement of animals within this larger pedigree allows the inference of a wide range of more distant and complex relationships, in light of ancestry in common with other macaques in the pedigree.
Stage 2: Genetic value analysis.
Using the comprehensive ONPRC pedigree, we first measure the average of all pairwise kinship coefficients for each living animal in the breeding colony with every other living animal in the colony4 according to the algorithm of Lange18 as implemented in the R package ‘kinship2’ (http://cran.r-project.org/web/packages/kinship2/index.html). The kinship coefficient measures the probability that alleles drawn at random from the same locus in each of 2 subjects will be identical by descent.7 The mean kinship of a subject to the rest of the colony can then be calculated as the average of all kinship coefficients between itself and every other living animal in the colony, including itself.4 Furthermore, a grand mean kinship can then be calculated from the distribution of all individual mean kinships. Because this distribution is expected to be approximately normal, measures of individual mean kinship for each candidate animal can be expressed in terms of standard deviates from this grand mean kinship, as z = (x−µ)/σ, that is, ‘z scores,’ in which x is the estimate of individual mean kinship, µ is the grand mean kinship, and σ is the standard deviation of the distribution of all individual mean kinship values. Therefore, z scores indicate the extent and direction to which each animal's individual mean kinship deviates from the grand mean kinship in the colony. An animal with a positive z score has an individual mean kinship that lies above the colony grand mean kinship, suggesting that the focus animal's genome is overrepresented in the colony, thus decreasing the focus animal's genetic value relative to that of animals with negative z scores. Conversely, an animal with a negative z score has an individual mean kinship that lies below the grand mean kinship, suggesting that this animal's genome is underrepresented in the colony and thereby increasing its genetic value.
After z scores based on mean kinship have been determined for each candidate macaque, measures of genome uniqueness are calculated using a ‘gene-drop’ simulation.19 A gene drop simulation assigns unique alleles to all founders in the current pedigree configuration and simulates the inheritance of these alleles throughout the pedigree according to Mendelian principles. Genome uniqueness is then calculated as the proportion of an arbitrary number of simulations in which a subject receives the only copy of a founder allele.4 Thus, genome uniqueness is an estimate of the probability that an animal possesses founder alleles that are rare and at risk of being lost from the population, due to their occurrence in only one or a few other animals. At the ONPRC, we use a threshold of 3 animals to calculate genome uniqueness to identify rare (and not just unique) genetic variation that may be in danger of being lost from the colony. Increasing the retention of rare alleles may be particularly relevant to the ONPRC colony, in which only approximately 30% of female macaques and 73% of adult males in a breeding group are estimated to produce offspring during any given year. The calculation of genome uniqueness and z scores based on individual mean kinship thus provides summary information on the expected genetic diversity represented by each candidate animal, in terms of both shared common ancestry and allelic diversity relative to the colony as a whole.
After z scores and genome uniqueness have been calculated, the macaques are ranked according to the following rules. First, imported animals without offspring are automatically ranked highest in genetic value. Then all animals with greater than 10% genome uniqueness are ordered first by genome uniqueness from largest to smallest value and then—within the same genome uniqueness— by z score from smallest to largest value. Next all remaining animals (those with 10% genome uniqueness or less) are ranked by z score from smallest to largest values. At this point, a threshold is drawn at animals with genome uniqueness 10% or less and whose z scores are 0.25 or greater. Animals ranking above this threshold are considered to be of high genetic value, whereas those below this threshold are not recommended for breeding.
Stage 3: Relatedness analysis and automated group formation.
After the macaques are ranked according to their genetic value, only animals lying above the threshold are considered for further analysis. During this stage, potential groups are formed on the basis of 1) the estimation of pairwise kinship between candidate group members and 2) any specified breeding-group criteria, including animal numbers, sex ratio, or age distribution. We have developed an algorithm that automatically parses the list of high-value animals into the desired number of groups, on the basis of relationships that must be excluded, the threshold value of within-group relatedness that may be tolerated, any relationships that can be ignored, and additional demographic criteria. We choose to exclude relatedness greater than second cousin between breeding males and between breeding males and females; however, we disregard any level of relatedness between breeding adults and animals ≤ 1 y of age and between females of any age. Given this information, the algorithm assigns randomly selected animals to hypothetical groups, with the constraint that they cannot be related at the specified threshold level or greater to any macaque already assigned to the group. This process is repeated 10,000 times to thoroughly sample the collection of possible groups that can be formed from the high-value set of candidate animals being considered. On completion, this algorithm returns the best set of groups, defined as the set with the largest minimum group size. The identification of multiple potential groups is an important result of this analysis, because it allows flexibility in the hands-on development of the final group, depending on behavioral conflicts or other constraints that may be difficult to predict. We have implemented all analyses described in Stages 2 and 3 in a user-friendly R-based software package, available upon request to the authors.
Simulation study.
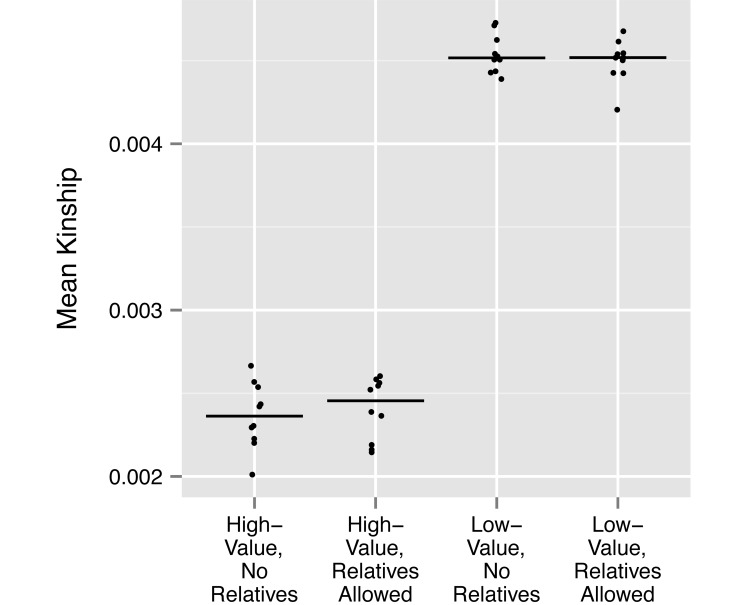
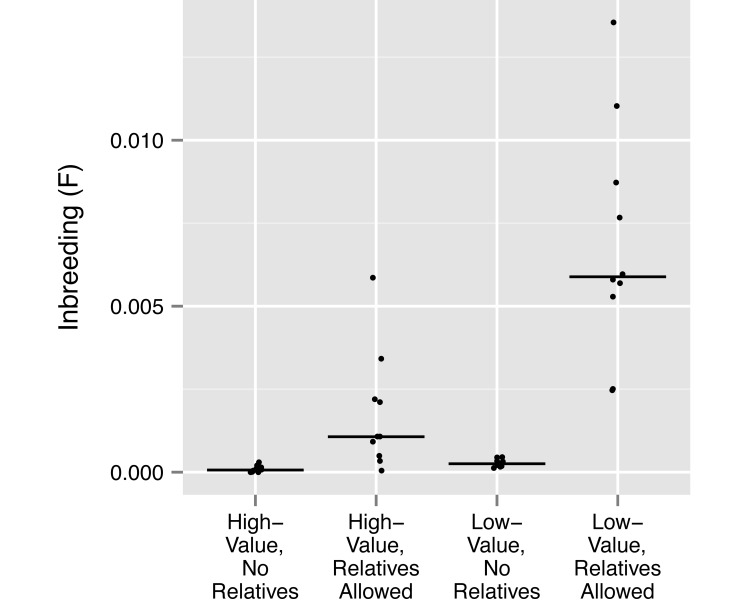
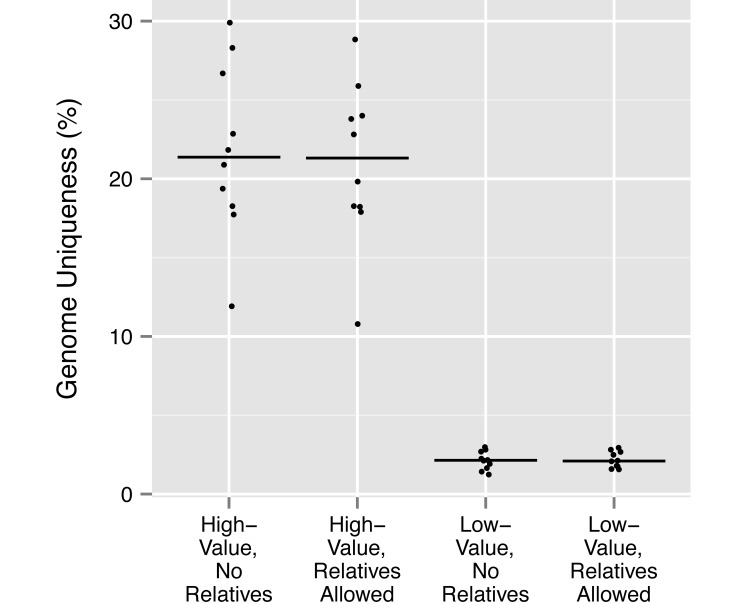
To explore the utility of our breeding group protocol beyond this single example, we also investigated the magnitude of differences in 1) mean kinship of the group with the rest of the colony, 2) mean group genome uniqueness, and 3) the mean inbreeding coefficient of group offspring (that is, the probability that any 2 alleles at a locus in the same animal are identical by descent7) between high- and low-value groups that either included or excluded relatives, according to a genetic-value analysis of the entire living ONPRC colony. To this end, we simulated sets of 10 replicate breeding groups for each combination of these 2 factors (that is, either high or low genetic value, and either including or excluding relatives). Each replicate group comprised 4 adult males and 20 adult females randomly selected from macaques either above or below our threshold in the genetic value rankings of the entire colony. Relationships above the level of 2nd cousin between adult males, between adult males and adult females or between all adults and juveniles (except between females) were either removed or retained as indicated; all other relatedness between group members was disregarded. For the calculation of mean group inbreeding, a single offspring was simulated from each unique combination of adult males and females within each replicate group. The mean values of each parameter were calculated for each replicate group and plotted, with the median of these values indicated (Figures 1 through 3).
Figure 1.
Mean group kinship with the rest of the colony for 10 replicate groups in each of 4 sets of conditions: high-value or low-value groups that either included or excluded relatedness as described in Materials and Methods. The median value within each set of 10 replicate groups is indicated with a horizontal line.
Figure 3.
Mean group inbreeding coefficient among offspring in 10 replicate groups in each of 4 sets of conditions: high-value or low-value groups that either included or excluded relatedness as described in Materials and Methods. Median value among the 10 replicate groups is indicated with a horizontal line.
In addition, we explored the effects of our protocol on mean group heterozygosity, using all available microsatellite marker information. To this end, we conducted simulations as described earlier and calculated the mean group heterozygosity for each simulated group, with the contribution of each marker weighted by the frequency of subjects in the group with genotype information available for that marker.
Results
The comprehensive ONPRC pedigree extends for 10 generations, includes approximately 22,000 animals both past and present, and provides the basis for all genetic analysis of macaques currently living at the center. In our example case involving 246 macaques, the vast majority (224 macaques, approximately 91%) of all candidate animals proposed had both parents assigned as a result of previous parentage analysis, with a few (16 macaques, approximately 6.5%) having parentage assigned for only 1 parent (the remaining 6 animals were imported from elsewhere and do not have assigned parents at the ONPRC). Of the 6 imported animals, 3 had not yet produced offspring; these 3 were automatically ranked highest in genetic value according to our ranking scheme. In addition, 42 animals were infants (that is, younger than 1 y) and thus were automatically assigned to the same group as their dam.
Ultimately, the genetic-value analysis of the remaining 204 candidate macaques identified 65 low-value animals that were removed from further consideration as potential breeders, leaving 139 animals ranked as having high genetic value. Relatedness among these 139 high-value macaques proved to be considerable, based on our threshold of 2nd-cousin relatedness or greater; however, ignoring relatedness between breeding-age females greatly reduced the numbers of pairwise relationships that exceeded this threshold. During automated group formation, despite our relatively simplistic algorithm, we found that the 2 best groups generated came very close to the specified parameters (Table 2). For example, these 2 groups included at least 4 to 7 males aged 6 y or older (3 to 4 breeding age males originally requested), 27 to 32 females aged 4 y or older (18 to 20 breeding age females originally requested), and 9 to 16 juveniles aged 1 to 4 y (1 to 30 juveniles originally requested). The ability to design feasible groups with somewhat larger numbers of animals than requested is helpful, because some animals ultimately will be removed due to behavioral incompatibilities or other nongenetic factors.
Table 2.
Initial candidate animal set and final breeding groups formed by using the described protocol
| Starting set of candidate macaques | Final membership, group 1 | Final membership, group 2 | |
| Total | 204 | 47 | 48 |
| Breeding-age macaques (no. male, no. female) | 21, 99 | 4, 27 | 7, 32 |
| Juvenile macaques (no. male, no. female) | 25, 59 | 0, 16 | 3, 6 |
| No. of genetically high-value macaques (median genome uniqueness, median mean kinship with colony) | 139 (12.60%, 0.0039) | 47 (14.60%, 0.0043) | 48 (12.15%, 0.0036) |
| No. of macaques with low genetic value (median genome uniqueness, median mean kinship with colony) | 65 (0.80%, 0.0066) | 0 | 0 |
The original request was for 2 groups, each containing 18 to 20 breeding-age female macaques older than 4 y (with their infants [younger than 1 y]), 3 or 4 breeding-age male animals 6 y or older, and 1 to 30 juveniles (age, 1 to 4 y) that will reach sexual maturity within the group.
The simulation study revealed that the median values of group mean kinship calculated for all 20 high-value groups (with relatives, 0.00246; without relatives, 0.00236) were approximately half of the median values for the 20 low-value groups (median, 0.00452 both for groups with and without relatives; Figure 1). Similarly, the median values of group mean genome uniqueness for all high-value groups (with relatives, 21.32%; without relatives, 21.37%) are approximately 10-fold higher than those for low-value groups (with relatives, 2.10%; without relatives, 2.15%; Figure 2). As expected, because the presence or absence of relatives within groups is not strongly related to mean kinship of group members to the rest of the colony, or to group genome uniqueness, differences between groups that included or excluded relatives within high-value or low-value conditions were either very small or absent altogether.
Figure 2.
Mean group genome uniqueness for 10 replicate groups in each of 4 sets of conditions: high-value or low-value groups that either included or excluded relatedness as described in Materials and Methods. The median value among each set of 10 replicate groups is indicated with a horizontal line.
However, because the presence (or absence) of relatives within groups does determine the level of inbreeding expected in any resulting offspring, it has a direct impact on the retention of genetic diversity in the colony as a whole. We found that groups that excluded relatives had lower median values of mean offspring inbreeding than did groups that included relatives, regardless of whether groups were of high or low genetic value (Figure 3); that is, for high-value and low-value groups that excluded relatives, the median F = 0.00007 and 0.00026, respectively; for high-value and low-value groups that included relatives, the median F = 0.00107 and 0.00589, respectively). Note that, as an estimate of the total effect expected from preferentially breeding high-value animals while excluding relatives, these groups had a median value of mean offspring inbreeding that was a full 2 orders of magnitude lower than that for low-value groups that did not exclude relatives (F = 0.00007 compared with 0.00589).
Finally, estimates of mean heterozygosity, as measured by our panel of microsatellite markers for each group and for each set of conditions, showed no appreciable differences either within or across groups or conditions.
Discussion
Compared with zoologic or other small-population settings, the complex social organization of NHP and the large size of NHP colonies that support biomedical research pose substantially different challenges to the implementation of genetic management strategies. For example, as many as 70% of the approximately 4000 rhesus macaques at the ONPRC may be housed in outdoor breeding corrals and other group enclosures, in which animals breed opportunistically. In these enclosures, insertion or removal of individual animals is not only logistically difficult but might extensively disrupt the group hierarchy, potentially leading to injury or death of group members. For this reason, most of the genetic management at the ONPRC occurs during routine development of the housing or breeding group, in contrast to the managed pairing of selected animals that may be more feasible in other settings. Furthermore, in addition to the genetic value of candidate breeders, numerous logistic and other factors must be considered in developing breeding group protocols for NHP, including the desired number of animals in the group, the desired sex ratio, the dominance rank and behavior of individual animals, the availability of animals, and the availability of physical space for developing the group. The need to simultaneously maximize genetic diversity in the face of these constraints and the lack of published protocols that provide practical guidance on these issues spurred us to develop the genetic management protocol we have described here.
In the absence of genome-wide genotype data that can be used to infer many types of relationship, the importance of accurate curation of pedigree data to successful genetic management in large NHP colonies cannot be overstated.3,4 To maximize the accuracy of pedigree data, colony managers should be aware of the potential sources of inaccurate or incomplete information that may be inherent to large NHP colonies. For example, at the ONPRC, the assignment of a dam to a specific offspring is initially recorded by animal care staff based on observation of animal behavior. Although dams can be assigned accurately by observation much of the time, infant swapping has been noted among rhesus macaques in captivity, and the parentage assignment of the observed dam to offspring should be confirmed using genotype data.27 Furthermore, because multimale breeding groups are preferred at the ONPRC, paternity is impossible to infer by observation alone, and there is usually little to no baseline information on the parentage assignment of sires to offspring in pedigree records. In these cases, genotype data are necessary to assign sires to offspring. Inaccurate pedigree data also can result from incorrect parentage assignment even when the assignment is based on genotype data, due to the limited genotyping performed in parentage analysis, the increased kinship or coancestry expected in semiisolated populations, and the presence of close relatives among potential parents—all of which are factors that are intrinsic to large captive NHP colonies. This inevitable increase in coancestry in NHP colonies can be exacerbated greatly by poor genetic management, that is, the preferential breeding of a small number of (usually) males. Increased coancestry will cause increased identical-by-descent allele-sharing among animals and may make it difficult to distinguish between a true parent and its close relatives when genotype information is limited, particularly when markers are linked. It is worth noting that probabilities of exclusion described in the literature in cases when one or both parents are unknown assume that all potential parents are unrelated, and expanded genotyping of animals that currently appear to be parent and offspring undoubtedly will prove this relationship to be wrong in some cases. Adherence to a rigorous set of decision rules for parentage analysis, such as those outlined here, is recommended to maximize the accuracy of pedigree information.
Pedigree information needs to be both correct and complete to produce an accurate ranking of an animal's genetic value to the colony. In practice, however, it is common for pedigrees to be neither correct nor complete, and these inaccuracies will affect estimates of kinship and genome uniqueness. For example, when relationships are indicated as correct in a pedigree but are incorrect in fact, an individual animal's mean kinship and genome uniqueness can be either over- or underestimated. However, when pedigree information is incomplete, such as when one or both parents are unknown but are likely to be related to others within the pedigree, either the offspring or the unknown parent(s) are treated as a new colony member during analysis; thus mean kinship with the colony will be underestimated and genome uniqueness will be overestimated for offspring. A lack of information on one or both parents is a common occurrence in extended pedigrees with multiple generations, particularly for older generations in which parentage assignment cannot be determined due to a lack of genotype data. In this case, genetic diversity will continue to be overestimated in more recent generations, even when complete information on parentage is available for these generations.
Despite the inevitable pedigree inaccuracies, the ranking of animals by genetic value based on the best information available can prove useful, not only for the design of new breeding groups but also in the selection of animals for research assignment or sale. In practice, at the ONPRC our threshold genetic value results in approximately one third of all macaques being ranked as of low value to the colony. Although these animals may not be recommended for breeding, they are ideal candidates for assignment to research or for sale to other research centers. In fact, an animal of low genetic value to one research center is likely to be of high genetic value to another. Summarizing the genetic value of each animal in terms of both mean kinship to the colony and genome uniqueness is essential, given that some animals may have genomes that combine alleles from an overrepresented lineage with alleles from an underrepresented lineage; in these cases, we prefer to rank these animals above others that may have lower mean kinship but that do not contain underrepresented alleles. In general, we prefer the risk of increasing background coancestry or kinship to the risk of losing a rare genotype in the colony. For this reason, we first rank macaques with the greatest genome uniqueness, followed by the z-score indicator of mean kinship within genome uniqueness. Once an animal's genome uniqueness falls below 10%, we then prefer to weight the risk of increased mean kinship to the colony more heavily than the comparatively low probability of losing rare alleles. Given the median 50% reduction in group mean kinship and 10-fold increase in group mean genome uniqueness for high-value groups compared with low-value groups in our simulation study (Figures 1 and 2), this ranking scheme appears to perform reasonably well in optimizing genetic diversity in the face of specific constraints.
The procedure we outline for ranking animals by genetic value can be applied to different breeding strategies or adjusted to incorporate a different threshold between high- and low-value animals, depending on the needs of individual centers. For example, the use of a ranking procedure for genetic value is not limited to multimale, multifemale grouping strategies. High-ranking single males and multiple females, or single males and single females, can also be selected from this list for harems or focused breeding pairs, respectively. In fact, the recalculation and selection of animals by genetic rank becomes increasingly critical with these alternative breeding strategies, as these practices can dramatically reduce colony genetic diversity if implemented naively, for example, when the same high value males are used among multiple harems or in repeated pairings. In addition, the threshold between high- and low-value animals can be adjusted in a way that better utilizes animal availability but that still maximizes the genetic value of available animals. If this adjustment is made, however, colony managers should remain mindful that any threshold is arbitrary and that breeding animals with higher mean kinship and lower genome uniqueness will always reduce colony genetic diversity relative to breeding those with lower mean kinship and higher genome uniqueness. Other adjustments to our ranking procedure are not expected to produce substantial changes in final group genetic diversity, including prioritizing according to a different scheme of mean kinship or z-score and genome uniqueness. This outcome is expected because, although mean kinship and genome uniqueness reflect different facets of genetic diversity, there is a strong relationship between these 2 measures, so that prioritizing by one will tend to maximize the other (see reference 3 for a detailed exploration of this relationship).
After the assignment of genetic value to candidates for a new breeding group, it is important to recognize that animals of high genetic value on the basis of lower mean kinship with the colony, may still be closely related to other high-value animals. Consequently, genetic diversity in the colony can still be greatly reduced when even high-value relatives are allowed to produce offspring. In particular, we avoid relatedness between breeding males and between breeding males and females, but we ignore relatedness between breeding adults and infants younger than 1 y and between females of any age. Ideally, a breeding group is maintained for several years before being disbanded, therefore ignoring all relationships between adults and infants is a pragmatic decision, given that infants are dependent on their dam for optimal social development and are the least likely to become sexually mature in their assigned group. However, these animals could become a genetic liability in the future if they are allowed to breed within their assigned group. Unlike infants, juveniles 1 to 4 y of age are expected to reach reproductive maturity before the group is disbanded, and for this reason, their relatedness to adults is considered. Although reports of free-ranging animals indicate that rhesus macaques become reproductively mature at the age of 3.5 to 5.5 y for females and around 6.5 y for males,8 parentage analysis at the ONPRC has revealed that male macaques can (albeit rarely) produce offspring at 2.5 y, similar to observations in other systems of early puberty induced by the captive environment.17,24 For this reason, we recommend the regular removal of males who were infants at group assignment and have since reached 3 y of age while in the group, given that their options for breeding with unrelated females might be limited or nonexistent. In general, depending on the group size and corresponding access of sexually mature males to unrelated females, it may be wise to disband smaller groups more frequently than larger groups to prevent inbreeding.
The second type of relatedness that we ignore is that between females of any age. This strategy permits a social structure that is consistent with wild-type social groups, which are composed of one or several maternal lineages with stable dominance hierarchies8,21,22 and which avoid inbreeding by male-biased dispersal from the natal group.20,28 From a practical standpoint, however, this practice allows us to further reduce the number of relationships that need to be accounted for during group formation. By excluding relatedness only among adult males, between adult males and adult females, and between adults and juveniles, we are able to form larger social groups than would be possible if we constrained relatedness between adult females as well. This compromise is necessary, given the finite number of group housing locations and the commitment of the ONPRC to social housing for as many animals as possible. That our strategy balances feasibility with the maintenance of genetic diversity can be seen in the order-of-magnitude decreases observed in the mean inbreeding coefficient expected among offspring when relatedness within groups is managed rather than ignored, regardless of whether a group is of high or low genetic value (Figure 3). However, it should be noted that the inbreeding coefficient expected among offspring in high-value groups that exclude relatives is still approximately 4-fold lower than that in low-value groups that also exclude relatives and approximately 84-fold lower than that in low-value groups that don't exclude relatives. These findings make it abundantly clear that managing relatedness in breeding groups is a critical component of an overall management strategy aimed at maximizing genetic diversity within the colony as a whole. In marked contrast to mean inbreeding coefficients, mean group heterozygosity, estimated using microsatellite genotypes across all experimental conditions, did not differ between groups, suggesting that these markers will be a poor predictor of the loss of genetic diversity over the short term. This finding is not unexpected, given the hypervariability of microsatellite markers, and other genetic markers may ultimately prove more useful for this purpose.
The maximization of genetic diversity in a large NHP breeding colony will always be constrained by the degree to which optimal genetic management protocols can be followed. In practice, breeding group formation is rarely based on genetic considerations alone. At the ONPRC, animal behavior concerns in particular tend to limit the formation of a breeding group that is optimal by genetic criteria. These concerns are not trivial, given that animal injury or death caused by other group members is a frequent outcome of behavioral incompatibilities within the group. However, the consequences of poor genetic management in NHP colonies will continue to erode the usefulness of this animal model to the very research it is intended to support, and at some point are likely to be irreversible. Future research focused on the development of a scoring system for individual behavioral traits or maladaptive behavior would greatly enhance the efficacy of breeding group formation and related genetic management protocols.
Acknowledgments
We gratefully acknowledge the ONPRC veterinary staff in the Division of Comparative Medicine for detailed information on social group housing at the ONPRC and the UC Davis Veterinary Genetics Laboratory for many years of genotyping ONPRC animals. This project was supported by the Office of the Director/Office of Research Infrastructure Programs (OD/ORIP) of the NIH (grant no. OD 011092). We also thank Betsy Ferguson (head of the ONPRC Primate Genetics Program), Samone Khoungsathiene, and Satyavathi Chinni for helpful analysis and discussion as this protocol evolved.
References
- 1.Allendorf FW, Luikart GH, Aitken SN. 2012. Conservation and the genetics of populations, 2nd ed. West Sussex (UK): John Wiley and Sons. [Google Scholar]
- 2.Andrade MC, Penedo MCT, Ward T, Silva VF, Bertolini LR, Roberts JA, Leite JPG, Cabello PH. 2004. Determination of genetic status in a closed colony of rhesus monkeys (Macaca mulatta). Primates 45:183–186. [DOI] [PubMed] [Google Scholar]
- 3.Ballou JD. 1997. Genetic and demographic modeling for animal colony and population management. ILAR J 38:69–75. [DOI] [PubMed] [Google Scholar]
- 4.Ballou JD, Lacy RC. 1995. Identifying genetically important individuals for management of genetic variation in pedigreed populations, p 77–111. In: Ballou JD, Gilpin M, Foose TJ. Population management for survival and recovery. New York (NY): Columbia University Press. [Google Scholar]
- 5.Dakin EE, Avise JC. 2004. Microsatellite null alleles in parentage analysis. Heredity (Edinb) 93:504–509. [DOI] [PubMed] [Google Scholar]
- 6.Ebeling M, Küng E, See A, Broger C, Steiner G, Berrera M, Heckel T, Iniguez L, Albert T, Schmucki R, Biller H, Singer T, Certa U. 2011. Genome-based analysis of the nonhuman primate Macaca fascicularis as a model for drug safety assessment. Genome Res 21:1746–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics, 4th ed. Essex (UK): Addison Wesley Longman. [Google Scholar]
- 8.Fooden J. 2000. Systematic review of the rhesus macaque, Macaca mulatta (Zimmermann, 1780). Chicago (IL): Field Museum of Natural History. Field Zool 96:1–180. [Google Scholar]
- 9.Frankham R. 1995. Conservation genetics. Annu Rev Genet 29:305–327. [DOI] [PubMed] [Google Scholar]
- 10.Garber RA, Morris JW. 1983. General equation for the average power of exclusion for genetic systems of n codominant alleles in 1-parent and in no-parent cases of disputed parentage, p 277–280. In: Walker RH. Inclusion probabilities in parentage testing. Arlington (VA): American Association of Blood Banks. [Google Scholar]
- 11.Ha JC, Robinette RL, Sackett GP. 1999. Social housing and pregnancy outcomes in captive pigtailed macaques. Am J Primatol 47:153–163. [DOI] [PubMed] [Google Scholar]
- 12.Jamieson A, Taylor SC. 2003. Comparisons of 3 probability formulae for parentage exclusion. Anim Genet 28:397–400. [DOI] [PubMed] [Google Scholar]
- 13.Jones AG, Ardren WR. 2003. Methods of parentage analysis in natural populations. Mol Ecol 12:2511–2523. [DOI] [PubMed] [Google Scholar]
- 14.Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106. [DOI] [PubMed] [Google Scholar]
- 15.Kanthaswamy S, Kou A, Satkoski J, Penedo MCT, Ward T, Ng J, Gill L, Lerche NW, Erickson BJA, Smith DG. 2010. Genetic characterization of specific pathogen-free (SPF) rhesus macaque (Macaca mulatta) populations at the California National Primate Research Center (CNPRC). Am J Primatol 72:587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura M, Crow JF. 2009. On the maximum avoidance of inbreeding. Genet Res 4:399–415. [Google Scholar]
- 17.King HD. 1939. Life processes in gray Norway rats during 14 years in captivity, p 1–77. In: Stockard CR, Evans HM. American anatomical memoirs, vol 17 Philadelphia (PA): The Wistar Institute of Anatomy and Biology. [Google Scholar]
- 18.Lange K. 2002. Mathematical and statistical methods for genetic analysis. New York (NY): Springer–Verlag. [Google Scholar]
- 19.MacCluer JW, VandeBerg JL, Read B, Ryder OA. 1986. Pedigree analysis by computer simulation. Zoo Biol 5:147–160. [Google Scholar]
- 20.Manson JH, Perry SE. 1993. Inbreeding avoidance in rhesus macaques: whose choice? Am J Phys Anthropol 90:335–344. [DOI] [PubMed] [Google Scholar]
- 21.Melnick DJ, Pearl MC. 1987. Cercopithecines in multimale groups: genetic diversity and population structure, p 121–134. In: Smuts BB, Cheney DL, Seyfarth RM, Wrangham RW, Struhsaker TT. Primate societies. Chicago (IL): The University of Chicago Press. [Google Scholar]
- 22.Melnick DJ, Pearl MC, Richard AF. 2005. Male migration and inbreeding avoidance in wild rhesus monkeys. Am J Primatol 7:229–243. [DOI] [PubMed] [Google Scholar]
- 23.Murphy WJ, Agarwala R, Schäffer AA, Stephens R, Smith C, Jr, Crumpler NJ, David VA, O'Brien SJ. 2005. A rhesus macaque radiation hybrid map and comparative analysis with the human genome. Genomics 86:383–395. [DOI] [PubMed] [Google Scholar]
- 24.O'Regan HJ, Kitchener AC. 2005. The effects of captivity on the morphology of captive, domesticated and feral mammals. Mamm Rev 35:215–230. [Google Scholar]
- 25.Poiley SM. 1960. A systematic method of breeder rotation for noninbred laboratory animal colonies. Proc Anim Care Panel 10:159–166. [Google Scholar]
- 26.Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, Johnson Z, Bergstrom M, Novakowski L, Nair P, Vinson A, Newman D, Heckman G, Cameron J. 2006. An initial linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics 87:30–38. [DOI] [PubMed] [Google Scholar]
- 27.Smith DG. 1994. Genetic heterogeneity in 5 captive specific pathogen-free groups of rhesus macaques. Lab Anim Sci 44:200–210. [PubMed] [Google Scholar]
- 28.Smith DG. 2005. Avoidance of close consanguineous inbreeding in captive groups of rhesus macaques. Am J Primatol 35:31–40. [DOI] [PubMed] [Google Scholar]
- 29.VandeBerg JL, Williams-Blangero S. 2011. Advantages and limitations of nonhuman primates as animal models in genetic research on complex diseases. J Med Primatol 26:113–119. [DOI] [PubMed] [Google Scholar]
- 30.Williams-Blangero S. 1993. Research-oriented genetic management of nonhuman primate colonies. Lab Anim Sci 43:535–540. [PubMed] [Google Scholar]
- 31.Williams-Blangero S, Eichberg JW, Dyke B. 2005. The genetic demography of a chimpanzee colony. Am J Primatol 27:73–83. [DOI] [PubMed] [Google Scholar]
- 32.Williams-Blangero S, VandeBerg JL, Dyke B. 2002. Genetic management of nonhuman primates. J Med Primatol 31:1–7. [DOI] [PubMed] [Google Scholar]