Summary
Histone variants play crucial roles in gene expression, genome integrity and chromosome segregation. However, to what extent histone variants control chromatin architecture remains largely unknown. We report genome-wide profiles of all four types of H2A variants in Arabidopsis and identify that the previously uncharacterized histone variant H2A.W specifically associates with heterochromatin. Genetic analyses show that H2A.W acts in synergy with the heterochromatic marks H3K9me2 and DNA methylation to maintain genome integrity. In vitro, H2A.W enhances chromatin condensation through a higher propensity to promote fiber-to-fiber interactions via its conserved C-terminal motif. In vivo, elimination of H2A.W causes decondensation of heterochromatin and conversely, ectopic expression of H2A.W promotes heterochromatin condensation. These results demonstrate that H2A.W plays critical roles in heterochromatin by promoting higher order chromatin condensation. Since motifs similar to the H2A.W C-terminal motif are present in other histone variants in other organisms, our findings impact our understanding of heterochromatin condensation in eukaryotes.
Introduction
All eukaryotic genomes are packaged into chromatin by nucleosomes, which consist of 146 base pairs of DNA wrapped around an octamer of histones H2A, H2B, H3 and H4 (Luger et al., 1997). Chromatin is organized into transcriptionally active euchromatin and more compact inactive heterochromatin. In somatic cells of Arabidopsis, heterochromatin can be visualized as dense structures called chromocentres by DAPI staining (Fransz et al., 2006). In Schizosaccharomyces pombe and mammals, heterochromatin is demarcated by histone H3 lysine 9 methylation (H3K9me3) (Jenuwein and Allis, 2001). Similarly, in Arabidopsis, heterochromatin is marked by H3K9me2, which is deposited by the histone methyltransferases KRYPTONITE (KYP), SUVH5 and 6 (Bernatavichute et al., 2008; Jackson et al., 2002; Malagnac et al., 2002). DNA methylation plays an important function in regulating heterochromatin (Law and Jacobsen, 2010). The plant specific DNA methyltransferase CHROMOMETHYLASE 3 (CMT3) binds to H3K9me2 and methylates cytosine residues at CHG sites (where H is A or C or T) (Du et al., 2012; Lindroth, 2001). In addition, DNA methylation in CG contexts is carried out by METHYLTRANSFERASE 1 (MET1) (Finnegan and Dennis, 1993; Law and Jacobsen, 2010). DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), and the recently discovered CHROMOMETHYLASE 2 (CMT2) perform methylation in CHH contexts (Haag and Pikaard, 2011; Zemach et al., 2013; Stroud et al., 2014). DNA methylation and H3K9me2 act together to silence genes and transposable elements (TEs) in Arabidopsis (Law and Jacobsen, 2010).
Histone variants carry out diverse chromatin functions including transcription, DNA repair, and chromosome segregation (Bönisch and Hake, 2012; Millar, 2013; Szenker et al., 2011; Talbert and Henikoff, 2010). The vast majority of known histone variants are homologs of H2A and H3 histones. A phylogenetic analysis of histone families supports the notion that genes encoding ancestral histone H3.3 and H2A.Z duplicated and evolved to encode histone variants with distinct properties (Talbert et al., 2012). In the case of H3, the cell cycle controlled H3.1 class evolved independently in most phyla, acquiring similar properties in plants and animals (Filipescu et al., 2013; Stroud et al., 2012; Wollmann et al., 2012). Similarly, the canonical H2A, thereafter referred to simply as “H2A”, as well as H2A.X, evolved independently from the ancestral H2A.Z in almost all eukaryotes (Talbert et al., 2012). In all eukaryotes, the centromeric H3 variant (cenH3) is functionally conserved, specifically recruited at the centromere and instrumental for kinetochore function (Rop et al., 2012; Talbert et al., 2008).
Genome-wide profiling of histone variants using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) revealed that some histone variants decorate specific functional regions of the genome (Talbert and Henikoff, 2010). In yeast and animals, H2A.Z is enriched at transcriptional start sites (TSS) or 5’ ends of genes (Guillemette et al., 2005; Li et al., 2005; Raisner et al., 2005). On the other hand, H3.3 is enriched over gene bodies with inclination towards the 3’ end of genes in Drosophila and humans (Goldberg et al., 2010; Mito et al., 2005). Histone variants also co-localize with posttranslational modifications on their own N-terminal tails. For example, H3.3 tends to be associated with H3K4me2/3 while H3.1 tends to be enriched at heterochromatin marked by H3K9me2 (Loyola and Almouzni, 2007). Although H2A.Z and H3.3 are predominantly linked to transcriptional activation, they are also associated with heterochromatin maintenance. In yeast, H2A.Z is enriched in subtelomeric regions and prevents spreading of telomeric heterochromatin into euchromatic regions (Meneghini et al., 2003). Similarly, in humans, H3.3 is localized to telomeric heterochromatic regions (Goldberg et al., 2010; Wong et al., 2009). In contrast, in Arabidopsis H2A.Z and H3.3 are excluded from heterochromatin. H2A.Z is enriched at the TSS of expressed genes and also enriched in gene bodies of response genes, which are differentially activated during development or stress (Aceituno et al., 2008; Coleman-Derr and Zilberman, 2012; Zilberman et al., 2008). Similarly, H3.3 enrichment is confined towards the 3’ end of genes and is correlated positively with gene expression (Stroud et al., 2012; Wollmann et al., 2012). Hence, histone variants H2A.Z and H3.3 appear to exclusively define euchromatin in Arabidopsis. This observation led us to question whether a specific combination of histone variants defines heterochromatin in plants, and if so what their impact might be chromatin organization.
To understand the roles of histone H2A variants in chromatin regulation in Arabidopsis, we generated a comprehensive localization map of all four types of H2A variants and found that they demarcate different features of the genome. Interestingly, heterochromatin was specifically marked by the previously uncharacterized H2A variant H2A.W. We find that heterochromatin specific localization of H2A.W does not depend on H3K9me2 or DNA methylation, and that H2A.W and DNA methylation cooperatively silence TEs through independent pathways. In vitro nucleosome array experiments suggest that H2A.W causes higher order chromatin condensation by promoting chromatin fiber-to-fiber interactions. Finally, our findings further demonstrate that H2A.W is both necessary and sufficient for heterochromatin condensation in vivo.
RESULTS
Genome-wide profiling of H2A variants identifies the heterochromatin specific H2A.W
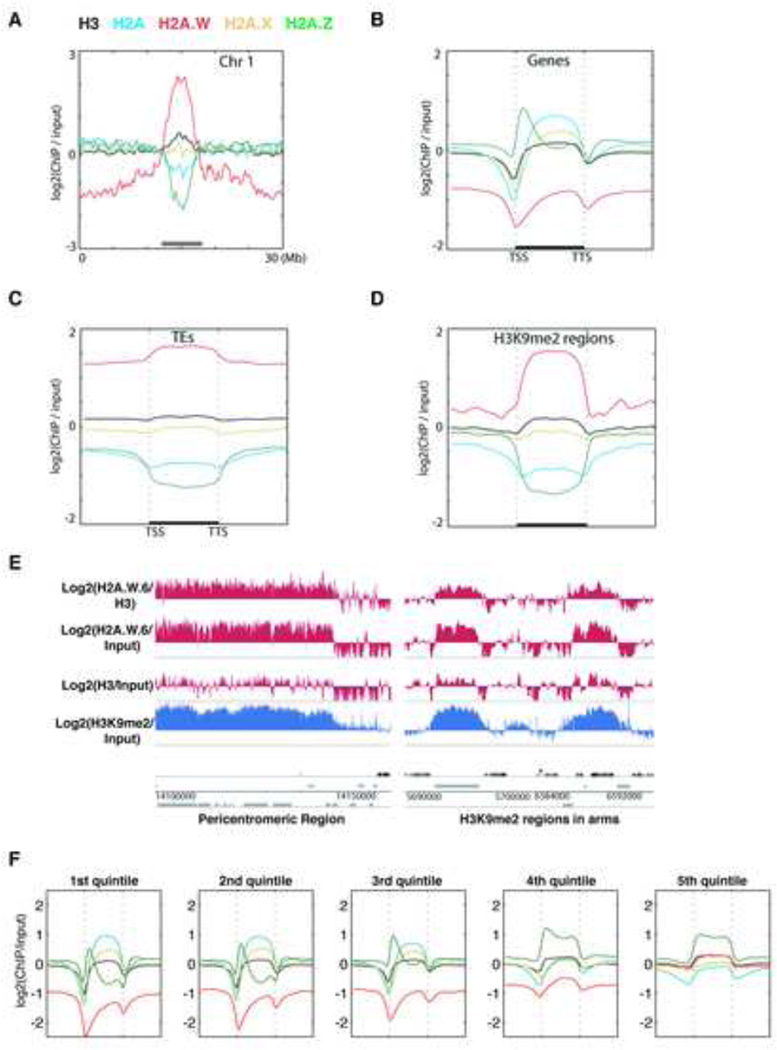
The Arabidopsis genome comprises thirteen genes encoding histone H2A variants, which are grouped into four classes according to C-terminal conserved motifs (Figure S1A) (Talbert et al., 2012). We obtained antibodies specific and representative of each H2A variant class (Figure S1B). We performed genome-wide profiling by chromatin immunoprecipitation for H2A variant-bound DNA followed by deep sequencing (ChIPseq) (Figures 1 and S1C). H2A.X covered the entire genome (Figures 1A and S1C). H2A showed a relative depletion at pericentromeric chromatin as well as over transposable elements (TEs) and islands of H3K9me2 present in chromosome arms outside pericentromeric heterochromatin (Figures 1A, 1C, 1D, 1E and S1C). Both H2A.X and H2A were enriched over gene bodies (Figure 1B). In contrast with the rather uniform profiles of H2A and H2A.X over gene bodies, H2A.Z was enriched specifically at the 5’ end of genes (Figure 1B) and was highly depleted over all heterochromatic features (Figures 1C and 1D), as reported previously (Coleman-Derr and Zilberman, 2012). In addition, we analyzed whether enrichment of each H2A variant over gene bodies depends on the level of gene expression. Comparing quintiles of genes defined by their expression level, we observed gradual depletion of H2A and H2A.X and in turn enrichment of H2A.Z when expression levels decreased (Figure 1F). Hence highly expressed genes are defined by a marked enrichment in H2A.Z and H2.A at the 5’end and 3’end respectively, whereas inactive genes are covered by H2A.Z.
Figure 1. The histone variant H2A.W is specifically localized to heterochromatin.
(A) Distribution of ChIP-seq reads along chromosome 1 for each class of H2A variants and total H3, relative to input DNA. The grey bar at the bottom indicates pericentromeric heterochromatin. (B) Average enrichment of each class of H2A variants and total H3 over genes delimited by a transcription start site (TSS) and a transcription termination site (TTS). (C) Average enrichment of each class of H2A variants and total H3 over transposable elements (TEs). (D) Average enrichment of each class of H2A variants and total H3 over defined H3K9me2 enriched regions in chromosomal arms. In (B to D) the middle region marked by the horizontal black bar represents the body of the element considered. The left and right flanking regions represent the 5’ upstream and the 3’ downstream regions, respectively. (E) Genome browser views of H2A.W.6 enrichment over pericentromeric chromatin and over a H3K9me2 enriched region in the arm of a chromosome. Black bars represent genes and grey bars represent TEs. The log2 ratios of H2A.W to input genomic DNA, and H2A.W to total H3 are shown. (F) Average enrichment of each class of H2A variants and total H3 over genes as defined in (B). Genes were grouped in quintiles defining highest (first quintile) to lowest (5th quintile) transcription levels in 10 day old seedlings. See also Figure S1.
Unlike all other H2A variants, H2A.W was strongly depleted from gene bodies (Figure 1B). H2A.W was specifically enriched in pericentromeric heterochromatin, TEs, and islands of H3K9me2 (Figures 1A–C, 1F and S1C). The Pearson correlation between H3K9me2 and H2A.W within TEs across the genome was 0.7, indicating a very high degree of overlap between H2A.W and H3K9me2. Overall we conclude that antagonistic localizations of H2A.Z and H2A.W distinguish active and inactive domains of chromatin.
H2A.W indexes heterochromatin
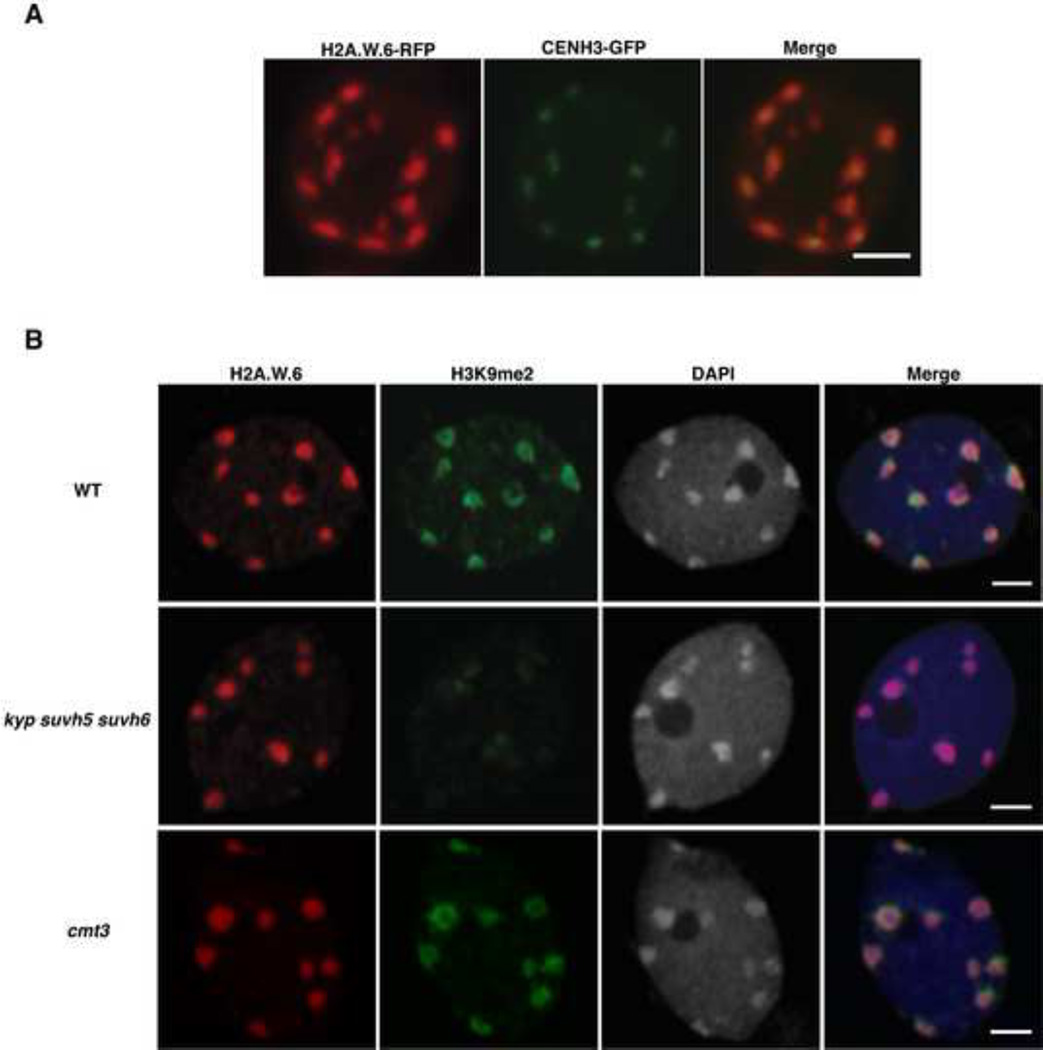
H2A.W carries an extended C-terminal tail with a conserved SPKK (in general T/SPXK) motif (Figure S1A) (Bönisch and Hake, 2012). The H2A.W variant class evolved and is highly conserved in seed bearing plants (i.e gymnosperms and the angiosperms) (Figure S2A). In Arabidopsis, the H2A.W family comprises three proteins H2A.W.6, H2A.W.7, and H2A.W.12, which are encoded by the genes HTA6, HTA7, and HTA12, respectively (Figure S1A). HTA6 is expressed at higher levels than the other two H2A.W homologs (Figure S2D) and was used for the ChIP-seq analyses described above. In order to investigate whether the properties of H2A.W.6 revealed by ChIP-seq analysis were representative of the entire H2A.W class, we analyzed the localization of each member of the H2A.W family in vivo. We fused the RED FLUORESCENT PROTEIN (RFP) to each gene encoding H2A.W and expressed the fusion protein under the control of its respective promoter in transgenic plants that were also expressing the CENTROMERIC HISTONE 3 (CenH3) fused to the GREEN FLUORESCENT PROTEIN (GFP). CenH3 is located at the centromere of each chromosome and is surrounded by pericentromeric chromatin that forms heterochromatin domains called chromocenters (Fang and Spector, 2005; Talbert et al., 2002; Fransz et al., 2002). Each H2A.W-RFP localized primarily to domains corresponding to the ten chromocenters, each containing a CENH3-GFP patch in its center (Figures 2A, S2B and S2C). H2A.W.7-RFP and H2A.W.12-RFP also colocalized with H3K9me2 (Figures S2E and S2F) as demonstrated for H2A.W.6 (Figures 2B WT row and 1D). In conclusion, all members of the H2A.W family are tightly associated with heterochromatin in somatic cells.
Figure 2. Localization of H2A.W does not depend on H3K9me2 and DNA methylation.
(A) Localization of H2A.W.6 with the centromeric histone CENH3. In vivo confocal section of a root nucleus of transgenic plants expressing pCENH3::CENH3- GFP and pH2A.W.6::H2A.W.6-RFP. (B) Confocal sections of leaf nuclei immunostained for H3K9me2 and H2A.W.6 with DAPI counterstaining in genetic backgrounds WT, kyp suvh5 suv6, and cmt3. Scale bars represent 2 µm. See also Figure S2.
Recruitment of H2A.W is independent of H3K9me2 or DNA methylation
The co-localization between H2A.W and H3K9me2 suggested a possible mechanistic link between the deposition of these two heterochromatic features. Preventing H3K9me2 deposition by mutating the three methyltransferases KYP, SUVH5, and SUVH6 did not significantly alter the pattern of H2A.W.6 (Figure 2B). Similarly, H2A.W.6 localization was not perturbed significantly by the absence of CMT3, the factor that methylates cytosine residues in CHG contexts at heterochromatic loci marked by H3K9me2 (Figure 2B). Since H2A.W localization does not depend on H3K9me2 or the associated CHG methylation, we tested the influence of other pathways responsible for DNA methylation. The loss of the de novo DNA methyltransferase DRM2 did not affect H2A.W nor heterochromatin organization (Figures S2G – S2I). In contrast, heterochromatin was partially dispersed in the absence of the CG methyltransferase MET1 or the chromatin remodeling protein DECREASED DNA METHYLATION 1 (DDM1) that contribute to maintenance of DNA methylation and silencing of heterochromatic loci (Figures S2G – S2I) (Vongs et al., 1993; Jeddeloh et al., 1999; Zemach et al., 2013). However, in spite of the change of heterochromatin condensation in met1 and ddm1 genetic backgrounds, H2A.W.6 was still co-localized with dispersed heterochromatin marked with H3K9me2 (Figures S2G – S2I). These results suggest that the recruitment or maintenance of H2A.W to heterochromatin depend neither on DNA methylation nor on H3K9me2 and that a mechanism independent from these markers of heterochromatin is responsible for H2A.W deposition. However, other unknown components of heterochromatin might participate in H2A.W localization.
Loss of H2A.W causes heterochromatin decondensation
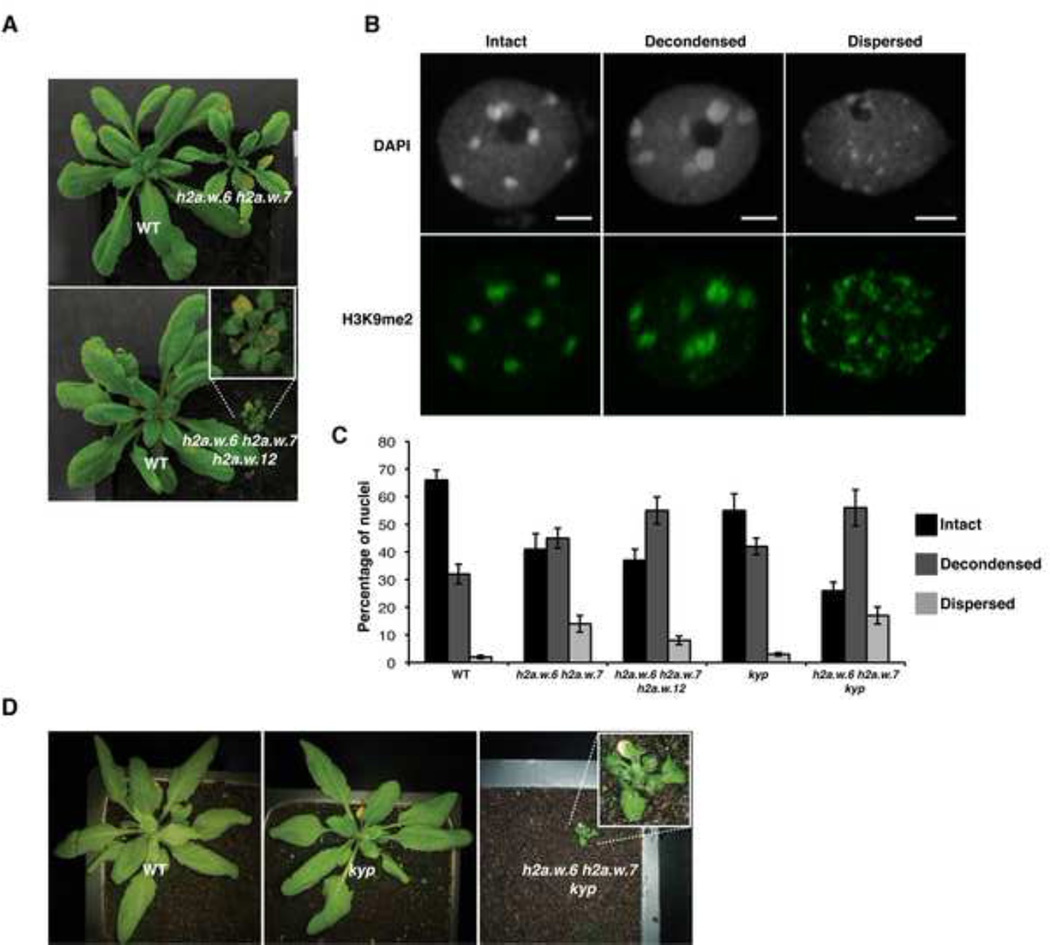
To study the function of H2A.W in vivo, we obtained knock-out lines for all three genes encoding H2A.W (Figure S3A). In single mutants we did not observe any obvious visible phenotype. Significant growth defects were observed in h2a.w.6 h2a.w.7 and h2a.w.6 h2a.w.12 double mutants and plant growth was severely affected in h2a.w.6 h2a.w.7 h2a.w.12 (h2a.w) triple mutants (Figure 3A). The strong effect of h2a.w.6 is likely due to its high level of expression among the three H2A.W encoding genes (Figure S2D). Triple mutant plants failed to flower and did not give rise to the next generation, suggesting that H2A.W is essential. Since H2A.W is specifically localized to heterochromatin, we analyzed heterochromatin structure in nuclei of h2a.w knock-out plants. In the double and triple mutants, a significant proportion of nuclei showed variable degrees of physical expansion of heterochromatin marked by H3K9me2 (Figure 3B) reaching in some nuclei a dispersed state in which chromocenters were no longer defined by condensed DAPI staining (Figure 3C). We conclude that H2A.W is required for heterochromatin condensation.
Figure 3. H2A.W is required for heterochromatin condensation.
(A) Morphological phenotypes observed in h2a.w double and triple mutants growing side by side with wild type (WT) plants. The triple mutant shown in the main panel is magnified in the inset. (B) Decondensed chromocentres observed in leaf nuclei stained with DAPI and immunostained against the heterochromatin marker H3K9me2. Scale bars represent 2 µm. (C) The bar chart represents percentages of nuclei belonging to each class of nuclei shown in B based on the degree of decondensation of chromocentres observed in DAPI staining. Error bars represent the standard deviation. (D) Morphological phenotype observed in the h2a.w.6 h2a.w.7 kyp mutant growing side by side with a WT plant. The triple mutant shown in the main panel is magnified in the inset. See also Figure S3.
Since heterochromatin was disorganized in h2a.w mutant, we analyzed the localization of the conserved heterochromatic mark H3K9me2 in h2a.w triple mutants. In this genetic background, H3K9me2 was still localized to DAPI rich regions even in nuclei with dispersed heterochromatic chromocenters, indicating that H3K9me2 deposition does not require H2A.W (Figure S3B). The independent colocalization of H2A.W and H3K9me2 prompted us to examine the genetic interaction between these two marks of heterochromatin. In the H3K9 methyltransferase mutant kyp (Figure S3C), H3K9me2 is reduced but does not show a significant effect on plant growth (Jackson et al., 2002) as SUVH5 and 6 also contribute to H3K9me2 (Bernatavicute et al., 2008; Malagnac et al., 2002). We found that simultaneous mutation of KYP and H2.A.W in h2a.w.6, h2a.w.7, kyp plants caused severe growth defects compared to growth defects observed in h2a.w.6, h2a.w.7 mutant plants (Figure 3D). Similarly, we also observed an increase in the proportion of nuclei with decondensed heterochromatin (Figure 3C). These results indicate that H2A.W and KYP participate in parallel pathways controlling heterochromatin condensation.
H2A.W represses transposable elements in synergy with CMT3-mediated CHG methylation
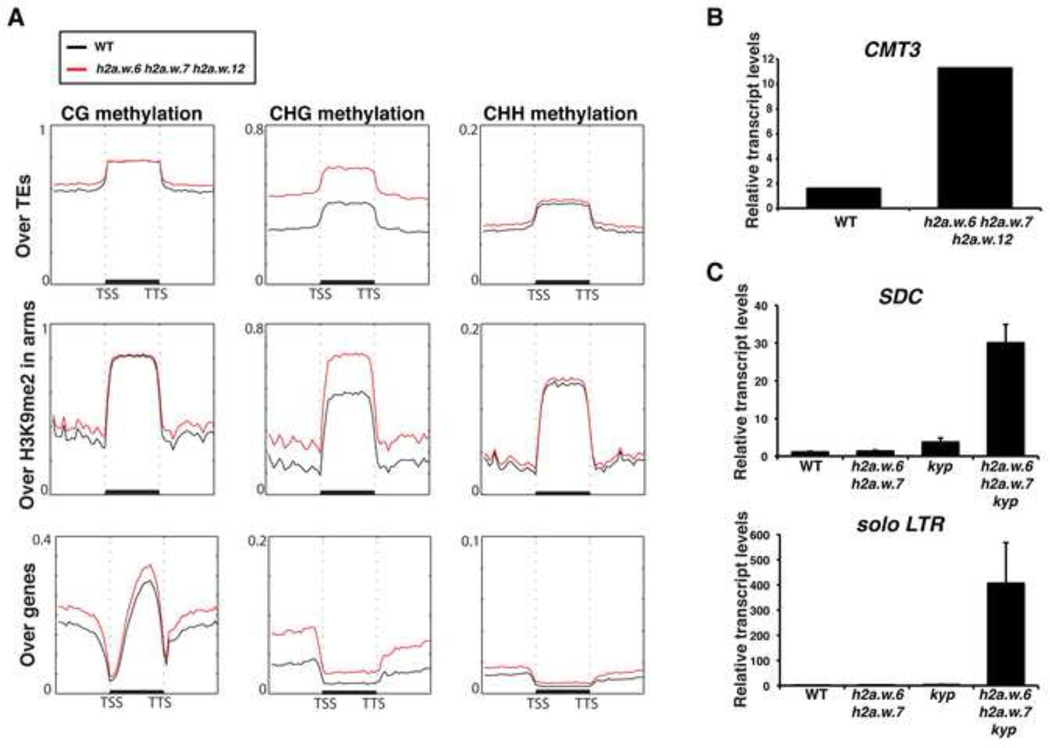
Decondensation of heterochromatin in mutants such as met1 has been previously shown to be associated with increased transcription of TEs and genes in the proximity of heterochromatic loci (Vaillant et al., 2008; Reinders et al., 2009; Law and Jacobsen, 2010). However, RNA sequencing (RNA-seq) analyses of the h2a.w triple mutant did not show a substantial impact on expression of TEs (Figure S4A). This suggests that in the absence of H2A.W, other silencing pathways might play compensatory roles in maintaining the transcriptional repressive status of TEs. Indeed, DNA methylation analysis by whole genome bisulfite sequencing (BS-seq) in the h2a.w triple mutant showed a significant increase in CHG methylation over TEs and regions enriched in H3K9me2, while DNA methylation over gene bodies was not affected (Figure 4A). A notable up-regulation of the expression of CMT3 encoding the primary CHG DNA methyltransferase likely accounts for the increase in CHG methylation (Figure 4B). DNA methylation at CG and CHH sites was not significantly affected, suggesting a specific association of H2A.W with CHG methylation (Figure 4A). These results suggested the possibility that increased CHG methylation may compensate for the lack of H2A.W, resulting in maintenance of the transcriptional repression of TEs. To test this hypothesis, we combined the h2a.w.6, h2a.w.7, and cmt3-11 null mutant alleles. This combination led to male and female gamete abortion (Figures S4B and S4C) and we were unable to obtain any triple homozygous plants out of 704 progeny segregating from the self fertilized triple heterozygous mutant, indicating that CMT3 and H2A.W pathways are synergistically required for viability. CMT3 methylates heterochromatic DNA by binding to H3K9me2 (Du et al., 2012), and consistently, CHG methylation is reduced in kyp mutants (Jackson et al., 2002; Stroud et al., 2013). Accordingly, the synergistic action of H2A.W and KYP was also observed on expression of solo LTR TE and the gene SDC, which were both dramatically increased in the h2a.w.6 h2a.w.7 kyp triple mutant (Figure 4C). These data suggests that genetic pathways of H2A.W and CHG methylation interact to ensure heterochromatin integrity.
Figure 4. H2A.W represses the transcription of heterochromatic elements in synergy with methylation mediated by CMT3.
(A) Metaplot analysis from whole genome bisulfite sequencing showing different types of methylation levels in WT and h2a.w triple mutant over major genomic features. Graphs were plotted as in Figure 1A. (B) Expression levels of CMT3 in h2a.w triple mutant in comparison to WT. Expression values were obtained from RNA-seq data. (C) Expression levels of SDC and solo LTR in h2a.w.6 h2a.w.7 kyp mutant compared with WT. The data from quantitative real-time RT-PCR was normalized to ACTIN7. Error bars represent standard deviation based on three independent biological replicates. See also Figure S4.
H2A.W promotes chromatin condensation through its conserved C-terminal motif in vitro
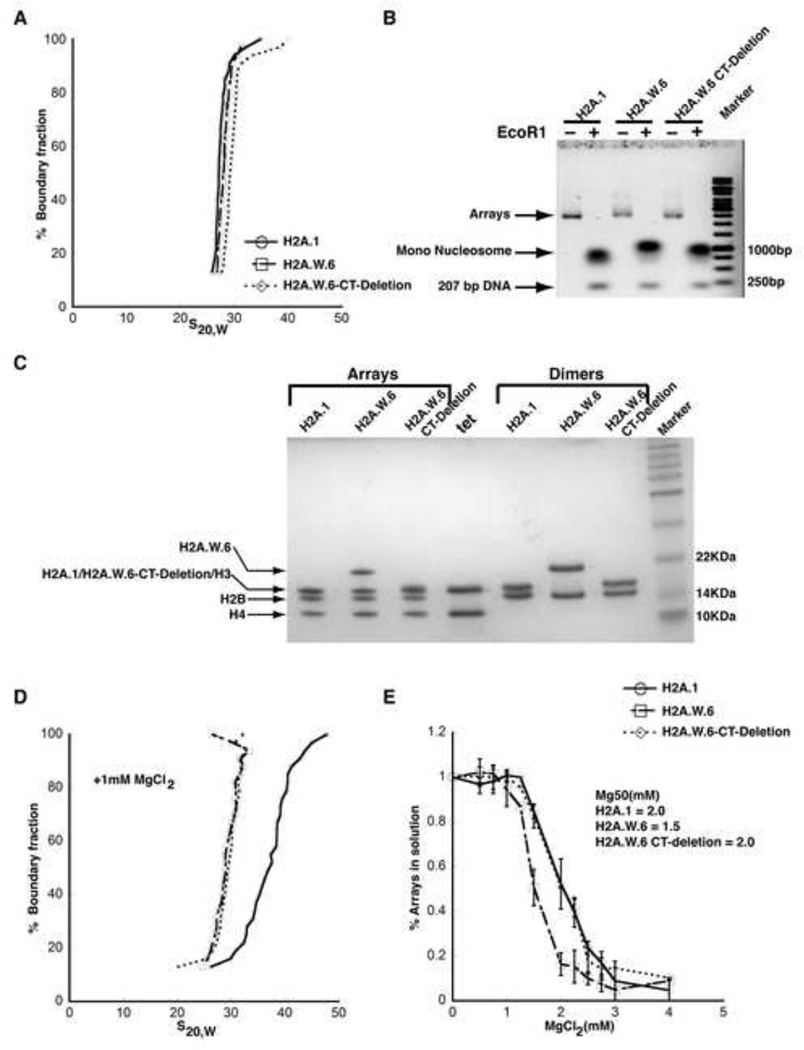
To further understand the potential mechanism of H2A.W in chromatin condensation, we tested whether H2A.W is able to cause condensation of nucleosome arrays in vitro. Chromatin condensation in vitro is composed of two independent structural transitions. Nucleosomes undergo short-range interactions with neighboring nucleosomes to form locally folded chromatin fibers. In addition, chromatin fibers self-associate to form supramolecular oligomers that share many of the properties of large-scale structure in condensed chromosomes. These two reversible structural transitions rely on different types of nucleosome-nucleosome interactions (Hansen, 2002), and can be studied in a model system by analysing the magnesium-dependent solution-state properties of defined 12-mer nucleosomal arrays in vitro. To understand the mechanism of H2A.W in chromatin condensation in vitro, we tested whether H2A.W affects self-association or folding. We assembled nucleosome arrays with Arabidopsis H2A.1, H2A.W.6, or a truncated version of H2A.W.6 (H2A.W-CT) that lacks the C-terminal tail containing the SPKK motif (Figure 5). Because folding and self-association depend on the number of nucleosomes per array, the three types of arrays were closely matched for nucleosome saturation. We ensured that all arrays contain on average 11-12 nucleosomes per DNA molecule, i.e. every nucleosome positioning sequence repeat is occupied (Figures 5A and 5B). We also verified equimolar distribution of all histone components by SDS-PAGE (Figure 5C). We then determined the extent of self-association as a function of salt concentration, and found that H2A.W.6 arrays engaged in oligomerization at significantly lower MgCl2 concentrations than H2A.1 or H2A.W.6-CT (Figure 5E). This suggests that H2A.W.6 promotes long-range nucleosome-nucleosome interactions in chromosomes, and that the C-terminal tail of H2A.W.6 contributes to this behaviour. Intriguingly, both arrays with H2A.W.6 and H2A.W.6-CT exhibited no signs of folding at 1 mM MgCl2, whereas arrays with canonical H2A.1 behave like other nucleosomal arrays and were moderately folded under these conditions (Figure 5D). Altogether, this suggests that H2A.W is able to enhance large-scale chromatin condensation by promoting long-range chromatin fiber-to-fiber interactions through its conserved C-terminal tail but is deficient in the short-range interactions that form the 30 nm fiber, suggesting a role in promoting highly condensed heterochromatin in vivo.
Figure 5. H2A.W causes chromatin condensation through chromatin fiber-fiber interactions promoted by its conserved C terminal tail in vitro.
(A) SV-AUC profile for 207-12 arrays assembled with H2A.1, H2A.W.6, and H2A.W.6-CT deletion indicates similar (and near-complete) saturation. (B) EcoRI digestion of nucleosome arrays to assay the degree of saturation of each array. Each array exhibits similar amounts of free 207 bp DNA confirming similar saturation of the arrays. (C) 1-2 µg of each array from (A) was run on a 15% SDS-PAGE to confirm equimolar distribution of histones. Tet indicates (H3-H4)2 tetramer. Note that H2A.1 and H2A.W.6-CT deletion runs in the same position as H3 on the gel. (D) The arrays were subjected to folding assay in the presence of 1 mM MgCl2. The H2A.1 containing control arrays show moderate folding (mid-point S ~40S), whereas both H2A.W.6 and H2A.W.6-CT deletion containing arrays do not undergo folding under these conditions. (E) Magnesium chloride-induced self-association assay showing that H2A.W.6 promotes inter-nucleosome array interactions. Arrays were incubated at the indicated concentrations of MgCl2, and the concentration of Mg2+ at which 50 % of the arrays have precipitated (Mg50) is listed in the figure (inset). Error bars are derived from two independent measurements (two preparations of reconstituted arrays).
H2A.W promotes chromatin condensation in vivo
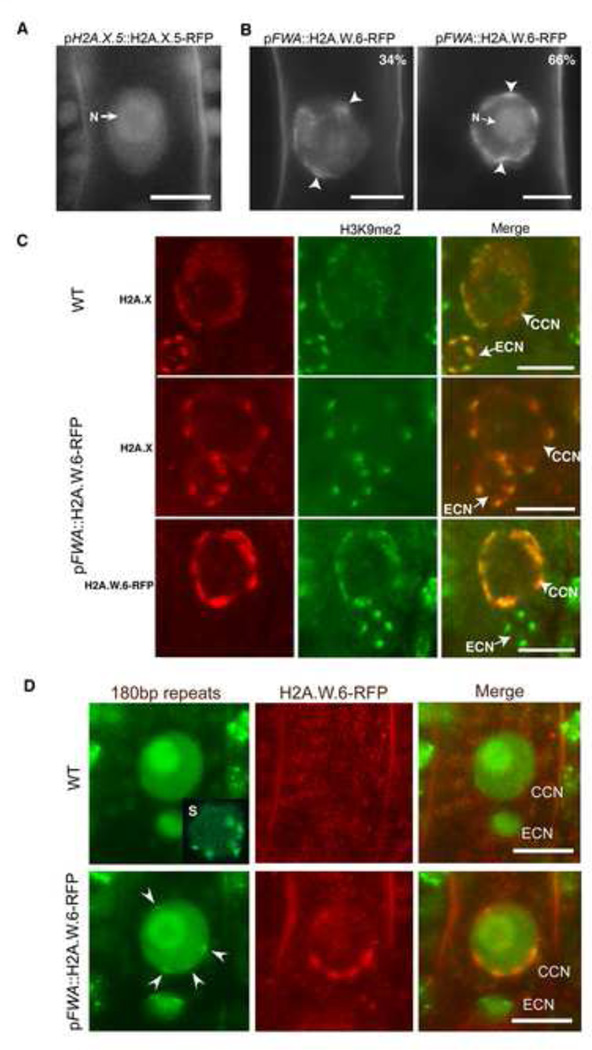
Since there was gain of DNA methylation upon loss of H2A.W in h2a.w triple mutants (Figure 4A), it was difficult to address directly whether H2A.W is sufficient to promote heterochromatin condensation in somatic cells. However, we observed that none of the genes encoding H2A.W was expressed in the central cell, one of the two female gametes of flowering plants (Figures S5A–S5C). The absence of H2A.W in this cell type correlated with the complete absence of condensed heterochromatic domains as indicated by the uniform distribution of H2A.X.5-RFP (Figure 6A), which marks equally heterochromatin and euchromatin in somatic cells (Figures 1A–1D). In order to test whether the lack of H2A.W was responsible for the absence of condensed heterochromatin in the central cell nucleus, we ectopically expressed H2A.W.6 using the promoter of FWA, which is specifically active in the central cell and the endosperm (Kinoshita et al., 2004). The chromatin of the central cell expressing pFWA-H2A.W.6-RFP showed domains of varying intensity with distinct zones of higher condensation (Figure 6B). We could distinguish two types of organization. A minor group of nuclei showed one or two strong patches of dense chromatin. The most representative class of central cell nuclei showed several patches of dense chromatin assembled in the periphery of the nucleus (Figure 6B), contrasting with the uniform wild-type pattern of H2A.X-RFP that marks both euchromatin and heterochromatin (Figure 6A). To test whether the assembled condensed domains are heterochromatic we immunostained whole ovules against H2A.X to report the general chromatin organization and against H3K9me2 to mark heterochromatin. We also used the POLYDACTYL ZINC FINGER (PZF) – GFP that binds to centromeric 180bp repeats in somatic cells (Lindhout et al., 2007). In the WT, chromocenters marked by chromatin condensation and H3K9me2 were clearly observed in the egg cell nucleus (Figures 6C) while the central cell chromatin organization was more diffuse with elongated patches of H3K9me2 lining the periphery of the nucleus (Figure 6C). In contrast, central cells expressing H2A.W.6-RFP clearly showed dense chromatin domains marked by H3K9me2, indicating that these domains are heterochromatic. In addition within these domains we detected a single dot of centromeric marker PZF-GFP (Figure 6D, lower panels), indicating that condensed chromatin domains caused by ectopic H2A.W are indeed pericentromeric heterochromatin foci. The absence of visible dots of PZF-GFP in WT central cell nuclei (Figure 6D, WT) was likely explained by the decondensed status of the pericentromeric and centromeric domains in the central cell and the absence of the cenH3 assembly reported previously (Ingouff et al., 2010). These observations indicate that H2A.W is sufficient to cause heterochromatin condensation in vivo.
Figure 6. H2A.W is sufficient to cause chromatin condensation in vivo.
(A) Uniform pattern of pH2A.X.5-H2A.X.5-RFP expressed in the central cell nucleus indicating the absence of defined chromocenters. N points the nucleolus. (B) Impact of ectopic expression of H2A.W.6-RFP in the central cell nucleus (CCN). We observe mild or severe chromatin condensation (arrowheads) and the proportion of each type of nuclei is indicated in the top right of each image. N points the nucleolus. (C) Impact of ectopic expression of H2A.W.6-RFP in the CCM on H2A.X and H3K9me2 immunolocalization. Top panel - The egg cell nucleus (ECN) shows well-defined chromocenters reported by H2A.X condensation and marked with H3K9me2 and serve as a reference point for comparison with the cc nucleus. Middle panel – Ectopic expression of H2A.W.6 in the CCN causes condensation of chromatin domains reported by H2A.X. Lower panel – These domains are marked with H3K9me2 (lower panel). (D) A CCN expressing PZFGFP that recognizes the centromeric 180bp repeats. Top panel – In the WT, PZF-GFP marks condensed centromeres in somatic cells (S, insert, a nucleus from root cells) but does not mark any condensed structure in the CCN. Bottom panel – In a central cell expressing pFWA::H2A.W.6-RFP arrowheads point the position of condensed chromatin domains which contain a dot of PZF-GFP marking the centromeric location. Scale bars in confocal sections represent 5 µm. See also Figure S5.
Discussion
Comprehensive genomic profiling showed that each H2A variant is characterized by a specific profile. In contrast to H2A, H2A.Z and H2A.W, the distribution of H2A.X is relatively ubiquitous, in keeping with the idea that H2A.X is involved in DNA repair (Downs et al., 2007; Soria et al., 2012). H2A uniformly marks gene bodies similar to that described for H3.1 (Stroud et al., 2012; Wollmann et al., 2012). Transcription of both H3.1 and H2A is highly coupled with S-phase (Talbert et al., 2012), suggesting that H2A and H3.1 are likely subject to common dynamics related to the loading of new nucleosomes over replicated DNA. However, unlike H3.1, H2A enrichment over gene bodies positively correlates with transcription with a gradual replacement by H2A.Z in genes expressed at low levels, suggesting that transcription is coupled to an exchange between H2A and H2A.Z over gene bodies. We confirmed that in highly expressed genes H2A.Z enrichment is confined to the 5’ end of the gene bodies, similar to that described in yeast, Drosophila and human (Zilberman et al., 2008). This enrichment appears to be a mirror image of H3.3 distribution, confined to the 3’ end of gene bodies (Stroud et al., 2012; Wollmann et al., 2012). Hence, the boundaries of gene bodies appear to be defined by differential enrichment of H2A, H2A.Z and H3.3 variants. In sharp contrast to euchromatin, heterochromatin is depleted of H2A and H2A.Z and is occupied nearly exclusively by H2A.W, suggesting that H2A variants restrict the deposition of each other over larger genomic domains defining euchromatin and heterochromatin. Therefore, combinatorial localizations of H2A and H3 variants define the main genomic features in Arabidopsis.
We show that H2A.W recruitment to chromatin is independent of the pathways that deposit heterochromatic marks H3K9me2 and DNA methylation. However, H2A.W acts together with these pathways to maintain heterochromatin condensation into higher order domains and to silence TEs. The lethal genetic interaction observed in our study suggests that H2A.W and H3K9me2/CHG methylation are absolutely essential for plant life, although each pathway on its own is dispensable. The lethality observed in mutants deficient for both pathways likely results from a lack of heterochromatin integrity, which could impact cell division. In response to H2A.W depletion, we also observe a cross talk between these pathways as both CMT3 expression and CHG methylation increase in heterochromatin.
In vitro micrococcal nuclease digestion data showed that nucleosomes containing H2A.W protect a longer stretch of DNA (162 bp) than nucleosomes containing H2A (146 bp) (Lindsey et al., 1991), suggesting that H2A.W prevents accessibility to DNA. Consistently, we find that H2A.W represses TE expression in synergistic action with H3K9me2. The acidic patch present in the histone core region of H2A affects the folding ability of nucleosome arrays (Kalashnikova et al., 2013). For example, euchromatic H2A.Z shows higher chromatin folding than H2A, as a consequence of its extended acidic patch (Fan et al., 2004). The acidic patch in H2A.W and canonical H2A are identical, and H2A.W shows a lower folding capacity than H2A, suggesting that other domains of H2A participate to chromatin folding. Our results show that H2A.W promotes chromatin condensation through long-range fiber-to-fiber interactions in vitro. This data is supported by the contrasting impacts of loss or gain of function of H2A.W in vivo on heterochromatin organization. We propose that H2A.W is necessary and sufficient to organize heterochromatin into higher order functional domains such as chromocenters, which are essential for kinetochore assembly and mitosis (Bian and Belmont, 2012; Luger et al., 2012).
H2A.W specific chromatin condensation properties rely on the extended C-terminal domain that contains the conserved SPKK motif. This is consistent with previous in vitro studies of the function of the SPKK motif in other histone variants (Khadake and Rao, 1997). The SPKK motif binds preferentially to the minor groove of DNA at A/T rich sites (Churchill and Suzuki, 1989), which are generally enriched in satellite repeats of heterochromatin (Brutlag, 1980). During evolution, the SPKK motif not only became associated with H2A.W in seed plants but was also recruited to other histones, which are associated with condensed chromatin, the linker H1 in most eukaryotes and the sea urchin testis specific H2B (Suzuki, 1989; Buttinelli et al., 1999; Poccia and Green, 1992). Motifs similar to SPKK are also present in the linker region of the vertebrate specific macroH2A that separates the core histone domain from the Cterminal macrodomain (Figure S6). MacroH2A is highly enriched on the X-chromosome and is linked to transcriptional repression (Costanzi and Pehrson, 1998; Gamble et al., 2009; Gaspar-Maia et al., 2013). In vitro, the linker region of macroH2A causes chromatin condensation (Muthurajan et al., 2011), and macroH2A devoid of the macrodomain stabilizes nucleosomes (Chakravarthy et al., 2012). However, how macroH2A affects chromatin architecture in vivo remains unclear. The similarity between H2A.W and macroH2A devoid of its macrodomain suggests that the properties of H2A.W described in this study may reveal conserved aspects of histone variant functions that may not only aid in our understanding of plant heterochromatin, but also shed light on mechanisms controlling higher-order heterochromatin condensation in many other eukaryotes.
Experimental procedures
Plant material and growth conditions
All mutants are in Columbia-0 (Col) ecotype. h2a.w.6 (SALK_024544.32), h2a.w.7 (GK_149G05), and h2a.w.12 (SAIL_667) T-DNA lines were obtained from ABRC at Ohio State University and NASC at The University of Nottingham, respectively. drm2-2 (SALK_150863.37.35), cmt3-11 (SALK_148381), kyp-6 (SALK_041474), suvh4,5,6 triple mutant (SALK_041474, Gabi_263C05, SAIL_1244_F04), cmt3-11 (SALK_148381), drm2-2 (SALK_150863), met1-3 and the EMS-induced SNP in ddm1-2. met1-3, ddm1-2, and kyp suvh5 suvh6 were previously described (Johnson et al., 2008; Saze et al., ; Vongs et al., 1993). The 180bp repeat reporter PDZ-GFP was a kind gift from Prof. B. van der Zaal. h2a.w.6 and h2a.w.7 T-DNA insertions were confirmed by PCR-based genotyping. Primer sequences are described in Table S1. Arabidopsis plants were grown under long day conditions i.e 16 hours light and 8 hours dark.
H2A variants ChIP-seq analyses
ChIP was performed as previously described (Wollmann et al., 2012) with few modifications. Two grams of seedlings were ground in liquid nitrogen and fixed in 1% formaldehyde. Sonicated chromatin was incubated overnight with either anti-H2A, anti- H2A.X, anti-H2A.W.6, anti-H2A.Z.9, and anti-H3 (ab1791, Abcam) antibodies or IgG (ab46540-1, Abcam). Immunoprecipitated DNA was treated with RNase A (Fermentas) and purified with the QIAquick purification kit (Qiagen). ChIP-seq libraries were generated as per manufacturer’s instructions (Illumina). Libraries were sequenced on a HiSeq2000 (Illumina). Reads were mapped to the TAIR8 genome and analysis performed was done as previously described (Stroud et al., 2012).
DNA methylation analyses
For genome-wide bisulfite sequencing (BS-seq), plants were grown under continuous light and samples were collected from 3-week-old plants. Genomic DNA was extracted using Plant DNeasy Mini kit (Qiagen). BS-seq libraries were generated, sequenced, and analyzed exactly as previously described (Stroud et al., 2013), except that reads were mapped to the TAIR8 genome.
Nucleosome array assembly
Nucleosomal arrays were reconstituted onto a DNA fragment that comprises of 12 repeats of 207 base pairs “601” positioning sequence(Lowary and Widom, 1998). A twostep reconstitution method was applied to assemble nucleosome arrays (Muthurajan et al., 2011). First, different amounts of (H3-H4)2 tetramers were titrated to DNA. The tetrasomes were reconstituted by step dialysis (4-6 hours each step) starting from 1 M NaCl to 0.75 M NaCl and finally 2.5 mM NaCl (in a buffer containing 10 mM Tris, pH 7.5 and 0.25 mM EDTA). The best ratio between H3-H4 and DNA for tetrasome formation was determined by sedimentation velocity AUC. Once the S20,w midpoint of reconstitution reached around 19S, the ratio of tetramer to DNA was fixed for the next step. Varying amounts of variant H2A-H2B dimers were added to the optimized the ratio of tetramer to DNA. The saturation levels of arrays were determined by sedimentation velocity AUC (SV-AUC). To reach a S20,w of 29S (characteristic of a fully saturated array), the ratio of dimer to tetrasome was around 2.0. Eco RI digestion also was be used to monitor the saturation status of nucleosome arrays as described previously(Muthurajan et al., 2011).
Self-association assay
Nucleosomal arrays (the equivalent of ~1 µg DNA) were combined with various concentrations of MgCl2 (from 0 to 5 mM). The mixtures were incubated at room temperature for 5 minutes, and then spun at 14,000g for 10 minutes. The percentage of arrays remaining in the supernatant was determined at 259 nm, and plotted against the concentration of MgCl2.
Folding assay by sedimentation velocity
Nucleosome arrays containing ~10 µg DNA was treated with 1 mM MgCl2. SV- AUC was applied to monitor the impact of MgCl2 on array folding. Sedimentation velocity experiments were done in a Beckman XL-A or XL-I ultracentrifuge at 20,000 rpm and 20 °C. The absorbance at 259 nm was scanned continuously with a redial increment of 0.003 cm. All results were processed and analyzed by UltraScanIII. By the improved van Holde-Weischet method in UltraScanIII, the diffusion-corrected integral distribution (G(s)) of sedimentation coefficients over the entire boundaries were analyzed and plotted.
Supplementary Material
Highlights.
Genomic features are marked by combination of H2A variants in Arabidopsis
Heterochromatin is specifically marked by H2A.W in correlation with H3K9me2
H2A.W is required for heterochromatin condensation and silencing
A conserved C-terminal motif of H2A.W promotes chromatin condensation
Acknowledgements
RY, SH, XN, and FB were funded by Temasek Lifesciences Laboratory. Sequencing was performed at the UCLA BSCRC BioSequencing Core facility. We thank Meredith Calvert for help with imaging, and Mahnaz Akhavan for Illumina sequencing. We thank Dr Maruyama for his comments and support with the graphical abstract. HS was supported by a Dissertation Year Fellowship from UCLA. XZ is a research fellow of Ruth L. Kirschstein National Research Service Award (F32GM096483-01). SF is a Special Fellow of the Leukemia & Lymphoma Society. Work in the Jacobsen lab was supported by NSF grant 1121245. KL, UM, and KZ are supported by NIH-GM067777. MG is supported by EMBO ALTF 986-2011 and HHMI. SEJ and KL are Investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
KZ, UMM, and KL performed experiments reported in Figures 5 and S5; SF and XZ performed the first series of ChIP-seq for H2A.W.6; SF performed genome-wide bisulfite sequencing; HS performed the RNA-seq and all bioinformatic analyses; SH participated in H2A variants ChIP; SH, TK and FB produced the data shown in Figures 6 and S6; XN performed preliminary transcriptome analyses; MG provided Figure S2H, RY performed all other experiments and analyses. FB conceived the initial project, which was developed with the help of a fruitful collaboration with SEJ lab. RY, FB, and SEJ wrote the manuscript with inputs from all other authors.
Accession numbers
All the sequencing data is deposited at GEO, can be found in following link. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=atmdiooajxiplid&acc=GSE50942
The data reported in this paper are available (accession number).
References
- Aceituno FF, Moseyko N, Rhee SY, Gutierrez RA. The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics. 2008;23:438. doi: 10.1186/1471-2164-9-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Current Opinion in Cell Biology. 2012;24:359–366. doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Research. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag DL. Molecular Arrangement and Evolution of Heterochromatic DNA. Annual Review of Genetics. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Buttinelli M, Panetta G, Rhodes D, Travers A. The role of histone H1 in chromatin condensation and transcriptional repression. Genetica. 1999;106:117–124. doi: 10.1023/a:1003745315540. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Patel A, Bowman GD. The basic linker of macroH2A stabilizes DNA at the entry/exit site of the nucleosome. Nucleic Acids Research. 2012;40:8285–8295. doi: 10.1093/nar/gks645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill MEA, Suzuki M. SPKK MOTIFS PREFER TO BIND TO DNA AT A/T-RICH SITES. Embo Journal. 1989;8:4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson J. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. Dual Binding of Chromomethylase Domains to H3K9me2-Containing Nucleosomes Directs DNA Methylation in Plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z Alters the Nucleosome Surface to Promote HP1α-Mediated Chromatin Fiber Folding. Molecular cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL. Centromere Positioning and Dynamics in Living Arabidopsis Plants. Molecular Biology of the Cell. 2005;16:5710–5718. doi: 10.1091/mbc.E05-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipescu D, Szenker E, Almouzni G. Developmental roles of histone H3 variants and their chaperones. Trends in Genetics. 2013;29:630–640. doi: 10.1016/j.tig.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a pitative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acid Research. 1993:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emante. Proc Natl Acad Sci USA. 2002;99:14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble M, Frizzell K, Yang C, Krishnakumar R, Kraus W. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Development. 2009;24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Adrian Leu N, Leroy G, Liu S, Costanzi C, Valle-Garcia D, Schaniel C, et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat Commun. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant Histone H2A.Z Is Globally Localized to the Promoters of Inactive Yeast Genes and Regulates Nucleosome Positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- Hansen JC. CONFORMATIONAL DYNAMICS OF THE CHROMATIN FIBER IN SOLUTION: Determinants, Mechanisms, and Functions. Annual Review of Biophysics and Biomolecular Structure. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Šoljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F. Zygotic Resetting of the HISTONE 3 Variant Repertoire Participates in Epigenetic Reprogramming in Arabidopsis. Current Biology. 2010;20:2137–2143. doi: 10.1016/j.cub.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the Histone Code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. Journal of The Royal Society Interface. 2013;10 doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadake JR, Rao MRS. Condensation of DNA and Chromatin by an SPKK-Containing Octapeptide Repeat Motif Present in the C-Terminus of Histone H1. Biochemistry. 1997;36:1041–1051. doi: 10.1021/bi961617p. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-Way Control of FWA Imprinting in Arabidopsis Endosperm by DNA Methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutiérrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhout BI, Fransz P, Tessadori F, Meckel T, Hooykaas PJ, van der Zaal BJ. Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nuclei Acid Research. 2007;35:e107. doi: 10.1093/nar/gkm618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Lindsey G, Orgeig S, Thompson P, Davies N, Maeder D. Extended C-terminal tail of wheat histone H2A interacts with DNA of the "linker" region. J Mol Biol. 1991;218:805–813. doi: 10.1016/0022-2836(91)90268-b. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Marking histone H3 variants: How, when and why? Trends in Biochemical Sciences. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved Histone Variant H2A.Z Protects Euchromatin from the Ectopic Spread of Silent Heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Millar CB. Organizing the genome with H2A histone variants. Biochemical Journal. 2013;449:567–579. doi: 10.1042/BJ20121646. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Muthurajan UM, McBryant SJ, Lu X, Hansen JC, Luger K. The Linker Region of MacroH2A Promotes Self-association of Nucleosomal Arrays. Journal of Biological Chemistry. 2011;286:23852–23864. doi: 10.1074/jbc.M111.244871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poccia DL, Green GR. Packaging and unpackaging the sea urchin sperm genome. Trends in Biochemical Sciences. 1992;17:223–227. doi: 10.1016/0968-0004(92)90382-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone Variant H2A.Z Marks the 5′ Ends of Both Active and Inactive Genes in Euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BB, Mirouze M, Mari-Ordonez A, Dapp M, Rozhon W, Bucher E, Thelier G, Paszkowski J. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rop V, Padeganeh A, Maddox P. CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma. 2012;121:527–538. doi: 10.1007/s00412-012-0386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo Sophie E, Almouzni G. Prime, Repair, Restore: The Active Role of Chromatin in the DNA Damage Response. Molecular cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive Analysis of Silencing Mutants Reveals Complex Regulation of the Arabidopsis Methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Otero S, Desvoyes B, Ramírez-Parra E, Jacobsen SE, Gutierrez C. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA. 2012;109:5370–5375. doi: 10.1073/pnas.1203145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. Embo Journal. 1989;8:797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P, Ahmad K, Almouzni G, Ausio J, Berger F, Bhalla P, Bonner W, Cande W, Chadwick B, Chan SW, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics & Chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P, Bayes J, Henikoff S. The Evolution of Centromeres and Kinetochores: A Two-Part Fugue. The Kinetochore: From Molecular Discoveries to Cancer Therapy. 2008:193–230. [Google Scholar]
- Talbert P, Henikoff S. Histone variants-ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- Talbert P, Masuelli R, Tyagi A, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Tutois S, Jasencakova Z, Douet J, Schubert I, Tourmente S. Hypomethylation and hypermethylation of the tandem repetitive 5S rRNA genes in Arabidopsis. Plant J. 2008;54:299–309. doi: 10.1111/j.1365-313X.2008.03413.x. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen R, Richards E. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wollmann H, Holec S, Alden K, Clarke ND, Jacques P-E, Berger F. Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Arabidopsis. Transcriptome. PLoS Genet. 2012;8:e1002658. doi: 10.1371/journal.pgen.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Research. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer Stacey L, Zilberman D. The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.