Abstract
Efficient, effective cage decontamination and the detection of infection are important to sustainable biosecurity within animal facilities. This study compared the efficacy of cage washing at 110 and 180 °F on preventing pathogen transmission. Soiled cages from mice infected with mouse parvovirus (MPV) and mouse hepatitis virus (MHV) were washed at 110 or 180 °F or were not washed. Sentinels from washed cages did not seroconvert to either virus, whereas sentinels in unwashed cages seroconverted to both agents. Soiled cages from mice harboring MPV, Helicobacter spp., Mycoplasma pulmonis, Syphacia obvelata, and Myocoptes musculinus were washed at 110 or 180 °F or were not washed. Sentinels from washed cages remained pathogen-free, whereas most sentinels in unwashed cages became infected with MPV and S. obvelata. Therefore washing at 110 or 180 °F is sufficient to decontaminate caging and prevent pathogen transmission. We then assessed whether PCR analysis of debris from the bedding disposal cabinet detected pathogens at the facility level. Samples were collected from the prefilter before and after the disposal of bedding from cages housing mice infected with both MPV and MHV. All samples collected before bedding disposal were negative for parvovirus and MHV, and all samples collected afterward were positive for these agents. Furthermore, all samples obtained from the prefilter before the disposal of bedding from multiply infected mice were pathogen-negative, and all those collected afterward were positive for parvovirus, M. pulmonis, S. obvelata, and Myocoptes musculinus. Therefore the debris on the prefilter of bedding-disposal cabinets is useful for pathogen screening.
Abbreviations: ABDC, animal bedding disposal cabinet; MAV, murine adenovirus K87; MHV, mouse hepatitis virus; MNV, murine norovirus; MPV, mouse parvovirus; MVM, minute virus of mice; MMBTU, million British thermal units; SW, Swiss Webster
The use of evidence-based standard operating procedures in animal resource centers is crucial to cost containment and sustainable energy use. Understandably, biosecurity is a major driver of procedures and processes in rodent facilities and permeates virtually all aspects of animal resource operations, making it necessary to balance the cost: benefit of detection, prevention, and control of infection. As an example, recent publications that suggested that mouse parvovirus (MPV) infection can be caused by MPV-contaminated grains that were not inactivated during their processing into pelleted rodent feed25,36,47 have led to changes in husbandry procedures including the use of autoclaved or irradiated food. Although the successful control and prevention of MPV infections in our facilities has been attributed to the practice of autoclaving mouse cages preassembled with bedding and food prior to their use in the facility, this procedure was not definitively proven to be the sole factor in MPV eradication.30 Because wash centers are historically the largest utilities consumer in animal facilities,15 this practice of autoclaving cages prior to use is not only labor-intensive but also energy-intensive, and the labor and energy consumption is amplified in facilities that lack a bulk autoclave and in which the cages must be transported to be autoclaved. In addition, during outbreaks with MPV and other infectious agents, our long-standing standard operating procedure for soiled cages is to autoclave them to inactivate infectious agents prior to removing the soiled bedding and cage washing. This labor- and energy-intensive practice of using autoclaving to decontaminate cages has historically been justified because MPV is a nonenveloped virus that is highly stable in the environment and difficult to inactivate.5,6,16,27,38,42,49 Despite the environmental stability of the virus, infections with MPV are difficult to detect because the amount of virus shed is low; transmission can be inefficient, resulting in inconsistent seroconversion of mice housed in the same cage; and PCR analysis can detect low levels of MPV DNA in the feces of mice that are unable to transmit virus to contact sentinels.4,30,31 We recently demonstrated that cage washing alone removed or inactivated MPV from 14 cages that had housed outbred mice acutely infected with MPV; these findings are not surprising when the inefficiencies of MPV transmission are considered.11 These initial results challenge the cost:benefit ratio of autoclaving cages as a means to decontaminate them prior to cage washing during an MPV outbreak. Our findings with MPV led us to question whether cage washing alone might be effective for decontaminating cages after exposure to other infectious agents that are stable in the environment, even if they are shed for longer periods of time or at higher levels than is MPV.
In our previous study,11 we showed that cage washing was effective at preventing fomite-based transmission of MPV by cage components, and we postulated that water temperature and detergent type contribute to MPV decontamination either directly through inactivation of the virus and/or indirectly through effective mechanical removal of organic waste and residual virus from the cage. The Guide for the Care and Use of Laboratory Animals24 states that “disinfection from the use of hot water alone is the result of the combined effect of temperature and the length of time at a given temperature” and that “effective disinfection can be achieved with wash and rinse water at 143–180 °F or more.” Theoretically, contact times to disinfect equipment at these temperatures would need to be 1800 s at 143 °F (61.7 °C) and 0.1 s at 180 °F (82.2 °C).46 Contact times of 6 s or more at 168 to 180 °F (75.6 to 82.2 °C) killed 3 types of bacteria (Pseudomonas aeruginosa, Salmonella cholerasuis, and Staphylococcus aureus) in one study,45 and contact times of 2 min or more at 160 °F (71.1 °C) killed 5 types of bacteria (Escherichia coli, Klebsiella pneumonia, Proteus mirabilis, Providencia rettgeri, and Staphylococcus epidermidis) in another.39 However, these 2 studies were performed by using hot water in a test tube and, therefore, detergent and mechanical spraying action that occur within rack washers was not a factor.
Traditionally, the temperature used for the wash and rinse water in our facilities has been 180 °F (82.2 °C) with a belt speed of 2 to 3 ft./min (0.6 to 0.9 m/min). We postulated that if the volume and force of the wash water, combined with detergents, consistently diluted or removed infectious agents to below the level necessary for the transmission of infection, then wash temperatures high enough to inactivate the agents would be unnecessary. Given the energy usage and infrastructure necessary to boost ‘domestic’ hot water to 180 °F, the purpose of this study was to compare the efficacy of cage washing by using the domestic hot-water temperature (110 °F [43.3 °C]) and the traditional steam-boosted wash temperature (180 °F [82.2 °C]) on preventing the transmission from contaminated caging of 3 of the most prevalent viral agents (MNV, MPV, MHV; prevalence, 32.6%,1.8%, and 1.6%, respectively ) and bacterial and parasitic agents (Helicobacter spp. and pinworms; prevalence,15.9% and 0.3%, respectively).35
Effective and efficient decontamination of caging goes hand-in-hand with effective and efficient detection of infection. Timely detection of infection is important to biosecurity, and environmental sampling is a promising adjunct to sentinel exposure programs for the early detection of infectious agents. PCR analysis of cage and rack components, including the outflow prefilter of ventilated racks, has been shown to be of use for several infectious agents including MPV, MHV, Helicobacter spp., and fur mites.10,26,31 The ability to reliably detect infectious agents from a site where soiled bedding debris is aerosolized and concentrated might provide an efficient adjunct for infectious agent screening. In the current study, we determined whether monitoring by PCR analysis of dust and debris collected from the animal bedding disposal cabinet (ABDC) prefilter could be used as an efficient adjunct to sentinel programs to screen for contamination by infectious agents.
Materials and Methods
Mice.
Female Swiss Webster mice (Crl:CFW [SW]; age, 4 to 6 wk) were obtained from Charles River Laboratories (Kingston, NY). Vendor reports indicated that mice were seronegative for ectromelia virus, murine rotavirus, lymphocytic choriomeningitis virus, MHV, MPV, minute virus of mice (MVM), murine norovirus (MNV), pneumonia virus of mice, reovirus, Sendai virus, Theiler encephalomyelitis virus, and Mycoplasma pulmonis and were free of bacterial and parasitic infections at the time of shipment. Random-bred female mice were obtained from 3 local pet stores (4 mice per store) to serve as sources of Helicobacter spp. and pinworms. Mice were housed in IVC (70 cages per rack) under positive pressure (ACE MicroVent, Allentown, NJ). Cages containing corncob bedding (Harlan Teklad, Indianapolis, IN), rodent chow (Global 2018S, Harlan Teklad) and nesting material (Cotton squares, Ancare, Bellmore, NY) were preassembled and autoclaved. Mice had unrestricted access to hyperchlorinated (4 to 6 ppm) water delivered by water bottle. The mice were housed and husbanded according to standard biocontainment procedures. The animal room had a negative pressure differential relative to the corridor, a 12:12-h light:dark cycle, 10 to 15 air changes hourly, room temperature of 22.2 ± 1.1 °C, and room humidity of 50% ± 10% and was used exclusively for this study. All animal care and experimental procedures were performed in an AAALAC-accredited animal facility, were approved by the Yale IACUC, and adverse events (described later) were reported to the IACUC. All animal care followed the Guide for the Care and Use of Laboratory Animals,24 and experimental procedure were in accordance with federal policies and guidelines governing the use of vertebrate animals.
PCR analysis.
Individual fecal pellets were collected from the anus of each mouse, and pools of 6 to 10 pellets were collected from soiled bedding. The nape, abdomen, and rump of mice were swabbed by using sterile Hydraflock swabs (Puritan, Guilford, ME) for detection of fur mites. Cage swabs were collected by swabbing the perimeter of the cage just above the bedding and then passing the swab through the soiled bedding in an X pattern from cage corner to cage corner to detect viruses, Helicobacter spp., and pinworms. Cecal samples were collected from mice after carbon dioxide euthanasia. All samples were frozen in 1.5-mL tubes at –20 °C prior to PCR analysis. DNA and RNA were extracted from samples by using DNeasy or RNeasy kits (Qiagen, Valencia, CA) according to the manufacturer's instructions. PCR assays were performed by using iTaq Universal SYBR Green kit (BioRad, Hercules, CA), and RT–PCR assays were performed by using iScript One-Step RT–PCR kit with SYBR Green (BioRad) and a thermocycler (CFX Connect, Biorad). Primers specific for cilia-associated respiratory bacillus, Helicobacter spp., MHV, parvovirus (MPV and MVM), murine adenovirus K87 (MAV), MNV, murine rotavirus, Myobia musculi, Myocoptes musculinus, Pasteurella pneumotropica, pinworms (Aspiculuris tetraptera and Syphacia obvelata), Pneumocystis murina, reovirus, and Theiler encephalomyelitis virus were used as previously described.12,20,21,44 RT–PCR analysis for lymphocytic choriomeningitis virus was performed by using the primers LCMVS710 (5′ AGG CTC AGA TGG CAA GAC C 3′) and LCMVS1641 (5′ GCC CAA ATG TTG TGA CAC TCT 3′). RT–PCR analysis for pneumonia virus of mice was performed by using PVM1616 (5′ CCC AAC ATG GAG GTC AAG CAG 3′) and PVM1987 (5′ GCA TTG CCA AGC ACA ACA CTG 3′). RT–PCR analysis for Sendai virus was performed by using SEN399 (5′ GGA GTA AAC GCC GAT GTC AAA 3′) and SEN1274 (5′ CCC TTG GCT GTA TCC GTC ACT 3′). Mycobacteria spp. PCR analysis used MYB404 (5′ TTT CTC GGA TTG ACG GTA GG 3′) and MYB1235 (5′ TGA GAC CGG CTT TAA AAG GA 3′). Mycoplasma pulmonis PCR analysis used MYPUL180 (5′ TTA GAT CGC ATG ATT TAG AT 3′) and MYPUL875 (5′ TGC GAG CAT ACT ACT CAG 3′).
Serology.
Cardiocentesis was performed after carbon dioxide overdose. Sera were tested in immunofluorescent antibody assays as previously described40 for antibodies to one or more of the following agents: ectromelia virus, lymphocytic choriomeningitis virus, MAV, MHV, MPV, MVM, murine rotavirus, pneumonia virus of mice, reovirus, Sendai virus, Theiler encephalomyelitis virus, and M. pulmonis.
Parasitology.
To detect S. obvelata, cellophane tape tests were performed by applying tape to the perianal region of the mouse. The adherent surface of the tape was placed onto a glass microscope slide, and slides were observed by using a microscope at 10× and 40× magnification. Slurries were made in 0.9% saline from pools of 6 to 10 feces collected from soiled bedding. A small amount of the slurry was placed on a glass microscope slide, a cover slip was placed on the sample, and the sample was examined for evidence of ova or adult endoparasites by microscopy at 10× and 40× magnification. Portions of the cecum and colon from each mouse were submitted for direct examination for intestinal parasites. Immediately after collection of the cecum and colon, approximately 0.5 mL of 0.9% saline was added to the sample in a culture dish and mixed with a wooden applicator. A small amount of the sample was placed on a glass microscope slide, cover-slipped, and examined for evidence of ova or adult endoparasites by microscopy at 10× and 40× magnification. Parasites were identified and speciated morphologically. The cecocolonic contents in the culture dish were examined again, at 24 h after the initial exam, under low-power (10×) microscopy for evidence of adult endoparasites. Fur plucked from the nape and rump of each mouse was placed in a drop of mineral oil on a microscope slide; a cover slip was placed on the sample; and samples were examined by using a microscope under low power (10× magnification) for evidence of fur-mite ova, larvae, and adults, which were identified and speciated morphologically.
Statistical analysis.
Two-tailed Fisher exact probability tests were performed by using an online statistics application (vassarstats.net). A P value of less than 0.05 denoted statistical significance.
Experimental viral infection study.
Unanesthetized 4-wk-old female SW index mice (n = 36) were inoculated orally with 300 ID50 of MPV1d (20 μL of a 10% spleen stock), 36 unanesthetized 4-wk-old female SW index mice were inoculated orally with 3000 ID50 of MHV-Y (20 μL of a 10% intestinal stock), and 36 unanesthetized 4-wk-old female SW index mice were inoculated orally with 100 ID50 of MNV-G8 (20 μL of a 10% colon stock). Mice were housed in 36 cages, with each cage containing one MPV-inoculated mouse, one MHV-inoculated mouse, and one MNV-inoculated mouse. On day 9 after inoculation, all inoculated (index) mice were removed from their respective soiled cages and were placed in clean cages (3 mice per cage) for an additional 2 wk to allow for seroconversion. Feces were collected from all index mice at 10 d after inoculation, and feces and blood was collected from index mice at 23 d to test for MPV-, MHV-, and MNV-specific nucleic acids in feces and antibodies in serum.
On day 9 after inoculation, soiled cage tops and bottoms were marked with heat-resistant ink. Each cage was swabbed, and PCR analysis was performed to confirm that all cages had detectable levels of viral nucleic acids on their surfaces prior to cage washing. Cages were immediately transported to the wash center. Prior to removing the soiled bedding from the cages, approximately 1-mL samples (n = 4) of dust and debris were collected from the prefilter of the ABDC (model NU607400, Labgard Class 1 Animal Bedding Disposal Cabinet, NuAire, Plymouth, MN) by using forceps. PCR analysis was performed on the 4 samples to determine whether infectious agents had previously been deposited on the prefilter. Soiled bedding was removed by ‘dumping’ the soiled bedding from the contaminated cages into the ABDC; cages were then scraped prior to loading into the tunnel washer (Better Built Metal Wash Tunnel Washer, Northwestern Systems, Delta, Canada), which had a belt speed of 2.4 ft/min (0.74 m/min). Cages were exposed to hot water in the wash and rinse cycles for a total of 4.6 min. The chemicals used in the tunnel washer were Enviro-Kleen 1500, GLPC7, and Acidulate 150 (Quip Labs, Wilmington, DE). After the soiled bedding from all 36 cages was dumped, 4 approximately 1-mL samples of dust and debris were collected from the prefilter of the ABDC, and PCR analysis was performed to determine whether detectable amounts of MPV, MHV, or MNV had been deposited on the prefilter during dumping of soiled bedding. Then, the first 16 cages, with filter tops and wire bars, were washed at a wash temperature of 110 °F (43.3 °C), which was confirmed by using a chromel–alumel type K thermocouple connected to a Meter (Fluke, Everett, WA). The thermocouple was located in the rinse tank, which stores and maintains the preset water temperature for this tunnel washer. Before the second set of 16 cages was washed, the steam system was activated, and the water in the tunnel washer tank was allowed to heat to 180 °F (82.2 °C). The wash temperature was confirmed to be 180 °F by using temperature-sensitive tape (Thermolabel, Paper Thermometer, Greenfield, NH), and then the second set of 16 cages was washed. The final 4 cages were not washed and served as positive-control cages to confirm that the amount of virus present on the soiled cages was sufficient to cause infection of naive sentinel mice. After cage washing at either 110 or 180 °F, cages were reassembled, transported back to the animal room, and placed on the IVC rack to dry for 2 h. The efficacy of cage sanitation was assessed by ATPase testing (Accupoint 2 Sanitation Monitoring System with surface samplers, Neogen, Lansing, MI).
Autoclaved food, autoclaved bedding, autoclaved nesting material, and an autoclaved water bottle filled with hyperchlorinated water were added to each washed cage, and then a 4-wk-old female SW sentinel mouse was placed into each cage. One week after the addition of sentinel mice to the cages (that is, 1 wk after exposure to the washed cages), feces were collected from sentinel mice for PCR analysis. At 2 wk after exposure to the washed cages, sentinel mice were moved to clean cages. At 3 wk after exposure to the washed cages, mice were euthanized, and feces and blood were collected for PCR analysis and serology, respectively.
Helicobacter spp. and pinworm contact-infection study.
The study design included a 2-stage approach to produce SW mice infected with Helicobacter spp. and pinworms because stocks of these infectious agents were not readily available. Initially, 12 random-bred female mice were obtained from 3 local pet stores (4 mice per store); 9 of these mice were housed individually. We added 4-wk-old female SW mice (n = 4 per cage; 36 SW mice total) to each of the 9 cages to initiate infection with Helicobacter spp. and pinworms, understanding that other infectious agents harbored by the pet-store mice would also be transmitted to the SW contact mice. The extra 3 pet-store mice were housed separately as replacements for pet-store mice that might become ill during the study. Feces from pet-store mice were tested for Helicobacter spp., pinworms, lymphocytic choriomeningitis virus, MAV, MHV, MNV, parvovirus, murine rotavirus, reovirus, and Theiler encephalomyelitis virus by PCR analysis on their arrival to our facility. Pet-store mice were rotated among the 9 cages of contact SW mice on days 5, 10, 15 and 20 to produce a more uniform infection profile than would be obtained if each cage of contact SW mice were exposed to only a single pet-store mouse. Cages were changed on days 10, 18, 30, 42, 51, 65, and 78. Two SW mice were found dead on day 7 in a cage in which a water bottle accident had occurred, and 2 of the ‘extra’ pet-store mice were added as replacements to that cage to restore the cage census to 5 mice (45 mice total: 11 pet-store mice and 34 SW contact mice). The remaining pet-store mouse was euthanized on day 20, when it was determined that it would not be needed as a replacement.
On days 22 and 23, feces were collected individually from pet-store and SW contact mice for PCR analysis, fecal pools were collected for direct examination, fur swabs were collected for PCR analysis and tape tests, and fur plucks were performed. Unexpected adverse events occurred between days 22 and 36. A total of 5 SW contact mice were found dead on days 23 and 29, despite careful monitoring, and 6 SW contact mice were euthanized because they were observed to be acutely dyspneic, dehydrated, and hunched or circling on days 20, 23, 25, and 26. On day 30, pinworm- and Helicobacter-infected mice (11 pet-store mice and the 23 remaining SW contact mice) were placed individually in 34 cages, and a second set of 68 naïve 4-wk-old female SW mice were distributed (2 per cage) among the 34 cages, resulting in 102 mice total at a density of 3 mice per cage. Ten more of the original SW contact mice were found dead on days 30 to 36, despite careful monitoring, and 3 more of the original SW contact mice were euthanized because they were noted to be acutely dyspneic, and dehydrated or hunched on day 35. Therefore, on day 37, the remaining 12 original contact SW mice were euthanized because of concerns that they also would develop similar acute clinical disease. The euthanasia or death of all 36 of the original SW contact mice left 23 of the 34 cages to be used to generate soiled bedding containing Helicobacter spp. and pinworms without an infected mouse, and the second set of 68 SW mice had not yet had sufficient time to acquire and shed both Helicobacter spp. and pinworms by day 37 (7 d after exposure to infected mice). Given concerns that exposing additional naive SW mice would result in clinical disease or mortality, we decided that no additional SW mice would be used. One of the 68 SW mice appeared runted and was euthanized on day 42. Therefore on day 42, the remaining 78 (11 pet-store and 67 SW mice from the second set of SW mice) animals were distributed among 39 clean cages (2 mice per cage) to generate soiled cages to be washed at 110 and 180 °F on day 51. Prior to cage washing (day 50), pools of feces were collected from each cage for PCR analysis to determine whether all of the cages contained mice that were infected with Helicobacter spp., were infested with pinworms, and were infected with other agents.
On day 51, 9 d after the last cage change and 21 d after exposure of the second set of 67 SW contact mice to infected mice, this cohort of contact-infected mice (11 pet-store mice and the second set of 67 SW mice) were moved to clean cages, and soiled cage tops and bottoms were marked with heat-resistant ink. Each cage was swabbed, and PCR analysis was performed to confirm that all cages had detectable levels of Helicobacter spp. and pinworm DNA on their surfaces prior to cage washing. Cages were transported to the wash center. Prior to removal of the soiled bedding, 3 approximately 1-mL samples of dust and debris and were collected from the prefilter of the ABDC and were submitted for PCR analysis to determine whether MAV, MHV, MNV, parvovirus, Helicobacter spp., pinworms or fur mites had previously been deposited on the prefilter. Soiled bedding was dumped into the ABDC, and cages were scraped prior to loading into the tunnel washer. After the bedding from all 39 cages was dumped, 10 approximately 1-mL samples of dust and debris were collected from the prefilter of the ABDC. PBS was added to 5 samples of dust and debris, and RNA and DNA were extracted from the debris–PBS mixture (indirect extraction). DNA was extracted from 5 samples of dust and debris directly after the addition of lysis buffer (direct extraction). PCR analysis to determine whether detectable amounts of infectious agents had been deposited on the prefilter during soiled bedding dumping was performed on all dust and debris samples.
Cages were washed in a Better Built Metal Wash Tunnel Washer with a belt speed of 2.4 ft./min (0.74 m/min) by using EnviroKleen 1500, GLPC7, and Acidulate 150. Cages were exposed to hot water in the wash and rinse cycles for a total of 4.6 min. The first 16 cages were washing by using a wash temperature of 110 °F, after which the steam system was activated and the water in the tank was heated to 180 °F. The wash temperature was confirmed to be 180 °F by using Thermolabel temperature-sensitive tape (Paper Thermometer), and then the second 16 cages were washed. The final 7 cages were not washed and served as positive-control cages to confirm that the amount of infectious agents present on the soiled cages was sufficient to infect the sentinel mice. Cages were reassembled and transported back to the animal room, where they were placed on the IVC rack to dry for 2 h. Autoclaved food, autoclaved bedding, autoclaved nesting material, and a clean water bottle were added to each cage, and a single 4-wk-old female SW sentinel mouse was placed in each cage.
On days 55 and 56, fecal pellets and fur swabs were collected from the 78 infected mice for PCR analysis, tape tests were performed and fur plucks were collected and analyzed. Then the mice were euthanized by CO2 asphyxiation, and blood was collected for serology and ceca were collected for direct observation and PCR.
On days 65 and 78 (days 14 and 27 after exposure to contaminated cages) sentinels were moved to clean cages. One month after exposure to the washed cages (on day 82), sentinel mice were euthanized, and feces and blood were collected for PCR and serology, respectively.
Results
Experimental viral infection study.
To determine the effect of water temperature during cage washing on the removal or inactivation of viruses, cages were washed when peak levels of virus were expected to be present in the cages. Specifically, 9 d after experimental inoculation of mice with MPV, MHV, and MNV, by which time a substantial amount of each virus should have accumulated in the soiled bedding and on the cage surfaces, 16 soiled cages were washed at 110 °F, and 16 soiled cages were washed at 180 °F. In addition, 4 cages from which the bedding was removed, the surfaces scraped to remove adhered bedding, but not washed were used to confirm that the amount of virus present on the soiled cage surface was sufficient to infect the sentinel mice (positive control).
Experimental viral infections of mice.
PCR analysis of feces collected individually from the 108 index mice, 10 d after experimental inoculation, detected MPV in 94% and MHV in 98% of the mice, indicating that most of the mice were actively shedding MPV and MHV when cages were washed (Table 1). In addition, at 23 d after inoculation, 97% of the index mice were seropositive for MPV (all 3 mice in one cage did not seroconvert to MPV), and all index mice were seropositive for MHV (Table 1). These data indicate that all mice inoculated with MHV became infected, and all but one mouse inoculated with MPV became infected; thereafter the inoculated mice infected their cage mates. Experimental infection with MNV was attempted but was unsuccessful.
Table 1.
PCR and serologic analysis for experimental virus infection: cages, index mice, and sentinel mice
| 180 °F wash | 110 °F wash | Unwashed | |
| 9 DPI MPV cage swab | 15/16 | 16/16 | 4/4 |
| 10 DPI MPV index feces | 44/48 | 45/47 | 12/12 |
| 23 DPI MPV index sera | 45/48 | 48/48 | 12/12 |
| 7 DPE MPV sentinel PCR | 0/16a | 0/16b | 3/4a,b |
| 21 DPE MPV sentinel sera | 0/16a | 0/16b | 3/4a,b |
| 9 DPI MHV cage swab | 16/16 | 16/16 | 4/4 |
| 10 DPI MHV index feces | 47/48 | 46/47 | 12/12 |
| 23 DPI MHV index sera | 45/48 | 48/48 | 12/12 |
| 7 DPE MHV sentinel PCR | 0/16a | 0/16b | 3/4a,b |
| 21 DPE MHV sentinel sera | 0/16a | 0/16b | 3/4a,b |
DPE, days postexposure to washed or unwashed cages; DPI, days postinoculation
Data are given as the number of samples positive for viral nucleic acids or antibodies / the number of samples tested.
Significant (P < 0.005) difference between unwashed and 180 °F washed cage groups.
Significant (P < 0.005) difference between unwashed and 110 °F washed cage groups
Testing of cages for viral contamination.
On day 9, 36 cages were swabbed, just prior to removal of the soiled bedding from the cages. MPV DNA was detected on all but one cage, and MHV RNA was detected on all cages (Table 1).
ABDC testing.
The presence of MPV and MHV in soiled bedding was confirmed by testing of dust and debris that was aerosolized during soiled bedding disposal and that was deposited on the ABDC prefilter. PCR-based testing of 4 samples of the dust and debris collected from the prefilter of the ABDC prior to soiled bedding disposal from the cages housing MPV- and MHV-infected mice, indicated the presence of MNV RNA and Helicobacter spp. DNA in all 4 samples but not MPV DNA or MHV RNA. This result was expected, because both MNV and Helicobacter spp. are endemic in this facility, whereas MPV and MHV are not. In contrast, PCR-based testing of 4 samples of the dust and debris present on the prefilter of the ABDC after soiled-bedding disposal from the cages housing MPV- and MHV-infected mice detected MHV and MNV RNA as well as MPV and Helicobacter spp. DNA. The detection of MHV RNA and MPV DNA in all postdumping samples indicates that the accumulation of MHV and MPV particles in aerosolized dust and debris from the 16 cages’ worth of soiled bedding was sufficient to be detectable by PCR.
Cage cleanliness and decontamination assessment.
The cleanliness of cages after washing at 110 or 180 °F was assessed objectively by measuring residual ATP levels on the cage bottom to determine whether washing at the lower temperature was less effective at removing organic material. The cage-wash temperature (110 °F compared with 180 °F) did not affect cage sanitation. Specifically, the ATP levels detected in the 2 groups of washed cages did not differ significantly, given that ATP levels of 1 to 64 (mean, 14) were detected in the 16 cages washed at 180 °F, and ATP levels of 1 to 99 (mean,16) were detected in 16 cages washed at 110 °F and was consistent with visual assessments in which no differences in cage cleanliness could be detected. In contrast, much higher ATP levels (7614; 42,358; greater than 99,999; and greater than 99,999) were detected on the 4 unwashed cages, which were visibly ‘dirty.’
Testing of SW sentinels exposed to washed and unwashed cages.
PCR analysis of feces collected from the 32 sentinel mice, 7 d after exposure to washed cages, were all negative for MPV DNA and MHV RNA, whereas PCR of feces collected from 3 of the 4 sentinel mice, 7 d after exposure to unwashed cages, were positive for MPV DNA and MHV RNA (Table 1). Similarly, all serologic samples from the 32 sentinel mice, 21 d after exposure to washed cages, were negative for MPV and MHV, whereas 3 of 4 sentinel mice, 21 d after exposure to unwashed cages, were MPV- and MHV-seropositive (Table 1). One of the unwashed cages did not transmit MPV or MHV, indicating that the viral load in this cage was below the transmission threshold. These results indicate that both 110 and 180 °F were effective at decontaminating soiled cages in which mice with active infections of MPV and MHV were housed.
Helicobacter spp. and pinworm contact-infection study.
The effect of water temperature during cage washing on decontamination of cages harboring common murine bacteria (Helicobacter spp.) and parasites (A. tetraptera and S. obvelata) of laboratory mice was determined. To generate the soiled cages to be washed, SW mice were infected with Helicobacter spp. and pinwormsby contact with pet-store mice. Then 16 soiled cages were washed at 180 °F, and 16 soiled cages were washed at 110 °F. The final 7 cages were dumped and scraped clean but not washed and therefore served as positive control cages to confirm that the amount of infectious agents present on the soiled cages was sufficient to infect the sentinel mice.
Infection and testing of mice used as sources for cage contamination.
Initial testing of pet-store mice.
Pet-store mice (n = 9) were tested, within 1 wk of arrival, by fecal and fur-swab PCR analysis and by visual observation of pools of feces. Infection with Helicobacter spp. was detected in all mice (H. bilis [n = 5 mice], H. typhlonius [n = 2], H. ganmani [n = 1], H. mastomyrinus [n = 1]). Coinfection with A. tetraptera and S. obvelata was detected in 2 mice, and infection with A. tetraptera alone was detected in 3 mice. In addition, pet-store mice also were infected with Myocoptes musculinus (n = 5 mice), Rodentolepis nana (n = 9), Hymenolepis diminuta (n = 5), lice (n = 2), MPV or MVM (n = 4), MHV (n = 5), and MAV (n = 3). Fecal PCR did not detect MNV, lymphocytic choriomeningitis virus, murine rotavirus, reovirus, or Theiler encephalomyelitis virus in the pet-store mice.
Antemortem testing of SW and pet-store mice.
PCR analysis of feces from individual mice (33 SW and 11 pet-store mice) on day 22 detected Helicobacter spp. DNA in 98% of mice, S. obvelata DNA in 93% of mice, and A. tetraptera DNA in 4.5% of mice. Fur-swab PCR analysis on day 23 detected Myocoptes musculinus DNA in 93% of mice and Myobia musculi DNA in 73% of mice. Fur-pluck observation on day 23 detected mite eggs on 36% of the mice and detected lice eggs on 41% of the mice.
Postmortem testing of SW mice exhibiting clinical disease.
PCR evaluation of lungs from the 21 SW mice that were euthanized between days 22 and 37 detected parvovirus in 100%, M. pulmonis in 82%, and Pasteurella pneumotropica in 23% of mice but not Pneumocystis murina,Mycobacteriaspp., cilia-associated respiratory bacillus, MHV, Sendai virus, or pneumonia virus of mice. Visual observation of cecal contents from the 12 SW mice euthanized on day 37 revealed all 12 of mice had S. obvelata, 5 had A. tetraptera, 5 had R. nana, 2 had Hymenolepis diminuta, and 2 had Entamoeba muris. PCR assessment of feces and cecal contents from the 12 SW mice euthanized on day 37 detected parvovirus and Helicobacter spp. in 100%, S. obvelata in 85%, MAV in 50%, and MHV in 41% of the mice.
Antemortem testing of SW and pet-store mice just prior to cage washing.
PCR analysis of fecal pools collected from each of the 39 soiled cages 1 d prior to cage washing on day 50, when 7 d of fecal accumulation was present in the cage, detected Helicobacter spp. DNA in 97% of cages, pinworm DNA in 46%, parvovirus DNA in 100%, MAV DNA in 5%, and MHV RNA in 77% (Table 2).
Table 2.
PCR analysis of pools of feces or swabs collected on the day prior to cage washing from cages housing Helicobacter spp. and pinworm contact-infection mice
| 180 °F wash | 110 °F wash | Unwashed | |
| Helicobacter fecal pool | 16/16 | 16/16 | 6/7 |
| Helicobacter cage swab | 14/16 | 14/16 | 5/7 |
| Pinworm fecal pool | 7/16 (7S/0A) | 10/16 (8S/2A) | 1/7 (1S/0A) |
| Pinworm cage swab | 11/16 (11S/0A) | 11/16 (11S/1A) | 5/7 (4S/1A) |
| Parvovirus fecal pool | 16/16 | 16/16 | 7/7 |
| Parvovirus cage swab | 15/16 | 14/16 | 7/7 |
| MAV fecal pool | 0/16 | 2/16 | 0/7 |
| MAV cage swab | 0/16 | 1/16 | 0/7 |
| MHV fecal pool | 16/16a,b | 11/16a | 3/7b |
| MHV cage swab | 0/16 | 2/16 | 0/7 |
Data are given as the number of samples positive for infectious agent nucleic acids / the number of samples tested (the number of. cages positive for Syphacia /the number of cages positive for Aspiculuris).
Significant (P < 0.05) difference between 110 °F and 180 °F washed-cage groups.
Significant difference between unwashed and 180F washed cage groups (P < 0.005).
Postmortem testing of contact-exposed SW and pet-store mice.
The 78 SW and pet-store mice were euthanized 4 to 5 d after cage washing on days 55 to 56, and serology, PCR, cecal observation, and tape tests were performed. Serology detected antibodies specific for MPV in 82%, MVM in 13%, MHV in 99%, MAV in 4%, and M. pulmonis in 61% of the mice (Table 3). PCR analysis performed on feces, ceca, or lungs detected nucleic acids specific for Helicobacter spp. in 97%, parvovirus in 26%, MHV in 47%, MAV in 7%, M. pulmonis in 44%, and S. obvelata in 92% of mice (Tables 3 and 4). Sequencing of Helicobacter PCR products revealed the presence of H. bilis, H. ganmani, and H. typhlonius. In addition, direct observation of cecal contents detected pinworms in 90% of mice, with S. obvelata detected in 76% of mice, A. tetraptera detected in 16% of mice, and pinworm larva of unknown species in 8% of mice (Table 4). In addition, cecal observation detected several other intestinal parasites (Entamoeba muris, 27%; trichomonads, 13%; R. nana, 13%; and Hymenolepis diminuta, 5%). PCR analyses of individual fecal pellets and tape tests were less sensitive than cecal methods (PCR analysis and observation) at detecting S. obvelata (P < 0.005; Table 4). Fur-swab PCR assay was more sensitive than fur pluck analysis for the detection of Myocoptes musculinus (P < 0.005; Table 4).
Table 3.
Serologic and PCR analysis of Helicobacter spp. and pinworm contact-infection mice 4 to 5 d after cage washing
| 180 °F wash | 110 °F wash | Unwashed | |
| MPV serology | 25/31 | 26/29 | 10/14 |
| MVM serology | 6/31 | 3/30 | 1/14 |
| Parvovirus fecal PCR | 9/31 | 7/28 | 3/14 |
| MHV serology | 30/30 | 30/30 | 13/14 |
| MHV fecal PCR | 16/31 | 15/28 | 3/14 |
| MAV serology | 0/31 | 3/30 | 0/14 |
| MAV fecal PCR | 1/31 | 4/28 | 0/14 |
| M. pulmonis serology | 19/30 | 18/30 | 8/14 |
| M. pulmonis lung PCR | 13/32 | 14/32 | 7/14 |
| Helicobacter cecal PCR | 31/32 | 31/32 | 14/14 |
| Helicobacter fecal PCR | 24/31 | 24/28 | 9/14 |
Data are given as the number of samples positive for infectious agent nucleic acids or antibodies / the number of samples tested
No significant difference was detected between groups for any agent
Table 4.
Testing for parasites in Helicobacter spp. and pinworm contact-infection mice 4 to 5 d after cage washing
| 180 °F wash | 110 °F wash | Unwashed | |
| Pinworm fecal PCR (Syphacia) | 8/31 | 7/28 | 1/14 |
| Pinworm cecal PCR (Syphacia) | 31/32 | 28/32 | 13/14 |
| Pinworm cecal observation | 20/24 (19S, 3A,1L) | 24/25 (18S, 6A,3L) | 12/13 (10S, 1A,1L) |
| Pinworm tape test (Syphacia) | 14/25 | 15/25 | 11/13 |
| Fur pluck observation (Myocoptes) | 13/24 | 15/25 | 7/13 |
| Fur swab PCR (Myocoptes) | 30/32 | 26/30 | 13/14 |
Data are given as the number of samples positive for parasites / the number of samples tested (the number of. mice positive for S. obvelata /the number of mice positive for A. tetraptera /the number of mice positive for pinworm larvae)
No significant difference was detected between groups for any agent
Testing of cages for bacterial, parasitic, and viral contamination.
We swabbed 39 cages on day 51, just prior to removal of soiled bedding from the cages. Helicobacter spp. DNA was detected on 85% of cages, pinworm DNA on 69%, and parvovirus DNA on 92% of cages (Table 2). MAV DNA was detected on a single cage and MHV RNA was detected on 2 cages in the 110 °F wash group but not on any of the cages in the 180 °F or unwashed cage groups (Table 2). The efficacy of fecal-pool PCR assay on day 50 (Table 2) and of cage-swab PCR analysis (Table 2) on day 51 was significantly different only for MHV (P < 0.005), with MHV detected less frequently on cages.
ABDC testing.
The presence of infectious agents in soiled bedding was confirmed by testing of dust and debris generated during the dumping of soiled bedding. Regarding indirect sample extraction, PCR-based testing of 3 samples of the dust and debris present on the prefilter of the ABDC prior to soiled bedding disposal from the cages housing infected mice indicated the presence of MNV RNA in 2 samples and Helicobacter spp. DNA in 3 samples but not MHV RNA or MAV, parvovirus, pinworm, or fur mite DNA. This result was expected, because both MNV and Helicobacter spp. are endemic in and are not excluded from the mouse colonies in this facility. PCR-based testing of 5 samples of the dust and debris present on the prefilter of the ABDC after dumping of soiled bedding from cages housing infected mice was performed by using the indirect extraction method to extract nucleic acids. Parvovirus DNA, MNV RNA, and Helicobacter spp. DNA were detected in all 5 samples, indicating that sufficient parvoviral particle accumulation on the prefilter occurred during soiled bedding dumping. MHV RNA and MAV DNA were not detected in any of the 5 samples collected from the ABDC after dumping of soiled bedding. Even though the majority of mice housed in cages were infested with pinworms and fur mites, DNA from these agents was not PCR-amplified from these prefilter samples. The 8 dust and debris samples (3 before and 5 after bedding disposal) had been extracted by adding 1 mL PBS to the 1.5-mL tube containing the debris, followed by adding 100 μL of the buffer–debris mixture to the lysis buffer (indirect extraction).
In an effort to determine whether direct extraction of the dust and debris on the prefilter (by adding the lysis buffer directly to the dust and debris sample) allowed for better detection of pinworms and mites on the prefilter, DNA was extracted from 5 samples of dust and debris by adding the lysis buffer directly to the sample. All 5 direct-extraction samples were positive for Helicobacter spp., parvovirus, M. pulmonis, S. obvelata, and Myocoptes musculinus DNA but not MAV DNA, indicating that direct extraction allowed for better detection of several infectious agents.
Sentinel testing data.
PCR assays of feces and ceca collected from the 32 sentinel mice exposed to cages washed at either 110 or 180 °F (31 d after exposure) were all negative for Helicobacter spp., pinworm, parvovirus, and MAV DNA and for MHV RNA (Table 5). Similarly, sera from the 32 sentinel mice exposed to washed cages (31 d after exposure) were all negative for MPV, MVM, MAV, and MHV antibodies (Table 5). In contrast, approximately half of the ceca from sentinel mice exposed to unwashed cages (31 d after exposure) were positive for parvovirus and S. obvelata DNA (Table 5). In contrast, parvovirus and S. obvelata DNA were detected in the feces of only one sentinel housed in an unwashed cage, underscoring the lower sensitivity of fecal PCR analysis to detect these 2 infectious agents (Table 5). Helicobacter spp., MVM, and MAV DNA and MHV RNA were not detected in the feces or ceca from sentinel mice exposed to unwashed cages (Table 5). Serology performed on the sentinel mice exposed to unwashed cages revealed a low level of infection by MVM and MHV and a higher level of infection by MPV (Table 5). Sera from all sentinels housed in washed or unwashed cages were negative for ectromelia virus, murine rotavirus, lymphocytic choriomeningitis virus, pneumonia virus of mice, reovirus, Sendai virus, Theiler encephalomyelitis virus, and M. pulmonis. Fur mites were not detected in any of the sentinels by fur pluck or by fur-swab PCR assay. The difference between the percentage of infected sentinels housed in washed compared with unwashed cages was significant (P < 0.005) only for MPV and pinworms, primarily because of inconsistent transmission of the other agents by unwashed (positive control) cages. No difference in transmission was seen between cages washed at 110 and 180 °F, indicating that both temperatures were effective at inactivating or removing MPV and pinworms.
Table 5.
Testing for Helicobacter and pinworms in contact-infection cage sentinels 31 d postexposure to cages
| 180 °F wash | 110 °F wash | Unwashed | |
| MPV serology | 0/16a | 0/16b | 5/7a,b |
| MVM serology | 0/16 | 0/16 | 1/7 |
| Parvovirus fecal PCR | 0/16 | 0/16 | 1/7 |
| Parvovirus cecal PCR | 0/16a | 0/16b | 4/7a,b |
| MVM fecal and cecal PCR | 0/16 | 0/16 | 0/7 |
| MHV serology | 0/16 | 0/16 | 1/7 |
| MHV fecal and cecal PCR | 0/16 | 0/16 | 0/7 |
| MAV serology | 0/16 | 0/16 | 0/7 |
| MAV fecal and cecal PCR | 0/16 | 0/16 | 0/7 |
| Helicobacter fecal PCR | 0/16 | 0/16 | 0/7 |
| Helicobacter cecal PCR | 0/16 | 0/16 | 0/7 |
| Pinworm fecal PCR | 0/16 | 0/16 | 1/7 |
| Pinworm cecal PCR | 0/16a | 0/16b | 5/7a,b |
| Pinworm cecal observation | 0/16a | 0/16b | 5/7a,b |
| Pinworm tape test | 0/16a | 0/16b | 5/7a,b |
Data are given as the number of samples positive for virus, Helicobacter spp., or S. obvelata / number of samples tested
Significant (P < 0.005) difference between unwashed and 180 °F washed cage groups
Significant (P < 0.005) difference between unwashed and 110 °F washed cage groups
Steam-use estimations.
Steam use was estimated for 2 representative tunnel washers to demonstrate potential cost savings of using 140 °F domestic hot water, compared with water at 180 to 200 °F, the industry standard. Because the steam is not currently metered directly at the washers, the manufacturer's data and operating schedule were used to estimate energy required to heat water.
Tunnel washer 1 (model TT27X22, Metalwash, Northwestern Systems) was manufactured in 1991. Calculations were based on washer operating time estimates of 6 h daily, 5 d each week, for 52 wk annually, and the manufacturer's water-use data of 12.5 gal/min (1,170,000 gal yearly). Steam is used to boost domestic hot water (approximately 140 °F) to a final rinse temperature of approximately 200 °F (∆T = 60 °F). We used 200 °F instead of 180 °F in the calculation because the control valve for this machine does not control to a specific set point; it controls according to steam flow, resulting in water within the wash and rinse tanks reaching more than 200 °F. The equation used to determine the steam energy (in 1 million British thermal units [MMBTU]) per year required to heat domestic hot water is:
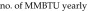 |
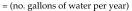 |
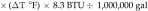 |
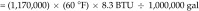 |
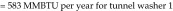 |
Note that additional steam is used to maintain approximately 50 gal of water within the wash tank and rinse tank at approximately 200 °F continuously (8760 h per year.) Although several reasonable engineering assumptions can be made to estimate the amount of steam use associated with maintaining the wash- and rinse-tank temperatures, it is more appropriate to meter this steam or solicit more detailed information from the manufacturer. No metering was performed in this analysis, so this additional steam use was not included in the savings estimates.
Tunnel washer 2 (model 3236LS, Getinge, Rochester NY) was manufactured in 2009. Calculations were based on washer operating-time estimates of 8 h daily, 5 d each week, 52 wk yearly (2080 h annually). Steam is used in the wash, initial rinse, final rinse, and dryer cycles at an average rate of 900 lb/h, according to the operating manual. Unlike those for tunnel washer 1, the manufacturer's data for this newer tunnel washer provided steam-supply specifications for life-cycle cost analysis. Therefore, steam is used at
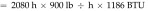 |
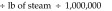 |
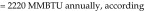 |
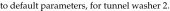 |
Cost-savings estimates from reduced steam use.
The potential savings is based on the estimated reduction in steam use and the current steam costs from the local utility ($14/MMBTU). For tunnel washer 1, the savings are based on the assumption that building-supplied domestic hot water would be used with no additional local steam heating. The potential savings is 583 MMBTU and US$8162 annually. Additional savings associated with ‘stand-by’ steam use and with energy costs to mitigate heat gain within the washing room can be assessed further. For tunnel washer 2, we assumed that steam use could be reduced by at least 75% with changes to the wash and rinse parameters. The total potential savings is 75% × 2220 MMBTU = 1665 MMBTU × $14/MMBTU = US$23,310 annually.
Discussion
Our hypothesis that cage washing alone at either 110 or 180 °F is sufficient to decontaminate soiled cages that had housed mice infected with viruses was confirmed. In the experimental infection study, 97% and 100% of cages had detectable levels of MPV and MHV, respectively, on their surfaces at the time of washing. MHV and MPV infection occurred in most sentinels exposed to the unwashed cages, whereas washing of cages at 110 °F or 180 °F resulted in inactivation or removal of MPV and MHV from all soiled cages that had housed mice acutely infected (9 d after inoculation) with these agents. In the second study, pet-store mice naturally infected with MPV infected most of the SW mice that were housed with them. Similar to the experimental MPV study, cage washing alone at either 110 or 180 °F was sufficient to inactivate or remove MPV from all soiled cages in the second study. The results of the MPV studies presented here combined with those of our previous study11 showed that none of the 46 washed cages (14 in previous study and 32 in current studies) transmitted MPV, whereas 8 of 11 of the unwashed cages in this study and all 14 of the unwashed cage bottoms in the previous study11 transmitted MPV. The ability of cage washing to remove or inactivate 2 very different viruses— MPV (a DNA virus that is very stable in the environment and difficult to inactivate)5,6,16,27,38,42,49 and MHV (an enveloped RNA virus that is less environmentally stable but is shed in high levels during acute infection3,9,20—suggests that cage washing is broadly effective at decontaminating cages that have housed mice infected with murine viruses.
Pinworms eggs, like MPV, are highly stable in the environment and difficult to inactivate with most disinfectants.13,32 Cage washing alone at either 110 or 180 °F inactivated or removed S. obvelata from all cages, suggesting that cage washing is effective at preventing infection with a wide range of murine infectious agents. Although more than half of the pet-store mice were infested with A. tetraptera, only 14% of SW mice became infested with A. tetraptera, and A. tetraptera DNA was detected on only 5% of cages prior to washing. In contrast, less than 25% of the pet-store mice were infested with S. obvelata, but more than 90% of the SW mice became infested with S. obvelata. This result is probably due to differences in the lifecycles of the 2 pinworm species. The duration of exposure of the second set of SW mice to the pet-store mice was only 21 d, to avoid the aforementioned clinical disease, which presumably was precipitated by M. pulmonis infection of the SW mice. This relatively short time frame favored S. obvelata infection, because S. obvelata eggs are released in a single burst of approximately 350 eggs, are infective 5 to 20 h after deposition on the perianal skin and hair, and can result in retrofection (migration of hatched larvae from the anus to the cecum of the mouse).7,41 In contrast, A. tetraptera eggs are released intermittently over a 3- to 4-wk period and are not infective for 5 to 8 d after excretion in the feces.2,23,34,41 In addition, A. tetraptera has a longer prepatent period (21 to 25 d) than does S. obvelata, which has a prepatent period of only 11 to 15 d.2,7,41
During the second study, the efficacies of several methods of pinworm detection were compared to determine the optimal sample to use for routine monitoring for pinworms in our facility. PCR and visual observation of cecal contents from SW mice were significantly better (Table 5, P < 0.01) at detecting pinworm infection (90% to 92%) than was testing of samples collected noninvasively (PCR of cage swabs, tape tests, PCR of fecal pools, and PCR of individual feces). These data agree with previous studies from our lab, which indicated that PCR and observation of cecal contents were equally effective at detecting pinworm infections.20 In light of our data, PCR of single fecal pellets collected directly from mice is not recommended as a reliable method for S. obvelata detection. The low sensitivity of the fecal PCR for Syphacia in our hands may be the result of inefficient DNA extraction, because others have suggested that inhibitors of pinworm PCR can be present in fecal DNA.17 Although only 22% of individual feces samples were positive for S. obvelata DNA on day 55 (Table 5), 37% of cages contained a mouse that was positive for S. obvelata DNA, indicating that most cages had only a single pinworm-positive mouse. Even though the pinworm PCR primers used were designed to amplify both S. obvelata and A. tetraptera, PCR assay of the cecal contents did not detect A. tetraptera, whereas observation of cecal contents detected A. tetraptera in 14% of the index mice. The majority of the SW contact mice infested with A. tetraptera (80%) were coinfested with S. obvelata, R. nana, Hymenolepis diminuta, Entamoeba muris, or trichomonads (or multiple parasites), whereas only 55% of mice infested with S. obvelata were also infested with these other parasites. Perhaps coinfection with these other parasites interfered with the amplification of A. tetraptera DNA but not S. obvelata DNA. Several studies have compared the various methods of pinworm detection, with divergent results.14,17,22 Further optimization of DNA extraction methods and amplification conditions is required before the sensitivities of the various pinworm-detection methods can be accurately compared in our laboratory.
Given that all 14 of the index mice housed in the unwashed cages were infected with Helicobacter spp. (Table 5) it is interesting to note that none of the sentinels placed in these cages became infected. This finding suggests that Helicobacter spp. are not readily transmitted by the residual waste present on unwashed cages. Several reports have shown efficient transmission of Helicobacter spp. to sentinel mice via exposure to soiled bedding,29,48 although the frequency of exposure and dose of soiled bedding were higher than those in our current study. Our results are more in line with 2 other studies, which reported that H. hepaticus, H. typhlonius, H. mastomyrinus, and H. muridarium were not transmitted to soiled-bedding sentinels.10,22
Although the contact-infection study was not intended to measure transmission of M. pulmonis, more than half of contact-exposed mice seroconverted to M. pulmonis, and lungs from almost half of the contact-exposed mice were positive for M. pulmonis DNA. We therefore assessed the transmission of M. pulmonis by soiled unwashed cages. Several mycoplasmal species, when dried on paper discs, have been stable for several weeks,33 and because lungs from 8 of the 14 contact-exposed mice in the unwashed cages were positive for M. pulmonis DNA, it would have been reasonable to expect that sentinels placed in unwashed cages would have become infected with M. pulmonis. But our results showed that M. pulmonis was not transmitted by unwashed cages. Our data are consistent with information presented in a recent study, which showed that M. pulmonis was not transmitted to soiled bedding or contact sentinels.22 It is important to note that although M. pulmonis was restricted to the lungs of infected mice and although M. pulmonis DNA was not detected on cage surfaces, at least one mycoplasmal species, which had 98% homology with M. moatsii19 and 99% homology with several uncultured bacterial species found in the jejunum of mice, was detected in the feces of most pet-store and SW mice and on cage surfaces. These findings underscore that testing for M. pulmonis in the environment by PCR assay should not be performed by using generic mycoplasmal primers.
Although Myocoptes musculinus was detected by PCR in 91% of the contact exposed pet-store and SW mice, it was not transmitted to sentinels housed in unwashed cages. These data are consistent with several previous studies, which have shown that soiled bedding transmission of fur mites to sentinels occurs infrequently.20,22,28 But 2 other studies37,43 have shown efficient transmission by soiled bedding to sentinel mice in 8 to 19 wk. The mice in the first study were infested with both Myobia musculi and Myocoptes musculinus, and transmission occurred to all cages, but the fur-mite species that was transmitted is unclear.37 In the second study, mice were housed in open-top cages that received soiled bedding from other open-top cages within the room that housed mice infested with Myobia musculi, and transmission occurred in 3 of 4 cages.43 To our knowledge, our current study is the first to evaluate fomite-based transmission of fur mites from mice infested with Myocoptes musculinus in the absence of Myobia musculi infestation.
The number and type of infectious agents present in pet-store mice and the potential for some of these agents to be spread by fomites prompt biosecurity concerns when animal care and laboratory personnel have pet-store mice as pets at home. Of particular concern is that these ‘clinically normal’ pet-store mice transmitted fatal disease to naive SW mice, albeit by direct contact. Nevertheless, these findings underscore the risk of pet-store mice and supports having a facility rodent-adoption program so that animal care and laboratory personnel wishing to acquire pet mice can officially do so through a pathogen-free source.
Early detection of an infectious agent can often lessen its overall effect on an animal program. Unfortunately, many routine sentinel-exposure protocols rely on periodic sampling of and exposure to soiled bedding within a housing rack, and therefore positive sentinel findings may represent an infection that began weeks to months previously. Environmental sampling is a promising adjunct to sentinel-exposure programs for early pathogen detection. Realizing that testing of individual ventilated cage rack filters is one environmental screening approach, we tested a complementary approach: the ability to reliably detect pathogens from an ABDC, a site where soiled bedding is concentrated and aerosolized. Because the ABDC serves large portions of a facility, the ability to reliably detect ‘excluded’ pathogens from the ABDC would allow for screening of large portions of a facility by using a few filter samples. Therefore, we assessed whether PCR-based testing of the accumulated dust-debris present on the prefilter of the ABDC after disposal of the soiled bedding from cages housing experimentally infected sentinel mice detected both MHV and MPV particles (diameter, 0.028 to 0.160 μm).1 Helicobacter DNA was detected in all samples of dust and debris present on the prefilter of the ABDC (both pre- and postdisposal samples from cages housing infected sentinel mice in both studies), indicating the accumulation of Helicobacter spp. (0.2 to 0.3 μm × 1.5 to 5 μm)18 during the routine dumping of cages from endemically infected cages within this animal facility.
In the contact-infection study, an initial PCR-based attempt to detect pinworms and fur mites in samples of debris from the dump station prefilter after the disposal of soiled bedding was unsuccessful when the DNA was extracted indirectly from the sample, even though the majority of mice housed in the represented cages were infested with pinworms and fur mites. We initially thought that the larger size of these parasites (pinworms eggs are 30 to 40 μm × 90 to 140 μm)41 led to inefficient aerosolization of these agents or parts of these agents during soiled bedding disposal. But a recent study showed that fur-mite DNA can be detected on swabs from the shelf of a horizontal manifold of an IVC rack housing a single cage of fur mite infested mice one week after placement of the cage on the rack indicating that aerosolization of detectable levels of debris containing fur mite components occurred.26 Follow-up testing of samples of prefilter dust and debris by using direct DNA extraction revealed that pinworms and fur mites were present in all of the samples of dust and debris. It seems that smaller agents such as viruses and Helicobacter spp. are readily washed from the debris; therefore indirect extraction resulted in a sufficient quantity of nucleic acids for detection by PCR analysis. In comparison, the larger infectious agents such as pinworms and fur mites were not readily transferred from the debris to the PBS during the indirect extraction procedure, possibly because these agents were too large to be easily drawn into the pipettor tip or because they were tightly associated with the debris (for example, fur mites attached to fur would not be transferred). Because debris samples collected prior to soiled-bedding dumping were extracted by using the indirect method, we perhaps would not have detected any pinworms or fur mites present on the prefilter because the predumping samples were extracted by using the indirect method. Because pinworms and fur mites have not been detected in the facilities served by this ABDC for over 9 mo, we concluded that the S. obvelata and Myocoptes musculinus DNA detected in the postdumping samples was deposited during dumping of the cages in this experiment.
PCR-based testing of the prefilter of the dump station, by using direct nucleic acid extraction, shows promise for murine infectious agents screening at the facility level when more than 10 cages housing mice infected with a virus, bacteria, or parasite are dumped. However, MHV and MAV, which were detected on only a few cages in the second study, were not detected in the debris present on the prefilter of the ABDC after soiled-bedding disposal, indicating that a threshold level of contaminated bedding needs to be present on the ABDC prefilter. Further studies to determine the sensitivity of this testing paradigm are needed.
Taken together, our studies demonstrate that cage decontamination does not require washing at 180 °F and that autoclaving of washed cages is unnecessary. Rather, cage washing at 110 °F with appropriate detergents and cycle times was sufficient to decontaminate cages that had housed mice infected or infested with common murine infectious agents— thereby representing a significant advancement to the ‘greening’ of vivarium operation. The effectiveness of washing at 110 °F is an important finding in the context of sustainability and the need for steam infrastructure. For example, the energy usage when 140 °F domestic hot water is used to wash caging rather than water steam boosted to 200 °F was calculated to result in a savings of 583 MMBTU when a 1991 Metalwash Tunnel Washer was operated for 1560 h annually, for a cost savings of approximately $8000 annually. Similarly, 1665 MMBTU can be saved by operating a newer but larger Getinge Tunnel Washer at 140 °F for 2080 h yearly, yielding a cost savings of approximately $23,000 each year. Additional energy savings might be achieved by scheduling cage washing to minimize the amount of time during which the tunnel washer is running but not actively washing caging and by factoring in the decrease in energy needed to mitigate the heat gain within the room so that room temperature and humidity are acceptable to personnel working in the wash facilities. Apart from the cost of the energy required to boost domestic hot water to 180 °F, the boiler required to supply the steam necessary to heat the volume of the water necessary may not be available in the building or nearby, resulting in significant increased construction or renovation costs. However, detailed manufacturer steam requirements are critical to understand the full range of savings associated with washer operating parameters, particularly requirements for stand-by modes maintaining temperature settings both during and after active use. Direct steam metering is not trivial in these settings but is required for more precise energy cost-savings estimates.
Acknowledgments
This work was supported by Grants for Laboratory Animal Science (GLAS) from AALAS. We thank Alison Faruolo for assistance with animal care and viral diagnostics and Fu-Chen Yang for assistance with parasitology. We thank Marco Garcia, Eric Georgelos, and Julie Paquette for assistance with cage washing and cage-wash energy calculations.
References
- 1.Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossman MG. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369–1381. [DOI] [PubMed] [Google Scholar]
- 2.Anya AO. 1966. Studies on the biology of some oxyurid nematodes. Part II. The hatching of eggs and development of Aspiculuris tetraptera Schulz, within the host. J Helminthol 40:261–268. [DOI] [PubMed] [Google Scholar]
- 3.Barthold SW, Beck DS, Smith AL. 1993. Enterotropic coronavirus (mouse hepatitis virus) in mice: influence of host age and strain on infection and disease. Lab Anim Sci 43:276–284. [PubMed] [Google Scholar]
- 4.Besselsen DG, Wagner AM, Loganbill JK. 2000. Effect of mouse strain and age on detection of mouse parvovirus 1 by use of serologic testing and polymerase chain reaction analysis. Comp Med 50:498–502. [PubMed] [Google Scholar]
- 5.Boschetti N, Niederhauser I, Kempf C, Stuhler A, Lower J, Blumel J. 2004. Different susceptibility of B19 virus and mice minute virus to low pH treatment. Transfusion 44:1079–1086. [DOI] [PubMed] [Google Scholar]
- 6.Boschetti N, Wyss K, Mischler A, Hostettler T, Kempf C. 2003. Stability of minute virus of mice against temperature and sodium hydroxide. Biologicals 31:181–185. [DOI] [PubMed] [Google Scholar]
- 7.Chan KF. 1952. Life-cycle studies on the nematode Syphacia obvelata. Am J Hyg 56:14–21. [DOI] [PubMed] [Google Scholar]
- 8.Compton SR. 2008. Prevention of murine norovirus infection in neonatal mice by fostering. J Am Assoc Lab Anim Sci 47: 25–30. [PMC free article] [PubMed] [Google Scholar]
- 9.Compton SR, Ball-Goodrich LJ, Paturzo FX, Macy JD. 2004. Transmission of enterotropic mouse hepatitis virus from immunocompetent and immunodeficient mice. Comp Med 54:29–35. [PubMed] [Google Scholar]
- 10.Compton SR, Homberger FR, Paturzo FX, Clark JM. 2004. Efficacy of 3 microbiological monitoring methods in a ventilated cage rack. Comp Med 54:382–392. [PubMed] [Google Scholar]
- 11.Compton SR, Paturzo FX, Smith PC, Macy JD. 2012. Transmission of mouse parvovirus by fomites. J Am Assoc Lab Anim Sci 51:775–780. [PMC free article] [PubMed] [Google Scholar]
- 12.Cundiff DD, Besch-Williford C, Hook RR, Jr, Franklin CL, Riley LK. 1994. Detection of cilia-associated respiratory bacillus by PCR. J Clin Microbiol 32:1930–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dix J, Astill J, Whelan G. 2004. Assessment of methods of destruction of Syphacia muris eggs. Lab Anim 38:11–16. [DOI] [PubMed] [Google Scholar]
- 14.Dole VS, Zaias J, Kyricopoulos-Cleasby DM, Banu LA, Waterman LL, Sanders K, Henderson KS. 2011. Comparison of traditional and PCR methods during screening for and confirmation of Aspiuluris tetraptera in a mouse facility. J Am Assoc Lab Anim Sci 50:904–909. [PMC free article] [PubMed] [Google Scholar]
- 15.Donaho JC. 2012. The green cage wash of 2012 and beyond. J Am Assoc Lab Anim Sci 51:648. [Google Scholar]
- 16.Eterpi M, McDonnell G, Thomas V. 2009. Disinfection efficacy against parvoviruses compared with reference viruses. J Hosp Infect 73:64–70. [DOI] [PubMed] [Google Scholar]
- 17.Feldman SH, Bowman SG. 2007. Molecular phylogeny of the pinworms of mice, rats, and rabbits, and its use to develop molecular-beacon assays for the detection of pinworms in mice. Lab Anim (NY) 36:43–50. [DOI] [PubMed] [Google Scholar]
- 18.Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, Collins MJ, Jr, Gorelick PL, Ward JM. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol 32:1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giebel J, Binder A, Kirchhoff H. 1990. Isolation of Mycoplasma moatsii from the intestine of wild Norway rats (Rattus norvegicus). Vet Microbiol 22:23–29. [DOI] [PubMed] [Google Scholar]
- 20.Grove KA, Smith PC, Booth CJ, Compton SR. 2012. Age-associated variability in susceptibility of Swiss Webster mice to MPV and other excluded murine pathogens. J Am Assoc Lab Anim Sci 51:789–796. [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashimoto N, Ueno M, Takakura A, Itoh T. 2007. A specific polymerase chain reaction based on the gyrB gene sequence and subsequent restriction fragment length polymorphism analysis of Pasteurella pneumotropica isolates from laboratory mice. J Am Assoc Lab Anim Sci 46:54–58. [PubMed] [Google Scholar]
- 22.Henderson KS, Perkins CL, Havens RB, Kelly ME, Francis BC, Dole VS, Shek WR. 2013. Efficacy of direct detection of pathogens in naturally infected mice by using a high-density PCR array. J Am Assoc Lab Anim Sci 52:763–772. [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh KY. 1952. The effect of the standard pinworm chemotherapeutic agents on the mouse pinworm Aspiculuris tetraptera. Am J Hyg 56:287–293. [PubMed] [Google Scholar]
- 24.Institute for Laboratory Animal Research 2011. Guide for the care and use of laboratory animals, 8th ed. Washington (DC): National Academies Press. [Google Scholar]
- 25.Janus LM, Bleich A. 2012. Coping with parvovirus infections in mice: health surveillance and control. Lab Anim 46:14–23. [DOI] [PubMed] [Google Scholar]
- 26.Jensen ES, Allen KP, Henderson KS, Szabo A, Thulin JD. 2013. PCR testing of a ventilated caging system to detect murine fur mites. J Am Assoc Lab Anim Sci 52:28–33. [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, Purdy GA, Riley LK, Livingston RS. 2007. Efficacy of disinfectants against MVM- and MNV-contaminated surfaces. J Am Assoc Lab Anim Sci 46:94. [Google Scholar]
- 28.Lindstrom KE, Carbone LG, Kellar DE, Mayorga MS, Wilkerson JD. 2011. Soiled-bedding sentinels for the detection of fur mites in mice. J Am Assoc Lab Anim Sci 50:54–60. [PMC free article] [PubMed] [Google Scholar]
- 29.Livingston RS, Riley LK, Besch-Williford CL, Hook RR, Jr, Franklin CL. 1998. Transmission of Helicobacter hepaticus infection to sentinel mice by contaminated bedding. Lab Anim Sci 48:291–293. [PubMed] [Google Scholar]
- 30.Macy JD, Cameron GA, Smith PC, Ferguson TA, Compton SR. 2011. Detection and control of mouse parvovirus. J Am Assoc Lab Anim Sci 50:516–522. [PMC free article] [PubMed] [Google Scholar]
- 31.Macy JD, Paturzo FX, Ball-Goodrich LJ, Compton SR. 2009. A PCR-based strategy for detection of mouse parvovirus. J Am Assoc Lab Anim Sci 48:263–267. [PMC free article] [PubMed] [Google Scholar]
- 32.Miyaji S, Kamiya M, Shikata J. 1988. Ovicidal effects of heat and disinfectants on Syphacia muris estimated by in vitro hatching. Jikken Dobutsu 37:399–404. [PubMed] [Google Scholar]
- 33.Nagatomo H, Takegahara Y, Sonoda T, Yamaguchi A, Uemura R, Hagiwara S, Sueyoshi M. 2001. Comparative studies of the persistence of animal mycoplasmas under different environmental conditions. Vet Microbiol 82:223–232. [DOI] [PubMed] [Google Scholar]
- 34.Phillipson RF. 1974. Intermittent egg release by Aspiculuris tetrapterain mice. Parasitology 69:207–213. [DOI] [PubMed] [Google Scholar]
- 35.Pritchett-Corning KR, Cosentino J, Clifford CB. 2009. Contemporary prevalence of infectious agents in laboratory mice and rats. Lab Anim 43:165–173. [DOI] [PubMed] [Google Scholar]
- 36.Reuter JD, Livingston R, Leblanc M. 2011. Management strategies for controlling endemic and seasonal mouse parvovirus infection in a barrier facility. Lab Anim (NY) 40:145–152. [DOI] [PubMed] [Google Scholar]
- 37.Ricart Arbona RJ, Lipman NS, Wolf FR. 2010. Treatment and eradication of murine fur mites. Part II. Diagnostic considerations. J Am Assoc Lab Anim Sci 49:583–587. [PMC free article] [PubMed] [Google Scholar]
- 38.Sauerbrei A, Wutzler P. 2009. Testing thermal resistance of viruses. Arch Virol 154:115–119. [DOI] [PubMed] [Google Scholar]
- 39.Silverman J, Chavannes J, Zwarun AA. 1981. Does 180 °F kill bacteria on rat and mouse cages? Lab Anim 10:67–68. [Google Scholar]
- 40.Smith AL, Singleton GR, Hansen GM, Shellam G. 1993. A serologic survey for viruses and Mycoplasma pulmonis among wild house mice (Mus domesticus) in southeastern Australia. J Wildl Dis 29:219–229. [DOI] [PubMed] [Google Scholar]
- 41.Taffs LF. 1976. Pinworm infections in laboratory rodents: a review. Lab Anim 10:1–13. [DOI] [PubMed] [Google Scholar]
- 42.Terpstra FG, van den Blink AE, Bos LM, Boots AG, Brinkhuis FH, Gijsen E, van Remmerden Y, Schuitemaker H, van't Wout AB. 2007. Resistance of surface-dried virus to common disinfection procedures. J Hosp Infect 66:332–338. [DOI] [PubMed] [Google Scholar]
- 43.Thigpen JE, Lebetkin EH, Dawes ML, Amyx HL, Caviness GF, Sawyer BA, Blackmore DE. 1989. The use of dirty bedding for detection of murine pathogens in sentinel mice. Lab Anim Sci 39:324–327. [PubMed] [Google Scholar]
- 44.Wakefield AE, Pixley F, Banerji S, Sinclair K, Miller R, Moxon E, Hopkin J. 1990. Detection of Pneumocystis carinii with DNA amplification. Lancet 336:451–453. [DOI] [PubMed] [Google Scholar]
- 45.Wardrip CL, Artwohl JE, Oswald J, Bennett BM. 2000. Verification of bacterial killing effects of cage wash time and temperature combinations using standard Penicylinder methods. Contemp Top Lab Anim Sci 39:9–12. [PubMed] [Google Scholar]
- 46.Wardrip CL, Artwohl J, Bennett B. 1994. A review of the role of temperature versus time in an effective cage-sanitation program. Contemp Top Lab Anim Sci 33:66–68. [PubMed] [Google Scholar]
- 47.Watson J. 2011. Unsterilized feed: the probable cause of sporadic mouse parvovirus outbreaks. J Am Assoc Lab Anim Sci 50:771–772. [PMC free article] [PubMed] [Google Scholar]
- 48.Whary MT, Cline JH, King AE, Hewes KM, Chojnacky D, Salvarrey A, Fox JG. 2000. Monitoring sentinel mice for Helicobacter hepaticus, H. rodentium, and H. bilis infection by use of polymerase chain reaction analysis and serologic testing. Comp Med 50:436–443. [PubMed] [Google Scholar]
- 49.Yang FC, Paturzo FX, Jacoby RO. 1995. Environmental stability and transmission of rat virus. Lab Anim Sci 45:140–144. [PubMed] [Google Scholar]


