Abstract
Background
Brahma-related gene 1 (BRG1) has been implicated in a variety of biological processes, and it has been found to be mutated or silenced in numerous cancers, including lung cancer. Recent reports have proposed BRG1 as a tumor suppressor, but its roles in cell proliferation and metastasis remain unknown. miR-148b functions as a tumor suppressor in non-small-cell lung cancer. However, the mechanism responsible for the downregulation of miR-148b in lung cancer is still elusive.
Methods
The expression of BRG1 and miR-148b was evaluated in lung cancer tissues and cells using quantitative real-time polymerase chain reaction. The effect of BRG1 on proliferation of lung cancer cells was investigated using MTT assay. Transwell and Western blot assays were used to analyze the effect of BRG1 on invasion and epithelial–mesenchymal transition (EMT), respectively. The target of miR-148b was ascertained using luciferase reporter assay. Chromatin immunoprecipitation (ChIP) assay was performed to analyze the relation of BRG1 and the promoter region of miR-148b.
Results
Restoration of BRG1 was demonstrated to inhibit cell proliferation, metastasis, and EMT in lung cancer cell lines. Furthermore, we found that miR-148b was positively regulated by BRG1. Additionally, we suggested that miR-148b suppressed cell proliferation, metastasis, and EMT in lung cancer cells by directly binging to 3′-untranslated region of WNT1, blocking the WNT1/β-catenin signaling pathway. ChIP assay showed that BRG1 bound to the promoter of miR-148b in A549 cells.
Conclusion
BRG1 positively regulated the expression of miR-148b, leading to inhibition of cell proliferation, metastasis, restraint of EMT, and inactivation of the WNT/β-catenin signaling pathway, which highlights potential therapeutic possibilities for the treatment of lung cancer.
Keywords: Brahma-related gene 1 (BRG1), proliferation, metastasis, miR-148b, WNT/β-catenin signaling pathway
Introduction
Mammalian switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complexes contain either BRG1 or BRM exclusively as the catalytic ATPase subunit that control the alteration of DNA–nucleosome structure and thus modulate target gene transcription.1,2 The loss of BRG1 subunit disrupts the complex and inactivates its function, which is associated with a variety of malignancies, including ovarian, renal, liver, and pediatric cancers.3,4 Brahma-related gene 1 (BRG1) was the fourth most frequently altered gene in non-small-cell lung cancer (NSCLC) cell lines and low BRG1 protein level have also been observed in clinical lung primary tumors.5–7 Restoration of wild-type BRG1 expression activated massive transcription in lung cancer cells and significantly inhibited invasion and growth of lung cancer cells orthotopically implanted in nude mice.6,8 However, the regulatory mechanism continues to be obscure.
MicroRNAs (miRNAs) are a class of single-stranded, small noncoding RNAs measuring 18–25 nucleotides in length that guide the cleavage of their target messenger RNAs (mRNAs) or bind to 3′-untranslated region (3′-UTR) of their recognition sequences of mRNA, causing either mRNA degradation or translational repression, thus effectively silencing their mRNA targets. Each miRNA potentially regulates numerous target mRNAs, and it is predicted that more than one-third of protein-coding genes are regulated by miRNAs.9,10 Recently, miRNAs have emerged as posttranscriptional regulators that are implicated in multiple cell biological processes, including cell differentiation, proliferation, metastasis, apoptosis, metabolism, tumorigenesis, and cancer progression.11,12 There is accumulating evidence that miRNAs show aberrant expressions in cancers and modulate cancer cell behaviors.13,14
miR-148b is frequently downregulated in several types of cancers, such as pancreatic cancer,15 gastric cancer,16 breast cancer,17 esophageal squamous cell carcinoma,18 and colorectal cancer.19 miR-148b functions as a new tumor suppressor via directly targeting specific protein-coding genes in these cancers. Moreover, miR-148b increases the radiosensitivity of non-Hodgkin’s lymphoma cells probably by enhancing ionizing radiation-induced apoptosis.20 In pancreatic cancer cell lines, upregulation of miR-148b returns DNA methylation to normal patterns and promotes re-expression of tumor suppressor genes, like BNIP3 and SPARC, through suppression of DNA methyltransferase-1 (DNMT-1) expression.21 miR-148b also plays a vital role in maintenance of a side population cells with cancer stem cell properties through regulating Neuropilin-1 in hepatocellular cancer.22 The latest report showed that miR-148b was downregulated in NSCLC, and aberrant miR-148b expression was significantly associated with the clinicopathological characteristics, including TNM stage, lymph node metastasis, distant metastasis, and worse overall survival rates.23 Liu et al24 found that miR-148b was downregulated in NSCLC cells, which resulted in carcinoembryonic antigen overexpression and disease progression in patients with NSCLC. These data show that miR-148b plays a critical role in regulating these physiological events, but the underlying mechanism accounting for the downregulation of miR-148b in human cancers, especially in lung cancer, remains unclear.
In this study, we confirmed that BRG1 and miR-148b were expressed at low levels in lung cancer tissues and cells and observed the correlation between BRG1 and miR-148b. Furthermore, we investigated the effects of restoration of BRG1 on the proliferation and metastasis of lung cancer cells and the potential mechanisms for lung cancer behavior affected by BRG1/miR-148b. These findings will help to better understand the roles of BRG1 and miR-148b in the progression of lung cancer.
Materials and methods
Clinical specimens
Surgically resected lung cancer tissues and matched adjacent normal lung tissues were collected from 29 lung cancer patients during operation at the Second Affiliated Hospital of Zhengzhou University. Exclusion criteria were presence of chemotherapy or radiotherapy prior to lung resection. All specimens were stored at −80°C until analysis. Written informed consent for the studies was obtained from all participants and their guardians. This study was approved by the Ethics Committee of the Second Affiliated Hospital of Zhengzhou University.
Cell culture and treatment
Human lung cancer cell lines and lung cell lines were obtained from the American Type Culture Collection (ATCC, Rock-ville, MD, USA). A549, H522, and H1299 were grown in Roswell Park Memorial Institute (RPMI) 1640 with 10% fetal calf serum, 5 mM L-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin, and maintained in a humidified incubator at 37°C containing 5% CO2. The normal lung cell lines (WI38 and MRC5) and the human bronchial epithelial cell line (Beas-2B) were cultured as described previously.25 For evaluation of epithelial–mesenchymal transition (EMT), cells were treated with 4 ng/mL of TGFβ1 (R&D Systems, Minneapolis, MN, USA) for 48 hours. Then the relative expression levels of E-cadherin, N-cadherin, and vimentin in cells were detected by using Western blot analysis.
Quantitative real-time PCR
Total mRNA from tissues and cells was extracted using Unizol Reagent (Biostar, Shanghai, People’s Republic of China) according to the manufacturer’s instructions. Total RNAs were reverse transcribed into cDNA using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). Expression level of miR-148b was quantified using a miRNA-specific TaqMan MiRNA Assay Kit (Applied Biosystems) according to the manufacturer’s instructions. For the amplification of BRG1 and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) mRNA, the primers were synthesized as follows: BRG1 (forward: 5′-CAT CAT CGT GCC TCT CTC AAC-3′; reverse: 5′-ACA CGC ACC TCG TTC TGC TG-3′) and GAPDH (forward: 5′-CAC GAT GGA GGG GCC GGA CTC ATC-3′; reverse: 5′-TAA AGA CCT CTA TGC CAA CAC AGT-3′). Relative levels of mRNA (amount of target gene normalized to GAPDH) were calculated using the comparative Ct method formula 2−∆∆Ct.
Chromatin immunoprecipitation assay
For the chromatin immunoprecipitation (ChIP) assay, chromatin was prepared from A549 cells. A549 cells were treated with 10% formaldehyde for 10 minutes at room temperature, washed with phosphate buffered saline (PBS), and resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 40 mM ethylenediaminetetraacetic acid (EDTA), 0.5% SDS). Chromatin was prepared according to standard protocols and sheared to fragments in a Bioruptor (Diagenode, Philadelphia, PA, USA) by three sonication cycles of 5 minutes. The sheared chromatin was then immunoprecipitated by using anti-BRG1 antibody (Abcam, Cambridge, UK) or control IgG (Sigma, St. Louis, MO, USA). After extensive washing and elution, crosslinks were reverted by adjusting NaCl concentrations and heat treatment. Precipitated DNA fragments were purified by using PCR purification kit (Qiagen, Valencia, CA, USA). Real-time PCR analysis was performed for BRG1 binding sites in miR-148b promoter by using the following primers: 5′-AGC GCC AGT GTT AAA GGC TA-3′ (forward) and 5′-TCC ATG GGG AAC AGA AGA AG-3′ (reverse).
Western blot analysis
Protein extraction and Western blot analysis were performed. Briefly, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes. After blocking, the filters were incubated with BRG1 (Abcam), Ki67 (Abcam), WNT1 (Cell Signaling Technology, Beverly, MA, USA), β-catenin (Abcam), c-myc (Cell Signaling Technology), E-cadherin (Santa Cruz Biotech, Santa Cruz, CA, USA), N-cadherin (Santa Cruz Biotech), vimentin (Santa Cruz Biotech), and GAPDH (Sigma, USA) antibodies at 4°C overnight. After washing and incubation with horseradish peroxidase-conjugated secondary antibodies for 1 hour, blots were visualized by using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA). Detection of GAPDH on the same membrane was used as a loading control.
Transfection
A549 and H522 cells (1×105 per well) were maintained in six-well plates and incubated for 24 hours. Cells were transfected with 50 nM of miR-148b mimics or control mimics (miR-control), 100 nM miR-148b inhibitors (anti-148b, with sequence of 5′-ACA AAG UUC UGU GAU GCA CUG A-3′) or control inhibitors (anti-ctrl), and 100 nM specific siRNA of the human BRG1 gene or control siRNA (si-control) with lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Transfection efficiency was evaluated by qRT-PCR or Western blot analysis. A549 cells were transfected with a retrovirus encoding shRNA sequence specific for WNT1 (shRNA-WNT1) or negative control sequence (shRNA-control). Approximately 10 µg of BRG1 expression plasmids (pcDNA3.1-3× Flag-BRG1) or empty vectors were transfected into A549 and H522 cells with Lipofectamine Plus (Invitrogen), according to the manufacturer’s instructions.
MTT assay
Following transfection with BRG1 vector (or cotransfection with BRG1 or control vector and anti-miR-148b mimics or anti-miR-control), MTT assay was used to assess relative cell viability of A549 and H522 cells, as described previously.26 The absorbance values were determined at a wavelength of 490 nm using a microplate spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
Cell migration assay
Cell migration was measured by using the Transwell assay. Briefly, A549 and H522 cells (2×104) transfected with vector containing BRG1 or control (or cotransfected with vector containing BRG1 or control and anti-miR-148b mimics or anti-miR-control) were seeded in serum-free medium in top chambers (BD Bioscience, Franklin Lakes, NJ, USA), whereas in the lower compartments Dulbecco’s Modified Eagle’s Medium containing 5% fetal bovine serum was added. After 20 hours of incubation, the nonmigrated cells in the top chambers were removed by a cotton swab. The number of A549 cells adhering to the lower membranes was counted after cells were fixed and stained with crystal violet.
Luciferase activity assay
A549 cells (2×104) in a 48-well plate were cotransfected with 10 nM miR-148b mimics or mimic control, 2 ng pRL-TK (Promega, Madison, WI, USA). Approximately 10 ng firefly luciferase reporter containing the wild-type or mutant 3′-UTR of WNT1 pRL-TK, which expresses Renilla luciferase, was used as the internal control in the dual-luciferase assay. Forty-eight hours after transfection, the luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
Statistics analysis
The data were expressed as the mean ± standard deviation from at least three independent experiments. Statistical significance between two groups was evaluated using Student’s t-test. All statistical tests were two sided, and a P-value <0.05 was considered to be statistically significant.
Results
BRG1 and miR-148b levels are significantly lower in lung cancer tissues and cell lines
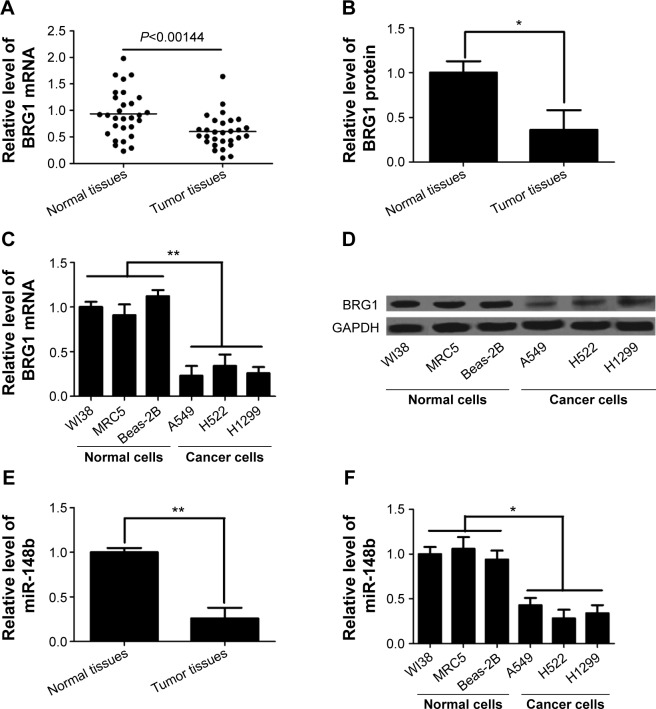
It has been reported that dysregulated expression of BRG1 is frequently present in lung cancer and that tumor progression and poor prognosis are greatly affected by downregulation of BRG1.27,28 Here, we confirmed the levels of BRG1 in lung cancer tissues and cell lines by using qRT-PCR and Western blot analysis. As shown in Figure 1A and B, the expression levels of BRG1 mRNA and protein in cancer lung tissues were obviously lower than those in matched adjacent normal lung tissues. In addition, lung cancer cell lines (A549, H522, and H1299) showed reduced BRG1 mRNA and protein levels compared with normal lung cells (WI38, MRC5, and Beas-2B; Figure 1C and D). Low expression of miR-148b has been found in NSCLC, and miR-148b functions as a tumor suppressor by directly targeting specific oncogenes.24 Here, we confirmed that the expression of miR-148b was decreased in lung cancer tissues and cell lines (Figure 1E and F). Collectively, these cumulative data verify that lower BRG1 and miR-148b expression levels frequently occur in human lung cancer.
Figure 1.
BRG1 and miR-148b are expressed at low levels in lung cancer tissues and cell lines.
Notes: (A) BRG1 mRNA level is lower in lung cancer tissues than that in adjacent normal lung tissues. (B) The expression level of BRG1 protein is decreased in lung cancer tissues compared to that in adjacent normal lung tissues. (C) BRG1 mRNA levels in lung cancer cell lines A549, H522, and H1299 are lower than those in lung cell lines WI38, MRC5, and Beas-2B. (D) BRG1 protein levels in lung cancer cell lines A549, H522, and H1299 are lower than those in lung cell lines WI38, MRC5, and Beas-2B. (E and F) miR-148b expression level is significantly decreased in lung cancer tissues and cells. *P<0.05, **P<0.01.
Abbreviations: BRG1, Brahma-related gene 1; mRNA, messenger RNA; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
BRG1 binds to miR-148b target gene promoter in lung cancer cells
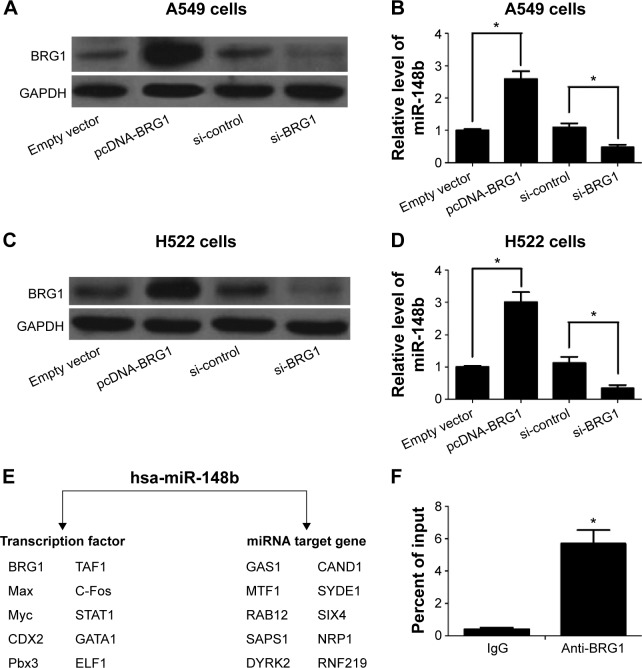
We further found that the expression of miR-148b correlates with the levels of BRG1 in A549 and H522 cells. As shown in Figure 2A–D, overexpression of BRG1 elevated the expression of miR-148b in A549 and H522 cells. In contrast, the expression levels of miR-148b were decreased when BRG1 was knocked down. The online platform ChIPBase shows that BRG1 is a predicted transcription factor of miR-148b (Figure 2E). To determine whether BRG1 binds to miR-148b promoter, ChIP assay was conducted using anti-BRG1 antibody in A549 cells. As shown in Figure 2F, BRG1 showed a high capacity to bind to the promoter region of miR-148b compared with control IgG.
Figure 2.
BRG1 binds to miR-148b target gene promoter in lung cancer cells.
Notes: (A) The change of BRG1 levels after treatment with pcDNA-BRG1 and si-BRG1 in A549 cells. (B) miR-148b levels are positively correlated with BRG1 expression levels in A549 cells. (C) The change of BRG1 levels after treatment with pcDNA-BRG1 and si-BRG1 in H522 cells. (D) miR-148b levels are positively correlated with BRG1 expression levels in H522 cells. (E) Online platform ChIPBase displays the integrated TF-miRNAs network of miR-148b. (F) ChIP assay shows that BRG1 binds to the promoter of miR-148b target gene in A549 cells. *P<0.01.
Abbreviations: BRG1, Brahma-related gene 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; si-control, control siRNA; ChIP, chromatin immunoprecipitation; TF, Transcription factor.
Restoration of BRG1 inhibits proliferation and metastasis of lung cancer cells
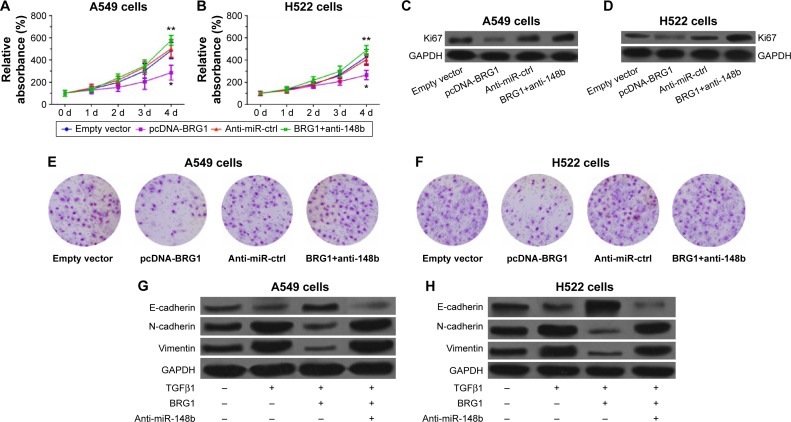
To demonstrate that restoration of BRG1 suppresses lung cancer development, we observed the effects of overexpression of BRG1 on lung cancer cells. As shown in Figure 3A and B, BRG1-overexpressing lung cancer cells had lower relative viability than the control group, but anti-miR-148b enhanced the cell viability. To confirm that BRG1 has an inhibitory effect on proliferation of cancer cells, we detected the expression of Ki67 in A549 and H522 cells by using Western blot analysis. The results showed that the Ki67 levels in BRG1-overexpressing lung cancer cells were significantly lower than those in the control groups, but downregulation of miR-148b elevated its levels (Figure 3C and D). We assessed the effects of BRG1 on metastasis of lung cancer in vitro by Transwell assay. As shown in Figure 3E and F, the number of cells transfected vector with BRG1 that migrated was reduced compared with the control, while knockdown of miR-148b could increased it. As EMT has been reported to facilitate tumor cell metastasis, we analyzed whether BRG1 functioned as an inhibitor of TGFβ1-induced EMT in A549 and H522 cells by detecting the expressions of E-cadherin, N-cadherin, and vimentin using Western blot analysis. The results indicated that after a 48-hour treatment of TGFβ1, the level of E-cadherin was decreased, while the levels of N-cadherin and vimentin were elevated, suggesting that TGFβ1 could induce EMT in lung cancer cells. Interestingly, we observed that overexpression of BRG1 could inhibit TGFβ1-induced EMT, but downregulation of miR-148b showed an opposite effect in A549 and H522 cells (Figure 3G and H). These data showed that restoration of BRG1 suppresses proliferation and metastasis of lung cancer cells and that miR-148b also functions as a tumor inhibitor.
Figure 3.
Restoration of BRG1 inhibits proliferation and metastasis of lung cancer cells but downregulation of miR-148b resists the effects.
Notes: (A and B) Restoration of BRG1 inhibits proliferation of A549 and H522 cells, respectively. Downregulation of miR-148b shows an opposite effect. (C and D) Overexpression of BRG1 reduces the expression levels of Ki67 in A549 and H522 cells, respectively, but downregulation of miR-148b elevates them. (E and F) Restoration of BRG1 suppresses the migration ability of A549 and H522 cells, respectively, which is resisted by downregulation of miR-148b. (G and H) TGFβ1 decreases the level of E-cadherin but increases the levels of N-cadherin and vimentin, enhancing EMT of A549 and H522 cell lines, respectively. Restoration of BRG1 suppresses TGFβ1-induced EMT, but downregulation of miR-148b reverses it. *P<0.01 versus the empty vector group; **P<0.01 versus the empty vector or the anti-miR-ctrl group. Empty vector: A549 or H522 cells were transfected with empty vectors. pcDNA-BRG1: A549 or H522 cells were transfected with vectors containing BRG1. Anti-miR-ctrl: Cells were transfected with control anti-miRNA mimics. BRG1+anti-148b: Cells were cotransfected with BRG1 vectors and anti-miR-148b mimics.
Abbreviations: BRG1, Brahma-related gene 1; EMT, epithelial–mesenchymal transition; d, days; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Anti-miR-ctrl, Anti-miR-control.
miR-148b suppresses lung cancer via regulating the WNT/β-catenin signaling pathway
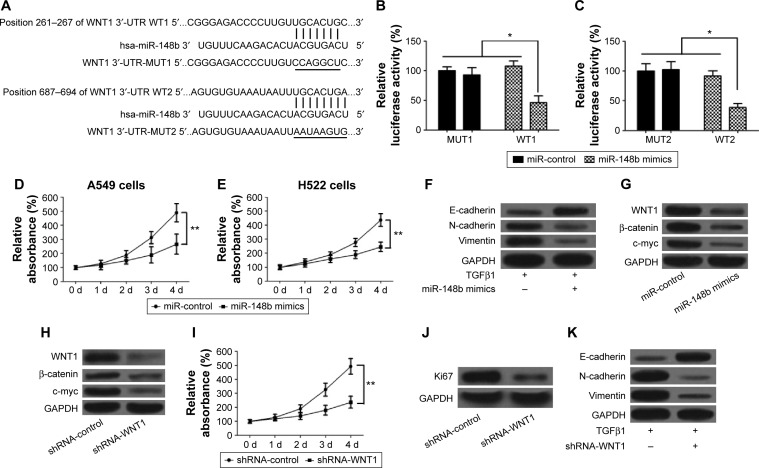
Increasing evidence showed that the WNT/β-catenin signaling pathway plays a vital role in the promotion of the EMT and the progression of cancer.29,30 We used the bioinformatic analysis (MiRanda and TargetScan) for prediction of miR-148b-binding sites. Two putative miR-148b complementary regions in the 3′-UTR of WNT1 mRNA were observed (Figure 4A). To confirm whether miR-148b regulates the expression of WNT1 via directly binding to WNT1 sequences, the reporter plasmids containing the wild-type or mutant 3′-UTR of WNT1 were constructed and then transfected into A549 cells along with pre-miR-148b or a precursor control, and then luciferase activity was detected. High expression of miR-148b significantly decreased reporter activity of wild-type WNT1 constructs but not mutant WNT1 (Figure 4B and C), suggesting that the 3′-UTR of WNT1 is responsible for miR-148b function. To confirm that miR-148b regulated the progress of lung cancer through inactivating the WNT/β-catenin signaling pathway, we first assessed the effects of miR-148b on lung cancer cells and the expression levels of WNT1, β-catenin, and c-myc. The results showed that upregulation of miR-148b suppressed proliferation of A549 and H522 cells compared with the miR-control group (Figure 4D and E). We also evaluated the effects of miR-148b on the EMT of lung cancer cells by detecting the expression levels of E-cadherin, N-cadherin, and vimentin. Our results showed that overexpression of miR-148b increased the level of E-cadherin but decreased the levels of N-cadherin and vimentin compared with the miR-control group (Figure 4F), which suggested that miR-148b could inhibit EMT of lung cancer. Furthermore, upregulation of miR-148b decreased the levels of WNT1, β-catenin, and c-myc in A549 and H522 cells (Figure 4G), which suggested that miR-148b has an inhibitory effect to the WNT/β-catenin signaling. In addition, we assessed the roles of the WNT/β-catenin signaling pathway in EMT and lung cancer. As shown in Figure 4H, knockdown of WNT1 significantly decreased the levels of WNT1, β-catenin, and c-myc. In addition, knockdown of WNT1 inhibited proliferation and the expression level of Ki67 (Figure 4I and J). Compared to the control group, knockdown of WNT1 elevated the level of E-cadherin but reduced the levels of N-cadherin and vimentin (Figure 4K), displaying a similar role to miR-148b. Taken together, these data revealed that miR-148b suppresses the promotion of EMT and the progression of lung cancer via inactivating the WNT/β-catenin signaling pathway.
Figure 4.
miR-148b inhibits proliferation and metastasis of lung cancer by directly regulating the WNT1/β-catenin signaling.
Notes: (A) The 3′-UTRs of WNT1 are bonded to miR-148b. (B and C) The wild types of WNT1 3′-UTR significantly inhibit the relative luciferase activity but not the mutants in A549 cells. (D and E) miR-148b suppresses proliferation of A549 and H522 cells, respectively. (F) miR-148b inhibits TGFβ1-induced EMT in A549 cells. (G) miR-148b inhibits the WNT1/β-catenin signaling pathway. (H) Knockdown of WNT1 blocks the WNT1/β-catenin signaling pathway, showing decreased levels of β-catenin and c-myc. (I) Knockdown of WNT1 suppresses proliferation of A549 cells. (J) Knockdown of WNT1 decreases the level of Ki67 in A549 cells. (K) Knockdown of WNT1 inhibits TGFβ1-induced EMT. *P<0.05, **P<0.01.
Abbreviations: EMT, epithelial–mesenchymal transition; 3′-UTR, 3′-untranslated region; d, days; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MUT, mutation; WT, wild type.
Discussion
Recent sequencing efforts have shown silence of BRG1 in multiple primary NSCLC clinical specimens and human lung tumor cell lines.31 In this study, BRG1 was confirmed to be significantly expressed at low levels in lung cancer tissues and cells. Furthermore, we validated that overexpression of BRG1 inhibited proliferation and metastasis of lung cancer cells and suppressed growth of xenografts in nude mice. Romero et al8 reported that BRG1-lacking cell lines from several cancer types were not sensitive to retinoic acid or glucocorticoid, while upregulation of BRG1 restored sensitivity. Additionally, restoration of BRG1 significantly suppressed invasion and progression of lung cancer cells orthotopically transplanted in nude mice. Another study showed that decreased BRG1 enhanced EMT and metastasis of colorectal cancer by regulating the miR-550a-5p/ring finger 43/WNT/β-catenin signaling pathway in vitro and in vivo.32 However, the effects of BRG1 on EMT and metastasis of lung cancer are unknown. In this study, we displayed that restoration of BRG1 suppressed EMT and metastasis of lung cancer cells by upregulation of miR-148b and inhibition of the WNT/β-catenin signaling pathway.
Overexpression of BRG1 promoted the transcriptional activity of miR-148b and silence of BRG1 by siRNA led to decreased miR-148b expression in this study, accounting for the low expression level of miR-148b, a new cancer suppressor, in lung cancer cells. BRG1 acts as a master regulator of the genome, for it transcriptionally regulates approximately 4.8% of the human genome, including genes that participate in plenty of signaling pathways.33 For example, BRG1 inhibits the transcriptional activity of miR-550a-5p promoter, and decreased BRG1 expression elevates miR-550a-5p expression level in colorectal cancer.33 BRG1 accelerates COUP-TFII expression via binding conserved regulatory regions to the COUP-TFII promoter and remodeling chromatin to make the promoter accessible to transcriptional machinery in vascular endothelial cells.34 In this study, we showed that BRG1 is a transcription factor of miR-148b using ChIP assay. Therefore, we deduced that BRG1 restoration transcriptionally regulated miR-148b, leading to inhibition of proliferation and metastasis of human lung cancer.
The WNT/β-catenin signaling pathway has been documented to play a critical role in the progression and promotion of EMT and cancer metastasis.35 Many studies have showed that WNT1, a mammary oncogene, is associated with mammary oncogenesis and that β-catenin is overexpressed in human cancers, which in turn promotes the expression of target genes such as E-cadherin and c-myc that are characterized by cancer development and poor clinical prognosis.36,37 Most recently, miR-148b was demonstrated to suppress cell proliferation and invasion by downregulating the expression of WNT1, β-catenin, and c-myc and upregulating E-cadherin expression in hepatocellular carcinoma.37 In breast cancer cell lines, miR-374a directly targets and suppresses numerous negative regulators of the WNT/β-catenin signaling cascade, including WIF1, PTEN, and WNT5A, promoting EMT and metastasis both in vitro and in vivo.35 In this study, we identified that miR-148b, regulated by BRG1, bound to two putative complementary regions in the 3′-UTR of WNT1 mRNA, inactivating the WNT/β-catenin signaling, resulting in inhibition of cell proliferation and metastasis in lung cancer cells. We also found that inactivation of the WNT/β-catenin signaling suppressed EMT of lung cancer cells. Previous studies have showed that EMT allows benign tumor cells to infiltrate surrounding tissues and metastasize to distant sites, facilitating the early stage of metastatic progression.38,39 Hence, our study revealed that inhibition of EMT by BRG1-depended miR-148b overexpression may contribute to decrease the metastatic capacity of lung cancer cells.
In conclusion, the gene encoding BRG1 was frequently downregulated in human lung cancer. Restoration of BRG1 suppressed cell proliferation and metastasis in lung cancer cells. Moreover, we identified that BRG1 positively regulated the expression of miR-148b and that miR-148b suppressed EMT of lung cancer cells by regulating the WNT/β-catenin signaling via directly binding to WNT1 sequences. The BRG1/miR-148b/WNT/β-catenin pathway provides potential therapeutic value for the treatment of lung cancer.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wiley MM, Muthukumar V, Griffin TM, Griffin CT. SWI/SNF chromatin-remodeling enzymes Brahma-related gene 1 (BRG1) and Brahma (BRM) are dispensable in multiple models of postnatal angiogenesis but are required for vascular integrity in infant mice. J Am Heart Assoc. 2015;4(4):e001972. doi: 10.1161/JAHA.115.001972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Li B, Li W, et al. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports. 2014;3(3):460–474. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orvis T, Hepperla A, Walter V, et al. BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res. 2014;74(22):6486–6498. doi: 10.1158/0008-5472.CAN-14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63(3):560–566. [PubMed] [Google Scholar]
- 5.Fukuoka J, Fujii T, Shih JH, et al. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10(13):4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- 6.Medina PP, Carretero J, Ballestar E, et al. Transcriptional targets of the chromatin-remodelling factor SMARCA4/BRG1 in lung cancer cells. Hum Mol Genet. 2005;14(7):973–982. doi: 10.1093/hmg/ddi091. [DOI] [PubMed] [Google Scholar]
- 7.Medina PP, Romero OA, Kohno T, et al. Frequent BRG1/SMARCA4–inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29(5):617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 8.Romero OA, Setien F, John S, et al. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med. 2012;4(7):603–616. doi: 10.1002/emmm.201200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Wang X, Wang H, et al. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer. 2012;48(16):3104–3111. doi: 10.1016/j.ejca.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Zhu X, Li Y, Shen H, et al. miR-137 inhibits the proliferation of lung cancer cells by targeting Cdc42 and Cdk6. FEBS Lett. 2013;587(1):73–81. doi: 10.1016/j.febslet.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Liu M, Lang N, Qiu M, et al. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer. 2011;128(6):1269–1279. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- 12.Kim HS, Lee KS, Bae HJ, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6(10):8089–8102. doi: 10.18632/oncotarget.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chresta CM, Schlicker A, Beran G, Cerillo G, Wessels LF, Orphanides G. Reciprocal expression of miR-125b and miR-192/miR-200 families define clinically relevant sub-types of colorectal cancer with differential sensitivity to EGFR and MEK targeted agents. Cancer Res. 2014;74(19 Suppl):5559. [Google Scholar]
- 14.Huang YC, Hung LY. Tumor suppressive miR-137 targets Aurora-A and leads to apoptosis in colorectal cancer cells. Cancer Res. 2014;74(19 Suppl):4344. [Google Scholar]
- 15.Zhao G, Zhang JG, Liu Y, et al. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol Cancer Ther. 2013;12(1):83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- 16.Song YX, Yue ZY, Wang ZN, et al. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10(1):1. doi: 10.1186/1476-4598-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cimino D, De Pittà C, Orso F, et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013;27(3):1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C, Wang C, Chen X, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56(12):1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 19.Song Y, Xu Y, Wang Z, et al. MicroRNA-148b suppresses cell growth by targeting cholecystokinin-2 receptor in colorectal cancer. Int J Cancer. 2012;131(5):1042–1051. doi: 10.1002/ijc.26485. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Liu GL, Liu SH, et al. MicroRNA-148b enhances the radio-sensitivity of non-Hodgkin’s Lymphoma cells by promoting radiation-induced apoptosis. J Radiat Res. 2012;53(4):516–525. doi: 10.1093/jrr/rrs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azizi M, Teimoori-Toolabi L, Arzanani MK, Azadmanesh K, Fard-Esfahani P, Zeinali S. MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther. 2014;15(4):419–427. doi: 10.4161/cbt.27630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Xu Y, Wei S, et al. microRNA-148b suppresses hepatic cancer stem cell by targeting neuropilin-1. Bios Rep. 2015;35(4):e00229. doi: 10.1042/BSR20150084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge H, Li B, Hu WX, et al. MicroRNA-148b is down-regulated in non-small cell lung cancer and associated with poor survival. Int J Clin Exp Pathol. 2015;8(1):800–805. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu GL, Liu X, Lv XB, Wang XP, Fang XS, Sang Y. miR-148b functions as a tumor suppressor in non-small cell lung cancer by targeting carcinoembryonic antigen (CEA) Int J Clin Exp Med. 2014;7(8):1990–1999. [PMC free article] [PubMed] [Google Scholar]
- 25.Tang SC, Sheu GT, Wong RH, et al. Expression of glutathione S-transferase M2 in stage I/II non-small cell lung cancer and alleviation of DNA damage exposure to benzo [a] pyrene. Toxicol Lett. 2010;192(3):316–323. doi: 10.1016/j.toxlet.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Xia J, Wu Z, Yu C, et al. miR-124 inhibits cell proliferation in gastric cancer through down-regulation of SPHK1. J Pathol. 2012;227(4):470–480. doi: 10.1002/path.4030. [DOI] [PubMed] [Google Scholar]
- 27.Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008;68(10):3689–3696. doi: 10.1158/0008-5472.CAN-07-6652. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe H, Mizutani T, Haraguchi T, et al. SWI/SNF complex is essential for NRSF-mediated suppression of neuronal genes in human nons-mall cell lung carcinoma cell lines. Oncogene. 2006;25(3):470–479. doi: 10.1038/sj.onc.1209068. [DOI] [PubMed] [Google Scholar]
- 29.Thu K, Radulovich N, Becker-Santos D, et al. SOX15 is a candidate tumor suppressor in pancreatic cancer with a potential role in Wnt/β-catenin signaling. Oncogene. 2014;33(3):279–288. doi: 10.1038/onc.2012.595. [DOI] [PubMed] [Google Scholar]
- 30.Kim WK, Oh J, Bae SY, et al. Antitumor activity of 2-hydroxycinnam-aldehyde against human colorectal cancer cells is mediated by the Wnt/β-catenin signaling pathway. Cancer Res. 2014;74(19 Suppl):4237. [Google Scholar]
- 31.Song S, Walter V, Karaca M, et al. Gene silencing associated with SWI/SNF complex loss during NSCLC development. Mol Cancer Res. 2014;12(4):560–570. doi: 10.1158/1541-7786.MCR-13-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, Fu Y, Yang X, et al. Brg-1 targeting of novel miR550a-5p/RNF43/Wnt signaling axis regulates colorectal cancer metastasis. Oncogene. 2015 May 11; doi: 10.1038/onc.2015.124. Epub. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Nemzow L, Chen H, Hu JJ, Gong F. Whole genome expression profiling shows that BRG1 transcriptionally regulates UV inducible genes and other novel targets in human cells. PloS One. 2014;9(8):e105764. doi: 10.1371/journal.pone.0105764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis RB, Curtis CD, Griffin CT. BRG1 promotes COUP-TFII expression and venous specification during embryonic vascular development. Development. 2013;140(6):1272–1281. doi: 10.1242/dev.087379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai J, Guan H, Fang L, et al. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123(2):566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G, Shukeir N, Potti A, et al. Up-regulation of Wnt-1 and β-catenin production in patients with advanced metastatic prostate carcinoma. Cancer. 2004;101(6):1345–1356. doi: 10.1002/cncr.20518. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Shi Y, Hong D, et al. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting Wnt/β-catenin pathway. Sci Rep. 2015;5:8087. doi: 10.1038/srep08087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, MacBeath G. A noncanonical frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell. 2014;159(4):844–856. doi: 10.1016/j.cell.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleeman JP, Thiery JP. SnapShot: the epithelial-mesenchymal transition. Cell. 2011;145(1):162.e1. doi: 10.1016/j.cell.2011.03.029. [DOI] [PubMed] [Google Scholar]